Background: Intron 2 of CNR1 gene contains multiple disease-associated SNPs.
Results: Allelic variants of a novel enhancer in CNR1 intron 2 affect its MAPK response in hypothalamus and hippocampus.
Conclusion: Alleles of enhancer may be functionally linked to obesity and addictive behavior.
Significance: Understanding the effects of CNR1 polymorphisms on gene regulation will accelerate understanding of human disease.
Keywords: Addiction, Cannabinoid Receptors, Cell Signaling, Gene Regulation, Genetic Polymorphism, Hypothalamus, Neurons, Obesity, Transcription Enhancers, Hippocampus
Abstract
Polymorphisms within intron 2 of the CNR1 gene, which encodes cannabinoid receptor 1 (CB1), have been associated with addiction, obesity, and brain volume deficits. We used comparative genomics to identify a polymorphic (rs9444584-C/T) sequence (ECR1) in intron 2 of the CNR1 gene that had been conserved for 310 million years. The C-allele of ECR1 (ECR1(C)) acted as an enhancer in hypothalamic and dorsal root ganglia cells and responded to MAPK activation through the MEKK pathway but not in hippocampal cells. However, ECR1(T) was significantly more active in hypothalamic and dorsal root ganglia cells but, significantly, and in contrast to ECR1(C), was highly active in hippocampal cells where it also responded strongly to activation of MAPK. Intriguingly, rs9444584 is in strong linkage disequilibrium with two other SNPs (rs9450898 (r2 = 0.841) and rs2023239 (r2 = 0.920)) that have been associated with addiction, obesity (rs2023239), and reduced fronto-temporal white matter volumes in schizophrenia patients as a result of cannabis misuse (rs9450898). Considering their high linkage disequilibrium and the increased response of ECR1(T) to MAPK signaling when compared with ECR1(C), it is possible that the functional effects of the different alleles of rs9444584 may play a role in the conditions associated with rs9450898 and rs2023239. Further analysis of the different alleles of ECR1 may lead to a greater understanding of the role of CNR1 gene misregulation in these conditions as well as chronic inflammatory pain.
Introduction
The cannabinoid receptor 1 (CB1)5 is a Gi/Go-protein-coupled receptor whose endogenous ligands include anandamide and 2-arachidonoyl glycerol (1, 2). CB1 is strongly expressed in a number of different regions of the central nervous system such as the hypothalamus, hippocampus, and dorsal root ganglia (DRG) where it is known to play a role in modulating appetite (3–5), cognition and memory (6–8), and inflammatory pain, respectively (9).
Because of the role of CB1 in these processes, there have been a number of genetic studies linking polymorphisms in and around the CNR1 gene to conditions such as cognitive decline, drug addiction, schizophrenia, obesity, and inflammatory pain. For example, one polymorphism that occurs within intron 2 of the CNR1 gene, rs2023239, has been associated with impulsivity (10), obesity (11), nicotine dependence (12), alcoholism (13), cannabis withdrawal and dependence (14, 15), substance addiction, and resistance to anti-depressive treatment (16). A second CNR1 intron 2 polymorphism, rs9450898, was associated with smaller fronto-temporal white matter volumes and greater schizophrenia risk due to cannabis misuse (17). Intriguingly, rs9450898 and rs2023239 are separated by only 2 kb and are in strong linkage disequilibrium (LD; r2 = 0.915). However, no mechanism has yet been discovered to explain the symptoms associated with rs9450898 or rs2023239.
We explored the hypothesis that polymorphic variation within CNR1 intron 2 might change the activity of unidentified cis-regulatory regions. We used comparative genomics to identify highly conserved functional elements within CNR1 intron 2 that might represent a cis-regulatory region. We then used molecular biology, primary cell culture, and pharmacology to isolate these sequences and to assess the effects of different alleles on the activity, tissue specificity, and signal transduction response of these polymorphic cis-regulatory regions. The significances of these results are discussed in the context of the role of the CB1 receptor in the hypothalamus, hippocampus, and dorsal root ganglia, and consequences for disease susceptibility are explored.
EXPERIMENTAL PROCEDURES
Bioinformatic Analysis
17-species vertebrate genome comparisons and detection of linkage disequilibrium was carried out using the Human (Homo sapiens) Genome Browser Gateway through the UCSC Genome Browser (see Fig. 1). Detection of transcription factor binding consensus sequences was carried out using the web-based program MATCH through BIOBASE (18). Prediction of the effects of rs9444584 on transcription factor-DNA binding was carried out using the newly developed RegSNP website. Linkage disequilibrium between SNPs was quantified using SNAP LD.
FIGURE 1.
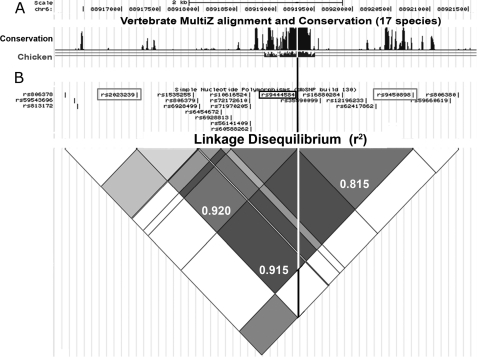
rs9444584 is in strong LD with rs92023230 and rs9450896 and occurs within a region of CNR1 intron 2 that has been conserved for 310 million years. A, a 17-species vertebrate alignment plot from the UCSC Genome Browser highlights the presence of a putative enhancer region conserved between birds and humans. The white line represents the location of rs944458 within this sequence. The top scale bar represents linear distance (2 kb), and the second scale bar represents coordinates in base pairs along the length of human chromosome 6. B, an LD heat map analysis of SNPs within CNR1 intron 2 derived from the UCSC browser demonstrating levels of LD (r2) between the different SNPs. This map is in linear register with the graph in A. Exact measurements of LD (as calculated by SNAP LD) are displayed as decimal fractions in the grey diamonds linking the three SNPs of interest (rs9444584, rs92023230, and rs9450896). The location of rs9444584 is highlighted using a black box and line. The locations of rs92023230 and rs9450896 are highlighted using gray boxes.
Plasmid Constructs
ECR1 was amplified from placental human DNA using the Expand high fidelity PCR kit and the following primers, ECR1F-TAAGCTAGGGCATGGGTGTG and ECR1R-TAGTGGAGAGGCAGGTTTGC (657 bp), as described in the manufacturer's instructions (Roche Applied Science), and cloned into pGEM-T Easy as described (Stratagene) to form pECR1(C)-GEMT-Easy. The correct amplification and orientation of ECR1(C) within the pGEM-T Easy vector was checked with restriction enzyme digestion and sequencing. The pLuc plasmid (renamed for clarity from pTAL-Luc, Clontech, see Fig. 2D) contains a herpes simplex virus-thymidine kinase minimal promoter that has a low transcriptional activity. pECR1(C) was produced by removing the ECR1(C) fragment from pECR1(C)-GEMT-Easy using NheI and SmaI and cloning into the ApaI (made blunt-ended using Klenow) and SpeI sites of the pLuc vector (see Fig. 2D). pECR1(T) was recreated by mutating the ECR1(C) sequence within pGEM-T Easy vector using QuikChange II site-directed mutagenesis kit (Stratagene) and the following primers, ECR1F-GAGAGTTCATTACTAATATGGCTTAGG and ECR1RCCTAAGCCATATTAGTAATGAACTCTC, to produce ECR1(T). ECR1(T) was then cloned into pLuc in the same way as pECR1(C) to produce pECR1(T) (see Fig. 2D).
FIGURE 2.
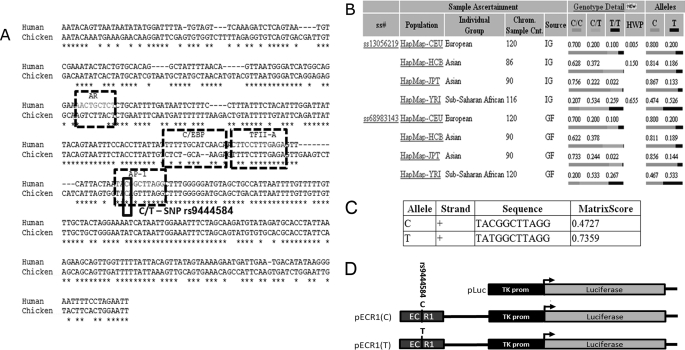
rs9444584 allele frequency varies greatly in different human populations and changes the predicted affinity of AP1 transcription factor. A, pairwise alignment of the human and chicken ECR1 sequences showing predicted transcription factor binding sites and allelic variants of rs944458. AR, androgen receptor; C/EBP, CCAAT enhancer-binding protein; TFII-A, transcription factor II A. B, table from the National Center for Biotechnology Information (NCBI) SNP web site showing population diversity of rs944458 in European, Asian, and sub-Saharan African populations. C, table generated by the RegSNP algorithm showing predicted relative binding affinities of each allele of rs944458 to the AP1 transcription factor. Matrix score and core score values represent the probability that AP1 will bind to each allelic binding site. D, diagrammatic representation of each of the plasmid constructs used in the current study (not to scale). Bent arrows represent the transcriptional start site of the luciferase gene. ss#, assigned submitter SNP ID, Chrm. Sample. Cnt, chromosome sample count, IC, individual Genotype, and GF, genotype frequency.
Primary Cell Culture
1–3-day-old rat neonates were humanely euthanized in accordance with current United Kingdom Home Office schedule 1 guidelines, and the hippocampal or hypothalamic regions were immediately dissected out in into DMEM/F12 (Invitrogen) under sterile conditions. The tissue was chopped into fine pieces and placed in 2 ml of trypsin/0.5 m EDTA for 6 min, and tissues were dissected as described previously (19). The tissue was then dissociated by gentle trituration in DMEM/F12 medium with a fire-polished glass pipette (∼1-mm diameter) and centrifuged at 5000 rpm for 2 min. This process was repeated, and cells were counted on a hemocytometer and plated out onto 24-well plates at a density of 5 × 104/cm2. Cultures were allowed to recover for 3 days at 37 °C in medium consisting of 90% DMEM/F12 media, 10% dialyzed FCS with penicillin/streptomycin until the majority of cells had formed neurite outgrowths. DRG cells were recovered and cultured as described previously (19, 20).
Cell Transfection
Magnetic transfection of plasmid constructs into primary lines was performed according to the manufacturer's instructions (OZ Biosciences). In summary, NeuroMag reagent was added to the plasmid preparation in the ratio of 3 μl/1 μg of plasmid DNA in the medium. The DNA-media-NeuroMag solution was mixed and incubated at room temperature for 15 min. The DNA-media-NeuroMag solution was then added to previously prepared single cell cultures in 24-well plates and incubated on a magnetofection magnetic plate for 15 min in a cell culture incubator at 37 °C and 5% CO2. Following transfection, cells were treated with 10 μm angiotensin II (Tocris) or angiotensin II plus a MEK kinase inhibitor (U0216, 1 μm, Tocris) a JNK II inhibitor (SP600125, 50 nm, Tocris), or a p38 kinase inhibitor (SB202190, 10 μm, Tocris) for 24 h.
Dual-Luciferase Assay
24 h after transfection and agonist/antagonist treatments, Dual-Luciferase assays were performed on lysates made from primary cell cultures according to the manufacturer's instructions (Promega). The Dual-Luciferase assay analysis was carried out on a GloMax 96 microplate luminometer (Promega) using 20 μl of cell lysate per well of a white 96-well plate.
Statistical Analysis
All experiments were repeated a minimum of three times on separate dates using separate groups of animals (n ≥ 3). Two-tailed Student's t tests or analysis of variance were used, where appropriate, to test the significance of data derived from primary cell cultures. Statistical analysis was done using Microsoft Excel.
RESULTS
A Highly Conserved Region within Intron 2 Contains an SNP in Strong LD with rs9450898 and rs2023239
Many novel cis-regulatory elements display high levels of evolutionary conservation because of their critical role in modulating the tissue-specific expression of genes (19–20, 22–30). Because intron 2 of the CNR1 gene contained a number of disease-associated SNPs (10–17), we asked whether intron 2 contained functional sequences such as cis-regulatory elements that could direct tissue-specific gene expression. Using comparative genomics, we identified a 402-bp region of high conservation (chr6:88919055–88919457) that had been conserved from the common ancestor of birds and humans (310 million years; Figs. 1A and 2A). Intriguingly, we found that this sequence contained a polymorphism (rs9444584; C/T) that was in strong LD with rs9450898 and rs2023239 (r2 = 0.815 and 0.920, respectively), which have been associated with different conditions in multiple association analyses (10–17). The two alleles of this 402-bp region of DNA were called ECR1(C) and ECR1(T).
Examination of the population diversity of these alleles demonstrated that in Eurasian populations, the T allele was comparatively rare and was only found in 13–20% of the alleles within the population (Fig. 2B). However, in sub-Saharan Africa, the frequency between C and T alleles was nearly equal (47 and 53%, respectively, Fig. 2B). Predictive software programs (Match and Transfac) were used to show the presence of androgen receptor (AR), CCAAT enhancer-binding protein (C/EBP), transcription factor II A (TFII-A), and activator protein-1 (AP1) binding sites within ECR1 (Fig. 2A). Using our newly developed RegSNP program, we were able to predict that ECR1(C) would have relatively low affinity for the AP-1 transcription factor (p-core binding = 0.448), whereas the ECR1(T) allele would have a significantly increased affinity (p-core binding = 0.736).
ECR1(C) Is Active within Primary Hypothalamic Cell Cultures and DRG but Not in Primary Hippocampal Cells
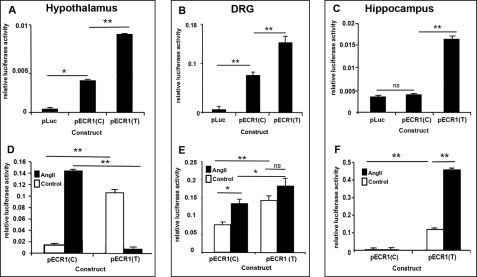
Because CNR1 is expressed in many different parts of the brain and peripheral nervous system such as the hypothalamus, hippocampus, and DRG, we explored the hypothesis that the most common allele, ECR1(C), acted as an enhancer of promoter activity within cells derived from specific regions of the brain. ECR1(C) was amplified using high fidelity PCR from human placental DNA and cloned into the pTAL-Luc vector (Clontech) that contains a TATA-like promoter (PTAL) region from the herpes simplex virus-thymidine kinase promoter linked to a luciferase reporter (Fig. 2D). This commercially available plasmid has been widely used to assay gene regulatory sequences in the past and will subsequently be referred to as pLuc. Because of the known function of CB1 in the hypothalamus, DRG, and hippocampus in appetite, cognition, and pain, respectively, we examined whether the ECR1(C) acted as an enhancer of promoter activity in primary cells derived from these different tissues. We also chose to use primary lines as the activity of many enhancer regions has been shown to be highly context-dependent (19–20, 22, 23, 25, 28–30). pLuc and pECR1(C) were magnetofected into primary cell cultures derived from the hypothalamus, hippocampus, or DRG, and after 24 h, Dual-Luciferase assays were performed. It was observed that ECR1(C) was able to act as an enhancer of promoter activity in hypothalamic and DRG cells (Fig. 3, A and B) but not in hippocampal cells (Fig. 3C).
FIGURE 3.
Allele-specific properties of ECR1 in different primary cell types and following stimulation of MAP kinase pathways. A–F, graphs showing the results of Dual-Luciferase analyses of primary cells derived from neonate rat hypothalamus (A and D), DRG (B and E), and hippocampus (C and F) following transfection with the different constructs shown in Fig. 2D and treatment with angiotensin II for 24 h (D–F; 10 μm). Cells were lysed, and relative luciferase activity was determined and normalized to an internal Renilla luciferase control (pRL-CMV). ns = not significant, *, p = ≤ 0.05, **, p = ≤ 0.01.
ECR1(C) Enhancer Activity Is Responsive to MAP Kinase Activation in Primary Hypothalamus- and DRG-derived Cells but Not in Hippocampal Cells
Angiotensin II (angiotensin) is a widely recognized inducer of many different MAP kinase pathways at concentrations ranging from 500 nm to 10 μm (31–33). We cultured different primary cells transfected with pECR1(C) in the presence of angiotensin II and demonstrated that ECR1(C) could respond to angiotensin in both primary DRG and hypothalamic cells (Fig. 3, A and B). However, we were unable to detect any significant up-regulation of ECR1(C) in hippocampus-derived cells.
Response of ECR1(C) to Angiotensin in Hypothalamic Cells Is Modulated by MEK Kinase and p38 Kinase Pathways
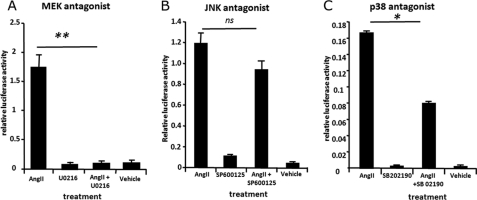
We explored which of the three major MAP kinase pathways were involved in mediating the effects of angiotensin on ECR1(C) by culturing pECR1(C)-transfected hypothalamic cells in the presence of angiotensin and an antagonist specific for the MEK kinase (U0216), JNK (SP6000125), or p38MAP (SB202190) kinase pathways in hypothalamic cells. The JNK kinase inhibitor had no significant effect on the ability of angiotensin to induce the activity of ECR1(C) (Fig. 4B). However, incubation with the p38 agonist halved the induction of ECR1(C) induced by angiotensin, and treatment with the MEK kinase inhibitor completely abolished the action of angiotensin (Fig. 4, A and C).
FIGURE 4.
ECR1(C) activity is controlled by MEK/p38 kinase signaling in hypothalamic neurons. Primary rat hypothalamic neurons were cultured for 3 days and then transfected with a pECR1(C) construct and treated with angiotensin II (AngII, 10 μm) in the presence/absence of MEK kinase inhibitor (U0216, 1 μm) (A), JNK II inhibitor (SP600125, 50 nm) (B), or p38 kinase inhibitor (SB 202190, 10 μm) (C). The control is the pECR1(C) construct treated with a respective vehicle. Cells were incubated with treatment for 24 h and then assayed for luciferase activity. Cells were lysed, and a relative luciferase measurement was determined and normalized to an internal Renilla luciferase control (pRL-CMV). ns = not significant, *, p = ≤ 0.05, **, p = ≤ 0.01.
ECR1(T) Allele Demonstrates Significant Differences from ECR1(C) as an Enhancer in Hippocampal Cell Cultures
Using bioinformatics, we predicted that ECR1(T) would bind the AP1 protein with a higher affinity (Fig. 2C). This is an interesting prediction in the context of MAP kinase signaling as AP1 activity is known to be modulated by MAP kinase pathways including JNK and MEK kinase (34). We used site-directed mutagenesis of the pECR1(C) plasmid to produce pECR1(T) (Fig. 2D). We compared pECR1(T) activity with that of pECR1(C) in hypothalamic, DRG, or hippocampal cell cultures. ECR1(T) demonstrated 50 and 40% higher enhancer activity in primary cells derived from hypothalamus and DRGs, respectively, than the ECR1(C) allele (Fig. 3, A and B). Significantly, and in strong contrast to ECR1(C), it was observed that ECR1(T) demonstrated potent enhancer activity in hippocampal neurons where ECR1(C) was inactive (Fig. 3C).
ECR1(T) Demonstrates Significant Allele-specific Differences from ECR1(C) in Its Response to Activation of MAP Kinases
We have demonstrated that ECR1(C) responded to MAP kinase activation in both hypothalamus-derived and DRG-derived primary cultures but not in hippocampal cultures (Fig. 3, D–F). However, in hippocampal cells treated with angiotensin, and in direct contrast to ECR1(C), activity of ECR1(T) was increased by over 300% when compared with the untreated sample (Fig. 3F). Surprisingly, although activation of MAP kinases in these hypothalamic cells increased the activity of ECR1(C) by nearly 10-fold, application of angiotensin to hypothalamic cultures transfected with pECR1(T) virtually inactivated ECR1(T) activity in these cells (Fig. 3D). Similarly, although treatment of DRG neurons with angiotensin increased activity of ECR1(C), no significant increase in ECR1(T) activity was observed (Fig. 3E).
DISCUSSION
Using a unique combination of in silico genetics, comparative genomics, molecular biology, and primary cell culture, we present evidence of a novel cis-regulatory region, ECR1(C), that is active in hypothalamus- and DRG-derived cells where it is responsive to MAP kinase signaling, but inactive in the hippocampus. However, the most significant finding from the current study is that, in contrast to ECR1(C) a second allele of ECR1 present in the population, ECR1(T), acts as a potent MAP kinase-inducible enhancer in hippocampus cells. These intriguing observations may have important implications for understanding disease because rs9444584 is in strong LD with two other polymorphisms that have been previously linked to diseases such as obesity, addiction, and reduced frontal brain volumes in schizophrenia as a result of cannabis misuse.
We explored the possibility that MAP kinase pathways were involved in controlling ECR1 as MAP kinase pathways in the hypothalamus and DRG have been shown to play a role in the modulation of appetite (35) and pain (36), processes in which the CB1 receptor is also involved (3–5). Furthermore, CB1 activation is known to trigger MAP kinase pathways (37). Thus, it is possible that one role for the ECR1(C) enhancer in the hypothalamus would be to allow a response to MAP kinase pathways, triggered by CB1 activation, to modulate CB1 levels in response to food availability. For example, MEK/ERK kinase pathways are induced by activation of CB1 in N1E-115 neuroblastoma cells, supporting the existence of a CB1-MAP kinase autocrine loop in certain groups of neurons (38). In keeping with these studies, we were able to demonstrate that the MAP kinase pathways responsible for the activity of ECR1(C) in hypothalamic cells were primarily the MEK kinase pathways with input from p38MAP kinases. These are interesting observations as the T allele of rs2023239, which is in strong LD with rs9444584, has been associated with higher body mass index in both Swiss obese subjects and Danish individuals (11). Considering the role of the CB1 gene in appetite in the hypothalamus and the triggering of MAP kinase signaling by CB1 activation, it can be hypothesized that increased sensitivity of ECR1(T) to MAP kinase signaling might play a role in perturbing CB1 expression in the hypothalamus, leading to changes in appetite and increased body mass index.
Because our algorithm predicted that AP1 binding affinity would be increased in ECR1(T), we were not surprised to observe that it became more responsive to MAP kinase activation in hypothalamic cells. However, we were surprised to observe such a clear difference in the ability of the ECR1(C) and ECR1(T) alleles to activate promoter activity in hippocampal cells. In addition, the ability of ECR1(T) to respond to MAP kinase activation in hippocampal cells, in clear contrast to the response of ECR1(C) in these cells, was also unexpected. These may be important observations as MAP kinase signaling, in the form of ERK-MEK kinase, in the hippocampus is involved in CB1-mediated neurogenesis and the synaptic integrity required for stable long term memory formation and cognition (39–42).
In addition, changes in the structure of the hippocampus and in plasticity of hippocampal neurons have been linked to cannabis-related psychosis (43–45). Intriguingly, rs94580898 is in strong LD with rs9444584 and has been associated with reductions in white matter in schizophrenia patients following cannabis overuse (17). The heightened sensitivity of ECR1(T) to MAP kinase signaling in the hippocampus in comparison with ECR1(C) may represent a possible mechanisms to explain the cognitive deficits and reduced white matter experienced by many schizophrenia sufferers and heavy marijuana users.
Consistent with the results of the current study, previous research has demonstrated that that CB1 receptor coupling to G-proteins differs between different brain regions and therefore depends on the cellular environment, i.e. it is context-dependent. One possible mechanism governing this context dependence is receptor coupling. For example, Breivogel et al. (46) demonstrated that receptor coupling efficacy is relatively higher in the hypothalamus than the hippocampus. From autoradiographic analysis, the relatively sparse cannabinoid receptors in the hypothalamus appeared to activate nearly as many G-proteins as in the receptor-dense hippocampus. These data imply that the CB1 receptor-mediated initiation of downstream signaling in the hypothalamus and hippocampus differ significantly and may have consequences for the initiation of MEK-mediated regulation of CNR1 expression in the hippocampus. This is in line with our results demonstrating regional difference in control of CNR1 expression via MAPK signaling. The findings have far reaching implications for the effects of cannabis in the brain as studies have demonstrated that the MEK pathway is implicated in regional differences in the development of tolerance to Δ9THC, the major psychoactive constitute of cannabis (47). Furthermore, cannabis smoking in adolescence is associated with an increased risk of psychosis, particularly in individuals with a family history of schizophrenia (48). Studies reveal that CB1 receptors in adolescent hippocampus have lower functional coupling to G-proteins and desensitize more slowly in response to THC treatment than those of adults (49). It could be speculated that polymorphisms in ECR1 may increase susceptibility of adults and adolescents to develop psychosis when exposed to high potency cannabis.
Further evidence for the possible harmful effects of ECR1(T) comes from the observation that following the migration of Eurasian ancestors out of Africa and into the Middle East 40,000 years ago, ECR1(T) was clearly selected against within these populations where the allelic frequency of ECR1(T) has been reduced from 50% in African populations to as little as 13% of the population in Europe and Asia. It may not be entirely coincidental that this reduction in the frequency of ECR1(T) occurred subsequent to the arrival of Eurasian ancestral populations in a region of the world where Cannabis sativa was endemic and in which it had been used for religious, medicinal, and recreational purposes for many thousands of years (50). Further support for this hypothesis comes from a number of studies that have reported a higher incidence of psychosis in individuals of African and Caribbean origin (21, 51–53), although the role of cannabis use in these studies was not fully explored.
A possible mechanism can be inferred if we consider that CB1 activation is known to trigger MAP kinase pathways (37) in neuronal cells and that the ECR1(T) allele responds more strongly to MAP kinase activation in hippocampal cells, where the ECR1(C) allele does not. Thus the current study raises the possibility of the presence of a potentially harmful CB1-MAP kinase-ECR1(T) autoregulatory loop in hippocampal neurons in a proportion of the human population that contributes to susceptibility to many of the conditions associated with rs9450898 and rs2023239 or the negative effects of high potency cannabis use.
This work was funded by the Tenovus Trust, the Wellcome Trust (080980/Z/06/Z), and the Medical Research Council (G0701003).
- CB1
- cannabinoid receptor 1
- DRG
- dorsal root ganglia
- LD
- linkage disequilibrium
- AP1
- activator protein-1.
REFERENCES
- 1. Iversen L. (2003) Cannabis and the brain. Brain 126, 1252–1270 [DOI] [PubMed] [Google Scholar]
- 2. López-Moreno J. A., González-Cuevas G., Moreno G., Navarro M. (2008) The pharmacology of the endocannabinoid system: functional and structural interactions with other neurotransmitter systems and their repercussions in behavioral addiction. Addict. Biol. 13, 160–187 [DOI] [PubMed] [Google Scholar]
- 3. Idelevich E., Kirch W., Schindler C. (2009) Current pharmacotherapeutic concepts for the treatment of obesity in adults. Ther. Adv. Cardiovasc. Dis. 3, 75–90 [DOI] [PubMed] [Google Scholar]
- 4. Akbas F., Gasteyger C., Sjödin A., Astrup A., Larsen T. M. (2009) A critical review of the cannabinoid receptor as a drug target for obesity management. Obes. Rev. 10, 58–67 [DOI] [PubMed] [Google Scholar]
- 5. Carr T. P., Jesch E. D., Brown A. W. (2008) Endocannabinoids, metabolic regulation, and the role of diet. Nutr. Res. 28, 641–650 [DOI] [PubMed] [Google Scholar]
- 6. Riedel G., Davies S. N. (2005) Cannabinoid function in learning, memory, and plasticity. Handb. Exp. Pharmacol. 445–477 [DOI] [PubMed] [Google Scholar]
- 7. Wilson R. I., Nicoll R. A. (2002) Endocannabinoid signaling in the brain. Science 296, 678–682 [DOI] [PubMed] [Google Scholar]
- 8. Pertwee R. G. (1999) Cannabis and cannabinoids: pharmacology and rationale for clinical use. Forsch. Komplementarmed. 6, Suppl. 3, 12–15 [DOI] [PubMed] [Google Scholar]
- 9. Talwar R., Potluri V. K. (2011) Cannabinoid 1 (CB1) receptor: pharmacology, role in pain, and recent developments in emerging CB1 agonists. CNS Neurol. Disord. Drug Targets 10, 536–544 [DOI] [PubMed] [Google Scholar]
- 10. Ehlers C. L., Slutske W. S., Lind P. A., Wilhelmsen K. C. (2007) Association between single nucleotide polymorphisms in the cannabinoid receptor gene (CNR1) and impulsivity in southwest California Indians. Twin Res. Hum. Genet. 10, 805–811 [DOI] [PubMed] [Google Scholar]
- 11. Benzinou M., Chèvre J. C., Ward K. J., Lecoeur C., Dina C., Lobbens S., Durand E., Delplanque J., Horber F. F., Heude B., Balkau B., Borch-Johnsen K., Jørgensen T., Hansen T., Pedersen O., Meyre D., Froguel P. (2008) Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum. Mol. Genet. 17, 1916–1921 [DOI] [PubMed] [Google Scholar]
- 12. Chen X., Williamson V. S., An S. S., Hettema J. M., Aggen S. H., Neale M. C., Kendler K. S. (2008) Cannabinoid receptor 1 gene association with nicotine dependence. Arch. Gen. Psychiatry 65, 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hutchison K. E., Haughey H., Niculescu M., Schacht J., Kaiser A., Stitzel J., Horton W. J., Filbey F. (2008) The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch. Gen. Psychiatry 65, 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haughey H. M., Marshall E., Schacht J. P., Louis A., Hutchison K. E. (2008) Marijuana withdrawal and craving: influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction 103, 1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filbey F. M., Schacht J. P., Myers U. S., Chavez R. S., Hutchison K. E. (2010) Individual and additive effects of the CNR1 and FAAH genes on brain response to marijuana cues. Neuropsychopharmacology 35, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dinu I. R., Popa S., Bîcu M., Moța E., Moța M. (2009) The implication of CNR1 gene's polymorphisms in the modulation of endocannabinoid system effects. Rom. J. Intern. Med. 47, 9–18 [PubMed] [Google Scholar]
- 17. Ho B. C., Wassink T. H., Ziebell S., Andreasen N. C. (2011) Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr. Res. 128, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., Kloos D. U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shanley L., Davidson S., Lear M., Thotakura A. K., McEwan I. J., Ross R. A., MacKenzie A. (2010) Long-range regulatory synergy is required to allow control of the TAC1 locus by MEK/ERK signaling in sensory neurons. Neurosignals 18, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shanley L., Lear M., Davidson S., Ross R., MacKenzie A. (2011) Evidence for regulatory diversity and auto-regulation at the TAC1 locus in sensory neurons. J. Neuroinflammation 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vanheusden K., Mulder C. L., van der Ende J., Selten J. P., van Lenthe F. J., Verhulst F. C., Mackenbach J. P. (2008) Associations between ethnicity and self-reported hallucinations in a population sample of young adults in The Netherlands. Psychol. Med. 38, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 22. Davidson S., Miller K. A., Dowell A., Gildea A., Mackenzie A. (2006) A remote and highly conserved enhancer supports amygdala-specific expression of the gene encoding the anxiogenic neuropeptide substance-P. Mol. Psychiatry 11, 410–421 [DOI] [PubMed] [Google Scholar]
- 23. Davidson S., Starkey A., MacKenzie A. (2009) Evidence of uneven selective pressure on different subsets of the conserved human genome: implications for the significance of intronic and intergenic DNA. BMC Genomics 10, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacKenzie A. (2010) GWA studies, gene regulation, and disease: pointers for the future of medical science? in Projects, Insight Publishers, http://www.projectsmagazine.eu.com/opinion/gwa_studies_gene_regulation_and_disease [Google Scholar]
- 25. Mackenzie A., Miller K. A., Collinson J. M. (2004) Is there a functional link between gene interdigitation and multispecies conservation of synteny blocks? Bioessays 26, 1217–1224 [DOI] [PubMed] [Google Scholar]
- 26. MacKenzie A., Payne C., Boyle S., Clarke A. R., Quinn J. P. (2000) The human preprotachykinin-A gene promoter has been highly conserved and can drive human-like marker gene expression in the adult mouse CNS. Mol. Cell. Neurosci. 16, 620–630 [DOI] [PubMed] [Google Scholar]
- 27. MacKenzie A., Purdie L., Davidson D., Collinson M., Hill R. E. (1997) Two enhancer domains control early aspects of the complex expression pattern of Msx1. Mech. Dev. 62, 29–40 [DOI] [PubMed] [Google Scholar]
- 28. Miller K. A., Barrow J., Collinson J. M., Davidson S., Lear M., Hill R. E., Mackenzie A. (2007) A highly conserved Wnt-dependent TCF4 binding site within the proximal enhancer of the anti-myogenic Msx1 gene supports expression within Pax3-expressing limb bud muscle precursor cells. Dev. Biol. 311, 665–678 [DOI] [PubMed] [Google Scholar]
- 29. Miller K. A., Davidson S., Liaros A., Barrow J., Lear M., Heine D., Hoppler S., MacKenzie A. (2008) Prediction and characterization of a highly conserved, remote, and cAMP-responsive enhancer that regulates Msx1 gene expression in cardiac neural crest and outflow tract. Dev. Biol. 317, 686–694 [DOI] [PubMed] [Google Scholar]
- 30. Davidson S., Lear M., Shanley L., Hing B., Baizan-Edge A., Herwig A., Quinn J. P., Breen G., McGuffin P., Starkey A., Barrett P., MacKenzie A. (2011) Differential activity by polymorphic variants of a remote enhancer that supports galanin expression in the hypothalamus and amygdala: implications for obesity, depression, and alcoholism. Neuropsychopharmacology 36, 2211–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takahashi E., Berk B. C. (1998) MAP kinases and vascular smooth muscle function. Acta Physiol. Scand. 164, 611–621 [DOI] [PubMed] [Google Scholar]
- 32. Schmitz U., Berk B. C. (1997) Angiotensin II signal transduction: stimulation of multiple mitogen-activated protein kinase pathways. Trends Endocrinol. Metab. 8, 261–266 [DOI] [PubMed] [Google Scholar]
- 33. Wolf G., Ziyadeh F. N. (1997) Renal tubular hypertrophy induced by angiotensin II. Semin. Nephrol. 17, 448–454 [PubMed] [Google Scholar]
- 34. Hipskind R. A., Bilbe G. (1998) MAP kinase signaling cascades and gene expression in osteoblasts. Front. Biosci. 3, d804–816 [DOI] [PubMed] [Google Scholar]
- 35. Jo Y. H., Chen Y. J., Chua S. C., Jr., Talmage D. A., Role L. W. (2005) Integration of endocannabinoid and leptin signaling in an appetite-related neural circuit. Neuron 48, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawasaki Y., Kohno T., Ji R. R. (2006) Different effects of opioid and cannabinoid receptor agonists on C-fiber-induced extracellular signal-regulated kinase activation in dorsal horn neurons in normal and spinal nerve-ligated rats. J. Pharmacol. Exp. Ther. 316, 601–607 [DOI] [PubMed] [Google Scholar]
- 37. Turu G., Hunyady L. (2010) Signal transduction of the CB1 cannabinoid receptor. J. Mol. Endocrinol. 44, 75–85 [DOI] [PubMed] [Google Scholar]
- 38. Graham E. S., Ball N., Scotter E. L., Narayan P., Dragunow M., Glass M. (2006) Induction of Krox-24 by endogenous cannabinoid type 1 receptors in Neuro2A cells is mediated by the MEK-ERK MAPK pathway and is suppressed by the phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 281, 29085–29095 [DOI] [PubMed] [Google Scholar]
- 39. Karanian D. A., Brown Q. B., Makriyannis A., Bahr B. A. (2005) Blocking cannabinoid activation of FAK and ERK1/2 compromises synaptic integrity in hippocampus. Eur. J. Pharmacol. 508, 47–56 [DOI] [PubMed] [Google Scholar]
- 40. Roth T. L., Sweatt J. D. (2008) Rhythms of memory. Nat. Neurosci. 11, 993–994 [DOI] [PubMed] [Google Scholar]
- 41. Miyamoto E. (2006) Molecular mechanism of neuronal plasticity: induction and maintenance of long-term potentiation in the hippocampus. J. Pharmacol. Sci. 100, 433–442 [DOI] [PubMed] [Google Scholar]
- 42. Waltereit R., Weller M. (2003) Signaling from cAMP/PKA to MAPK and synaptic plasticity. Mol. Neurobiol. 27, 99–106 [DOI] [PubMed] [Google Scholar]
- 43. Kristensen F. W. (1994) [Cannabis and psychoses]. Ugeskr. Laeger 156, 2875–2878, 2881 [PubMed] [Google Scholar]
- 44. Port R. L., Seybold K. S. (1995) Hippocampal synaptic plasticity as a biological substrate underlying episodic psychosis. Biol. Psychiatry 37, 318–324 [DOI] [PubMed] [Google Scholar]
- 45. Ashtari M., Avants B., Cyckowski L., Cervellione K. L., Roofeh D., Cook P., Gee J., Sevy S., Kumra S. (2011) Medial temporal structures and memory functions in adolescents with heavy cannabis use. J. Psychiatr Res. 45, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Breivogel C. S., Sim L. J., Childers S. R. (1997) Regional differences in cannabinoid receptor/G-protein coupling in rat brain. J. Pharmacol. Exp. Ther. 282, 1632–1642 [PubMed] [Google Scholar]
- 47. Rubino T., Forlani G., Viganò D., Zippel R., Parolaro D. (2005) Ras/ERK signaling in cannabinoid tolerance: from behavior to cellular aspects. J. Neurochem. 93, 984–991 [DOI] [PubMed] [Google Scholar]
- 48. Henquet C., Di Forti M., Morrison P., Kuepper R., Murray R. M. (2008) Gene-environment interplay between cannabis and psychosis. Schizophr. Bull. 34, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moore J. H., Asselbergs F. W., Williams S. M. (2010) Bioinformatics challenges for genome-wide association studies. Bioinformatics 26, 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Touw M. (1981) The religious and medicinal uses of cannabis in China, India, and Tibet. J. Psychoactive Drugs 13, 23–34 [DOI] [PubMed] [Google Scholar]
- 51. Zolkowska K., Cantor-Graae E., McNeil T. F. (2001) Increased rates of psychosis among immigrants to Sweden: is migration a risk factor for psychosis? Psychol. Med. 31, 669–678 [DOI] [PubMed] [Google Scholar]
- 52. Reeves S. J., Sauer J., Stewart R., Granger A., Howard R. J. (2001) Increased first-contact rates for very-late-onset schizophrenia-like psychosis in African- and Caribbean-born elders. Br. J. Psychiatry 179, 172–174 [DOI] [PubMed] [Google Scholar]
- 53. Veling W., Selten J. P., Susser E., Laan W., Mackenbach J. P., Hoek H. W. (2007) Discrimination and the incidence of psychotic disorders among ethnic minorities in The Netherlands. Int. J. Epidemiol. 36, 761–768 [DOI] [PubMed] [Google Scholar]