Background: Salmonella effector protein SopB binds host Rho GTPase Cdc42.
Results: Structural characterization of SopB with Cdc42 reveals that SopB contains a CRIB-like motif and contacts Cdc42 switch regions.
Conclusion: SopB structurally mimics contacts between Cdc42 switch regions and human guanine dissociation inhibitor (GDI) and slows nucleotide exchange.
Significance: This is the first structure of SopB and provides further insight into Salmonella regulation of Cdc42.
Keywords: Bacterial Pathogenesis, GTPase, Microbiology, Signaling, Structural Biology, X-ray Crystallography
Abstract
SopB is a type III secreted Salmonella effector protein with phosphoinositide phosphatase activity and a distinct GTPase binding domain. The latter interacts with host Cdc42, an essential Rho GTPase that regulates critical events in eukaryotic cytoskeleton organization and membrane trafficking. Structural and biochemical analysis of the SopB GTPase binding domain in complex with Cdc42 shows for the first time that SopB structurally and functionally mimics a host guanine nucleotide dissociation inhibitor (GDI) by contacting key residues in the regulatory switch regions of Cdc42 and slowing Cdc42 nucleotide exchange.
Introduction
Salmonella enterica is a Gram-negative bacterium that causes typhoid fever and gastroenteritis. Typhoid fever resulted in ∼22 million illnesses and 200,000 deaths during 2000 (1), whereas Salmonella-induced gastroenteritis results in an estimated 93.8 million illnesses and 155,000 deaths each year (2). Such numbers emphasize the importance of understanding the molecular mechanisms of Salmonella pathogenesis. Salmonella gains entry into human host phagocytic and intestinal epithelial cells by secreting virulence effector proteins into the host cytoplasm via a type III secretion system (T3SS)3 (3). Inside the host cell, Salmonella delivers additional effector proteins via a second distinct T3SS and creates a replication niche termed the Salmonella-containing vacuole (SCV).
SopB (also known as SigD) is a T3SS secreted Salmonella effector protein that contains a GTPase binding domain (residues 117–168) and a phosphoinositide phosphatase domain (residues 357–561). Upon T3SS-mediated delivery of SopB into host cells, the phosphoinositide phosphatase domain promotes host membrane fission by hydrolysis of phosphatidylinositol 4,5-bisphosphate, a process that facilitates bacterial entry (4). Subsequently, SopB is multi-monoubiquitinated and translocated to the SCV (5, 6), where SopB phosphatase activity is proposed to help prevent lysosomal degradation of the bacteria by reducing the negative surface charge of the SCV (7).
The potential function of the SopB GTPase binding domain was suggested when phosphatase-deficient SopB R468A expressed in yeast interfered with actin dynamics (8), cell cycle progression, and MAP kinase signaling through direct interaction with yeast Cdc42 (9). Cdc42 is an essential Rho GTPase that regulates cytoskeleton organization and membrane trafficking during cell motility, proliferation, and cytokinesis in eukaryotes (10, 11). Active Cdc42 (GTP-bound) and inactive Cdc42 (GDP-bound) differ in conformation in the flexible regulatory regions, switch I and II (Fig. 1A) (12). Downstream host effector proteins such as p21-activated kinase (PAK) serine/threonine kinases (13) and the Wiscott-Aldrich syndrome protein (WASP) (14) recognize and bind the switch regions of active Cdc42, and this interaction subsequently alters the signaling activity of the host effector (Fig. 1B) (12). The interaction of the Salmonella SopB virulence effector with Cdc42 was delineated by stable isotope labeling of amino acids in cell culture (SILAC) mass spectrometric analysis in mammalian cells with the resulting Cdc42 binding region (residues 117–168) shown to down-regulate Cdc42-dependent signaling (9, 15). SopB binds specifically to Cdc42 and no other related GTPases (15). SopB binds both active and inactive Cdc42, and this interaction is important for translocation of SopB to the SCV (16). Furthermore, disrupting interaction between SopB and Cdc42 during a Salmonella invasion assay reduces the efficiency of Salmonella replication within the SCV (16).
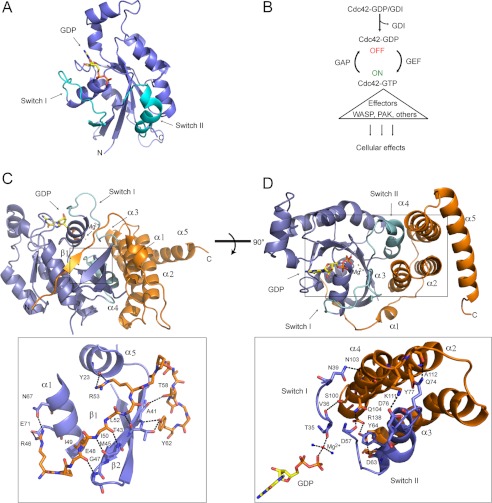
FIGURE 1.
Overview of Cdc42 (blue) structure and interaction with the GTPase binding domain of SopB (orange). A, ribbon structure of GDP-bound Cdc42 (PDB 1AN0). The regulatory switch regions of Cdc42 (cyan) interact with host effector proteins. B, Cdc42 activity regulation by GEFs, GAPs and GDIs. Active GTP-bound Cdc42 turns on numerous effector proteins to regulate cytoskeleton organization, cell cycle control, cellular trafficking, and proliferation. C, N-terminal strand of SopB (labeled as β1; orange) forming an intermolecular β-sheet with β2 of Cdc42 (blue) and contacting Cdc42 α1 and α5. Bound GDP nucleotide (yellow) is shown in stick representation, and magnesium is represented by a red star. Interacting amino acids are shown as sticks in the inset below. D, SopB helices α2, α3, and α4 inserting between switch I and II of Cdc42. SopB residues and Cdc42 switch residues that interact are shown as sticks in the inset below. A magnesium ion (red sphere) coordinates GDP nucleotide (yellow; shown in stick representation) and two water molecules (blue spheres).
Cdc42 is also targeted by Salmonella effector proteins SopE (and its paralogue SopE2) and SptP. SopE/E2 mimic eukaryotic guanine exchange factors (GEFs) to catalyze the exchange of GDP for GTP and activate Cdc42 and Rac1, a closely related Rho GTPase, to drive actin cytoskeleton assembly (Fig. 1B) (17, 18). SptP inactivates Cdc42 and Rac1 by mimicking a GTPase-activating protein (GAP) and catalyzing hydrolysis of GTP to GDP by Cdc42 and Rac1 (Fig. 1B) (19). The structures of SopE in complex with Cdc42 (20) and SptP in complex with Rac1 (21) have been solved, but a lack of structural information for SopB has impeded analysis and understanding of its interaction with and modulation of Cdc42.
Here, we present the crystal structure of the N-terminal GTPase binding domain of SopB in complex with its host target, Cdc42. This structure shows that SopB contacts Cdc42 by mimicking a eukaryotic CRIB (for Cdc42 and Rac-interactive binding)-like motif, which was previously unidentified due to a lack of sequence conservation. This is the first time a bacterial effector protein has been shown to contact a Rho GTPase through a CRIB-like motif. Furthermore, our structure shows that SopB mimics key interactions between Cdc42 and host guanine dissociation inhibitor (GDI), a regulatory protein that sequesters Cdc42 in the inactive state by preventing exchange of GDP for GTP. In vitro nucleotide exchange assays confirm that the N-terminal domain of SopB slows intrinsic Cdc42 nucleotide exchange as well as Cdc42 nucleotide exchange catalyzed by the Salmonella GEF, SopE.
EXPERIMENTAL PROCEDURES
Isothermal Titration Calorimetry (ITC)
ITC was done using an iTC200 (MicroCal GE Healthcare). Cdc42 and SopB(29–181) were each expressed in Escherichia coli BL21 (λDE3) cells and purified from cell lysate by injection over a 1-ml His-trap column (GE Healthcare) and a Superdex 75 26/60 column (GE Healthcare). Protein samples were dialyzed into 20 mm Hepes, pH 7.5, with 50 mm NaCl. The concentrations of SopB(29–181) and Cdc42 were determined by Bradford assay. SopB(29–181) was concentrated to 774 μm, and 19 aliquots of 1 μl were injected into the ITC cell containing 94.7 μm Cdc42. Titration experiments were repeated in triplicate. The dissociation constant was determined by fitting the raw data to a bimolecular interaction model.
Static Light Scattering
SopB(29–181)-Cdc42 complex was expressed and purified (described below) and concentrated to 2 mg/ml. The protein complex (100-μl volume) was injected over a Superdex 75 10/300 GL column (GE Healthcare) and analyzed by miniDAWN multiangle static light scattering device (Wyatt Technologies) and ASTRA program.
Analysis of SopB and Cdc42 Interaction by Gel Filtration
SopB(29–181), SopB I49A(29–181), and Cdc42 were expressed and purified (as described below). Protein was concentrated to 2 mg/ml and injected over a Superdex 75 10/30 column (GE Healthcare) for gel filtration analysis. To determine the effect of the SopB I49A point mutation on interaction with Cdc42, WT SopB(29–181) and SopB I49A(29–181) (2 mg/ml) were incubated with Cdc42 (2 mg/ml) on ice for 1 h and then injected over a Superdex 75 10/30 column for gel filtration analysis.
Protein Cloning, Purification, and Crystallization
Cdc42 with a deletion of seven C-terminal amino acids (Cdc42(1–183)) was amplified from Homo sapiens DNA and cloned into pET28 vector (Novagen) with restriction sites NdeI and SacI. Residues 29–181 of SopB were amplified from Salmonella typhimurium DNA and cloned into pET21 vector (Novagen) with restriction sites NdeI and Sac1. Expression constructs pET28 with N-terminal His6-tagged Cdc42(1–183) and pET21 SopB(29–181) were co-transformed into E. coli BL21 (λDE3) cells. Cells were grown in LB medium with 50 μg/ml kanamycin and 100 μg/ml ampicillin to an A600 of 0.6, and expression of Cdc42 and SopB(29–181) was induced with 1 mm IPTG for 20 h at 20 °C. Cells were harvested by centrifugation, resuspended in buffer with 20 mm Hepes, pH 7.5, 300 mm NaCl, and 50 mm imidazole and lysed by French press. The cell suspension was centrifuged at 45,000 rpm for 1 h, and supernatant was filtered and injected onto a 1-ml His-trap column. The column was washed with buffer (20 mm Hepes, pH 7.5, 300 mm NaCl, 50 mm imidazole), and His-tagged Cdc42 and bound SopB(29–181) were eluted with elution buffer (20 mm Hepes, pH 7.5, 300 mm NaCl, 500 mm imidazole). Fractions containing the protein complex were concentrated and further purified by gel filtration with a Superdex 75 26/60 column in 20 mm Hepes, pH 7.5, 300 mm NaCl. The final yield of protein was 100 mg of purified SopB(29–181) and Cdc42 complex for 2 liters of bacterial culture. Protein was concentrated to 12 mg/ml for crystallization trials. The SopB(29–181) and Cdc42 complex was crystallized using the sitting drop vapor diffusion method by mixing 0.5 μl of 12 mg/ml protein solution with 0.5 μl of well solution, 0.2 m sodium chloride, 0.1 m phosphate-citrate, pH 4.2, 20% (w/v) PEG 8000 (JCSG+ suite, Qiagen). The crystals were diamond-shaped and appeared after 14 days at 4 °C.
Data Collection, Structure Determination, and Refinement
Crystals were cryoprotected in a solution of mother liquor with 20% glycerol and flash frozen in liquid nitrogen. Crystals were screened at the CMCF-2 of the Canadian Light Source and diffracted to 2.35-Å resolution. The crystals were space group P43212 and had cell dimensions of 106.7 × 106.7 × 87.5 Å. Data were reduced and scaled using MOSFLM (22) and SCALA (23). The phases of the SopB(29–181)-Cdc42 complex were solved by molecular replacement using Phaser_MR (24), with the search model Cdc42-GDP (PDB code 1AN0). COOT (25) was used to perform model building and the model was refined with REFMAC5 (26) from the CCP4 suite and Phenix (27). Ligand and ion atoms were refined at an occupancy of one. The structure was analyzed using MolProbity (28) with 96.7, 3.3, and 0% of residues in the favorable, allowed, and disallowed regions of the Ramachandran plot. The SopB-Cdc42 structure was deposited in the Protein Data Bank (4DID). Figures were generated by PyMOL (29).
Cdc42 Nucleotide Exchange Assay
Cdc42 nucleotide exchange assay was adapted from Ref 30. Cdc42 was preloaded with a fluorescent GDP analog, N-methanylanthraniloyl (Mant)-GDP, by incubating 100 μm Cdc42 with 500 μm Mant-GDP in buffer (20 mm Tris pH 8.0, 150 mm NaCl, 5 mm EDTA, 1 mm DTT) at 37 °C for 30 min. The reaction was transferred to ice and stopped by addition of 10 mm MgCl2. A desalting column (GE Healthcare) was used to remove free Mant-GDP and exchange the Mant-GDP-loaded Cdc42 into reaction buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 5 mm MgCl2). Mant-GDP-loaded Cdc42 was diluted with reaction buffer at a final concentration of 2.5 μm. To initiate nucleotide exchange, unlabeled GTP was added to Mant-GDP-loaded Cdc42 at a final concentration of 500 μm. The intrinsic exchange rate was measured by exciting Mant-GDP at 355 nm and monitoring emission at 448 nm. The fluorescence intensity at 448 nm decreases as Mant-GDP is released from Cdc42 into solution. To determine the effect of SopB on nucleotide exchange rate, 2.5 μm Mant-GDP-loaded Cdc42 was incubated with a 1.5 m or 3 m excess of SopB(29–181) in buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 5 mm MgCl2) for 10 min on ice. The exchange reaction was initiated by addition of 500 μm GTP and measured as described above. Each reaction was repeated in three independent experiments, and the mean value of relative fluorescence was plotted versus time. S.D. was calculated in Microsoft Excel and is represented by error bars. The SopE-catalyzed nucleotide exchange reactions were prepared and measured as described above. GTP was added to reaction mixture, and measurements were recorded for 30 s. The spectrophotometer was paused as SopE was added to the reaction at a final concentration of 25 nm, and measurements resumed immediately after.
RESULTS
Biochemical Analysis and Crystallization of Bacterial SopB and Host Cdc42 Complex
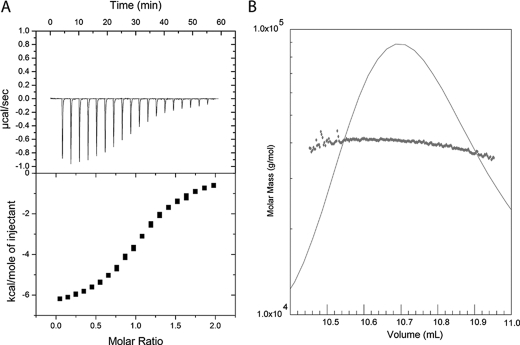
To determine how the SopB N-terminal domain binds Cdc42 to down-regulate activity, SopB(29–181) in complex with Cdc42 bound to GDP (Kd of 6 μm ± 2 μm as determined by ITC analysis; Fig. 2A) was co-purified for crystallization. SopB(29–181) and Cdc42 formed a 1:1 complex (as determined by static light scattering; Fig. 2B) and yielded crystals that diffracted to 2.35-Å resolution (one complex in the asymmetric unit; Table 1). Density could be observed for residues 45–171 of SopB and residues 1–179 of Cdc42 as well as its GDP ligand. SopB consists of a β-strand (β1) that forms an intermolecular β-sheet with Cdc42 and five α-helices (α1–5), three of which (α2, α3, and α4) contact Cdc42 switch I and II regions (Fig. 1, C and D). The interface of SopB and Cdc42 forms an area of ∼1770 Å2 as calculated by PISA (31).
FIGURE 2.
Analysis of SopB(29–181) binding to Cdc42 by ITC and light scattering. A, SopB(29–181) was concentrated to 774 μm, and 1-μl aliquots were injected into 94.7 μm Cdc42. The dissociation constant (Kd) 6 μm ± 2 μm was calculated as the mean value of the Kd determined. Experiments were repeated a minimum of three times and error is represented as S.D. B, purified SopB(29–181)-Cdc42 complex was analyzed by injection over a Superdex 75 10/300 GL column followed by static light scattering (Wyatt Technologies). The complex eluted from the column in a monodisperse peak and a molecular mass of ∼42 kDa, corresponding to a 1:1 binding stoichiometry.
TABLE 1.
Data collection and refinement statistics
| Crystal parameters | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 107.00, 107.00, 87.75 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Data collection | |
| Resolution (Å) | 45.7-2.35 |
| Completeness (%) | 99.9 (100)a |
| Redundancy | 14.5 (14.7) |
| Rmerge | 0.081 (0.578) |
| I/σI | 18.8 (4.7) |
| Refinement statistics | |
| Resolution (Å) | 45.7-2.35 |
| Rwork/Rfree | 0.2133/0.2522 |
| No. reflections | 405,282 |
| No. atoms | |
| Protein | 2,391 |
| Ligand/ion | 28/1 |
| Water | 116 |
| B-factors (Å2) | |
| Protein | 45.0 |
| Ligand/ion | 35.5/29.8 |
| Water | 49.5 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.0230 |
| Bond angles (°) | 1.9055 |
| Ramachandran valuesb | |
| Favored (%) | 96.7 |
| Allowed (%) | 3.3 |
a Values in parentheses are for highest resolution shell.
b Calculated by MolProbity (33).
SopB Contacts Host Cdc42 by Mimicking Eukaryotic CRIB-like Intermolecular β-Sheet
SopB residues 48–52 and Cdc42 β2 form an intermolecular β-sheet that extends the three-stranded β-sheet of Cdc42 by an additional strand (Fig. 1C). SopB residues flanking the intermolecular β-strand interact with Cdc42 α5 and α1 (Fig. 1C). Specifically, SopB residue Arg-46 side chain forms hydrogen bonds with side chains of Cdc42 α5 residues Asn-167 and Glu-171. Additionally, hydrophobic interactions are formed between SopB residues Pro-47 and Ile-49 and Cdc42 residue Leu-174 in α5. SopB residue Arg-53 forms a hydrogen bond with Cdc42 residue Tyr-23 in α1. Furthermore, SopB residues 58–62 form a short α-helix (α1) that packs against the intermolecular β-sheet such that SopB residues Thr-58 and Tyr-62 hydrogen bond with Cdc42 residues Ala-41 and Thr-43, respectively.
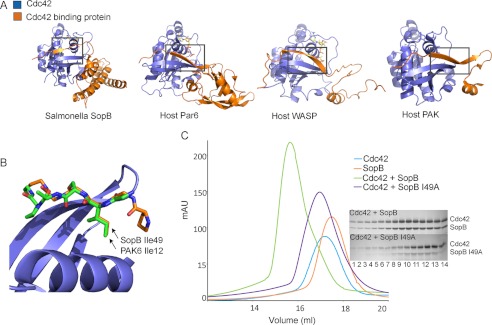
Eukaryotic effector proteins such as PAK, which couples activation of Cdc42 to cytoskeletal function, and cell migration (32) form an intermolecular β-sheet with Cdc42 β2 and contact Cdc42 α1 and α5 through a CRIB motif with the conserved sequence ISXP(X)2–4FXHXXHV (Fig. 3A) (13). The SopB region (resides 46–53) that forms the intermolecular β-sheet contacts the same regions of Cdc42 (β2, α1, and α5) as the PAK CRIB motif. The canonical CRIB motif is not conserved in SopB except for residue Ile-49, which remarkably superimposes closely with the PAK residue Ile-12 (Fig. 3B). In both the eukaryotic PAK and Salmonella SopB, this Ile residue contacts Cdc42 Ile-46 and additional hydrophobic residues contained within Cdc42 α5. As such, SopB is the first example of a bacterial effector protein that mimics a host CRIB-like interaction to contact and modulate the action of a mammalian Rho GTPase, despite lacking the identifying CRIB sequence motif. These data indicate a key structural role of the Ile residue in the context of Cdc42 binding. This observation is supported by earlier site-specific mutagenesis studies (33) of an I75N mutant within the eukaryotic αPAK CRIB that binds Cdc42 with ∼3-fold less affinity as well as our gel filtration analysis of an I49A point mutant of SopB that disrupts interaction with Cdc42 (Fig. 3C).
FIGURE 3.
SopB contacts Cdc42 through a CRIB-like motif that contains an Ile residue conserved with eukaryotic Cdc42 effector proteins. A, eukaryotic Cdc42 effector proteins Par6 (1NF3), WASP (1CEE), and PAK6 (2ODB) contain a CRIB domain that forms a intermolecular β-sheet with Cdc42 β2. B, SopB (orange) lacks a conserved CRIB motif except for residue Ile-49, which superimposes with the CRIB Ile residue of eukaryotic effector proteins such as PAK6 Ile-12 (green; PDB 2ODB) to contact Cdc42 (blue). C, gel filtration analysis of binding of SopB (residues 29–181) to Cdc42 after mutagenesis of a SopB Ile residue (I49A) that is conserved with the CRIB Ile of eukaryotic Cdc42-binding effector proteins is shown. SopB I49A with Cdc42 (purple) elutes later on the gel filtration profile than WT SopB with Cdc42 (green), indicating that Ile-49 is important for interaction with Cdc42. Corresponding elution fractions from WT SopB + Cdc42 (top) and SopB I49A + Cdc42 (bottom) are shown beside the gel filtration profile.
SopB Contacts Key Cdc42 Regulatory Switch Residues to Mimic a Rho GDI
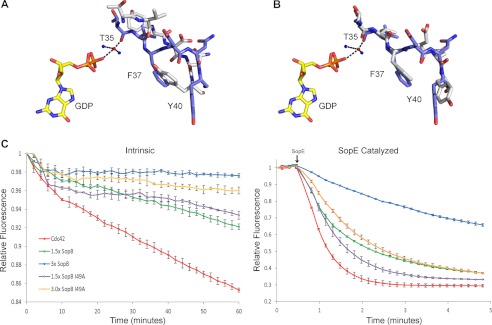
SopB helices (α2, α3, and α4) insert between Cdc42 regulatory loop regions, switch I and II (residues 26–45 and 59–74, respectively) (Fig. 1D). When our structure of Cdc42 in complex with GDP and SopB is superimposed with that of Cdc42 in complex with GDP (PDB 1AN0), the overall structure of Cdc42 is similar (average r.m.s.d. of 1.5 Å on 179 Cα atoms). However, our comparison shows that there is a localized change in conformation of the Cdc42 switch I (r.m.s.d. of 2.5 Å Cα atoms of residues 26–45) and that it is the SopB interaction with Cdc42 Val-36 that is predominantly responsible (Fig. 1D; Fig. 4A). Significantly, our observed conformation of Cdc42 switch I in complex with GDP and Salmonella SopB is similar to that of Cdc42 switch I in complex with GDP and human Rho GDI, an inactivator of Cdc42 activity (Fig. 1B). At switch I, the main contact between Cdc42 and SopB is a hydrogen bond between the hydroxyl group of SopB residue Ser-100 and the main chain carbonyl of Cdc42 residue Val-36 and van der Waals interactions between SopB residue Val-97 and the side chain atoms of Cdc42 Thr-35. In addition, the main chain carbonyl of Cdc42 switch I residue Thr-35 and the terminal phosphate group of GDP are coordinated by magnesium. Similarly, in the human Rho GDI, a serine residue contacts Cdc42 Val-36, and the main chain carbonyl of Cdc42 Thr-35 and the terminal phosphate group of GDP are coordinated by magnesium (34). Additional observations from our data show that in the presence of SopB, Cdc42 switch I residue Phe-37, which is solvent-exposed in Cdc42-GDP, is reoriented to a hydrophobic pocket that Tyr-40 occupies in the Cdc42-GDP structure (Fig. 4A). This flip of Phe-37 also occurs when Rho GDI is bound to Cdc42 (Fig. 4B) (34). Collectively, our structure shows that Salmonella SopB mimics several contacts between host Rho GDI and Cdc42 to reposition Cdc42 switch I residues such that Thr-35 forms a coordination network with magnesium and GDP, an interaction that prevents dissociation of bound nucleotide from Cdc42 (34, 35).
FIGURE 4.
When Cdc42 is bound to SopB the conformation of Cdc42 switch I residues aligns with the conformation of Cdc42 switch I residues when bound to human Rho GDI, and the GTP exchange activity of Cdc42 is down-regulated. A, alignment of Cdc42 switch I (blue) when bound to SopB and Cdc42 switch I (gray) in the absence of SopB (PDB 1AN0). Both Cdc42 structures contain GDP (yellow). B, alignment of Cdc42 switch I (blue) when bound to SopB with Cdc42 switch I (gray) when bound to Rho GDI (PDB 1DOA). C, intrinsic Cdc42 nucleotide exchange monitored by measuring change in emitted fluorescence when Mant-GTP is exchanged for unlabeled GTP. SopB was added in excess to the nucleotide exchange assay, and the change in fluorescence was measured. SopE-catalyzed Cdc42 nucleotide exchange was measured as for intrinsic Cdc42 nucleotide exchange. The experiment was repeated with SopB I49A. The arrow indicates the addition of SopE to the reaction mix. Experiments were repeated in triplicate, and error bars represent S.E.
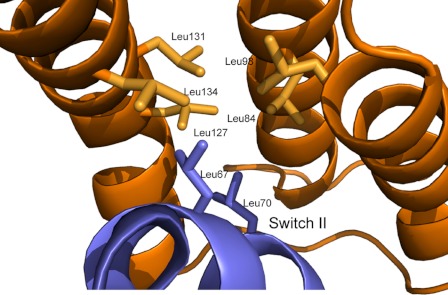
Our structural data also show that SopB makes extensive contacts with Cdc42 switch II; however, the conformation of switch II in our structure is similar to that of Cdc42 in complex with GDP (r.m.s.d. 0.6 Å on Cα atoms of residues 59–74), suggesting that SopB uses switch II as an anchoring point to further facilitate binding. SopB α2, α3, and α4 form a helical bundle with a hydrophobic groove composed of several leucine residues (Leu-84, Leu-98, Leu-127, Leu-131, Leu-134) that contact Cdc42 switch II residues Leu-67 and Leu-70 (Fig. 5). SopB mutant L84P has previously been shown to disrupt SopB binding to Cdc42 (16). These interactions are complemented by additional polar interactions (Fig. 1D). Like host Rho GDI (25) and the GDI domain of the Yersinia effector YpkA (35), two SopB residues, Arg-138 and Asp-95, hydrogen bond to the hydroxyl group of switch II residue Tyr-64. Two salt bridges are formed between SopB residues Lys-111 and Arg-138 and switch II residues Asp-76 and Arg-138, respectively. SopB residue Gln-104 contacts switch II residue Asp-57, which is also coordinated in Rac1 by YpkA.
FIGURE 5.
SopB leucine residues (orange) form van der Waals contacts with Cdc42 switch II residues Leu-67 and Leu-70 (blue). A similar set of interactions is also observed when eukaryotic Rho GDI (PDB 1DOA) binds Cdc42.
SopB Prevents Cdc42 Nucleotide Exchange in Vitro
To support our observation that SopB structurally mimics Rho GDI, we used an in vitro nucleotide exchange assay to test whether SopB(29–181) can slow dissociation of bound GDP from Cdc42 and subsequent exchange for GTP (Fig. 4C). Our data show that SopB significantly decreases the rate of in vitro intrinsic Cdc42 nucleotide exchange in a dose-dependent manner. In addition, we tested whether SopB slows nucleotide exchange in the presence of the potent Salmonella GEF, SopE. As seen with the intrinsic nucleotide exchange reaction, SopB slows the rate of Cdc42 nucleotide exchange catalyzed by SopE (Fig. 4C). To test whether the SopB mutant I49A can still inhibit Cdc42 nucleotide exchange, we repeated the intrinsic and catalyzed Cdc42 exchange assay with SopB I49A. SopB I49A could inhibit nucleotide exchange at levels similar to wild-type SopB for the intrinsic exchange assay. However, in the presence of the potent GEF SopE, the SopB I49A mutation significantly decreased the ability of SopB to inhibit nucleotide exchange.
DISCUSSION
Rho GTPases regulate a multitude of cellular processes such as cytoskeleton organization, cell division, and cell motility (36). Members of the Rho GTPase family (for example Cdc42, Rac1, and RhoA) are low molecular weight proteins containing two flexible regulatory switch regions, termed switch I and II, that bind and hydrolyze GTP. As the name implies, they function as molecular switches, which are active in the GTP-bound state and inactive in the GDP-bound state (36). Cycling between the active and inactive state causes a change in conformation at the switch regions critical to function. In the active state, Rho GTPases interact with effector proteins, such as kinases, lipases, and oxidases to alter their activity (12). The activity of Rho GTPases is regulated by GEFs, which catalyze exchange of GDP for GTP to activate the GTPase; GAPs, which catalyze hydrolysis of GTP to inactivate the GTPase; and GDIs, which prevent dissociation of bound nucleotide to prevent reactivation of the GTPase (36).
Because Rho GTPases play a fundamental role in many cellular processes, they are common targets of bacterial effector proteins and toxins. Whereas bacterial toxins often inhibit or activate Rho GTPases by irreversible covalent modification (37), Salmonella uses a T3SS to deliver multiple effector proteins to the host cell that target specific Rho GTPases and modify their activity in a reversible and temporally regulated (38) manner. This sophisticated mechanism of Rho GTPase regulation by Salmonella allows the bacterium to manipulate the activity of Rho GTPases at different times after infection and exploit the resulting cellular effect for efficient entry into the host cell (38). Whereas the interaction between Salmonella effectors SopE and SptP and Rho GTPases Cdc42 and Rac1 have been structurally characterized (20, 21), there has been no structural information of the Salmonella effector SopB and its interaction with Cdc42.
Here, we have presented the first structure of the Cdc42 binding domain of SopB in complex with Cdc42. In this model, SopB contacts key Cdc42 switch I and II residues that are also contacted by host Rho GDI (34). The most notable interaction in this region is a hydrogen bond between SopB Ser-100 and the main chain carbonyl of Cdc42 Val-36. This hydrogen bond positions the main chain carbonyl of the neighboring Cdc42 residue Thr-35 to coordinate a magnesium ion, which is also coordinated by the terminal phosphate group of the bound GDP nucleotide. This coordination network, which is similar to that seen in the complex between Rho GDI and Cdc42 (34), serves to stabilize the bound GDP nucleotide and prevent exchange for GTP. In vitro nucleotide exchange assays confirm that the GTPase binding domain of SopB can slow intrinsic and SopE catalyzed Cdc42 nucleotide exchange. The GDI-like activity of the GTPase binding domain of SopB is consistent with the observation that phosphatase-deficient SopB can down-regulate Cdc42 signaling in a yeast model system (9).
Like Salmonella, Yersinia also encodes a T3SS effector protein, YpkA, which mimics a host GDI (35). The C-terminal GDI domain of YpkA binds to Rac1 and RhoA, but not Cdc42 (16). Although there is no sequence similarity between the GDI domains of SopB and YpkA, both interact with Rho GTPases by inserting α-helices between switch I and II and specifically contacting switch I residues Val-36 (SopB), and Thr-35 (YpkA) so that the magnesium ion coordinates the main chain carbonyl of Thr-35 and the terminal phosphate of GDP.
One unexpected feature of the SopB and Cdc42 interaction is the CRIB-like domain of SopB that forms an intermolecular β-sheet with Cdc42. This region of the complex closely resembles interactions between Cdc42 and eukaryotic effector proteins that contain a CRIB motif, such as PAK (13) and WASP (13), but was not predicted because of a lack of sequence similarity between this region of SopB and the canonical CRIB motif. However, SopB contains a isoleucine residue that superimposes with a conserved CRIB isoleucine of eukaryotic effector proteins. This isoleucine is positioned within a hydrophobic pocket on the surface of Cdc42. The SopB and Cdc42 CRIB-like interaction is not conserved with YpkA (35) because YpkA does not form an intermolecular β-sheet with β2 of Rac1 or RhoA. This is the first known example of a bacterial effector protein that mimics a CRIB-like domain to contact Cdc42.
In conclusion, our structure shows that SopB contacts Cdc42 with a eukaryotic CRIB-like motif and slows in vitro Cdc42 nucleotide exchange by structurally mimicking key contacts made between human Rho GDI and Cdc42. The discovery of the GDI-like function of SopB demonstrates that Salmonella uses a GEF (17, 18), GAP (19), and GDI to contact Cdc42, resulting in sophisticated regulation of host Cdc42 signaling. This is the first time that a pathogenic bacterium has been shown to mimic all three eukaryotic regulators of small GTPases.
Acknowledgments
We thank M. Vuckovic for cloning of protein constructs; T. Spreter for help with multiangle light scattering; and R. Gruninger for helpful discussion. We thank the Canadian Light Source staff at the CMCF-2 beamline for synchrotron data collection.
This work was supported by the Canadian Institute for Health Research, the Canadian Foundation of Innovation, and the British Columbia Knowledge Development Fund. G. P. and L. J. W. are Michael Smith Foundation for Health Research (MSFHR) postdoctoral fellows.
The atomic coordinates and structure factors (code 4DID) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- T3SS
- type III secretion system
- CRIB
- Cdc42 and Rac-interactive binding
- GAP
- GTPase-activating protein
- GDI
- guanine dissociation inhibitor
- GEF
- guanine exchange factor
- ITC
- isothermal titration calorimetry
- Mant-GDP
- N-methanylanthraniloyl-GDP
- PAK
- p21-activated kinase
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- SCV
- Salmonella-containing vacuole
- WASP
- Wiscott-Aldrich syndrome protein
- SILAC
- stable isotope labeling of amino acids in cell culture.
REFERENCES
- 1. Crump J. A., Luby S. P., Mintz E. D. (2004) The global burden of typhoid fever. Bull. World Health Organ. 82, 346–353 [PMC free article] [PubMed] [Google Scholar]
- 2. Majowicz S. E., Musto J., Scallan E., Angulo F. J., Kirk M., O'Brien S. J., Jones T. F., Fazil A., Hoekstra R. M. (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889 [DOI] [PubMed] [Google Scholar]
- 3. Ibarra J. A., Steele-Mortimer O. (2009) Salmonella—the ultimate insider: Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11, 1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Terebiznik M. R., Vieira O. V., Marcus S. L., Slade A., Yip C. M., Trimble W. S., Meyer T., Finlay B. B., Grinstein S. (2002) Elimination of host cell PtdIns(4,5)P2 by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 4, 766–773 [DOI] [PubMed] [Google Scholar]
- 5. Patel J. C., Hueffer K., Lam T. T., Galán J. E. (2009) Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knodler L. A., Winfree S., Drecktrah D., Ireland R., Steele-Mortimer O. (2009) Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell. Microbiol. 11, 1652–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bakowski M. A., Braun V., Lam G. Y., Yeung T., Heo W. D., Meyer T., Finlay B. B., Grinstein S., Brumell J. H. (2010) The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 7, 453–462 [DOI] [PubMed] [Google Scholar]
- 8. Alemán A., Rodríguez-Escudero I., Mallo G. V., Cid V. J., Molina M., Rotger R. (2005) The amino-terminal non-catalytic region of Salmonella typhimurium SigD affects actin organization in yeast and mammalian cells. Cell. Microbiol. 7, 1432–1446 [DOI] [PubMed] [Google Scholar]
- 9. Rodríguez-Escudero I., Rotger R., Cid V. J., Molina M. (2006) Inhibition of Cdc42-dependent signalling in Saccharomyces cerevisiae by phosphatase-dead SigD/SopB from Salmonella typhimurium. Microbiology 152, 3437–3452 [DOI] [PubMed] [Google Scholar]
- 10. Sinha S., Yang W. (2008) Cellular signalling for activation of Rho GTPase Cdc42. Cell. Signal. 20, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 11. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 23, 509–514 [DOI] [PubMed] [Google Scholar]
- 12. Bishop A. L., Hall A. (2000) Rho GTPases and their effector proteins. Biochem. J. 348, 241–255 [PMC free article] [PubMed] [Google Scholar]
- 13. Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., Lowe P. N., Laue E. D. (2000) Structure of Cdc42 bound to the GTPase binding domain of PAK. Nature 7, 384–388 [DOI] [PubMed] [Google Scholar]
- 14. Abdul-Manan N., Aghazadeh B., Liu G. A., Majumdar A., Ouerfelli O., Siminovitch K. A., Rosen M. K. (1999) Structure of Cdc42 in complex with the GTPase-binding domain of the Wiskott-Aldrich syndrome protein. Nature 399, 379–383 [DOI] [PubMed] [Google Scholar]
- 15. Rogers L. D., Kristensen A. R., Boyle E. C., Robinson D. P., Ly R. T., Finlay B. B., Foster L. J. (2008) Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J. Proteomics 71, 97–108 [DOI] [PubMed] [Google Scholar]
- 16. Rodriguez-Escudero I., Ferrer N. L., Rotger R., Cid V. J., Molina M. (2011) Interaction of the Salmonella typhimurium effector protein SopB with host cell Cdc42 is involved in intracellular replication. Mol. Microbiol. 80, 1220–1240 [DOI] [PubMed] [Google Scholar]
- 17. Hardt W. D., Chen L. M., Schuebel K. E., Bustelo X. R., Galán J. E. (1998) S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826 [DOI] [PubMed] [Google Scholar]
- 18. Stender S., Friebel A., Linder S., Rohde M., Mirold S., Hardt W. D. (2000) Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36, 1206–1221 [DOI] [PubMed] [Google Scholar]
- 19. Fu Y., Galán J. E. (1999) A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401, 293–297 [DOI] [PubMed] [Google Scholar]
- 20. Buchwald G., Friebel A., Galán J. E., Hardt W. D., Wittinghofer A., Scheffzek K. (2002) Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stebbins C. E., Galán J. E. (2000) Modulation of host signalling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol. Cell 6, 1449–1460 [DOI] [PubMed] [Google Scholar]
- 22. Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, 26 [Google Scholar]
- 23. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) COOT: model building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. (1997) Application of maximum likelihood refinement. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) Collaborative Computational Project, Number 4 Newsletter 42 [Google Scholar]
- 28. Chen V. B., Arendall W. B., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 30. Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., Roy C. R. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide exchange factor. Nat. Cell Biol. 8, 971–977 [DOI] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32. Eswaran J., Soundararajan M., Kumar R., Knapp S. (2008) UnPaking the class differences among p21-activated kinases. Trends Biochem. Sci. 33, 394–403 [DOI] [PubMed] [Google Scholar]
- 33. Zhao Z., Manser E., Chen X., Chong C., Leung T., Lim L. (1998) A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18, 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffman G. R., Nassar N., Cerione R. A. (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with multifunctional regulator GDI. Cell 100, 345–356 [DOI] [PubMed] [Google Scholar]
- 35. Prehna G., Ivanov M. I., Bliska J. B., Stebbins C. E. (2006) Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell 126, 869–880 [DOI] [PubMed] [Google Scholar]
- 36. Jaffe A., Hall A. (2005) Rho GTPases: biochemistry and biology. Annu. Rev. Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 37. Aktories K. (2011) Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 9, 487–498 [DOI] [PubMed] [Google Scholar]
- 38. Kubori T., Galán J. E. (2003) Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115, 333–342 [DOI] [PubMed] [Google Scholar]
- 39. Dukuzumuremyi J. M., Rosqvist R., Hallberg B., Akerström B., Wolf-Watz H., Schesser K. (2000) The Yersinia protein kinase A is a host factor inducible RhoA/Rac-binding virulence factor. J. Biol. Chem. 275, 35281–35290 [DOI] [PubMed] [Google Scholar]