The characterization of modifier genes can provide insights into disease pathways and identify novel therapeutic targets. This study identifies Mtap1a as a modifier gene of photoreceptor loss in Tulp1 and Tub mutant mice, which are models of retinal degeneration.
Abstract
Purpose.
To identify genes that modify photoreceptor cell loss in the retinas of homozygous Tulp1tm1Pjn and Tubtub mice, which exhibit juvenile retinitis pigmentosa.
Methods.
Modifier loci were identified by genetic quantitative trait locus analysis. F2 Tulp1tm1Pjn/tm1Pjn mutant mice from a B6-Tulp1tm1Pjn/tm1Pjn × AKR/J intercross were genotyped with a panel of single nucleotide polymorphism markers and phenotyped by histology for photoreceptor nuclei remaining at 9 weeks of age. Genotype and phenotype data were correlated and examined with Pseudomarker 2.02 using 128 imputations to map modifier loci. Thresholds for the 63%, 10%, 5%, and 1% significance levels were obtained from 100 permutations. A significant, protective candidate modifier was identified by bioinformatic analysis and confirmed by crossing transgenic mice bearing a protective allele of this gene with Tulp1- and Tub-deficient mice.
Results.
A significant, protective modifier locus on chromosome 2 and a suggestive locus on chromosome 13 that increases photoreceptor loss were identified in a B6-Tulp1tm1Pjn/tm1Pjn × AKR/J intercross. The chromosome 2 locus mapped near Mtap1a, which encodes a protein associated with microtubule-based intracellular transport and synapse function. The protective Mtap1a129P2/OlaHsd allele was shown to reduce photoreceptor loss in both Tulp1tm1Pjn/tm1Pjn and Tubtub/tub mice.
Conclusions.
It was demonstrated that the gene Mtap1a, which modifies hearing loss in Tubtub/tub mice, also modifies retinal degeneration in Tubtub/tub and Tulp1tm1Pjn/tm1Pjn mice. These results suggest that functionally nonredundant members of the TULP family (TUB and TULP1) share a common functional interaction with MTAP1A.
Retinal degenerative diseases are highly variable in onset, severity, and rate of progression. Retinitis pigmentosa (RP), with an incidence of roughly 1:4000 in the United States, is a prominent example of this variability.1 RP consists of a heterogeneous group of retinal dystrophies characterized by rod and cone photoreceptor cell degeneration, usually through apoptotic cell death. Clinical features of the disease include night blindness, which is often accompanied by a gradual loss of peripheral visual field and eventual loss of central vision. Variation in the clinical presentation of the disease is partly explained by the existence of >48 RP genes, each with multiple alleles that may cause substantial differences in disease phenotype (Retinal Information Network, http://www.sph.uth.tmc.edu/Retnet/).1 However, variation has been documented even among family members of similar age with identical RP mutations, suggesting contributions from other genetic or environmental factors.2–12 Modifier genes, which influence the phenotypic expression of genes in monogenic and multigenic traits, are a potential source of variation in such cases.13 Identifying modifier genes in RP may help to explain variation in disease presentation and therapeutic response, improve understanding of functional pathways that underlie the RP phenotype, and reveal important targets for clinical intervention.
Our interest in RP gene modifiers arose from studying two members of the tubby-like protein (TULP)14 gene family that are associated with retinal degeneration. The TULP gene family consists of four members: Tub, Tulp1, Tulp2, and Tulp3. Homozygous mutations of Tub (Tubtub) and Tulp1 in mice cause increased apoptotic photoreceptor cell death and early-onset retinal degeneration; Tub mutants also exhibit cochlear degeneration and late-onset obesity.15–19 In humans, a disruption in Tulp1 has been identified as the causative factor of RP type 14, a rare, autosomal recessive form of RP that is genetically heterogeneous.20–23 Indeed, 23 pathogenic TULP1 mutations were identified from the literature in a recent study of human patients with RP or Leber congenital amaurosis.24 Much evidence suggests that TUB and TULP1 function in vesicular transport in neurons, because disruption of either Tub or Tulp1 leads to abnormal accumulation of vesicles in the photoreceptor matrix at the level of the inner segments,25,26 and deficiency of Tulp1 results in structural and functional defects at the photoreceptor synapse.27,28 These defects are thought to be responsible for the observed photoreceptor cell apoptosis. However, other studies point to a role of the TULP family14 in insulin signaling,29 transcriptional regulation,30,31 and phagocytosis.32,33 Thus, it remains to be determined which of the proposed functions of TUB and TULP1 are of greatest relevance in the healthy and diseased retina.
To gain insight into how disruptions in TULP family members lead to retinal degeneration, we previously performed a quantitative trait locus (QTL) analysis by outcrossing B6.Cg-Tubtub mutant mice to strain AKR/J, and assessed the progression of photoreceptor degeneration in F2 progeny.34 In homozygous tub F2 mice, a suggestive protective locus (P < 0.05) from AKR/J was identified on chromosome 2. Likewise, here, in a similar cross with Tulp1tm1Pjn/tm1Pjn mutant mice, a significant locus (P < 0.01) from AKR/J is identified in the same region on chromosome 2. In both studies, the photoreceptor protective locus on chromosome 2 overlaps the moth1 (modifier of tubby hearing1) locus, which our laboratory had previously identified as Mtap1a.35,36
Crossing the Tulp1tm1Pjn/tm1Pjn and Tubtub/tub mutant mice to transgenic mice carrying a 129P2/OlaHsd Mtap1amoth1-r allele resulted in a significant protective effect (P < 0.0005 and P < 0.04, respectively) against photoreceptor loss in the F2 generation in transgene-positive (tg+) Tulp1tm1Pjn/tm1Pjn and B6.Cg-Tubtub/tub mutant mice. These results indicate that, although Tulp1 and Tub are not functionally redundant, they both exhibit a genetic interaction with Mtap1a. Because MTAP1A is implicated in cytoskeletal processes at the neuronal synapse,37 these findings further support the hypothesis that TULP family members are involved in synapse maintenance36 or architecture.28,38
Methods
Experimental Animals
All mice were treated in accordance with the Animal Care and Use Committees at each of the contributing institutions and in compliance with the Association for Research in Vision and Ophthalmology (ARVO) statement for ethical care and use of animals.
Mouse Production and Mapping
Mutant mice from an intercross of homozygous B6.129 × 1-Tulp1tm1Pjn× AKR/J were genotyped by PCR to identify Tulp1tm1Pjn homozygotes (designated as Tulp1tm1Pjn/tm1Pjn mice elsewhere in this study). The eyes from 9-week-old F2 Tulp1tm1Pjn/tm1Pjn intercross mice were processed for histologic observation and scored for the loss of photoreceptor nuclei (1, mild; 2, moderate; 3, severe). Mice were genotyped (KBiosciences, Beverly, MA) with a panel of single nucleotide polymorphism (SNP) markers spaced at approximate 20-cM intervals. The genotype and phenotype data were analyzed with Pseudomarker 2.0239 using 128 imputations. One hundred permutations were performed to determine thresholds for the 63%, 10%, 5%, and 1% significance levels.
To confirm the chromosome 2 QTL, additional F2 Tulp1tm1Pjn/tm1Pjn mutant mice were collected, and follow-up genotyping on these mice combined with the initial cohort was performed in a fashion similar to that described by Danciger and colleagues.40 Briefly, DNA was genotyped with simple sequence length polymorphic (SSLP) markers at 10-cM intervals in chromosomal regions, demonstrating possible QTL loci, and analyzed using Pseudomarker 2.02 as described earlier.
Microscopy
Whole eyes were enucleated, fixed in 37.5% methanol, 12.5% glacial acetic acid in phosphate buffered saline, embedded in paraffin, and sectioned at 6-μm intervals. The sections were mounted on commercial slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA) and stained with hematoxylin and eosin. Sections were analyzed by brightfield illumination on a light microscope (Leitz DMRO; Leica Microsystems, Buffalo Grove, IL) and images were collected with a commercial software package (FireCam software; Leica Microsystems). Brightness and contrast of images were optimized with a digital imaging package (Photoshop 7.0; Adobe Systems, San Jose, CA).
Quantification of Retinal Degeneration
To quantify the degree of retinal degeneration, we followed a previously published protocol.35 The section in which the optic nerve was at its greatest width was selected from serial sections, and carefully photographed. For a quantitative measure of retinal degeneration the number of rows of nuclei contained within the outer nuclear layer (ONL) were counted at three points within a 200-μm square near the optic nerve head and the values were averaged. Because the thickness of the inner nuclear layer (INL) is not affected by the Tulp1 mutation,34 INL thickness was measured to identify oblique sections, which were subsequently removed from the data set.
Results
QTL Analysis of Genetic Modifiers of Tulp1tm1Pjn/tm1Pjn Retinal Degeneration
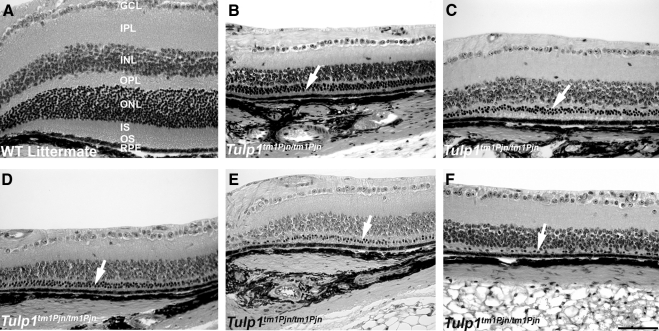
To detect genetic modifiers of Tulp1tm1Pjn/tm1Pjn retinal degeneration, we first conducted pilot studies of F2 Tulp1tm1Pjn/tm1Pjn mutant mice from a B6-Tulp1tm1Pjn/tm1Pjn × AKR/J intercross to identify an age at which the greatest range of ONL phenotypes could be detected. The ONL phenotype of the oldest mice examined, 21 weeks of age, consisted of only one or two rows of photoreceptor nuclei and exhibited little variation (data not shown). Thus, 9-week-old mice were selected for QTL analysis. A total of 72 age-matched F2 Tulp1tm1Pjn/tm1Pjn mutant mice from a B6-Tulp1tm1Pjn/tm1Pjn × AKR/J intercross were identified by PCR genotyping and the eyes processed for histologic analysis. Although the retinas of wild-type and heterozygous mice were arranged in orderly layers (Fig. 1A) with a prominent INL and ONL, the retinas of Tulp1tm1Pjn/tm1Pjn × AKR/J mutant mice were abnormal, with a marked reduction in the number of nuclei in the ONL, indicating photoreceptor cell loss (Figs. 1B–F, arrows). Strikingly, the number of ONL nuclei among these animals varied considerably. This variation in ONL nuclei was specific for homozygous Tulp1tm1Pjn/tm1Pjn mice and was not observed in randomly sampled retinas from heterozygous or wild-type littermates (data not shown).
Figure 1.
Variation of ONL thickness in Tulp1-deficient mice is variable in a segregating genetic background. Wildtype (WT) F2 progeny from a B6-Tulp1tm1Pjn/tm1Pjn × AKR/J intercross showed normal thickness and arrangement of retinal layers (A). GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; IS, inner segment; OS, outer segment; RPE, retinal pigment epithelium. F2 Tulp1tm1Pjn/tm1Pjn progeny showed attenuated ONL, IS, and OS layers, with considerable variability in the ONL appearance (B–F). Arrows: ONL layer in Tulp1-deficient progeny. Bar: 50 μm.
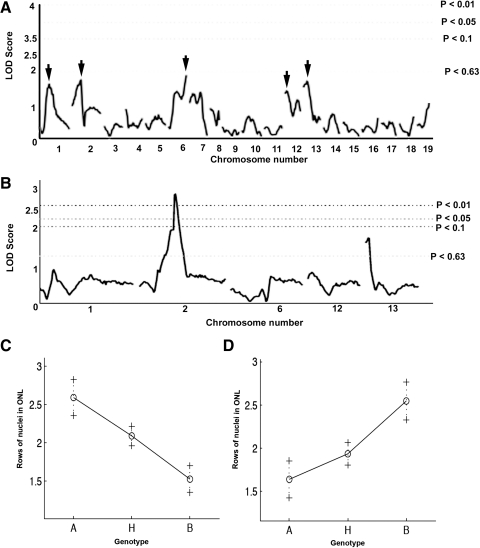
Photoreceptor cell loss in each mutant mouse was scored by assigning a value based on the distribution of photoreceptor nuclei remaining (1, mild; 2, moderate; 3, severe loss) and the animals were genotyped with SNP markers. Although QTL analysis identified no statistically significant modifier loci with the first cohort of 72 animals, peaks on chromosomes 1, 2, 6, 12, and 13 nearly reached the significance level for suggestive loci (P < 0.63; n = 72; Fig. 2A). Therefore, 44 additional mutant mice were collected, and SSLP markers were tested at 10-cM intervals across chromosomes 1, 2, 6, 12, and 13 for all samples combined (n = 116 mice). Analysis of the combined data indicated a significant modifier locus on chromosome 2 (P < 0.01) and a suggestive locus on chromosome 13 (P < 0.63) (Fig. 2B). The locus on chromosome 2 slowed the rate of photoreceptor cell degeneration (Fig. 2C), whereas the locus on chromosome 13 enhanced the rate of photoreceptor cell loss (Fig. 2D).
Figure 2.
QTL analysis revealed a significant modifier locus on chromosome 2 and a suggestive modifier locus on chromosome 13. Analysis of 72 F2 mice at an approximately 20-cM SNP interval yielded five nearly suggestive loci (A). Analysis of an additional 44 F2 mice at an approximately 10-cM SNP interval revealed a significant modifier on chromosome 2 and a suggestive modifier on chromosome 13 (B). Effect plots of homozygous (A, AKR/J; B, B6) and heterozygous (H) phenotypes indicated a protective and sensitizing effect of the modifiers on chromosomes 2 and 13, respectively (C, D). LOD: log of the odds ratio.
Identification of Candidate Modifier Genes on Chromosome 2
The region contained under the highest peak on chromosome 2 comprised an interval of approximately 45 Mb (115 to 160 Mb). We expected modifier genes within this interval to show nonsynonymous coding sequence differences between the strain backgrounds that were intercrossed, C57BL/6J and AKR/J. Since this interval contained too many genes to analyze individually, candidate quantitative trait genes (QTGs) were identified by a “virtual” approach using bioinformatic tools as described.40 Searching the critical interval with the PosMed virtual positional cloning database (http://omicspace.riken.jp/PosMed/) and the keyword “retina” produced a list of 161 genes known to be expressed or function in the retina. To further narrow the list of candidate genes, each gene from the PosMed list was evaluated as to whether mutant alleles were associated with an abnormal eye phenotype using the MGI Phenotypes database (http://www.informatics.jax.org/phenotypes.shtml) and PubMed listings (http://www.ncbi.nlm.nih.gov/pubmed/). Candidate genes known to influence eye development, degeneration, or function were selected, yielding 15 candidate genes (Table 1). Additionally, the Mtap1a gene, which we have found to modify cochlear and possibly retinal degeneration in homozygous Tubtub/tub mice, was identified within the critical region.
Table 1.
Candidate QTGs Identified on AKR/J Chromosome 2
| Gene* | Position | GO Molecular Function† | Mutant Eye Phenotype‡ |
|---|---|---|---|
| Meis2 | 115 Mb | Sequence-specific DNA binding transcription factor activity (GO:0003700) | Developmental eye defects |
| Tyro3 | 119 Mb | Transmembrane receptor protein tyrosine kinase activity (GO:0004714) | Vision/eye; retinal degeneration |
| Mtap1a§ | 121 Mb | Actin binding (GO:0003779) | Possible modifier of tub/tub retinal degeneration |
| Nphp1 | 127 Mb | Protein binding (GO:0005515) | Vision/eye; retinal degeneration |
| Mertk | 128 Mb | Transmembrane receptor protein tyrosine kinase activity (GO:0004714) | Vision/eye; retinal degeneration |
| Bcl2l11 | 128 Mb | Microtubule binding (GO:0008017) | Increased retinal thickness |
| Pank2 | 131 Mb | Pantothenate kinase activity (GO:0004594) | Vision/eye; retinal degeneration |
| Plcb4 | 135 Mb | Mitogen-activated protein kinase binding (GO:0051019) | Vision/eye; reduced visual processing |
| Jag1 | 136 Mb | Notch binding (GO:0005112) | Vision/eye; developmental eye defects |
| Mkks | 136 Mb | ATP binding (GO:0005524) | Vision/eye; retinal degeneration |
| Bfsp1 | 143 Mb | Structural constituent of eye lens (GO:0005212) | Vision/eye; cataract |
| Vsx1 | 150 Mb | Sequence-specific DNA binding transcription factor activity (GO:0003700) | Vision/eye; impaired retinal electrophysiology |
| E2f1 | 154 Mb | Sequence-specific DNA binding transcription factor activity (GO:0003700) | Vision/eye; defects |
| Gss | 155 Mb | Glutathione synthase activity (GO:0004363) | Seasonal sensitivity to light |
| Src | 157 Mb | Nonmembrane spanning protein tyrosine kinase activity (GO:0004715) | Vision/eye; Glaucoma |
| Rbl1 | 157 Mb | Transcription factor binding (GO:0008134) | Vision/eye; retinoblastoma |
Candidate genes within the modifier locus were identified from PosMed, MGI Phenotypes, and PubMed databases as described in the text.
When multiple gene ontology (GO) molecular functions were found, a single informative molecular function was chosen, with the corresponding GO term accession number given in parentheses.
The existence of mutant alleles affecting vision or the eye (Vision/eye) was determined with MGI Phenotypes. Other evidence for an ocular phenotype was obtained from PubMed listings.
Nonsynonymous coding difference between strains.
Comparison of the C57BL/6J and AKR/J alleles in the MGI Strains, SNPs, and Polymorphisms database (http://www.informatics.jax.org/strainsSNPs.shtml) gave no result for the Vsx1 and E2f1 alleles. Of the other candidate genes in the critical region, 13 showed no allelic differences between the two strains or showed sequence changes that were limited to introns or synonymous coding changes. Mertk, which has been suggested to interact with Tub and Tulpl,33 was among the genes that showed no coding sequence changes. By contrast, the C57BL/6J and AKR/J alleles of Mtap1a differ at multiple nucleotides corresponding to nonsynonymous coding changes,36 suggesting that it is the modifier gene on chromosome 2.
An Allele of Mtap1a Modifies Retinal Degeneration in Tulp1tm1Pjn/tm1Pjn Mutant Mice
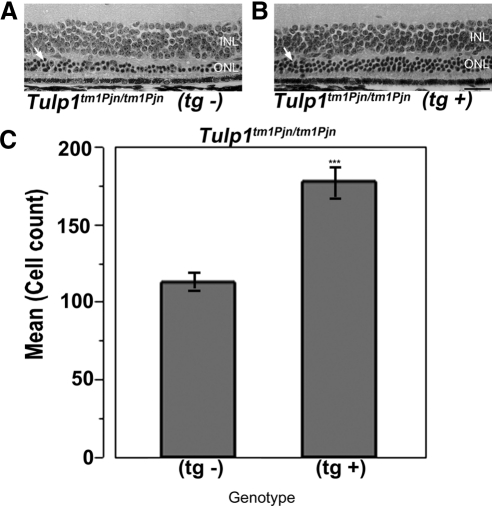
Our laboratory has previously reported a modifier of Tub hearing (moth1) on chromosome 2 in strains AKR/J, CAST/Ei, and 129P2/OlaHsd.35 Subsequent fine mapping and sequencing identified multiple polymorphisms in the microtubule-associated protein 1A (Mtap1a) gene. A C57BL/6J mouse carrying a transgenic Mtap1amoth1-r allele from the 129P2/OlaHsd strain (Mtap1a transgene) was mated to Tubtub/tub mice and, subsequently, the Mtap1a transgene was found to rescue hearing loss in Tubtub/tub mice.35 To test whether the Mtap1a transgene also attenuates photoreceptor cell loss in Tulp1tm1Pjn/tm1Pjn mice, Tulp1tm1Pjn/tm1 Pjn mutant mice were mated to Mtap1a transgene positive mice, and the F1s were intercrossed to generate F2 progeny. Retinas of F2 Tulp1tm1Pjn/tm1Pjn mice with the Mtap1a transgene (tg+) and without the Mtap1a transgene (tg−) were compared histologically at 9 weeks of age (Figs. 3A, 3B). The number of ONL nuclei remaining in the central retina of Tulp1tm1Pjn/tm1Pjn mutant mice with and without the Mtap1a transgene were counted. The average number of photoreceptor nuclei remaining in mutant retinas lacking the Mtap1a transgene averaged 112 ± 7.3 (Fig. 3A), whereas the number of photoreceptor nuclei remaining in the mutant retinas carrying the Mtap1a transgene averaged 177 ± 12.2 (Fig. 3B). The degree of photoreceptor cell loss was significantly decreased in mutant mice carrying the Mtap1a transgene (P < 0.0005, Student's t-test, n = 6 each) (Fig. 3C).
Figure 3.
Mtap1a modifies photoreceptor cell loss in Tulp1 mutant retinas. Retinal micrographs reveal fewer ONL nuclei Tulp1tm1Pjn/tm1Pjn mice lacking a transgenic copy of the 129P2/OlaHsd Mtap1amoth1-r gene (tg−, A) than in those containing the transgene (tg+, B). Retinal layers are labeled as in Figure 1A. Bar: 50 μm. ONL nuclei counts (mean ± SE) indicate a significantly higher number of photoreceptor cells in tg+ mice (C). Asterisks: P < 0.0005.
Mtap1a Modifies Retinal Degeneration in Tubtub/tub Mutant Mice
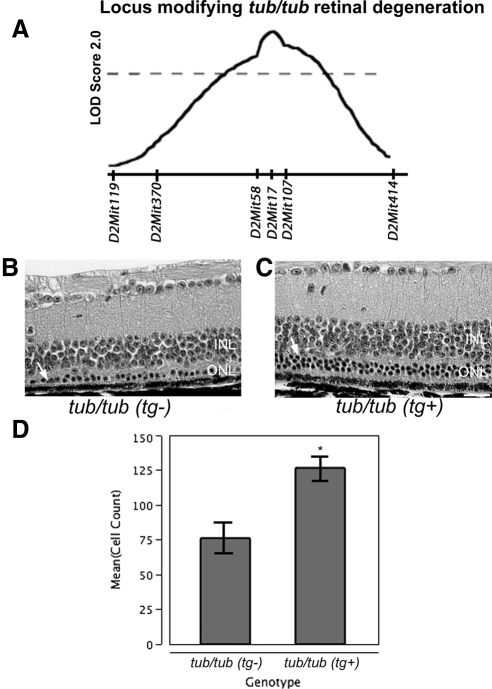
Mtap1a maps to a region located at 121 Mb on mouse chromosome 2, near the peak location (D2Mit17, 122 Mb) of a suggestive QTL for retinal degeneration in an intercross of B6.Cg-tub/tub × AKR/J mice34 (Fig. 4A). Due to the ability of the 129P2/OlaHsd Mtap1amoth1-r transgene to slow the rate of photoreceptor loss in Tulp1tm1Pjn/tm1Pjn mutant mice, we investigated whether the same allele of Mtap1a was capable of modifying retinal degeneration in homozygous Tubtub/tub mutant mice as well. Mtap1a transgenic mice were intercrossed with B6.Cg-tub/tub mice and the retinas of the F2 generation were examined. Retinas of B6.Cg-tub/tub mice with and without the Mtap1a transgene were compared histologically at 22 weeks of age. The later time point was selected because retinal degeneration in B6.Cg-tub/tub is slower than that in Tulp1tm1Pjn/tm1Pjn mutant mice. The numbers of ONL nuclei remaining in the central retina of B6.Cg-tub/tub mice with and without the Mtap1a transgene were counted. The average number of photoreceptor nuclei remaining in mutant retinas lacking the Mtap1a transgene averaged 76 ± 18.2 (Fig. 4B), whereas the number of photoreceptor nuclei remaining in mutant retinas carrying the Mtap1a transgene averaged 126 ± 14.4 (Fig. 4C). Statistical analysis indicated the degree of photoreceptor cell loss was markedly decreased in the mutant B6.Cg-tub/tub mice carrying the Mtap1a transgene (P < 0.04, Student's t-test, n = 5 each) (Fig. 4D).
Figure 4.
Mtap1a modifies photoreceptor cell loss in Tubtub/tub retinas (A). Previous QTL studies suggested a retinal-protective locus on chromosome 2, similar to findings with Tulp1 (reprinted with permission from the International Society for Eye Research/Elsevier Publishers34). Retinal micrographs reveal fewer ONL nuclei in Tubtub/tub mice lacking a transgenic copy of the 129P2/OlaHsd Mtap1amoth1-r gene (tg−, B) than in those containing the transgene (tg+, C). Retinal layers are labeled as in Figure 1A. Bar: 50 μm. ONL nuclei counts (mean ± SE) indicate a significantly higher number of photoreceptor cells in tg+ mice (D). Asterisk: P < 0.04.
Discussion
This study has identified a significant locus on AKR/J chromosome 2 that modifies the progression of retinal degeneration in Tulp1tm1Pjn/tm1Pjn mice. Mating of Tulp1tm1Pjn/tm1Pjn mice to mice carrying an Mtap1a transgene derived from the 129P2/OlaHsd strain indicated that the protective gene within the chromosome 2 locus was Mtap1a. We demonstrated a similar protective effect of the 129P2/OlaHsd Mtap1amoth1-r allele on photoreceptor loss in Tubtub/tub mice. Our results indicating that Mtap1a is a genetic modifier of both Tulp1 and Tub raise the possibility that two members of the TULP protein family (TULP1 and TUB) share a common functional interaction with MTAP1A. This interaction may be a characteristic feature of sensory neurons because the protective allele of Mtap1a modulates the Tubtub/tub mutant phenotype in both the ear35 and retina.
We combined QTL analysis and bioinformatic techniques to identify candidate genetic modifiers. Although QTL analysis is a powerful tool for identifying large chromosomal regions that influence a given phenotype, the identification of individual genes within the QTL that specifically influence the phenotype of interest is rare. This is inherent to this type of analysis because, as the region of interest becomes smaller, the number of crossovers observed within the region become correspondingly infrequent. Moreover, the imperfect phenotype–genotype correspondence of a QTL dictates that quantitative statistical analysis must be performed to define the most likely candidate regions. The expense to generate, house, and genotype enough mice to obtain sufficient crossovers to delineate the critical region required to narrow the candidate gene pool to a single or a few genes is often prohibitive. Even well-designed studies using large numbers of mice with closely spaced genotypes can fail to minimize the critical region to a manageable number of candidate genes. For this reason, we chose to filter our results through the PosMed database as reported by Danciger and colleagues,40 and to further filter the results through reports of phenotypic relevance of these genes to retinal development, degeneration, and function and by the presence of nonsynonymous coding SNPs. The filtering process allowed us to test a subset of biologically relevant genes with known retinal associations within the critical interval for sequence changes. The success of this combined QTL and bioinformatics approach is expected to improve as genetic tools such as the Collaborative Cross and Diversity Outbred mice become available for high-resolution genetic mapping,41–43 and as the annotations and quality of bioinformatics search engines increase.
Although the modifier locus on chromosome 13 identified in our study did not reach statistical significance, the data raise the possibility of an intriguing modifier gene that enhances the loss of photoreceptor cells. By filtering genes with the same PosMed and bioinformatic criteria we used for chromosome 2 and by screening the results for nonsynonomous coding changes between C57BL/6J and AKR/J (data not shown), we have tentatively identified Lyst as a potential candidate gene in the critical interval of chromosome 13. LYST is a widely expressed protein involved in vesicular trafficking, particularly in the biogenesis of lysosomes and related organelles.44 Lyst mutations in beige and gray mice exhibit lysosomal abnormalities within the retinal pigment epithelium,45,46 and LYST mutations in humans cause Chédiak–Higashi syndrome,44 a systemic disease with ocular manifestations that include a progressive visual loss consistent with retinal degeneration.47,48 A spontaneous mouse mutant of the Lyst gene is available (Lystbg-J), which might be used to test for modifier effects on Tulp1 and Tub retinal degeneration.
Our finding that Mtap1 is a genetic modifier of both Tulp1 and Tub supports the hypothesis that TUB and TULP1 function in synaptic maintenance or architecture, possibly reflecting their role in vesicular trafficking throughout the photoreceptor cell.14,25–28,36,49 MTAP1A (also known as MAP1A) is predominantly found at the synapses of adult neurons, where it associates with both filamentous-actin and microtubules, presumably integrating these two components of the cytoskeleton.37 We previously showed that MTAP1A interacts with DLG4 (previously known as PSD95),36 a PDZ-domain protein that organizes transmembrane signaling proteins and the cytoskeleton at postsynaptic membranes. In the mouse retina, DLG4 localizes to the presynaptic termini of rod and cone photoreceptor cells.50 Interestingly, TULP1 has also been shown to interact with dynamin-1, a neuron-specific GTPase that has roles in endocytosis, vesicle formation, vesicular movement at the trans-Golgi network, and plasma membrane and vesicle recycling at neuronal synapses.38 TULP1 colocalizes with this protein at the photoreceptor synapse and inner segment,38 and with the actin cytoskeleton of photoreceptor cells.51 Tub- and Tulp1-deficient mice mislocalize rhodopsin into extracellular vesicles and alter the cellular distribution of arrestin and transducin, consistent with a trafficking defect.25,26,49 Tulp1 mice also show striking defects in the normal distribution of photoreceptor synaptic proteins.27,28 Together with our observations, these findings suggest a working model in which MTAP1, TUB, and TULP1 in conjunction with components of the cytoskeleton are necessary for maintaining the architecture of the photoreceptor synapse and/or vesicular trafficking at the synapse and inner segment.
Acknowledgments
The authors thank Gareth Howell and Kenneth Johnson for careful review of the manuscript and Jeanie Hansen for expert technical assistance.
Footnotes
Supported in part by National Institutes of Health Grant 5R01DK085441 (JKN) and a Cancer Center CORE Grant 5P30CA034196.
Disclosure: D.M. Maddox, None; S. Ikeda, None; A. Ikeda, None; W. Zhang, None; M.P. Krebs, None; P.M. Nishina, None; J.K. Naggert, None
References
- 1. Ayuso C, Millan JM. Retinitis pigmentosa and allied conditions today: a paradigm of translational research (Abstract). Genome Med. 2010;2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. To K, Adamian M, Dryja TP, Berson EL. Histopathologic study of variation in severity of retinitis pigmentosa due to the dominant rhodopsin mutation Pro23His. Am J Ophthalmol. 2002;134:290–293 [DOI] [PubMed] [Google Scholar]
- 3. Berson EL, Grimsby JL, Adams SM, et al. Clinical features and mutations in patients with dominant retinitis pigmentosa-1 (RP1). Invest Ophthalmol Vis Sci. 2001;42:2217–2224 [PubMed] [Google Scholar]
- 4. Berson EL, Rosner B, Sandberg MA, Dryja TP. Ocular findings in patients with autosomal dominant retinitis pigmentosa and a rhodopsin gene defect (Pro-23-His). Arch Ophthalmol. 1991;109:92–101 [DOI] [PubMed] [Google Scholar]
- 5. Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Dryja TP. Ocular findings in patients with autosomal dominant retinitis pigmentosa and rhodopsin, proline-347-leucine. Am J Ophthalmol. 1991;111:614–623 [DOI] [PubMed] [Google Scholar]
- 6. Jacobson SG, Cideciyan AV, Iannaccone A, et al. Disease expression of RP1 mutations causing autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:1898–1908 [PubMed] [Google Scholar]
- 7. Passerini I, Sodi A, Giambene B, Menchini U, Torricelli F. Phenotypic intrafamilial variability associated with S212G mutation in the RDS/peripherin gene. Eur J Ophthalmol. 2007;17:1000–1003 [DOI] [PubMed] [Google Scholar]
- 8. Paunescu K, Preising MN, Janke B, Wissinger B, Lorenz B. Genotype-phenotype correlation in a German family with a novel complex CRX mutation extending the open reading frame. Ophthalmology. 2007;114:1348–1357 [DOI] [PubMed] [Google Scholar]
- 9. Ayuso C, Trujillo MJ, Robledo M, et al. Novel rhodopsin mutation in an autosomal dominant retinitis pigmentosa family: phenotypic variation in both heterozygote and homozygote Val137Met mutant patients. Hum Genet. 1996;98:51–54 [DOI] [PubMed] [Google Scholar]
- 10. Chang W, Ding Q, Tang Z, et al. A novel de novo frameshift mutation of RPGR ORF15 is associated with X-linked retinitis pigmentosa in a Chinese family. Mol Vis. 2007;13:1548–1554 [PubMed] [Google Scholar]
- 11. Bandah-Rozenfeld D, Mizrahi-Meissonnier L, Farhy C, et al. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2010;87:382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langmann T, Di Gioia SA, Rau I, et al. Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa. Am J Hum Genet. 2010;87:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124:357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukhopadhyay S, Jackson PK. The tubby family proteins (Abstract). Genome Biol. 2011;12:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coleman DL, Eicher EM. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered. 1990;81:424–427 [DOI] [PubMed] [Google Scholar]
- 16. Kleyn PW, Fan W, Kovats SG, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290 [DOI] [PubMed] [Google Scholar]
- 17. Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538 [DOI] [PubMed] [Google Scholar]
- 18. Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Invest Ophthalmol Vis Sci. 1999;40:2706–2712 [PubMed] [Google Scholar]
- 19. Ikeda S, Shiva N, Ikeda A, et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet. 2000;9:155–163 [DOI] [PubMed] [Google Scholar]
- 20. Hagstrom SA, North MA, Nishina PL, Berson EL, Dryja TP. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176 [DOI] [PubMed] [Google Scholar]
- 21. Banerjee P, Kleyn PW, Knowles JA, et al. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet. 1998;18:177–179 [DOI] [PubMed] [Google Scholar]
- 22. Paloma E, Hjelmqvist L, Bayes M, et al. Novel mutations in the TULP1 gene causing autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:656–659 [PubMed] [Google Scholar]
- 23. Gu S, Lennon A, Li Y, et al. Tubby-like protein-1 mutations in autosomal recessive retinitis pigmentosa. Lancet. 1998;351:1103–1104 [DOI] [PubMed] [Google Scholar]
- 24. Abbasi AH, Garzozi HJ, Ben-Yosef T. A novel splice-site mutation of TULP1 underlies severe early-onset retinitis pigmentosa in a consanguineous Israeli Muslim Arab family. Mol Vis. 2008;14:675–682 [PMC free article] [PubMed] [Google Scholar]
- 25. Hagstrom SA, Adamian M, Scimeca M, Pawlyk BS, Yue G, Li T. A role for the Tubby-like protein 1 in rhodopsin transport. Invest Ophthalmol Vis Sci. 2001;42:1955–1962 [PubMed] [Google Scholar]
- 26. Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci. 1999;40:2795–2802 [PubMed] [Google Scholar]
- 27. Grossman GH, Pauer GJ, Narendra U, Peachey NS, Hagstrom SA. Early synaptic defects in tulp1−/− mice. Invest Ophthalmol Vis Sci. 2009;50:3074–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grossman GH, Pauer GJT, Narendra U, Hagstrom SA. Tubby-like protein 1 (Tulp1) is required for normal photoreceptor synaptic development. Adv Exp Med Biol. 2010;664:89–96 [DOI] [PubMed] [Google Scholar]
- 29. Kapeller R, Moriarty A, Strauss A, et al. Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J Biol Chem. 1999;274:24980–24986 [DOI] [PubMed] [Google Scholar]
- 30. Bateman A, Finn RD, Sims PJ, Wiedmer T, Biegert A, Soding J. Phospholipid scramblases and Tubby-like proteins belong to a new superfamily of membrane tethered transcription factors. Bioinformatics. 2009;25:159–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125 [DOI] [PubMed] [Google Scholar]
- 32. Caberoy NB, Maiguel D, Kim Y, Li W. Identification of tubby and tubby-like protein 1 as eat-me signals by phage display. Exp Cell Res. 2010;316:245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caberoy NB, Zhou Y, Li W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010;29:3898–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ikeda A, Naggert JK, Nishina PM. Genetic modification of retinal degeneration in tubby mice. Exp Eye Res. 2002;74:455–461 [DOI] [PubMed] [Google Scholar]
- 35. Ikeda A, Zheng QY, Rosenstiel P, et al. Genetic modification of hearing in tubby mice: evidence for the existence of a major gene (moth1) which protects tubby mice from hearing loss. Hum Mol Genet. 1999;8:1761–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ikeda A, Zheng QY, Zuberi AR, Johnson KR, Naggert JK, Nishina PM. Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet. 2002;30:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins (Review). Genome Biol. 2006;7:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xi Q, Pauer GJ, Ball SL, et al. Interaction between the photoreceptor-specific tubby-like protein 1 and the neuronal-specific GTPase dynamin-1. Invest Ophthalmol Vis Sci. 2007;48:2837–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goring HH, Terwilliger JD. Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters. Am J Hum Genet. 2000;66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Danciger M, Ogando D, Yang H, et al. Genetic modifiers of retinal degeneration in the rd3 mouse. Invest Ophthalmol Vis Sci. 2008;49:2863–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chesler EJ, Miller DR, Branstetter LR, et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137 [DOI] [PubMed] [Google Scholar]
- 43. Iraqi FA, Churchill G, Mott R. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm Genome. 2008;19:379–381 [DOI] [PubMed] [Google Scholar]
- 44. Kaplan J, De Domenico I, Ward DM. Chediak-Higashi syndrome. Curr Opin Hematol. 2008;15:22–29 [DOI] [PubMed] [Google Scholar]
- 45. Robison WG, Jr, Kuwabara T, Cogan DG. Lysosomes and melanin granules of the retinal pigment epithelium in a mouse model of the Chédiak–Higashi syndrome. Invest Ophthalmol. 1975;14:312–317 [PubMed] [Google Scholar]
- 46. Runkel F, Bussow H, Seburn KL, et al. Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25. Mamm Genome. 2006;17:203–210 [DOI] [PubMed] [Google Scholar]
- 47. BenEzra D, Mengistu F, Cividalli G, Weizman Z, Merin S, Auerbach E. Chediak-Higashi syndrome: ocular findings. J Pediatr Ophthalmol Strabismus. 1980;17:68–74 [DOI] [PubMed] [Google Scholar]
- 48. Sayanagi K, Fujikado T, Onodera T, Tano Y. Chediak-Higashi syndrome with progressive visual loss. Jpn J Ophthalmol. 2003;47:304–306 [DOI] [PubMed] [Google Scholar]
- 49. Kong L, Li F, Soleman CE, et al. Bright cyclic light accelerates photoreceptor cell degeneration in tubby mice. Neurobiol Dis. 2006;21:468–477 [DOI] [PubMed] [Google Scholar]
- 50. Koulen P, Fletcher EL, Craven SE, Bredt DS, Wassle H. Immunocytochemical localization of the postsynaptic density protein PSD-95 in the mammalian retina. J Neurosci. 1998;18:10136–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xi Q, Pauer GJ, Marmorstein AD, Crabb JW, Hagstrom SA. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest Ophthalmol Vis Sci. 2005;46:4754–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]