Background: Oxygen is required for three enzymatic steps in tetrapyrrole biosynthesis.
Results: A transcriptional regulator gene was found by genome resequencing of a mutant of Synechocystis sp. PCC 6803.
Discussion: Sll1512 (ChlR) is a MarR-type transcriptional regulator involved in induction of genes in response to low-oxygen conditions.
Significance: Maintaining the chlorophyll level constant under hypoxia is important for cyanobacteria to survive.
Keywords: Cyanobacteria, Genome Structure, Hypoxia, Photosynthetic Pigments, Transcription Regulation, Chlorophyll, Tetrapyrrole
Abstract
Oxygen is required for three enzyme reactions in chlorophyll and bilin biosynthesis pathways: coproporphyrinogen III oxidase (HemF), heme oxygenase (HO1), and Mg-protoporphyrin IX monomethylester cyclase (ChlAI). The cyanobacterium Synechocystis sp. PCC 6803 has alternative enzymes, HemN, HO2, and ChlAII, to supply chlorophyll/bilins even under low-oxygen environments. The three genes form an operon, chlAII-ho2-hemN, that is induced in response to low-oxygen conditions to bypass the oxygen-dependent reactions. Here we identified a transcriptional regulator for the induction of the operon in response to low-oxygen conditions. A pseudorevertant, Δho1R, was isolated from a HO1-lacking mutant Δho1 that is lethal under aerobic conditions. Δho1R grew well even under aerobic conditions. In Δho1R, HO2 that is induced only under low-oxygen conditions was anomalously expressed under aerobic conditions to complement the loss of HO1. A G-to-C transversion in sll1512 causing the amino acid change from aspartate 35 to histidine was identified as the relevant mutation by resequencing of the Δho1R genome. Sll1512 is a MarR-type transcriptional regulator. An sll1512-lacking mutant grew poorly under low-oxygen conditions with a remarked decrease in Chl content that would be caused by the suppressed induction of the chlAII and hemN genes in Chl biosynthesis under low-oxygen conditions. These results demonstrated that Sll1512 is an activator in response to low-oxygen environments and that the D35H variant becomes a constitutive activator. This hypothesis was supported by a gel shift assay showing that the Sll1512-D35H variant binds to the DNA fragment upstream of the operon. We propose to name sll1512 chlR.
Introduction
Cyanobacteria are photosynthetic prokaryotes that adapt and thrive in a wide variety of environments on the Earth (1). They perform oxygenic photosynthesis like plants by extracting electrons from water with generation of oxygen as a byproduct. Cyanobacteria also use oxygen for respiration to produce energy in the dark (2). The oxygen levels greatly change in the field environment from hyperoxic to almost anaerobic conditions (3, 4). Even in aerobic environments, cyanobacterial cells are exposed to anaerobic conditions in the night on a daily basis because oxygen is quickly consumed by respiration in the cells (5, 6). Thus, adaptation of anaerobiosis is important for cyanobacteria to survive.
Chl and bilin are important tetrapyrrole pigments in photosynthesis. Their biosynthetic pathways from glutamate to protoporphyrin IX is the same as that of heme (7, 8). The Chl-specific pathway starts from the insertion of a magnesium ion into protoporphyrin IX by Mg-chelatase. Protoporphyrin IX is converted to heme by the insertion of ferrous ion catalyzed by ferrochelatase. Bilin biosynthesis starts from the oxygen-dependent cleavage of heme catalyzed by heme oxygenase (HO)2. Photosynthetic organisms, including cyanobacteria, should have elaborate mechanisms to regulate the biosynthetic pathways of Chl and bilin to avoid photooxidative damage caused by the accumulation of free pigments and their intermediates. However, in plants, the mechanisms largely remains to be solved while some plausible mechanisms are proposed (9, 10).
In the biosynthetic pathways of Chl and bilin pigments, there are at least three oxygen-dependent reactions (7, 8). One is coproporphyrinogen III oxidation catalyzed by oxygen-dependent coproporphyrinogen III oxidase. In this reaction, oxygen is required for oxidative decarboxylation (11). The second is the heme cleavage reaction to form biliverdin IXα catalyzed by HO (12, 13). Third is the fifth ring (E-ring) formation of chlorophyll, catalyzed by Mg-protoporphyrin IX monomethylester cyclase (ChlAI) (14). In this reaction, Mg-protoporphyrin IX monomethylester is converted to protochlorophyllide through oxygenation of the C13-methylpropionate and ring closure reactions with the incorporation an oxygen atom as the oxo group at the C131 position. In the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis 6803), these oxygen-dependent reactions are bypassed by three sets of enzymes (12). An analogous coproporphyrinogen III oxidase, HemN, catalyzes the reaction in an oxygen-independent manner instead of HemF (11). Low-oxygen-specific isoforms, HO2 and ChlAII, operate together with the constitutive isoforms, HO1 and ChlAI, respectively (12, 14). The three low-oxygen-specific genes form a small operon, chlAII-ho2-hemN, and are induced in response to low-oxygen conditions. This operon is also conserved in another cyanobacterium Synechococcus sp. PCC 7002 (Synechococcus 7002) and the transcription of the operon is also induced by low-oxygen conditions as well as Synechocystis 6803 (15). However, the low-oxygen induction mechanism is still elusive in cyanobacteria. Recently Summerfield et al. (16) reported that Hik31-histidine kinase plays a role as a negative regulator for expression of some genes in response to low oxygen in Synechocystis 6803. However, because the expression profile of the chlAII-ho2-hemN operon is not altered in the ΔHik31 mutant, this operon is regulated by a Hik31-independent pathway that has remained unknown.
Here we report a MarR-type transcriptional regulator Sll1512 involved in the low-oxygen induction of the chlAII-ho2-hemN operon through genome resequencing of a mutant with a Next Generation Sequencer SOLiD. Sll1512 would play a role as an activator for transcription of the operon in response to low-oxygen environments.
EXPERIMENTAL PROCEDURES
Cyanobacterial Strains and Growth Conditions
Synechocystis sp. PCC 6803 and its derivative strains used in this study were cultivated in BG-11 medium as described (12). To prepare cells grown under aerobic and low-oxygen conditions, we incubated agar plates in air and in an anaerobic jar (BBL GasPak anaerobic systems, BD Biosciences; gas-generating kit anaerobic system, Oxoid, Basingstoke, Hants, UK), respectively. Agar plates under both conditions were illuminated at light intensity of 50 μmolphoton m−2s−1 (FLR40SW, Hitachi, Tokyo, Japan).
In the process of genome resequencing, our wild-type “YF” was found to have a genome sequence closer to PCC strains (PCC-N and PCC-P; 17) rather than GT strains (GT-Kazusa, GT-S, and GT-I 17,18) because three SNPs in the YF genome were identical to those of PCC strains (supplemental Table S1).
Determination of Chl and PC Contents
The concentration of Chl in the 90% methanol extracts from the cells was determined by absorption at 665 nm (model V-550, JASCO, Hachioji, Japan) (19). The amount of Chl was normalized by the optical density at 730 nm (A730; UV1700, Shimadzu, Kyoto, Japan) of the cell suspension used for the extraction. PC contents were determined as described (12). The protein concentration was determined by a dye-binding assay (protein assay, Bio-Rad) using bovine serum albumin as a standard.
Preparation of RNA and RT-PCR
Cells were grown on the agar plates for 7 days, collected, and harvested by centrifugation at 4 °C. RNA preparation and the following RT-PCR procedures were described previously (14). The isolated total RNA was used for the synthesis of cDNA with ReverTra Ace (Toyobo, Osaka, Japan) and reverse primers for RT-PCR (supplemental Table S2) according to the manufacturer's manual. Thus, obtained cDNA fraction was used as the template for PCR amplification with the specific primers (supplemental Table S2).
Solid DNA Sequencing
Sequencing runs were done using cycled ligation sequencing on a SOLiD analyzer (Invitrogen). Ten micrograms of purified genomic DNA of wild-type, Δho1, and Δho1R were sheared into ∼125-bp short fragments with the Covaris S2 system (Covaris, Woburn, MA). The sheared DNA fragments were blunted by Next NR enzyme (New England Biolabs, Beverly, MA) and then purified using a Pure Link PCR purification kit (Invitrogen). The purified DNA fragments were ligated to short SOLiD P1 and P2 adapters (supplemental Table S2) by T4 Ligase (Invitrogen). Adapter-ligated DNA was purified using the Pure Link PCR purification kit. These DNA fragments were nick-translated and amplified by PCR (cycle number 3, supplemental Table S2) and then purified using the Pure Link PCR purification kit. The purified PCR products were separated by SOLiD size selection gel (2% agarose gel, Invitrogen) and collected the fragments corresponding 150- to 200-bp DNA. These DNA libraries were quantified by quantitative PCR using a SOLiD TaqMan probe.
In preparation for sequencing, the DNA fragments were clonally amplified by emulsion PCR using beads with P1 primer covalently attached to the surface. Emulsions were broken with butanol, and emulsion PCR beads were enriched by hybridization with P2-coated capture beads (Life Technology). These beads were added dTs at the 3′ end in the presence of terminal transferase to connect on the slide glass. About 60 million beads were deposited onto one-fourth of a glass surface of a 25 mm × 75 mm SOLiD slide. The slide was loaded onto a SOLiD instrument, and the 50-base sequences were obtained according to the manufacturer's protocol.
SNP Analysis
DNA sequence data (csfasta) were analyzed using Takeru II (NABE International, Tsukuba, Japan). Sequence data were mapped to the genome sequence of Synechocystis 6803 obtained from KEGG using “Map Data” of BioScope (v. 1.2). From them, we gained SNPs data using SNPs (supplemental Fig. S2).
Transformation Assay for Identification of SNPs
The sll1512 (2,065 bp) and slr0887 (2,098 bp) fragments containing an SNP site were amplified by PCR using the genomic DNA of Δho1 and Δho1R by a standard thermal cycle (KOD-plus DNA polymerase, Toyobo). The amplified DNA fragments were separated by 1% agarose gel and purified by Wizard SV gel and PCR clean-up kit (Promega, Madison, WI). During preparation of the sll1512 fragment, we found another nonspecific fragment with almost the same size as the sll1512 fragment. We removed it by digesting a mixture of these fragments with BstXI. The resultant 1,942-bp fragment was purified by agarose gel electrophoresis. The cell suspension of Δho1 (300 μl, A730 = 1) was spread on BG-11-agar plate containing kanamycin. The DNA fragments (2–60 ng/μl, 5 μl) were spotted on the plate. The plate was incubated under low-oxygen conditions for 3 days and then transferred under aerobic conditions for 2 weeks.
Construction of Plasmids for sll1512 Disruption and Transformation
Plasmids for targeted gene disruption were constructed as follows (Fig. 3A, supplemental Table S2). A DNA fragment (2,338 bp) containing sll1512 was amplified by PCR from genomic DNA of Synechocystis 6803 with the primer pairs (supplemental Table S2) by a standard thermal cycle (KOD-plus DNA polymerase). The amplified DNA fragments were cloned into the BamHI site of pUC118. The recombinant plasmid carrying sll1512 was digested by HpaI and BsmI (Fig. 3A). A spectinomycin-resistant cartridge amplified from pJN3 (20) was inserted into the blunted HpaI and BsmI sites to construct the sll1512-disrupted plasmid. Synechocystis 6803 was transformed with the plasmid constructed as described (12).
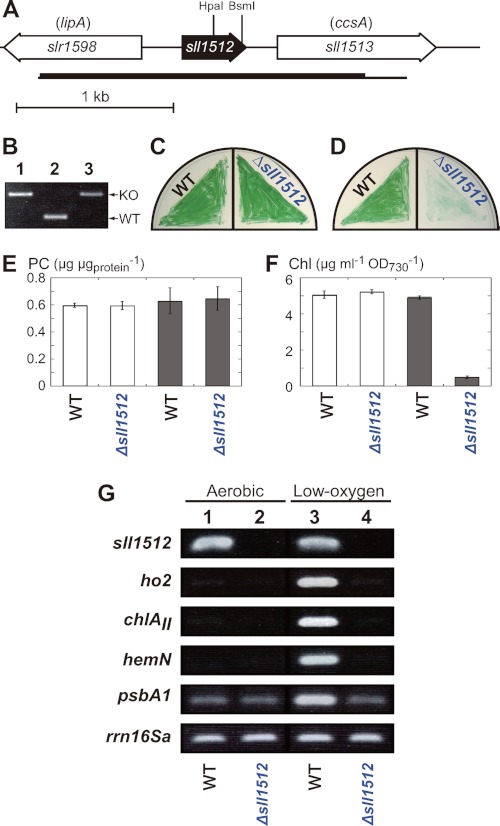
FIGURE 3.
Characterization of Δsll1512. A, gene arrangement of sll1512 in the genome of Synechocystis 6803. The chromosomal region amplified by PCR to construct plasmids for targeted gene disruption is shown by a horizontal thin bar. The region between the HpaI and BsmI sites was replaced with a spectinomycin resistance cartridge in the Δsll1512 mutant. B, colony PCR analysis to confirm the gene replacement in Δsll1512. A chromosomal region (a horizontal thick bar in A) was amplified by PCR from single colonies on the selective agar plates (lane 3). Longer and shorter fragments correspond to the mutant (KO) and the WT copies of sll1512, respectively. As controls, the mutant (lane 1) and the wild-type (lane 2) fragments were amplified from the plasmid used for transformation and the wild-type genome, respectively. C and D, comparison of photoautotrophic growth of the wild-type and Δsll1512. Cells were cultivated under aerobic (C) and low-oxygen (D) conditions for 5 days. E and F, comparison of PC (E) and Chl (F) contents in the wild-type and Δsll1512. PC (E) and Chl (F) were extracted from cells grown photoautotrophically under aerobic (white bars) and low-oxygen (gray bars) conditions on the agar plates for 7 days. The data are mean ± S.D. for three independent experiments. G, RT-PCR analysis for the transcript levels of sll1512, ho2, chlAII, hemN, and psbA1. Total RNA was isolated from the wild-type (lanes 1 and 3) and Δsll1512 (lanes 2 and 4) cells. Aerobic samples (lanes 1 and 2) were grown under low-oxygen conditions for 5 days and then transferred under aerobic conditions and grown for 2 days. Low-oxygen samples (lanes 3 and 4) were grown for 7 days under low-oxygen conditions. The housekeeping gene rrn16Sa was used as a control. Cycle numbers of RT-PCR for sll1512 was 25, and those for the other genes are the same as in Fig. 1.
Overexpression of Sll1512 in Escherichia coli and Purification
The plasmids for overexpression of Sll1512 (wild type) and the D35H variant were constructed as follows. The BstXI-digested 1,942-bp fragments from Δho1 and Δho1R that were prepared for the transformation assay were used as the template to amplify the coding regions of Sll1512 and Sll1512-D35H, respectively, by PCR in a standard thermal cycle (supplemental Table S2). After digestion with BsaI, the amplified DNA fragments were cloned into the BsaI site of pASK-IBA5plus (IBA, Göttingen, Germany) to form pASK5-Sll1512 and pASK5-Sll1512D35H. The N-terminal sequence of Strep-tagged Sll1512 is MASWSHPQFEKGA followed by the second amino acid residue (Glu) of the native sequence. E. coli JM105 transformants harboring the plasmids were cultivated in LB (1% Trypton, 0.5% Yeast extract, 1% NaCl) medium (5 ml) containing 100 μg/ml ampicillin for 6 h with vigorous shaking at 37 °C. Overexpression of the protein was induced upon the addition of anhydrotetracycline (final 200 ng/ml) 2 h after inoculation. E. coli cells harvested by centrifugation were washed in 1 ml of 0.9% (w/v) NaCl, resuspended in lysis buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.1 mm PMSF) and disrupted by sonication (Sonifier model 250; Branson, Danbury, CT). The soluble fractions were obtained by centrifugation of the sonicates (15,000 rpm, 4 °C, 10 min). Affinity purification with Strep-Tactin was carried out as described (21).
Gel Shift Assay
Two DNA fragments upstream of chlAII and psbA1 (312 bp from the position −1 to −312 of chlAII, 297 bp from the position −1 to −297 of psbA1) were amplified by PCR from genomic DNA of Synechocystis 6803 (supplemental Table S2). DNA fragments were labeled at both termini with 32P and used as the probe for gel retardation assays. Gel retardation assays were performed essentially as described (22). Proteins were incubated with probes (0.1 nm) in 20 μl of binding buffer (20 mm HEPES-KOH, pH 8.0, 20 mm Tris-HCl, 1 mm EDTA, 1 mm DTT, 60 mm KCl, 12% glycerol, 0.1 mg ml−1 BSA, 0.05 mg ml−1 poly(deoxyinosinic-deoxycytidylic) acid sodium salt) for 20 min at 30 °C. The mixtures were separated by a native 4% polyacrylamide gel. Gels were dried, and the signals were detected by autoradiography.
RESULTS
Isolation of a Pseudorevertant Δho1R and Characterization
Previously, we isolated a ho1-deficient mutant, Δho1, that cannot survive under aerobic conditions but can grow under low-oxygen conditions (12). A single colony survived on an agar plate incubated under aerobic conditions where all other Δho1 cells died. We picked up the colony as a pseudorevertant “Δho1R” for further study. We confirmed that Δho1R could grow under aerobic conditions where the original strain Δho1 could not (Fig. 1A). The growth of Δho1R under low-oxygen conditions was equal to that of the wild type (Fig. 1B).
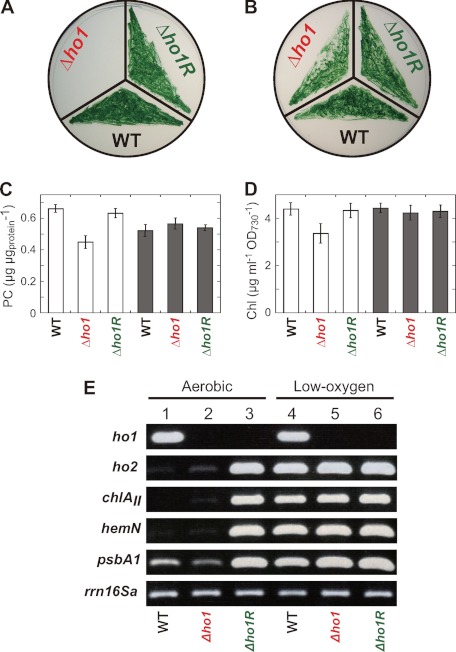
FIGURE 1.
Characterization of a pseudorevertant Δho1R. Comparison of photoautotrophic growth (A and B) and PC (C) and Chl (D) contents. Cells grew under aerobic (A) and low-oxygen (B) conditions for 7 days. PC (C) and Chl (D) were extracted from cells grown under low-oxygen conditions for 5 days followed by 2-day aerobic incubation (white bars) and from cells grown under low-oxygen conditions on the agar plates for 7 days (gray bars). The data are mean ± S.D. for three independent experiments. E, RT-PCR analysis for the transcript levels of ho1, ho2, chlAII, hemN, and psbA1. Total RNA was isolated from the wild-type (lanes 1 and 4), Δho1 (lanes 2 and 5), and Δho1R (lanes 3 and 6) cells. Aerobic samples (lanes 1-3) were grown under low-oxygen conditions for 5 days and then transferred under aerobic conditions and grown for 2 days. Low-oxygen samples (lanes 4-6) were grown under low-oxygen conditions for 7 days. The housekeeping gene rrn16Sa was used as a control. Cycle numbers of RT-PCR were 6 and 25 for rrn16Sa and the others, respectively.
PC and Chl contents in Δho1R were determined (Fig. 1, C and D). PC is a dominant phycobilisome protein of Synechocystis 6803. The PC and Chl contents in aerobically incubated Δho1 were significantly decreased as shown previously (12). The levels of PC and Chl in Δho1R were the same as those of the wild type under aerobic conditions.
After we confirmed the absence of ho1 in Δho1R, we examined the transcript levels of the low-oxygen inducible operon consisting of chlAII, ho2, and hemN in Δho1R (11, 12, 14) by RT-PCR (Fig. 1E). Surprisingly, the ho2 transcript level in aerobically grown Δho1R was almost comparable with that in Δho1R grown under low-oxygen conditions (Fig. 1E, lane 3), which was in contrast to the expression profiles of ho2 in the wild type and Δho1. This change in the transcript levels was also observed in the chlAII and hemN. In Δho1R, all three genes in the operon were expressed in a constitutive manner in both aerobic and low-oxygen conditions. This result suggested that the constitutive expression of ho2 restores the defect of ho1 to allow growth under aerobic conditions. This is consistent with the observation of the ho2 overexpression by an artificial promoter restored the Δho1 growth defect (12). Summerfield et al. (23) reported five gene clusters, including the chlAII operon, that are induced in response to low-oxygen conditions in Synechocystis 6803. We examined whether the expression patterns of 10 representative genes (chlG, por, ctpA, hoxE, flv4, psbA1, ppc, ppsA, cpcG2, and atpF) are altered in Δho1R by RT-PCR (supplemental Fig. S1 and Fig. 1E). The mRNA levels of most genes except for flv4 and psbA1 did not change significantly between wild-type cells grown under aerobic and low-oxygen conditions, and the expression profile in Δho1R was almost the same as that in the wild type. The flv4 transcript was not detected in any cells grown under low-oxygen conditions. psbA1 encoding an isoform of the D1 protein (D1′) of photosystem II showed a clear low-oxygen induction profile in the wild type and Δho1, and the profile was changed to a constitutive one in Δho1R like the chlAII operon (Fig. 1E).
Genome Resequencing of Δho1R and Identification of the Mutation Site
To identify the relevant mutation(s) responsible for the growth restoration of Δho1R, we carried out whole genome resequencing of Δho1R, Δho1, and the wild type by a next-generation sequencer, SOLiD. Comparison with the Genome Database of Cyanobacteria, CyanoBase, revealed 12 SNPs that were common in all the three strains (supplemental Table S1). Six of these 12 SNPs were recently reported to be sequence errors in the data base (18), three are identical SNPs found in PCC strains (17), and the remaining three appear to be unique to our wild-type strain YF. In addition to the common 12 SNPs, we found two SNPs unique to Δho1R. One was a G-to-C transversion (at 462,452) in sll1512 that causes an amino acid substitution from Asp-35 to His, and the other was a C-to-A transversion (at 1,153,565) in slr0887 causing an amino acid replacement of Pro-92 to Thr (supplemental Fig. S2 and Table S1).
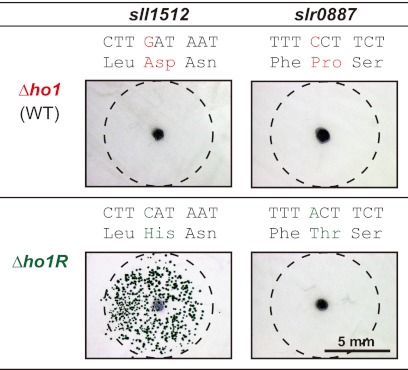
We performed a transformation analysis to identify which of the two Δho1R-specific SNPs is responsible for the growth recovery of Δho1. DNA fragments of about 2 kb containing the SNPs in sll1512 and slr0887 were amplified by PCR from the genomes of Δho1 and Δho1R to transform Δho1. Many colonies appeared on only one sector transformed with the sll1512 fragment from Δho1R, whereas no colonies appeared on the other three sectors (Fig. 2). This result clearly indicated that the D35H mutation in sll1512 is responsible for the recovery of growth in Δho1 under aerobic conditions.
FIGURE 2.
The transformation analysis to identify the mutation site important for Δho1R. Left and right panels show the results of the transformation assay with DNA fragments containing sll1512 and slr0887, respectively. DNA fragments amplified from the genome of Δho1 and Δho1R were used in the top and bottom panels, respectively. We confirmed that Δho1 carries the wild-type copies of sll1512 and slr0887. The black dots at the center indicate the points where DNA fragments were spotted (100 ng), and the dashed circles indicate the area where the DNA solutions were spread. The sequences above the pictures show the mutation sites in the Δho1R. Red and green characters correspond to wild-type and Δho1R sequences, respectively.
Isolation of an sll1512-lacking Mutant and Phenotype Analysis
sll1512 encodes a protein with 135 amino acid residues (Fig. 3A). Although Sll1512 is annotated as a hypothetical protein in the cyanobacterial genome databases CYORF and CyanoBase, they suggested that 11 cyanobacteria carry probable orthologs of sll1512 (supplemental Fig. S3). A BLAST search indicated that the amino acid sequence of Sll1512 shows significant similarity to those of a MarR family transcriptional regulator from a variety of prokaryotes. Among MarR family proteins with known functions HxlR from Bacillus subtilis showed the highest similarity score except for many cyanobacterial homologues (supplemental Fig. S3). B. subtilis HxlR activates the expression of the hxlAB operon for formaldehyde fixation in response to the presence of formaldehyde (24).
To elucidate the physiological function of Sll1512 in Synechocystis 6803, we isolated a mutant, Δsll1512, that lacked the sll1512 gene (Fig. 3, A and B). Δsll1512 grew poorly under low-oxygen conditions, although it grew as vigorously as the wild type under aerobic conditions (Fig. 3, C and D). We determined the contents of PC and Chl (E and F). The PC content of the Δsll1512 cells was the same as that of the wild type under both aerobic and low-oxygen conditions (Fig. 3E). On the other hand, the Chl content of Δsll1512 grown under low-oxygen conditions was greatly decreased to about 10% of the wild type, whereas the Chl content of aerobically grown cells was the same as that of the wild type (Fig. 3F). These results indicated that Sll1512 plays an important role for growth under low-oxygen conditions. The significant growth retardation under low-oxygen conditions in Δsll1512 would be caused by the drastic decrease in Chl.
We examined the effects of the sll1512-defeciency on the transcript levels of the chlAII-ho2-hemN operon (Fig. 3G). Interestingly, the transcripts of all the three genes in the operon were not detected at all in the Δsll1512 cells grown even under low-oxygen conditions. The transcript for psbA1 was detected in a low level in both wild-type and Δsll1512 cells grown aerobically. Although the transcript level was greatly increased in the wild-type cells grown under low-oxygen conditions, it was kept low in the Δsll1512 cells grown under low-oxygen conditions. These results suggested that Sll1512 is essential for the transcriptional activation of these genes under low-oxygen conditions. The transcript profile is in contrast to the constitutive expression of these genes in the Δho1R. In Δho1R an Sll1512-D35H variant would behave as a constitutively active form resulting in the aberrant expression of the chlAII-ho2-hemN operon and psbA1 under aerobic conditions.
Gel Shift Assay of Sll1512 and the D35H Variant
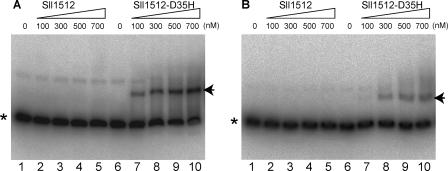
To confirm the DNA binding ability of Sll1512, we performed a gel shift assay using two DNA fragments of the 312-bp chlAII and 297-bp psbA1 promoters and Sll1512 or the D35H variant (Fig. 4). The genes encoding Sll1512 and the D35H variant were overexpressed in E. coli, and the proteins (theoretical molecular mass, 16.4 kDa) were purified to almost homogeneity by affinity chromatography (supplemental Fig. S4A). There was no specific gel shift with the wild-type Sll1512 protein even at the highest concentration (700 nm) in either fragment. In contrast, an obvious mobility shift in both promoter fragments was detected in the assay with the D35H variant. Because all procedures, from crude extract preparation and protein purification to the assay, were carried out under normal aerobic conditions, the Sll1512 protein would be kept inactive without binding to the operator sequence. The binding features of Sll1512 and the D35H variant were consistent with our hypothesis (Fig. 5), in which the D35H variant is a constitutively active form to bind to the operator region under either aerobic or low-oxygen conditions.
FIGURE 4.
Gel mobility shift assays showing retardation of the 32P-labeled DNA fragments of chlAII (A) and psbA1 (B) promoters by Sll1512 and Sll1512-D35H in polyacrylamide gel. Purified Sll1512 (lanes 1–5) and Sll1512-D35H (lanes 6–10) were added to the reaction mixtures to give the indicated concentrations of protein. Formed DNA-protein complexes are shown by the arrows, and asterisks indicate free probes.
FIGURE 5.
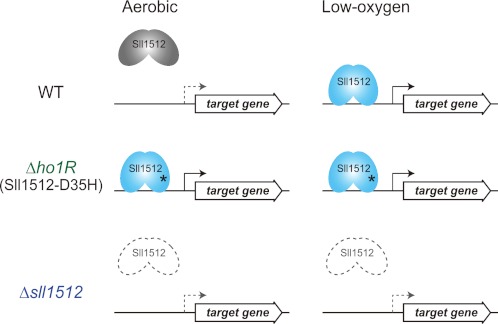
A working model of Sll1512 (ChlR) in Synechocystis 6803. In the wild-type cells, Sll1512 is maintained as an inactive form (gray) under aerobic conditions, so the target gene is not expressed (dotted gray arrows). When cells are exposed to low-oxygen conditions, Sll1512 is converted to an active form (light blue), and the target gene is transcribed (solid arrows). In the pseudorevertant Δho1R, Sll1512 is fixed as an active form even under aerobic conditions because of the point mutation of D35H (asterisk), and the target gene is constitutively transcribed. In Δsll1512, there is no Sll1512 in the cell, and the target gene is not transcribed.
DISCUSSION
The MarR family is one of the major transcriptional regulator families in prokaryotes and regulates a wide variety of cellular processes such as antibiotic resistance; virulence; oxidative stress; and environmental sensing of aromatic compounds, formaldehyde, and organic hydrogen peroxides (25, 26). In this work, we identified a MarR-family transcriptional regulator for the low-oxygen induction of genes for Chl and bilin biosynthesis in cyanobacteria for the first time. We propose to name it sll1512 “chlR.”
Although cyanobacteria evolve oxygen by photosynthesis in the light like plants, they face hypoxia in a diurnal cycle because the oxygen level drops quickly after the onset of night (6). Many cyanobacteria also thrive in anoxic habitats such as hypolimnia, lake sediments, and soil (27). The transcriptional activator ChlR enables the cyanobacterium to produce Chl and bilin pigments even under low-oxygen environments. D1′ encoded by psbA1 is a special D1 isoform that is expressed only under low-oxygen conditions. In the field environment low-oxygen conditions are often associated with a high concentration of H2S damaging to PSII. D1′ is thought to help maintain PSII by rapid turnover of damaged conventional D1 isoforms under H2S-rich conditions (23). Thus, it is reasonable that the psbA1 is coregulated with the operon for Chl biosynthesis by ChlR.
MarR (multiple antibiotic resistance regulator) family regulatory proteins are small proteins consisting of 120- to 150-amino acid residues and form a homodimer to recognize specific palindromic nucleotide sequences upstream of the target genes (25). In most cases, MarR proteins function as repressors. The binding of a MarR regulatory protein on the operator region inhibits the transcription of the target genes by RNA polymerase, resulting in repression. Some MarR family members play a role as an activator. Such MarR members bind to a region close to the operator to activate the RNA polymerase to stimulate the transcription. We concluded that ChlR is an activator-type MarR member on the basis of the result of RT-PCR in Δsll1512 (Fig. 3G).
The chlAII-ho2-hemN operon is conserved in Synechococcus 7002, and the expression is induced under low-oxygen conditions (15). Interestingly, a MarR-type transcriptional regulator (SYNPCC7002_A1993) is encoded just upstream of the operon in the opposite direction, which is a typical gene arrangement of MarR transcriptional regulator and the target genes. Because SYNPCC7002_A1993 showed about 67% identity to ChlR (supplemental Fig. S3), it may play a role as an activator of the operon in response to low-oxygen conditions like ChlR in Synechocystis 6803. We compared the upstream sequences of the chlAII operon and the psbA1 gene from Synechocystis 6803 and the chlAII operon from Synechococcus 7002 to find a consensus motif that is recognized by ChlR. A palindromic sequence of TT(A/C)CC-N3/4-GG(T/A)AA was found to be conserved commonly in the three regions (supplemental Fig. S5). This motif would be a binding site for ChlR. This consensus sequence is very similar to that (TTAC-N5-GTAA) of CatR, which is a repressor of the catDE operon encoding catechol 2,3-dioxygenase for quinone resistance in B. subtilis (28).
Crystal structures of many MarR family members have been reported in reduced, ligand-bound, and DNA-bound forms, starting with the crystal structure of MarR from E. coli (29), showing a common architecture with a domain for DNA binding called winged helix-turn-helix fold (25). Some MarR family members, including HxlR, constitute a group of regulatory proteins for oxidative stress responses. OhrRs in B. subtilis and Xanthomonas campestris have been most extensively studied as MarR members involved in oxidative stress sensing (30, 31). OhrR is a repressor of the gene ohrA encoding a thiol peroxidase that degrades organic hydroperoxides. They commonly possess oxidation-sensitive Cys residues in the N- and/or C-terminal region(s) by which they are divided into two subtypes, one-Cys-type and two-Cys-type (31). OhrR of B. subtilis represents a one-Cys-type regulator. Organic hydroperoxides react with a commonly conserved single Cys in N-terminal part, giving rise to Cys sulfenic acid (Cys-SOH) followed by rapid S-thiolation, resulting in dissociation from the operator to induce the ohrA gene expression (32). A representative of two-Cys-type is OhrR of X. campestris (31). The oxidation of a Cys (Cys-22) in the N-terminal part leads to the reaction with the other Cys in the C-terminal part (Cys-127′) in the other protomer of the dimer, forming a disulfide bond (31). This S-S bond formation inactivates OhrR activity as a repressor by attenuation of the DNA binding ability.
ChlR appears to be a one-Cys-type regulator because only one Cys (Cys-25) of five Cys is conserved among all 12 cyanobacterial homologues (supplemental Fig. S3). In our hypothesis, ChlR may have a modified Cys like sulfenic acid or S-thiolated found in OhrR under aerobic conditions, and the modified ChlR would be activated by the conversion of the modified Cys to reduced Cys with free thiol upon exposure to low-oxygen environments. Because the wild-type ChlR did not bind to DNA (Fig. 4), the ChlR protein purified from E. coli would be an inactive form with the modified Cys residue. We performed a gel shift assay with the wild-type ChlR in the presence of dithiothreitol (10 mm) or sodium dithionite (10 mm). However, no detectable mobility shift was detected (supplemental Fig. S4B). How the inactive ChlR is converted to an active form that binds to DNA like the D35H variant remains unknown.
In the crystal structure of OhrR from B. subtilis, the active Cys-15 is located adjacent to two Tyr residues (Tyr-29′ and Tyr-40′) and Met-25′ that are from the other protomer (33). In the sequence alignment, Asp-35 of ChlR corresponds to Met-25 of OhrR (supplemental Fig. S3), suggesting that Asp-35′ is located in the vicinity of the probable redox active Cys-25 in ChlR. It is possible that the substitution of Asp-35 with His affects the conformation of ChlR to be kept as the active DNA-binding form even though Cys-25 is modified under aerobic conditions or that the Cys-25 modification itself is inhibited in ChlR-D35H, resulting in the constitutive active form.
Δsll1512 showed a severe growth defect and drastic decrease of Chl content under low-oxygen conditions (Fig. 3, D and F). In contrast, the individual mutants of chlAII, ho2 and hemN, which we isolated previously, showed only slight growth retardation with no such significant decrease in the Chl contents (11, 12, 14). The poor growth phenotype of Δsll1512 would be caused by the defect of some other unidentified genes under low-oxygen conditions. Further study is needed to identify the whole set of the ChlR regulon.
Microarray analysis under low-oxygen conditions in Synechocystis 6803 has been reported (16, 23). Summerfield et al. (16) found that Hik31 histidine kinase is involved in negatively regulating gene expression of a large set of genes in response to low oxygen. Genes regulated by the Hik31-depedent system include subunits of photosystems I and II, phycobiliproteins, chaperons, and ATPase subunits. The other low-oxygen inducible genes, including chlAII, ho2, hemN, and psbA1, are regulated in a Hik31-independent system that has remained unknown until this work. Our results clearly demonstrated that ChlR is one of the key players in the Hik31-independent pathway. However, except for the four genes, there were a number of genes up-regulated by low-oxygen conditions such as chlG, por, ctpA, hoxE, and flv4 (16). In our experiments, the up-regulation of these genes was not detected (supplemental Fig. S1). This discrepancy may be attributed to a difference in culture conditions. Although Summerfield et al. (16) observed the difference in transcript levels after a 1-h exposure to low-oxygen conditions of aerobically grown cells, we compared the transcript levels in cells grown under low-oxygen conditions (7 days) with those in cells after 2-day exposure to aerobic conditions. Our results, taken together with Summerfield et al. (16), suggest that the Hik31-independent regulatory system consists of more than two pathways, including the ChlR pathway. Further research is needed to elucidate the complex regulatory system to allow fine-tuning of the cellular metabolism in response to hypoxia.
Supplementary Material
Acknowledgments
We thank Kazuma Uesaka and Yuto Hiraide for technical help in analyzing SOLiD data and Haruki Yamamoto for technical help in using the anaerobic techniques. We also thank Kazuki Terauchi for valuable discussions.
This work was supported by Grants-in-Aid for Scientific Research 19570036 and 20200063 from Japan Society for the Promotion of Science (JSPS), by the Advanced Low Carbon Technology Research and Development Program (ALCA), and by Precursory Research for Embryonic Science and Technology (PRESTO) of the Japan Science and Technology Agency (JST).

This article contains supplemental Figs. S1–S5 and Tables S1 and S2
- HO
- heme oxygenase
- Chl
- chlorophyll
- PC
- phycocyanin
- PCC
- Pasteur culture collection
- GT
- glucose-tolerant
- KEGG
- Kyoto encyclopedia of genes and genomes.
REFERENCES
- 1. Stal L. J., Zehr J. P. (2008) in The Cyanobacteria: Molecular Biology, Genomics and Evolution (Herrero A., Flores E., eds) pp. 423–446, Caister Academic Press, Norfolk, UK [Google Scholar]
- 2. Bernroitner M., Zamocky M., Pairer M., Furtmüller P. G., Peschek G. A., Obinger C. (2008) Heme-copper oxidases and their electron donors in cyanobacterial respiratory electron transport. Chem. Biodivers. 5, 1927–1961 [DOI] [PubMed] [Google Scholar]
- 3. Keeley J.F. (1988) Photosynthesis in Quillworts, or why are some aquatic plants similar to cacti? Plants Today 1, 127–132 [Google Scholar]
- 4. Jørgensen B.B., Revsbech N.P., Blackburn T.H., Cohen Y. (1979) Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl. Environ. Microbiol. 38, 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stal L. J. (1995) Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131, 1–32 [DOI] [PubMed] [Google Scholar]
- 6. Steunou A.S., Bhaya D., Bateson M. M., Melendrez M. C., Ward D. M., Brecht E., Peters J. W., Kühl M., Grossman A. (2006) In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. U.S.A. 103, 2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chew A. G., Bryant D. A. (2007) Chlorophyll biosynthesis in bacteria. The origins of structural and functional diversity. Annu. Rev. Microbiol. 61, 113–129 [DOI] [PubMed] [Google Scholar]
- 8. Masuda T., Fujita Y. (2008) Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 7, 1131–1149 [DOI] [PubMed] [Google Scholar]
- 9. Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A. G., Tanaka A., Terry M. J. (2010) The cell biology of tetrapyrroles. A life and death struggle. Trends Plant Sci. 15, 488–498 [DOI] [PubMed] [Google Scholar]
- 10. Stenbaek A., Jensen P. E. (2010) Redox regulation of chlorophyll biosynthesis. Phytochemistry 71, 853–859 [DOI] [PubMed] [Google Scholar]
- 11. Goto T., Aoki R., Minamizaki K., Fujita Y. (2010) Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 51, 650–663 [DOI] [PubMed] [Google Scholar]
- 12. Aoki R., Goto T., Fujita Y. (2011) A heme oxygenase isoform is essential for aerobic growth in the cyanobacterium Synechocystis sp. PCC 6803. Modes of differential operation of two isoforms/enzymes to adapt to low oxygen environments in cyanobacteria. Plant Cell Physiol. 52, 1744–1756 [DOI] [PubMed] [Google Scholar]
- 13. Yilmaz M., Kang I., Beale S. I. (2010) Heme oxygenase 2 of the cyanobacterium Synechocystis sp. PCC 6803 is induced under a microaerobic atmosphere and is required for microaerobic growth at high light intensity. Photosynth. Res. 103, 47–59 [DOI] [PubMed] [Google Scholar]
- 14. Minamizaki K., Mizoguchi T., Goto T., Tamiaki H., Fujita Y. (2008) Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 2684–2692 [DOI] [PubMed] [Google Scholar]
- 15. Ludwig M, Bryant D. A. (2011) Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by Next-Gen (SOLiDtm) sequencing of cDNA. Front. Microbiol. 2, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Summerfield T. C., Nagarajan S., Sherman L. (2011) Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 157, 301–312 [DOI] [PubMed] [Google Scholar]
- 17. Kanesaki Y., Shiwa Y., Tajima N., Suzuki M., Watanabe S., Sato N., Ikeuchi M., Yoshikawa H. (2012) Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tajima N., Sato S., Maruyama F., Kaneko T., Sasaki N. V., Kurokawa K., Ohta H., Kanesaki Y., Yoshikawa H., Tabata S., Ikeuchi M., Sato N. (2011) Genomic structure of the cyanobacterium Synechocystis sp. PCC 6803 strain GT-S. DNA Res. 18, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamazaki S., Nomata J., Fujita Y. (2006) Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol. 142, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nomata J., Mizoguchi T., Tamiaki H., Fujita Y. (2006) A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis. Reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281, 15021–15028 [DOI] [PubMed] [Google Scholar]
- 21. Yamamoto H., Nomata J., Fuita Y. (2008) Functional expression of nitrogenase-like protochlorophyllide reductase from Rhodobacter capsulatus in Escherichia coli. Photochem. Photobiol. Sci. 7, 1238–1242 [DOI] [PubMed] [Google Scholar]
- 22. Nishimura T., Takahashi Y., Yamaguchi O., Suzuki H., Maeda S., Omata T. (2008) Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol. Microbiol. 68, 98–109 [DOI] [PubMed] [Google Scholar]
- 23. Summerfield T. C., Toepel J., Sherman L. A. (2008) Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 47, 12939–12941 [DOI] [PubMed] [Google Scholar]
- 24. Yurimoto H., Hirai R., Matsuno N., Yasueda H., Kato N., Sakai Y. (2005) HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol. Microbiol. 57, 511–519 [DOI] [PubMed] [Google Scholar]
- 25. Perera I. C., Grove A. (2010) Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell. Biol. 2, 243–254 [DOI] [PubMed] [Google Scholar]
- 26. Ellison D. W., Miller V. L. (2006) Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159 [DOI] [PubMed] [Google Scholar]
- 27. Stal L., Moezelaar R. (1997) Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21, 179–211 [Google Scholar]
- 28. Chi B. K., Kobayashi K., Albrecht D., Hecker M., Antelmann H. (2010) The paralogous MarR/DUF24-family repressors YodB and CatR control expression of the catechol dioxygenase CatE in Bacillus subtilis. J. Bacteriol. 192, 4571–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. (2001) The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8, 710–714 [DOI] [PubMed] [Google Scholar]
- 30. Fuangthong M., Helmann J. D. (2002) The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U.S.A. 99, 6690–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Panmanee W., Vattanaviboon P., Poole L. B., Mongkolsuk S. (2006) Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188, 1389–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. W., Soonsanga S., Helmann J. D. (2007) A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005) Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20, 131–141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.