Background: Anti-factor D antibody blocks a rate-limiting step in the alternative complement pathway.
Results: The structure of anti-factor D in complex with factor D provides the molecular basis of complement inhibition.
Conclusion: Anti-factor D binds to the factor D exosite and inhibits alternative pathway complement activation.
Significance: Targeting the exosite on proteases could have great potential for antibody therapies.
Keywords: Antibody Engineering, Complement System, Convertases, Crystal Structure, Immunology
Abstract
By virtue of its amplifying property, the alternative complement pathway has been implicated in a number of inflammatory diseases and constitutes an attractive therapeutic target. An anti-factor D Fab fragment (AFD) was generated to inhibit the alternative complement pathway in advanced dry age-related macular degeneration. AFD potently prevented factor D (FD)-mediated proteolytic activation of its macromolecular substrate C3bB, but not proteolysis of a small synthetic substrate, indicating that AFD did not block access of the substrate to the catalytic site. The crystal structures of AFD in complex with human and cynomolgus FD (at 2.4 and 2.3 Å, respectively) revealed the molecular details of the inhibitory mechanism. The structures show that the AFD-binding site includes surface loops of FD that form part of the FD exosite. Thus, AFD inhibits FD proteolytic function by interfering with macromolecular substrate access rather than by inhibiting FD catalysis, providing the molecular basis of AFD-mediated inhibition of a rate-limiting step in the alternative complement pathway.
Introduction
Complement activation is an innate defense mechanism that, when uncontrolled, leads to inflammation and local tissue damage. Both gain- and loss-of-function SNPs in genes of the alternative complement pathway control risk of age-related macular degeneration. The presence of SNPs in the complement pathway is further associated with increased plasma levels of complement proteins Ba, C3a, C5a, and factor D (FD)3 and with the incidence of intermediate and advanced age-related macular degeneration (1–4). Because of a strong genetic and pathophysiological link between alternative complement pathway activation and age-related macular degeneration, the past years have seen an increase in the number of therapeutics that target the complement pathways in this disease. Inhibitors that target complement pathway convertases are currently in preclinical development or in clinical trials (5).
FD is responsible for conversion of the alternative pathway proconvertases C3bB and C3b2B to form the active C3 convertase C3bBb or the C5 convertase C3b2Bb. Moreover, FD is a rate-limiting enzyme of the alternative complement pathway and has the lowest concentration in plasma among all complement proteins (6). Therefore, targeting FD with a neutralizing anti-FD antibody is a promising therapeutic strategy to inhibit alternative complement activation for treatment of age-related macular degeneration. In addition to inhibiting the alternative pathway, an anti-FD antibody indirectly also blocks classical and mannose-binding lectin pathways, as amplification of these pathways depends on alternative pathway convertase activation (7, 8). The low concentration of FD in blood is maintained by an extremely rapid catabolic rate (6, 9, 10). Systemic treatment with blocking anti-FD antibodies would require very high dosing to overcome the high turnover rate (11). Therefore, we generated an anti-FD Fab fragment (AFD) for inhibition of local complement activation in the eye.
AFD is an Escherichia coli-expressed, humanized Fab fragment derived from monoclonal antibody (mAb) 166-32 (11–13). mAb 166-32 inhibits complement activation in serum from human and non-human primates and has been shown to reduce complement and leukocyte activation in a baboon model of cardiopulmonary bypass surgery (14). mAb 166-32 binds human and cynomolgus FD and is a potent and selective inhibitor of the alternative complement pathway in human and non-human primate sera (13). Humanization was performed by grafting the complementarity-determining regions of mAb 166-32 on human germ line framework regions. Minimal framework changes were required in the variable regions to retain high-affinity binding of AFD to FD. So far, it is unclear how AFD inhibits FD activation.
The structural, biophysical, and biochemical studies described herein provide a detailed insight into the molecular mechanism by which AFD interferes with FD-dependent complement activation. AFD binds to an exosite region that is remote from the catalytic center of FD. As a result, AFD does not impair catalysis of a small synthetic substrate, but it potently inhibits binding and activation of the macromolecular substrate C3bB, leading to efficient attenuation of alternative complement pathway activation and its amplification.
EXPERIMENTAL PROCEDURES
Antibodies
Hybridoma 8E2 was generated by immunizing mice with recombinant human FD using a previously described immunization method (15). Fab fragments were generated by papain digestion as described by the manufacturer (Pierce). To generate AFD, mAb 166-32 (12) was humanized on a human kappa I subgroup variable domain for the light chain, with one mouse framework residue carried over (Val at residue 4). The heavy chain was humanized on a human VH7 subgroup variable domain with one mouse framework residue included (Pro at residue 9). The N-terminal Gln was changed to Glu to remove potential for pyro-Gln formation. The Fab fragment of an antibody directed against hepsin was used as a control (control Fab) (16).
C3 Convertase and Hemolysis Assay
The convertase assay mixture consisted of 0.1% gelatin Veronal buffer and 10 mm MgCl2 with complement proteins FD (0.125 μm), factor B (FB) (0.5 μm), and C3b (0.5 μm) and antibodies (AFD, 8E2, and control Fab). C3b was generated as described (15). Ten μl of FD (0.5 μm) and 10 μl of AFD, 8E2, or control Fab (2 μm or a concentration range) were mixed for 15 min. Ten μl of FB (2 μm) and 10 μl of C3b (2 μm) were added to the FD/AFD mixture and incubated for 30 min at 37 °C. Forty μl of Laemmli buffer was added to stop the reaction. Samples were boiled for 5 min and run on a Novex 4–20% Tris/glycine-polyacrylamide gel for 1.5 h at 125 mV (SeeBlue2 molecular mass marker). Gels were stained for 1 h with SimplyBlue SafeStain, washed overnight with double-distilled water, and dried between Cellophane. The hemolysis assay was performed as described (17).
Biacore Analysis
Binding analysis was performed on a Biacore 3000 instrument. C3b was amine-coupled to a CM5 chip following the manufacturer's recommendation. The CM5 chip was activated with N-hydroxysuccinimide and N-ethyl-N′-(dimethylaminopropyl) carbodiimide (flow rate of 5 μl/min, 30 μl). C3b (50 μg/ml) was flowed at 5 μl/min and 20 μl to achieve 7300 final resonance units. FB, FD, AFD, and 8E2 Fab fragment proteins and antibodies were buffer-exchanged using a HiTrap desalting column (GE Healthcare) in assay buffer (Veronal buffer, 1 mm NiCl2, and 0.05% surfactant P-20). Binding assays used the “co-inject” mode in the Biacore instrument. One μm FB was injected (flow rate of 30 μl/min, 90 μl), followed by co-injecting a mixture of 1 μm FB and FD(S195A) (2-fold dilutions, FD(S195A) concentration range of 15.6 nm to 4.0 μm, flow rate of 30 μl/min, 90 μl), and then allowed to dissociate in assay buffer for 5 min. The Biacore chip was regenerated by three 1-min washes with 3 m NaCl in 50 mm sodium acetate (pH 5.0) and re-equilibrated for 5 min with assay buffer. To determine the effect of AFD on binding to the proconvertase, the same C3b-coated CM5 chip was used. FB, FD, AFD, and 8E2 Fab fragment proteins and antibodies were buffer-exchanged using a HiTrap desalting column in assay buffer. Binding assays used the “co-inject” mode. One μm FB was injected (flow rate of 30 μl/min, 90 μl), followed by co-injecting a mixture of 1 μm FB, 1 μm FD, and different concentrations of antibodies (flow rate of 30 μl/min, 90 μl), and then allowed to dissociate in assay buffer for 5 min. The chip was regenerated with three 1-min washes with 3 m NaCl in 50 mm sodium acetate (pH 5.0) and re-equilibrated for 5 min with buffer.
FD Expression and Purification
Cynomolgus pro-FD was cloned using a cynomolgus lung cDNA library and primers directed to the 5′- and 3′-ends of human FD. The cDNAs of the entire pro-FD open reading frames of human and cynomolgus FD were cloned into a pRK expression vector and expressed in CHO cells. An affinity column was prepared by coupling anti-FD Ab 8E2 to Affi-Gel 10 (Bio-Rad). Unreacted sites were blocked with ethanolamine. The harvested transient supernatants were applied to the 8E2 affinity column, washed with PBS, eluted with 0.1 m acetic acid, neutralized with 1.5 m Tris (pH 8.6), and dialyzed into 20 mm HEPES (pH 7.2) and 200 mm NaCl. Characterization of the purified proteins confirmed intact, monomeric, >98% pure FD with the signal peptide and propeptide fully processed. The final protein samples were in 10 mm HEPES (pH 7.2) and 140 mm NaCl. AFD was provided in lyophilized form and reconstituted with water. The resulting solution was 50 mg/ml protein in 40 mm histidine hydrochloride, 20 mm sodium chloride, 180 mm sucrose, and 0.04% (w/v) polysorbate 20 (pH 5.4).
Crystallization
Human FD protein and AFD were mixed at a 1:1 ratio and purified on a Superdex 200 column pre-equilibrated with 20 mm HEPES (pH 7.2) and 200 mm NaCl. The peak fractions containing the complex were pooled, concentrated to 30 mg/ml, and used in a crystallization trial. Crystals were grown at 4 °C using the sitting drop vapor diffusion method. The crystallization buffer containing 0.1 m Tris-HCl (pH 8.5), 0.2 m ammonium phosphate, 50% 2-methyl-2,4-pentanediol, and 0.01 m hexamine cobalt(III) chloride was mixed in an equal volume with the protein solution. Crystals appeared after 6 days and belonged to space group P43212. The crystals were flash-frozen in liquid nitrogen. A 2.4 Å data set was collected at Stanford Synchrotron Radiation Lightsource on beamline 9-2. The cynomolgus FD-AFD complex was purified following the same protocol as described above. Crystals used in the structure determination were grown at 19 °C in 0.1 m MES (pH 6.5), 25% PEG monomethyl ether 550, 0.01 m zinc sulfate, and 3% 6-aminohexanoic acid using the sitting drop vapor diffusion method and mixed with an equal volume of protein solution (20 mg/ml) and mother liquor. The crystals appeared after 1 day and belonged to space group C2. The crystals were dipped in artificial mother liquor containing 10% glycerol and flash-frozen in liquid nitrogen. A 2.1Å data set was collected at Stanford Synchrotron Radiation Lightsource beamline 9-2. Structures were solved using the program Phaser (18), the model was manually improved using the program Coot, and refinement was done with REFMAC and BUSTER (19). The buried surface in the AFD-FD complex (supplemental Table S2) was determined by first calculating the surface area of each residue in the two structures (FD and AFD) separately and then calculating the surface that is accessible in the complex. The difference shows how much of the surface of each residue is buried in the interface. Figures were made using the program PyMOL (39).
RESULTS
AFD Inhibits Proconvertase Activation, but Not Esterolytic Activity
AFD efficiently blocked FB cleavage and the formation of a C3 convertase (Fig. 1, A and B). AFD also inhibited amplification of the complement response in human serum (Fig. 1C). In contrast, 8E2, a Fab fragment of an antibody that binds to FD, did not inhibit FD proteolytic activity even at the highest concentration tested.
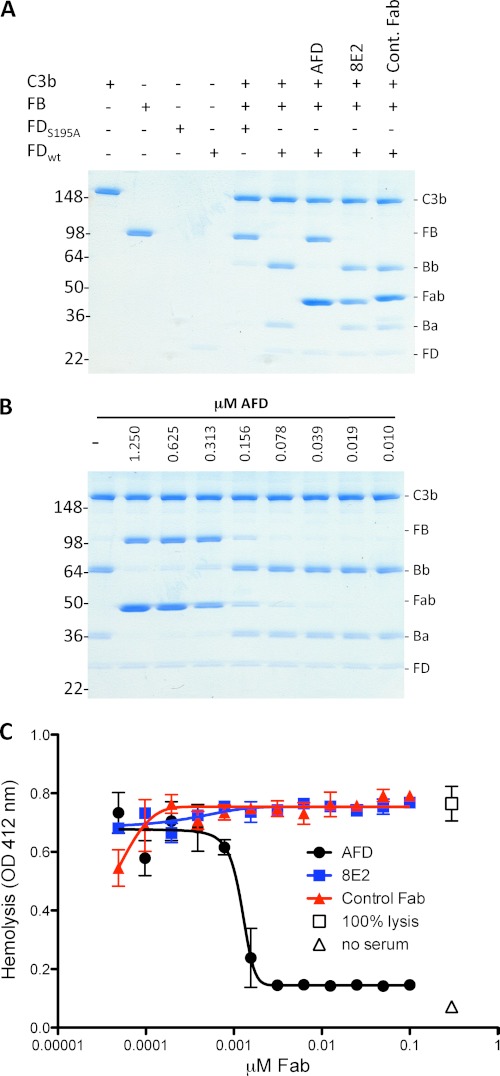
FIGURE 1.
AFD blocks activation of C3bB proconvertase. A, SDS-PAGE of proconvertase formation in the presence of catalytically inactive FD (FD(S195A)) and wild-type FD and the effects of blocking (AFD) or non-blocking (8E2) FD antibodies or anti-hepsin (control) Fab fragments. B, AFD fully inhibited C3 proconvertase activation (indicated by the absence of Ba and Bb formation) at a 1:2 molar ratio of AFD to FD. Convertase formation was measured by conversion of FB to C3Ba and C3Bb. C, AFD blocked hemolysis in an assay selective for the alternative pathway. Maximal blockade of hemolysis was achieved at a 1:1 molar ratio of AFD to FD.
AFD did not decrease the proteolytic conversion of the thioester substrate thiobenzyl benzyloxycarbonyl-l-Lysinate (Z-lys-SBzl) ester by FD (20), indicating that FD thioesterolytic activity is not inhibited by AFD (supplemental Fig. S1). Rather, AFD enhanced the kinetics of the enzymatic reaction, whereas antibody 8E2 did not affect the enzymatic activity of FD. This indicates that AFD does not prevent small substrates from accessing the FD catalytic site. The unexpected increase in thioesterolytic activity following AFD binding maybe explained by stabilization of the active site, facilitating proteolysis of small substrates, but further analysis needs to be performed to confirm this. Thus, AFD binds to an epitope on FD that is implicated in proteolytic conversion of its macromolecular substrate C3bB, in human serum.
AFD Blocks Binding of FD to C3bB
Because AFD did not affect access of small substrates to the FD active site but effectively inhibited proteolysis of its macromolecular substrate, we employed surface plasmon resonance to determine whether AFD blocks alternative pathway complement activation by interfering with FD binding to C3bB. To establish binding of FD to C3bB, a constant amount of FB was injected onto immobilized C3b. This was followed by injection of a dilution series of FD(S195A), which is unable to cleave FB (Fig. 2A). FD did not bind C3b directly (supplemental Fig. S2), in line with previous observations (21). The KD of FD(S195A) binding to C3bB is 720 nm (Fig. 2B), which is in reasonable agreement with the affinity reported previously (315 nm) (21). To determine the effect of AFD on binding, a constant amount of FD(S195A) was premixed with a dilution series of AFD or control Ab 8E2 before binding to C3bB was measured. AFD, but not 8E2, inhibited binding of FD(S195A) to C3bB in a concentration-dependent manner (Fig. 2C). Full inhibition of binding was achieved at a ∼1:1 molar ratio of AFD to FD(S195A).
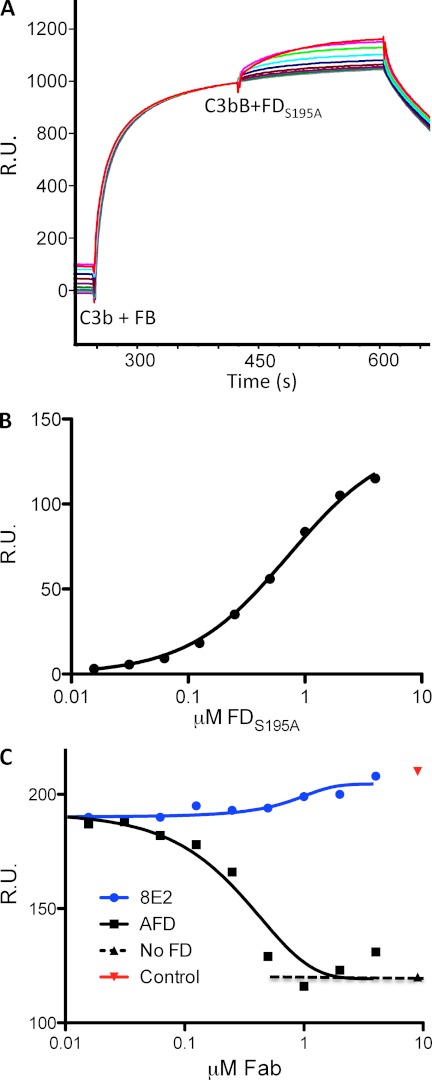
FIGURE 2.
AFD blocks binding of FD to C3bB proconvertase. A, binding of FD to a preformed C3bB complex as shown by surface plasmon resonance. A fixed concentration of FB (1 μm) was injected over immobilized C3b, followed by a dilution series of enzymatically inactive FD (FD(S195A), 2-fold dilution series of 4.0 μm to 15.6 nm). B, the binding affinity of FD for C3bB (720 nm) was measured by detecting the response (y axis) to increasing concentrations of FD(S195A) and a fixed concentration of FB (1 μm) flowed over immobilized C3b. Steady-state analysis of the binding data was used to derive dissociation constants for FD binding to C3bB. C, AFD blocked binding of FD to the C3bB complex. Fixed concentrations of FB (1 μm) and FD(S195A) (1 μm) and increasing concentrations of AFD or 8E2 were injected over immobilized C3b. The IC50 value averaged over three separate experiments is 0.43 ± 0.20 μm (mean ± S.D.) with full blockade at 0.82 ± 0.18 μm, which indicates that AFD blocks proconvertase activation at an ∼1:1 molar ratio of AFD to FD. R.U., resonance units.
Crystal Structure of FD in Complex with AFD
The structures of human and cynomolgus FD in complex with AFD were solved at 2.4 and 2.3 Å resolution, respectively (Fig. 3A and supplemental Fig. S3, A–C, and Table S1). The two symmetry-related FD molecules in either of the Fab complexes superimposed with a root mean square deviation of 0.5 Å for the 228 common Cα positions. Similarly, human FD in complex with AFD superimposed well onto the cynomolgus FD-AFD complex with a root mean square deviation ranging from 0.5 to 0.75 Å for all Cα positions, with the largest differences occurring around the poorly ordered catalytic residue His-57 and the inhibitory loop (residues 212–218; chymotrypsin numbering has been used throughout this work) (supplemental Fig. S6B). Comparison of the FD-AFD complexes with published structures of FD illustrates that no conformational changes are required in FD to allow binding of AFD and that the structure of FD in the FD-AFD complex is similar to the overall structures of native FD (Protein Data Bank codes 1HFD (22) and 1DSU (23)) and diisopropyl fluorophosphate-inhibited FD (code 1DFP) (24, 25).
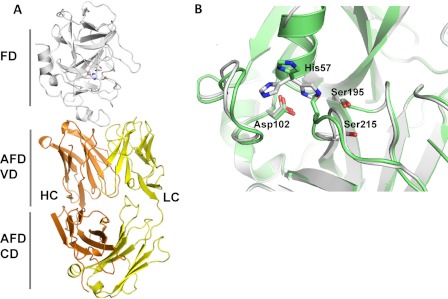
FIGURE 3.
Crystal structure of human FD in complex with AFD. A, the structure of one of the two FD-AFD complexes in the asymmetric unit is shown as a ribbon diagram. FD is shown in white, and the AFD LC and HC are shown in yellow and orange, respectively. VD, variable domain; CD, constant domain. B, close-up of the FD catalytic triad His-57, Asp-102, and Ser-195. The structures of human FD (green) and cynomolgus FD (white) are overlaid. His-57 is shown in three different positions as found in different structures. Ser-215 of the self-inhibitory loop is also indicated.
The FD-AFD binding interface is >20 Å away from the catalytic center, and the interaction of AFD with FD does not change the conformation of the active site pocket or of the oxyanion hole. In the structure of cynomolgus FD (Fig. 3B and supplemental Fig. S3C), the catalytic triad residues His-57, Asp-102, and Ser-195 are in a catalytically incompetent conformation, with the side chain of His-57 pointing away from the active site and the inhibitory loop (residues 212–218) adopting a non-competent conformation as seen in the structures of free FD (22, 23). In contrast, the inhibitory loop in human FD is rearranged compared with free FD and adopts a competent conformation similar to the one observed in the complex between C3b, FB, and FD (21). However, the side chain of His-57 is poorly defined in the electron density and was modeled in two different conformations, only one of which is catalytically competent (Fig. 3B). These differences in residues of the rather flexible active site underscore the lack of an induced conformation by binding of AFD, and it is apparent that AFD can bind the active as well as the inactive conformation of FD.
Structural Basis for High Affinity of AFD for Human and Cynomolgus FD
The interface between FD and AFD is virtually identical in all four complexes; we describe here the interface between chain A of human FD and AFD. A total of 1649 Å2 of solvent-accessible surface is buried in the interface between human FD and AFD: 901 Å2 buried on FD, 485 Å2 on the heavy chain (HC) of AFD, and the remaining 263 Å2 on the light chain (LC) of AFD (supplemental Table S2). AFD is in contact with residues 129A–132, the loop spanning residues 164–178, and Arg-223 and Lys-223A of FD (Fig. 4A). The bulk of the interaction (∼80% of the buried surface) is due to interactions involving the 170 loop of FD. Most prominently, the 170 loop of FD is “sandwiched” between the LC and HC of AFD with Arg-170A of FD deeply buried between the two chains. Arg-170A forms numerous potential hydrogen bonds and charged interactions with Glu-97 and Glu-99 of the HC and with Tyr-36 and Asn-34 of the LC (Fig. 4B and supplemental Fig. S4). Additional charged interactions are formed between Lys-223A of FD and the side chains of Asp-30, Asp-32, and Asp-92 of the LC of AFD (Fig. 4B). The interaction between Arg-170A and the LC as well as HC residues in AFD likely explains the high affinity (KD < 10 pm by surface plasmon resonance) (supplemental Fig. S5, A and B) of AFD for FD. There are nine residue differences between human and cynomolgus FD, all of which, with one exception, are located in regions that are distant from the AFD-binding interface (supplemental Fig. S6, A and B). Residue 178 is the exception, with the Glu-178 side chain of human FD engaged in a weak hydrogen bond interaction with HC Thr-28. In comparison, Gln-178 of cynomolgus FD is engaged in a hydrogen bond interaction with HC Asn-31. These minor differences likely explain the lower affinity of AFD for cynomolgus FD compared with human FD (supplemental Fig. S5B).
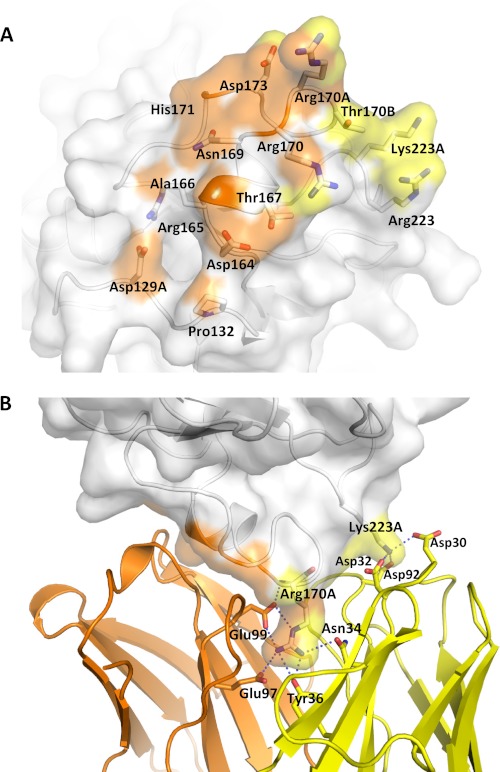
FIGURE 4.
Interface between AFD and FD. A, FD (transparent surface) interacts with the AFD HC mainly through residues in the 170 loop (orange) and interacts with both the HC and LC through residues in the 220 loop (yellow). B, Arg-170A of FD forms six hydrogen bonds with LC and HC residues of AFD. Potential hydrogen bonds are formed between Lys-223A of FD and Asp-30 and Asp-32 of the AFD LC and between Arg-170A of FD and Tyr-36 and Asn-34 of the AFD LC as well Glu-97 and Glu-99 of the AFD HC.
Major Steric Clash Prevents FD-AFD Binding to C3bB
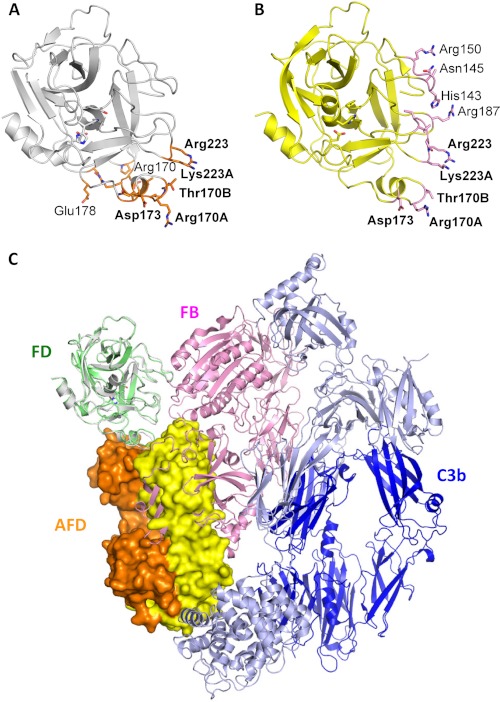
Recently, the crystal structure of FD bound to the proconvertase C3bB was reported (21). In this complex, FD forms contacts with FB in the C3bBD complex via four loops, including residues 145–149, 169–173, 185–188, and 220–224, collectively defining the FD exosite (Fig. 5, A and B) (21). Upon binding to C3bB, FD exhibits conformational changes that increase serine protease activity (21). These include structural changes in the self-inhibitory loop and the flipping of the side chain of His-57 to induce an active conformation of the catalytic triad. There are five residues in FD that establish contacts with both AFD and C3bB: Arg-170A, Thr-170B, Asp-173, Arg-223, and Lys-223A. Superimposition of AFD in complex with FD onto the structure of FD bound to C3bB (C3bBD) shows that steric clashes prevent access of FD to C3bB when bound to AFD (Fig. 5C). Hence, the structures clearly confirm the mechanism of action by which AFD inhibits the activation of alternative pathway convertase C3bB by FD.
FIGURE 5.
AFD sterically prevents FD from binding to C3bB proconvertase. A, residues in FD (white ribbon representation, from the FD-AFD complex) in contact with AFD (orange sticks). B, residues in FD (yellow ribbon representation, from the C3bBD complex) in contact with FB (pink sticks). C, modeling of a major steric clash between FB (pink) and the AFD HC (orange) and LC (yellow). Dark blue indicates the C3b β-chain, and light blue indicates the C3b α-chain.
DISCUSSION
Here, we have described the molecular mechanism by which AFD, a promising therapeutic antibody, inhibits alternative pathway complement activation. The specificity and proteolytic activity of FD for its substrate FB are determined primarily by its exosite-mediated binding to the open form of FB (21). Upon binding of AFD to the exosite, FD is not able to cleave FB in C3bB to generate an active C3Bb convertase and amplify the generation of complement effector molecules in serum. AFD is a potent inhibitor of the alternative complement pathway, and the FD-AFD co-structure provides a molecular explanation for AFD activity. Although the size of the surface area on FD that is buried by AFD (901 Å) is typical for an Ab-protein interaction (26), a key substrate-binding loop on FD is deeply buried in the interface wedged between the AFD HC and LC, allowing for formation of multiple hydrogen bonds and charged interactions between FD and AFD. Hence, the high potency of AFD can be explained by the high binding affinity of AFD for FD, which is >70,000-fold higher than the binding affinity of FD for its substrate C3bB.
AFD binding to the exosite loop distant from the catalytic site can fully explain the mechanism by which AFD inhibits proteolytic activity, but not thioesterolytic activity. The fact that AFD binding to FD increases thioesterolytic activity for a small substrate indicates that subtle conformational changes may take place in the active site configuration that cannot be defined in the structure. An allosteric effect of AFD may explain why human FD is partly in the active conformation in the complex with AFD. However, interactions of FD with the open form of FB are required to induce full activation of FD (21), and AFD prevents any of these interactions from taking place.
Previous crystal structures of Fab-serine protease complexes defined two distinct mechanisms by which antibodies block catalytic activity (27). The antibodies either insert complementarity-determining region loops into the catalytic cleft to prevent access of the scissile peptide (steric inhibition) or impose structural changes at the S2–S4 sites by an allosteric mechanism (27–29). Both mechanisms are competitive in nature and result in inhibition of small synthetic as well as macromolecular substrate proteolysis. The mechanism identified herein is different in that AFD binds to the FD exosite region to inhibit processing of the macromolecular substrate FB, but not of a small synthetic substrate. The latter finding that AFD did not inhibit small substrate cleavage is consistent with the absence of conformational changes at the catalytic cleft by AFD. Instead, AFD binds to two (170 and 220 loops) of the four FD surface loops that define the FD exosite (21) and thereby completely prevents access of the open FB form to productively interact with FD. Therefore, inhibition of the exosite interaction by AFD defines a third molecular mechanism by which antibodies interfere with catalysis of trypsin-like serine proteases.
Preventing substrates from interacting productively with complement convertases has been adopted by pathogens as a strategy to evade complement attack. Staphylococcus aureus inhibitors SCIN-A and SCIN-B bind to a functionally important site on C3b, entrapping the convertase in an inactive state (30–32). The extracellular fibrinogen-binding molecule of S. aureus acts as both an allosteric and a competitive inhibitor by inducing conformational changes in C3b that prevent formation of an active C3 convertase (33). An example of a complement inhibitor that targets the substrate rather than the convertase is the tick C5 inhibitor OmCI, which binds to the C345C domain of C5, preventing interaction of C5 with its convertase (34, 35). Steric blockade of enzyme-substrate interactions has also been described for therapeutic inhibitors of complement activation. Compstatin, a C3-binding peptide, sterically inhibits formation of the complement convertases (36, 37). S77, a C3b selective Ab, blocks alternative pathway convertase activation by inhibiting binding of FB to C3b (38). Finally, complement receptor CRIg inhibits complement activation by preventing binding of C5 to the C5 convertase (17). Thus, steric inhibition is a potent strategy to inhibit complement convertases. In sum, this study demonstrates that a high-affinity antibody directed to the FD exosite can block a low-affinity enzyme-substrate interaction required for complement activation and amplification.
Supplementary Material
Acknowledgments
We thank Michael Fung, Charles Winter, Charles Morgan, Laura DeForge, Kelly Loyet, Bernd Wranik, Jeremy Good, and Lizette Embuscado for valuable contributions.
All authors were employees at Genentech, Inc., a for-profit institution, while the studies were performed.

This article contains supplemental “Methods,” Figs. S1–S6, and Tables S1 and S2.
The atomic coordinates and structure factors (codes 4D9R and 4D9Q) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FD
- factor D
- AFD
- anti-FD Fab fragment
- mAb
- monoclonal antibody
- FB
- factor B
- HC
- heavy chain
- LC
- light chain.
REFERENCES
- 1. Hecker L. A., Edwards A. O., Ryu E., Tosakulwong N., Baratz K. H., Brown W. L., Charbel Issa P., Scholl H. P., Pollok-Kopp B., Schmid-Kubista K. E., Bailey K. R., Oppermann M. (2010) Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum. Mol. Genet. 19, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scholl H. P., Charbel Issa P., Walier M., Janzer S., Pollok-Kopp B., Börncke F., Fritsche L. G., Chong N. V., Fimmers R., Wienker T., Holz F. G., Weber B. H., Oppermann M. (2008) Systemic complement activation in age-related macular degeneration. PLoS ONE 3, e2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reynolds R., Hartnett M. E., Atkinson J. P., Giclas P. C., Rosner B., Seddon J. M. (2009) Plasma complement components and activation fragments: associations with age-related macular degeneration genotypes and phenotypes. Invest. Ophthalmol. Vis. Sci. 50, 5818–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stanton C. M., Yates J. R., den Hollander A. I., Seddon J. M., Swaroop A., Stambolian D., Fauser S., Hoyng C., Yu Y., Atsuhiro K., Branham K., Othman M., Chen W., Kortvely E., Chalmers K., Hayward C., Moore A. T., Dhillon B., Ueffing M., Wright A. F. (2011) Complement factor D in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 52, 8828–8834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ricklin D., Lambris J. D. (2007) Complement-targeted therapeutics. Nat. Biotechnol. 25, 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Volanakis J. E., Barnum S. R., Giddens M., Galla J. H. (1985) Renal filtration and catabolism of complement protein D. N. Engl. J. Med. 312, 395–399 [DOI] [PubMed] [Google Scholar]
- 7. Harboe M., Ulvund G., Vien L., Fung M., Mollnes T. E. (2004) The quantitative role of alternative pathway amplification in classical pathway-induced terminal complement activation. Clin. Exp. Immunol. 138, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lesavre P. H., Müller-Eberhard H. J. (1978) Mechanism of action of factor D of the alternative complement pathway. J. Exp. Med. 148, 1498–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barnum S. R., Niemann M. A., Kearney J. F., Volanakis J. E. (1984) Quantitation of complement factor D in human serum by a solid-phase radioimmunoassay. J. Immunol. Methods 67, 303–309 [DOI] [PubMed] [Google Scholar]
- 10. Sanders P. W., Volanakis J. E., Rostand S. G., Galla J. H. (1986) Human complement protein D catabolism by the rat kidney. J. Clin. Invest. 77, 1299–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ündar A., Eichstaedt H. C., Clubb F. J., Jr., Fung M., Lu M., Bigley J. E., Vaughn W. K., Fraser C. D., Jr. (2002) Novel anti-factor D monoclonal antibody inhibits complement and leukocyte activation in a baboon model of cardiopulmonary bypass. Ann. Thorac. Surg. 74, 355–362 [DOI] [PubMed] [Google Scholar]
- 12. Fung M., Loubser P. G., Undar A., Mueller M., Sun C., Sun W. N., Vaughn W. K., Fraser C. D., Jr. (2001) Inhibition of complement, neutrophil, and platelet activation by an anti-factor D monoclonal antibody in simulated cardiopulmonary bypass circuits. J. Thorac. Cardiovasc. Surg. 122, 113–122 [DOI] [PubMed] [Google Scholar]
- 13. Tanhehco E. J., Kilgore K. S., Liff D. A., Murphy K. L., Fung M. S., Sun W. N., Sun C., Lucchesi B. R. (1999) The anti-factor D antibody, mAb 166-32, inhibits the alternative pathway of the human complement system. Transplant. Proc. 31, 2168–2171 [DOI] [PubMed] [Google Scholar]
- 14. Ündar A., Fraser C. D., Jr. (2002) Anti-factor D monoclonal antibody, pulsatile flow, and cardiotomy suction during cardiopulmonary bypass. Eur. J. Cardiothorac. Surg. 22, 330–331 [DOI] [PubMed] [Google Scholar]
- 15. Helmy K. Y., Katschke K. J., Jr., Gorgani N. N., Kljavin N. M., Elliott J. M., Diehl L., Scales S. J., Ghilardi N., van Lookeren Campagne M. (2006) CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell 124, 915–927 [DOI] [PubMed] [Google Scholar]
- 16. Ganesan R., Kolumam G. A., Lin S. J., Xie M. H., Santell L., Wu T. D., Lazarus R. A., Chaudhuri A., Kirchhofer D. (2011) Proteolytic activation of pro-macrophage-stimulating protein by hepsin. Mol. Cancer Res. 9, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 17. Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. (2006) Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444, 217–220 [DOI] [PubMed] [Google Scholar]
- 18. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta. Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 20. Kim S., Narayana S. V., Volanakis J. E. (1995) Catalytic role of a surface loop of the complement serine protease factor D. J. Immunol. 154, 6073–6079 [PubMed] [Google Scholar]
- 21. Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R. S., Lambris J. D., Gros P. (2010) Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 330, 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jing H., Babu Y. S., Moore D., Kilpatrick J. M., Liu X. Y., Volanakis J. E., Narayana S. V. (1998) Structures of native and complexed complement factor D: implications of the atypical His-57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. J. Mol. Biol. 282, 1061–1081 [DOI] [PubMed] [Google Scholar]
- 23. Narayana S. V., Carson M., el-Kabbani O., Kilpatrick J. M., Moore D., Chen X., Bugg C. E., Volanakis J. E., DeLucas L. J. (1994) Structure of human factor D. A complement system protein at 2.0 Å resolution. J. Mol. Biol. 235, 695–708 [DOI] [PubMed] [Google Scholar]
- 24. Cole L. B., Chu N., Kilpatrick J. M., Volanakis J. E., Narayana S. V., Babu Y. S. (1997) Structure of diisopropyl fluorophosphate-inhibited factor D. Acta Crystallogr. D. Biol. Crystallogr. 53, 143–150 [DOI] [PubMed] [Google Scholar]
- 25. Cole L. B., Kilpatrick J. M., Chu N., Babu Y. S. (1998) Structure of 3,4-dichloroisocoumarin-inhibited factor D. Acta. Crystallogr. D. Biol. Crystallogr. 54, 711–717 [DOI] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27. Ganesan R., Eigenbrot C., Kirchhofer D. (2010) Structural and mechanistic insight into how antibodies inhibit serine proteases. Biochem. J. 430, 179–189 [DOI] [PubMed] [Google Scholar]
- 28. Farady C. J., Sun J., Darragh M. R., Miller S. M., Craik C. S. (2007) The mechanism of inhibition of antibody-based inhibitors of membrane-type serine protease 1 (MT-SP1). J. Mol. Biol. 369, 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y., Eigenbrot C., Liang W. C., Stawicki S., Shia S., Fan B., Ganesan R., Lipari M. T., Kirchhofer D. (2007) Structural insight into distinct mechanisms of protease inhibition by antibodies. Proc. Natl. Acad. Sci. U.S.A. 104, 19784–19789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ricklin D., Tzekou A., Garcia B. L., Hammel M., McWhorter W. J., Sfyroera G., Wu Y. Q., Holers V. M., Herbert A. P., Barlow P. N., Geisbrecht B. V., Lambris J. D. (2009) A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J. Immunol. 183, 2565–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rooijakkers S. H., Wu J., Ruyken M., van Domselaar R., Planken K. L., Tzekou A., Ricklin D., Lambris J. D., Janssen B. J., van Strijp J. A., Gros P. (2009) Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 10, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia B. L., Summers B. J., Lin Z., Ramyar K. X., Ricklin D., Kamath D. V., Fu Z. Q., Lambris J. D., Geisbrecht B. V. (2012) Diversity in the C3b convertase contact residues and tertiary structures of the staphylococcal complement inhibitor (SCIN) protein family. J. Biol. Chem. 287, 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H., Ricklin D., Hammel M., Garcia B. L., McWhorter W. J., Sfyroera G., Wu Y. Q., Tzekou A., Li S., Geisbrecht B. V., Woods V. L., Jr., Lambris J. D. (2010) Allosteric inhibition of complement function by a staphylococcal immune evasion protein. Proc. Natl. Acad. Sci. U.S.A. 107, 17621–17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fredslund F., Laursen N. S., Roversi P., Jenner L., Oliveira C. L., Pedersen J. S., Nunn M. A., Lea S. M., Discipio R., Sottrup-Jensen L., Andersen G. R. (2008) Structure of and influence of a tick complement inhibitor on human complement component 5. Nat. Immunol. 9, 753–760 [DOI] [PubMed] [Google Scholar]
- 35. Roversi P., Lissina O., Johnson S., Ahmat N., Paesen G. C., Ploss K., Boland W., Nunn M. A., Lea S. M. (2007) The structure of OmCI, a novel lipocalin inhibitor of the complement system. J. Mol. Biol. 369, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Janssen B. J., Halff E. F., Lambris J. D., Gros P. (2007) Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J. Biol. Chem. 282, 29241–29247 [DOI] [PubMed] [Google Scholar]
- 37. Ricklin D., Lambris J. D. (2008) Compstatin: a complement inhibitor on its way to clinical application. Adv. Exp. Med. Biol. 632, 273–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katschke K. J., Jr., Stawicki S., Yin J., Steffek M., Xi H., Sturgeon L., Hass P. E., Loyet K. M., Deforge L., Wu Y., van Lookeren Campagne M., Wiesmann C. (2009) Structural and functional analysis of a C3b-specific antibody that selectively inhibits the alternative pathway of complement. J. Biol. Chem. 284, 10473–10479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.4, Schrödinger, LLC, San Diego, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.