Background: Although J-proteins stimulate ATPase activity of Hsp70s, the additional physiological functions are still elusive.
Results: Besides its J-dependent functions, Dph4, a type III J-protein, exhibits redox activity and acts as electron carrier attributed to its iron-binding property.
Conclusion: 3D structure and multiple functions of Dph4 expand the functional repertoire of the J-protein family.
Significance: J-proteins have additional co-evolved specialized functions, previously not well appreciated.
Keywords: Heat Shock Protein, Molecular Chaperone, NMR, Protein Folding, Protein Synthesis, Diphthamide, Hsp70, J-protein, Iron Binding, Protein Metal-Ion Interaction
Abstract
J-proteins are obligate cochaperones of Hsp70s and stimulate their ATPase activity via the J-domain. Although the functions of J-proteins have been well understood in the context of Hsp70s, their additional co-evolved “physiological functions” are still elusive. We report here the solution structure and mechanism of novel iron-mediated functional roles of human Dph4, a type III J-protein playing a vital role in diphthamide biosynthesis and normal development. The NMR structure of Dph4 reveals two domains: a conserved J-domain and a CSL-domain connected via a flexible linker-helix. The linker-helix modulates the conformational flexibility between the two domains, regulating thereby the protein function. Dph4 exhibits a unique ability to bind iron in tetrahedral coordination geometry through cysteines of its CSL-domain. The oxidized Fe-Dph4 shows characteristic UV-visible and electron paramagnetic resonance spectral properties similar to rubredoxins. Iron-bound Dph4 (Fe-Dph4) also undergoes oligomerization, thus potentially functioning as a transient “iron storage protein,” thereby regulating the intracellular iron homeostasis. Remarkably, Fe-Dph4 exhibits vital redox and electron carrier activity, which is critical for important metabolic reactions, including diphthamide biosynthesis. Further, we observed that Fe-Dph4 is conformationally better poised to perform Hsp70-dependent functions, thus underlining the significance of iron binding in Dph4. Yeast Jjj3, a functional ortholog of human Dph4 also shows a similar iron-binding property, indicating the conserved nature of iron sequestration across species. Taken together, our findings provide invaluable evidence in favor of additional co-evolved specialized functions of J-proteins, previously not well appreciated.
Introduction
J-protein and Hsp705 genes have proliferated during evolution, with cellular compartments having multiple J-proteins and Hsp70s. The functional specificity of Hsp70 is determined by J-protein cochaperones, which are structurally highly diverse group of proteins co-localized in each compartment (1–3). Many Hsp70s and J-protein partners have coevolved to function together as a “molecular machine” with Hsp70 being the core component. The J-proteins sequester and deliver unfolded proteins to Hsp70 in the ATP-bound state and catalytically stimulate the ATPase activity of Hsp70 thereby, stabilizing the interactions with client proteins in ADP-bound state (1, 4). The nucleotide exchange factors regulate the exchange of nucleotides from Hsp70s, thus operating the chaperone cycle in connection to various physiological functions, including de novo protein folding, protein translocation across the membranes, and degradation (2, 4, 5).
J-proteins are polymorphic multidomain proteins identified by the presence of signature J-domain region essential for the stimulation of ATPase activity of Hsp70s. Based on domain organization, J-proteins are historically classified into three types (6). The class I and II J-proteins closely resemble Escherichia coli DnaJ that are ubiquitous in nature, share common functions, and are essential for maintaining the cellular protein homeostasis. On the contrary, class III J-proteins are present in large numbers and consist of heterogeneous structural organization outside the J-domain. As a consequence, they are involved in multiple cellular functions (1, 7).
A member of class III J-protein has been identified to play a critical role in diphthamide (DPH) biosynthesis, which is one of the important conserved cellular processes in the eukaryotic system (8). In the mammalian system, DPH is formed by unique post-translational modification of the histidine (His715) in elongation factor-2. DPH serves as an opportunistic target site for the diphtheria toxin (DT)-mediated ADP-ribosylation, thus abrogating protein translation. Of the five known proteins implicated in DPH biosynthesis, Dph4 has been identified as a class III J-protein consisting of a predicted N-terminal J-domain and a CSL-domain at the C terminus. In the mouse model system, the dph4 splice site mutant has been associated with multiple phenotypes, including retarded growth and late developmental defects (9). Interestingly, JJJ3, an ortholog of mammalian DPH4 does not exhibit a growth phenotype upon deletion but shows sensitivity toward DT overexpression in yeast (10). The CSL-domain of Jjj3 has been found critical for conferring the DT sensitivity, whereas the J-domain is dispensable, thus highlighting the central importance of the regions outside of the J-domain in DPH biosynthesis (10).
Our report uncovers the first structure of full-length human type III J-protein, Dph4, and reveals multiple insights into its cellular functions. We present here novel iron-mediated functional roles of Dph4 not known previously. Besides functioning with Hsp70, our findings establish several J-domain-independent functions of J-proteins, including redox and electron carrier activity and a role in the diphthamide biosynthesis.
EXPERIMENTAL PROCEDURES
Plasmids, Yeast Strains, and Genetic Techniques
The open reading frames (ORFs) encoding full-length DPH4, KTI11, HSC70, and HSPA1A were PCR-amplified from a HeLa cell cDNA library (Stratagene) using sequence-specific primers. For purification, wild type DPH4, yeast JJJ3, DPH4(1–76), DPH4(1–92), and DPH4(94–149) were cloned into pET3a vector, whereas HSC70 and HSPA1A were cloned into pRSFDuet-1 vector with the insertion of six histidine codons at the C terminus. The point mutant, dph4C139S was generated by QuikChange protocol using high fidelity Pfu Turbo DNA polymerase (Stratagene). For fusion protein purification, the entire ORF of DPH4 was cloned into the glutathione S-transferase (GST)-expressing vector pGEX-KG at the C terminus using EcoRI and XhoI restriction sites.
Yeast JJJ3 and human DPH4 ORFs were subcloned into pRS424 yeast expression vector under GPD promoter and transformed to Δjjj3 strain as described previously (10). The strains were separately co-transformed with pLMY101 plasmid encoding a diphtheria toxin under the galactose-inducible promoter (GAL-DT) and selected on dropout medium (Trp−). In vivo diphthamide biosynthesis was monitored by growth of yeast cells on conditional expression of GAL-DT (11).
Expression and Protein Purification from E. coli and Yeast
All proteins except Dph4(1–76), dph4C139S, HspA1A, and Hsc70 were overexpressed in E. coli Rosetta strain at 30 °C using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at A600 of 0.6 for 4 h. The induction of dph4C139S and Dph4(1–76) was carried out for 12 h in E. coli Rosetta strain at 18 °C. HspA1A and Hsc70 proteins were overexpressed in E. coli BL21 (DE3) strain. The cells were grown in M9 minimal medium supplemented with vitamins and either 1 mm ZnSO4·7H2O or 200 μm FeCl3 as per requirement. For the purification of His6-tagged proteins, the cells were resuspended in lysis buffer A (20 mm Tris-Cl, pH 7.5, 20 mm imidazole, 200 mm NaCl, 5% glycerol, and 1 mm DTT) containing protease inhibitor mixture and incubated with 0.2 mg/ml lysozyme at 4 °C for 1 h. The sample was gently lysed with 0.2% sodium deoxycholate (USB) followed by DNase I (10 μg/ml) treatment in the presence of 5 mm MgCl2 for 15 min. The cell lysate was prepared by sonication and clarified by centrifuging at 14,000 rpm for 30 min at 4 °C. The soluble supernatant was incubated with pre-equilibrated Ni2+-NTA-Sepharose beads (GE Healthcare) for 2 h at 4 °C. Unbound proteins were removed by extensive washing with buffer A, followed by a wash with buffer A containing 0.5 m NaCl. Nonspecific contaminants were removed using buffer B (20 mm Tris-Cl, pH 7.5, 30 mm imidazole, 200 mm NaCl, 1 mm ATP, 5 mm MgCl2, and 5% glycerol). The proteins were eluted with buffer A containing 250 mm imidazole. For purification of HspA1A and Hsc70 proteins, buffer A and B containing HEPES-KOH (pH 7.5) was used instead of Tris-Cl, pH 7.5.
The GST-tagged proteins were isolated using glutathione-Sepharose beads (GE Healthcare). The GST-Dph4 fusion protein was purified using lysis buffer C (20 mm Tris-Cl, pH 7.5, 200 mm NaCl, and 5% glycerol) followed by a wash with buffer D (20 mm Tris-Cl, pH 7.5, 500 mm NaCl, and 5% glycerol). The untagged Dph4 protein was obtained from GST-Dph4 fusion protein by cleaving the GST tag using thrombin as per the manufacturer's instructions (Novagen). The proteins used for UV-visible and fluorescence analysis were dialyzed against buffer X (20 mm Tris-Cl, pH 7.5, 200 mm NaCl, 10% glycerol, and 1 mm DTT). The apo-form of Dph4 was obtained by overnight treatment of Zn-Dph4 protein preparation with 10 mm EDTA and then extensive dialysis against buffer X.
For purification from yeast cells, Δjjj3 cells transformed with His6-tagged DPH4 in pRS424 vector were grown in Trp− medium at 30 °C supplemented with 50 μm FeCl3 and harvested at A600 of 1.3. Cells pelleted from 3.5 liters of culture were dissolved in buffer E (100 mm KH2PO4, 1.2 m sorbitol, pH 7.4) and treated with zymolyase (2 mg/g of pellet) for 45 min. The cells were then lysed by homogenization in buffer F (100 mm Tris-Cl, pH 7.4, and 0.6 m sorbitol). The soluble supernatant was incubated with pre-equilibrated Ni2+-NTA-Sepharose beads for 2 h at 4 °C. Nonspecific protein contaminants were removed by extensive washing with buffer A followed by wash with buffer A containing 0.5 m NaCl and buffer B. The proteins were eluted with buffer A containing 250 mm imidazole.
NMR Spectroscopy
Samples for NMR studies (full-length Dph4, Dph4(1–76), Dph4(1–92), and Dph4(93–149)) were prepared as mentioned above but supplemented with 4 g/liter d-[13C]glucose and 1 g/liter 15NH4Cl (Cambridge Isotope Laboratories, Inc.) as the sole sources of carbon and nitrogen, respectively. The following Dph4 samples were prepared with different isotope labeling schemes: uniform 15N-labeled, doubly labeled (uniform 13C- and 15N-labeled), and fractionally (10%) 13C-labeled. NMR experiments were performed with 0.5 ml of ∼1 mm protein samples in 20 mm Tris-Cl buffer, pH 7.5, containing 80 mm NaCl, constituted either in 99.9% 2H2O or in a mixed solvent of 90% H2O and 10% 2H2O.
The details of NMR experiments, sequence-specific assignments, and various constraints used in structure calculations are described in the supplemental Experimental Procedures.
UV-visible Spectroscopy
All absorption spectra for iron reduction assays were recorded on a PerkinElmer Lambda 35 UV-visible spectrometer equipped with the Julabo MB-5A temperature control unit. All of the samples used for the analysis were prepared in a buffer X and purged with N2 gas to maintain anaerobic conditions. All reduction reactions were initialized by the addition of NADH. The initial delay in recording the spectra was manually adjusted in the calculations for rate determination. The rate constants were calculated using SigmaPlot 9.0 (Systat Software, Inc.) by fitting the curve into y = y0 + a(1 − exp(−bx)), where b is the rate constant.
Miscellaneous Methods
Molecular dynamics (MD) simulations are detailed in the supplemental Experimental Procedures. EPR spectra were recorded on a Bruker EMX X-band spectrometer at 9.42 GHz in the quartz tube under standard conditions. Iron staining using 3,5-diaminobenzoic acid dihydrochloride (Sigma-Aldrich) and ATPase assays were carried out as described previously (12, 13). Fluorescence analysis was performed using a Varian Cary Eclipse fluorescence spectrophotometer. The protein samples were prepared in buffer X and purged with N2 gas before analysis. Immunoblot analysis was carried out by using the ECL system (PerkinElmer Life Sciences) according to the manufacturer's instructions. The thermal unfolding using circular dichroism and partial tryptic digestion were carried out as previously described (14).
RESULTS
Differential Metal Binding in Human Dph4
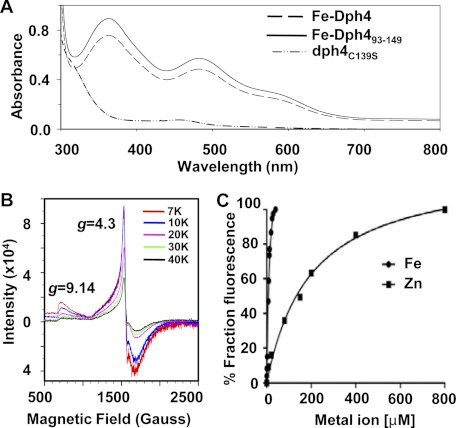
The primary sequence analysis reveals the presence of a N-terminal J-domain (residues 1–76) and a metal binding CSL-domain (residues 93–149) at the C terminus separated by long linker region (residues 77–92). Strikingly, the purified His6-tagged protein from E. coli in rich medium was found to be dark brown in color (supplemental Fig. S1A). The protein preparations were found to be greater than 98% homogeneous as analyzed on SDS-PAGE (supplemental Fig. S1B). UV-visible spectral analysis of purified protein revealed two prominent peaks in the visible region (365 and 485 nm) and a shoulder peak at 570 nm (Fig. 1A). Interestingly, these spectral properties are similar to proteins containing the classical rubredoxin fold, which are known to bind iron as a metal ion (15). On the contrary, CSL motif-containing proteins are reported to bind only zinc (16). To address this further, the protein was purified from E. coli selectively grown on M9 minimal medium supplemented with iron (for Fe-Dph4) or zinc (Zn-Dph4) salts. The metal ion analysis by atomic absorption spectroscopy revealed 0.8:1 and 0.9:1 stoichiometric binding of iron and zinc to Dph4, respectively. These results clearly demonstrate that both metal ions can be independently sequestered by Dph4, depending on growth conditions used for expression in E. coli. The spectral properties of Dph4 purified from iron-supplemented minimal medium were found to be identical to Dph4 produced from rich medium (supplemental Fig. S1C). Dph4 purified from zinc-supplemented minimal medium was found colorless and devoid of any absorption in the visible region (supplemental Fig. S1C). To eliminate any possibility of interference of the His6 tag in iron binding, we compared the absorption spectrum of untagged protein purified from a minimal medium supplemented with iron salts. Both His6-tagged and untagged Dph4 protein showed an identical UV-visible spectrum, indicating that Dph4 possesses the intrinsic ability to bind iron in solution (supplemental Fig. S1D). Additionally, the UV-visible spectrum recorded on purified isolated Dph4(93–149) fragment was found to be identical with full-length protein, thus confirming the binding site of iron in the CSL-domain (Fig. 1A).
FIGURE 1.
Analysis of iron binding in Dph4. A, UV-visible absorption spectra of 200 μm Fe-Dph4, Fe-Dph4(93–149), and dph4C139S. B, EPR spectra of 0.6 mm Fe-Dph4 in solution recorded at different temperatures. The observed resonances are marked for their g values. C, titration of apo-Dph4 (2 μm) with Fe2+ as (NH4)2Fe(SO4)2·6H2O (in 200 μm ascorbic acid) and Zn2+ as ZnSO4·7H2O. The change in Trp fluorescence intensity (342 nm) of apo-Dph4 was monitored.
It has been reported previously that in CSL motif-containing proteins, zinc coordinates spatially with 4 Cys residues in a tetrahedral geometry (16). To confirm that cysteines of the CSL-domain in Dph4 coordinate with iron, we prepared a point mutation by replacing Cys-139 with Ser. Interestingly, the purified dph4C139S showed a significant reduction in absorption in the UV-visible region in comparison with wild type, underlining the loss of iron binding ability by dph4C139S (Fig. 1A). To elucidate the geometry of iron coordination in the CSL-domain of Dph4, we have utilized electron paramagnetic resonance spectroscopy (EPR) (Fig. 1B). The resonances at g ∼4.3 and g ∼9.14 are characteristic of high spin Fe3+ ion in tetrahedral geometry as reported previously for oxidized rubredoxins (17). Furthermore, both of the signals were found to be sensitive to temperature, similar to rubredoxins (18) (Fig. 1B). Taken together, our results provide the first evidence showing unique iron-binding properties associated with human J-protein Dph4 besides its ability to bind zinc in solution.
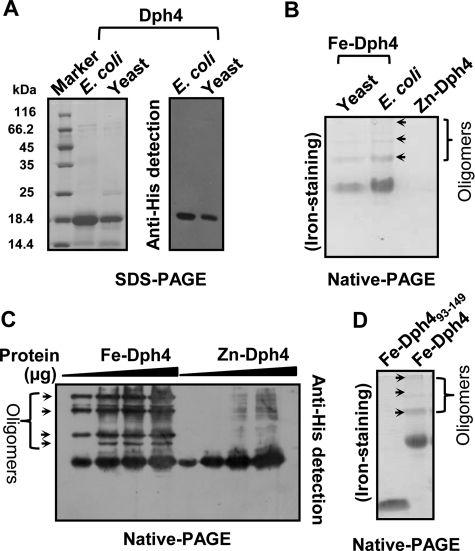
The binding affinities of zinc and iron for Dph4 in solution were compared by monitoring fractional changes in intrinsic tryptophan (Trp) fluorescence of apo-Dph4 upon titration with zinc or iron metal salts. The observed dissociation constants for iron and zinc were 12.5 ± 1.9 and 199.5 ± 25.2 μm respectively (Fig. 1C). A 16-fold enhanced binding of iron to Dph4 as compared with zinc suggests that Dph4 exists predominantly in iron-bound form under physiological conditions. To demonstrate the iron sequestration property in the eukaryotic physiological context, His6-tagged Dph4 protein was purified from Saccharomyces cerevisiae using pRS424-GPD vector. The expression was confirmed by immunodetection using anti-His antibodies (Fig. 2A). The iron binding was confirmed by using iron-specific staining after separating the proteins on a native PAGE (10). Dph4 purified from yeast showed strong reactivity toward iron-specific staining (Fig. 2B). Fe-Dph4 and Zn-Dph4 purified from E. coli served as positive and negative controls, respectively. Hence, our result conclusively proves that iron sequestration by Dph4 is a very specific process in the eukaryotic system.
FIGURE 2.
Oligomerization in Fe-Dph4. A, immunodetection of Dph4 purified from yeast. Purified protein was separated on SDS-PAGE and immunodetected by Western blotting using anti-His antibody. B, iron-specific staining of Dph4 purified from E. coli and yeast separated on native PAGE. C, Western blot of Fe-Dph4 and Zn-Dph4 separated on native-PAGE and immunodetected by anti-His antibody. Increasing amounts of protein (4–20 μg) were loaded as indicated. D, iron staining of Dph4(93–149) purified from E. coli separated on native PAGE. Oligomers are marked with arrows.
Iron Induces Oligomerization of Dph4
The native PAGE of Fe-Dph4 showed the presence of additional higher molecular weight bands, which were positive for iron staining (Fig. 2B). These oligomeric forms were also detected in the gel filtration analysis of purified Fe-Dph4 (supplemental Fig. S1E). We analyzed the oligomers in the protein preparations of both Zn- and Fe-Dph4. Increasing concentrations of Fe-Dph4 and Zn-Dph4 were loaded on native PAGE and immunodetected with anti-His antibody (Fig. 2C). Minimal higher order oligomers were detected in Zn-Dph4 as compared with Fe-Dph4 even at higher protein concentrations, indicating that preferential binding of iron to Dph4 promotes oligomerization. Upon quantification, nearly 50% of the Fe-Dph4 protein sample was found to be oligomeric as analyzed from both gel filtration and Western blot (Fig. 2C and supplemental Fig. S1E). To understand the region of protein required for the oligomerization, we tested the oligomerization status of purified Fe-bound isolated CSL-domain (Fig. 2D). The absence of detectable iron-bound oligomer in Dph4(93–149) suggests that iron-induced oligomerization is a property of full-length protein, where both domains are required to form higher order structures.
Fe-Dph4 Has Enhanced Ability to Stimulate ATPase Activity of Hsp70s
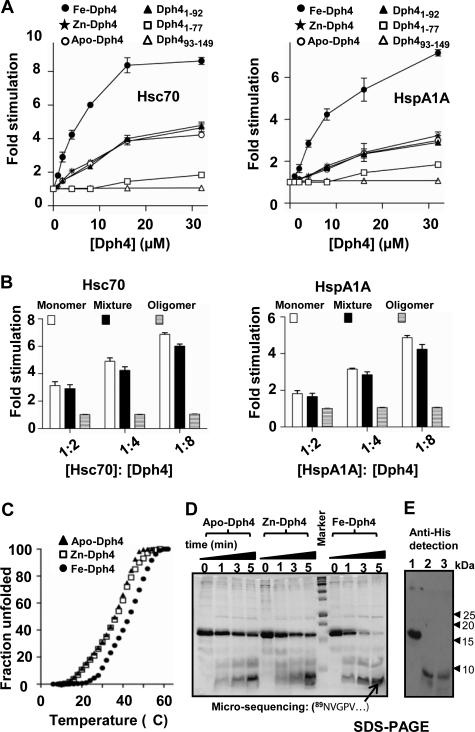
J-proteins are known to stimulate the ATPase activity of Hsp70s (1–5). To investigate whether Zn-Dph4 and Fe-Dph4 retain the ability to stimulate Hsp70s, we assayed for in vitro ATPase stimulation activity. Due to prevalent cytosolic localization of Dph4, we analyzed for the ability to stimulate Hsc70 and Hsp70A1A partners (19). The complexes of Hsc70-[α-32P]ATP and HspA1A-[α-32P]ATP were isolated, and the rate of ATP hydrolysis was measured under the single turnover conditions in the presence of Zn-Dph4 and Fe-Dph4. The basal rates of ATP hydrolysis by Hsc70 and HspA1A were estimated to be 0.16 and 0.07 min−1, respectively, which are comparable with values reported previously (20). Upon the addition of increased molar ratios of Zn-Dph4 and Fe-Dph4 to Hsp70s, a robust stimulation in the ATPase activity of Hsc70 and HspA1A was observed (Fig. 3A). These results demonstrate that Dph4 could potentially function in vivo as a J-protein cochaperone in both zinc- and iron-bound states. Strikingly, Fe-Dph4 displayed a significantly enhanced ability to stimulate the ATPase activity of Hsc70 and HspA1A as compared with Zn-Dph4 under similar conditions. Such elevated stimulation is attributable to overall conformational differences between Fe- and Zn-Dph4.
FIGURE 3.
ATPase assays of Dph4. A, stimulation of Hsc70 (left) and HspA1A (right) by apo-Dph4, Zn-Dph4, Fe-Dph4, Dph4(1–76), Dph4(1–92), and Fe-Dph4(93–149). The preformed radiolabeled Hsc70-ATP (1 μm) and Hsp70A1A-ATP (1 μm) complexes were incubated with increasing concentrations of Fe-Dph4 or Zn-Dph4. ATP hydrolysis was monitored under single turnover conditions at different time intervals at 25 °C, and rates of hydrolysis were calculated for different concentrations. -Fold stimulation was calculated by setting the intrinsic ATP hydrolysis rate as 1. Error bars are derived from two independent sets of experiments. B, stimulation of Hsc70 (left) and HspA1A (right) by monomer and oligomers of Fe-Dph4. C, thermal denaturation curve of apo-, Zn-, and Fe-Dph4 using circular dichroism. The fractional change in ellipticity at 222 nm was monitored. D, trypsin digestion of Fe-, Zn-, and apo-Dph4. The digestion was analyzed at different time points as indicated. The marked band was subjected to N-terminal microsequencing. The sequence is indicated in the figure. E, anti-His immunoblot of trypsin-digested Fe-Dph4. Lane 1, Fe-Dph4; lane 2, Fe-Dph4 after 10 min of trypsin digestion; lane 3, Dph4(93–149).
To test the conformational differences, Fe- and Zn-Dph4 were subjected to thermal denaturation using circular dichroism. As illustrated in Fig. 3C, Fe-Dph4 exhibited enhanced stability by 5 °C as compared with Zn-Dph4, suggesting plausible conformational differences between Zn- and Fe-Dph4. Both Zn-Dph4 and apo-Dph4 (used as a control) exhibited similar thermal stability. To probe further the conformational differences, we utilized the proteolytic susceptibility assay using trypsin treatment. As illustrated in Fig. 3D, all forms of proteins, including apo-, Fe-, and Zn-Dph4, showed a similar pattern of tryptic digestion. However, Fe-Dph4 exhibited higher susceptibility for tryptic cleavage (Fig. 3D). As indicated in Fig. 3E, one of the stable fragments showed mobility equal to the CSL-domain (Dph4(93–149)), which was confirmed by immunodetection using anti-His antibody. The cleavage site was mapped to Arg-88 of the exposed linker-helix by N-terminal micro-sequencing. Notably, apo- and Zn-Dph4 possess similar conformations in solution, as evident from tryptic digestion. On the other hand, the linker-helix is well exposed for tryptic cleavage by adopting a favorable conformation in the Fe-Dph4.
To determine the minimal region essential for such enhanced ATPase stimulation in Fe-Dph4 form, we generated truncation mutants. As illustrated in Fig. 3A, the J-domain alone (Dph4(1–76)) and the CSL-domain (Fe-Dph4(93–149)) were incapable of significant stimulation even at higher concentrations. Strikingly, the addition of a linker-helix to the J-domain (Dph4(1–92)) resulted in significantly enhanced stimulation. These observations are consistent with those reported previously, where the isolated J-domain was found incapable of cochaperone activity, with the additional linker sequence rendering the protein functional (10, 21). Intriguingly, Dph4(1–92) showed stimulation equivalent to full-length apo- and Zn-Dph4 (Fig. 3A), suggesting that the linker-helix plays a critical role in modulating the conformational dynamics of the J-domain. Next, to rule out the possibility of iron-mediated oligomerization enhancing the ATPase activity, we isolated monomeric and oligomeric forms of Fe-Dph4 (see the supplemental Experimental Procedures) and subsequently analyzed them for their stimulation activity. Surprisingly, the oligomeric form of Fe-Dph4 failed to stimulate Hsp70 ATPase activity in comparison with the monomeric state (Fig. 3B and supplemental Fig. S1F). Based on these observations, we conclude that the linker-helix together with the CSL-domain in Fe-Dph4 renders a conformation suitable for enhanced stimulation of ATPase activity of Hsp70s.
NMR-derived Structure of Dph4
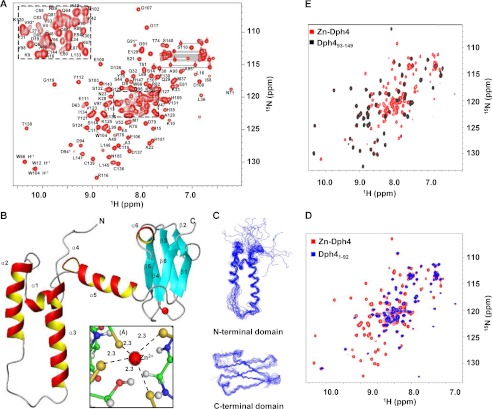
To gain further insight into the mechanistic aspects of iron-mediated functional roles of Dph4, we used NMR spectroscopy to elucidate the structure of Dph4. The two-dimensional 15N-1H HSQC spectrum of Zn-Dph4 is shown in Fig. 4A. Supplemental Fig. S1G shows the overlay of two-dimensional 15N-1H HSQC of both metal forms of Dph4. Due to the paramagnetic effect of Fe3+, many of the resonances of Fe-Dph4 were broadened, with the remaining peaks matching closely with those of Zn-Dph4 (22). The 18-kDa Zn2+-bound full-length Dph4 was a monomer under NMR sample conditions, as indicated by its average correlation time of 10 ± 0.5 ns at 298 K measured using 15N T1ρ and T2 relaxation rates (23). As described in the supplemental Experimental Procedures, >90% of resonances (excluding residues belonging to the His6 tag and 15N resonances of Pro) were assigned sequence-specifically. Doubling of peaks was observed for residues 91–95, indicative of two conformations in slow exchange on the NMR time scale.
FIGURE 4.
Structural analysis of full-length Zn-Dph4. A, two-dimensional 15N-1H HSQC spectrum of Zn-Dph4 acquired at 1H resonance frequency of 700 MHz at 298 K. Sequence-specific resonance assignments are indicated by labels depicting the single-letter code for the amino acid followed by the residue number. Side chain amide signals of asparagines and glutamines are shown connected by horizontal lines. Peaks from residues undergoing conformational exchange are marked by asterisks. The overlapped region of the spectrum is expanded in the inset. B, ribbon diagram of representative structure of Zn-Dph4, Zn2+ (red) in tetrahedral coordination with Sγ (gold) (inset). The average Zn–Sγ distance is marked with dotted lines. C, superimposition of the 20 best structures of N-terminal J-domain (top) and C-terminal CSL-domain (bottom). D, overlay of two-dimensional 15N-1H HSQC spectrum of full-length Zn-Dph4 and Dph4(1–92). E, overlay of two-dimensional 15N-1H HSQC spectrum of Zn-Dph4 and Dph4(93–149).
The structure having the least residual target function is shown in Fig. 4B. The structural statistics for the family of 20 Dph4 structures are presented in Table 1. The solution structure of Dph4 (PDB code 2L6L) reveals a multidomain structure comprising of an N-terminal J-domain (residues 1–76) and a C-terminal CSL-domain (residues 93–149) connected by a flexible helical linker (residues 77–92). Fig. 4C shows the superposition of 20 NMR derived conformers in the individual N- and C-terminal domains. Although each domain exhibits structural heterogeneity when aligned with respect to the other (supplemental Fig. S2A), residues within each domain converge with a low r.m.s. deviation of 1.18 ± 0.41 and 0.62 ± 0.14 Å, respectively, for ordered residues (Table 1). Notably, the two-dimensional 15N-1H HSQC spectrum recorded on Dph4(1–92) and Dph4(93–149) nearly matched with their counterparts in full-length Dph4 (Fig. 4, D and E). However, Dph4(1–76) fails to maintain a similar fold in the absence of linker-helix (supplemental Fig. S2B). This suggests that the two domains are independently folding units and that the linker maintains the structural integrity of the J-domain.
TABLE 1.
Structural statistics of 20 conformers of Dph4
| Total restraints | 1761 |
| Distance restraints (upper limit) | |
| All | 1558 |
| NOE intraresidue | 478 |
| Short range | 461 |
| Medium range | 271 |
| Long range | 223 |
| Hydrogen bonds | 116 |
| Zinc distance restraints | 9 |
| Dihedral angle constraints (°) | |
| ϕ | 104 |
| ψ | 99 |
| Mean CYANA target function | 2.13 ± 0.92 |
| Ramachandran plot statistics: all residues (selected residuesa) (%) | |
| Most favored regions | 74.7 (86.3) |
| Additionally allowed regions | 22.1 (13.5) |
| Generously allowed regions | 2.2 (0.2) |
| Disallowed regions | 1.0 (0.0) |
| r.m.s. deviation values (Å)b | |
| N-terminal domain | |
| Residues 11–72 | 1.53 ± 0.40 |
| Helix 1 (residues 11–16) | 0.27 ± 0.15 |
| Helix 2 (residues 24–38) | 0.39 ± 0.17 |
| Helix 3 (residues 50–68) | 0.52 ± 0.20 |
| Linker (residues 76–93) | |
| Helix 5 (residues 77–88) | 0.15 ± 0.06 |
| C-terminal domain | |
| Residues 94–149 | 1.05 ± 0.28 |
| Ordered residues (95–98, 103–105, 110–114, 120–124, 127–130, 133–135, and 142–146) | 0.62 ± 0.14 |
a Selected residues were as follows: 12–38, 50–69, 71–74, 77–90, 95–117, 120–137, and 142–146. Ramachandran plot statistics were obtained using PROCHECK. The residues observed in disallowed regions (1%) belonged to loop regions.
b r.m.s. deviations were evaluated using MOLMOL.
The canonical J-domain comprising residues 1–76 is characterized by four helices, as commonly observed in the three-dimensional structures of J-domain reported previously (24). The α-helices span residues Asp11–Leu16 (α1), Ile24–Tyr38 (α2), Gly50–Ile68 (α3), and Glu72–Lys75 (α4). The J-domain is followed by a long linker (residues 77–92) forming helix α5 (Glu77–Arg88). The C-terminal CSL-domain comprising residues 93–149 and containing the CSL zinc-binding motif (16, 25) consists of a three-stranded β-sandwich with one sheet comprising two parallel strands: (i) β1 (Ala95–Tyr98) and (ii) β6 (Ile142–Leu146) and one anti-parallel strand: β5 (Leu133–Ser135). The second sheet in the β-sandwich is comprised of strands β2 (Ser103–Asn105), β3 (Ser110–Ser114), and β4 (Lys120–Ser124) running anti-parallel to each other. The two β-sheets are separated by a short stretch α-helix α6 (Glu127–Glu130). All α-helices and β-strands have been identified by characteristic NOE patterns (26). The four cysteine residues binding Zn2+ in Dph4 (Cys115, Cys117, Cys136, and Cys139; see the supplemental Experimental Procedures) form a tetrahedral coordination sphere with an average Zn2+–Sγ distance of 2.30 ± 0.05 Å and Sγ–Sγ distance of 3.75 ± 0.14 Å.
The local backbone r.m.s. deviation for the family of 20 structures is plotted in supplemental Fig. S2D along with the values of S2 computed using TALOS+ (27). The most well defined regions of the protein consist of α-helices and β-sheets, followed by the Zn2+-binding loops. The S2 profile calculated using the backbone chemical shift matches that of the local r.m.s. deviation, indicating that regions with high r.m.s. deviation may also exhibit high flexibility.
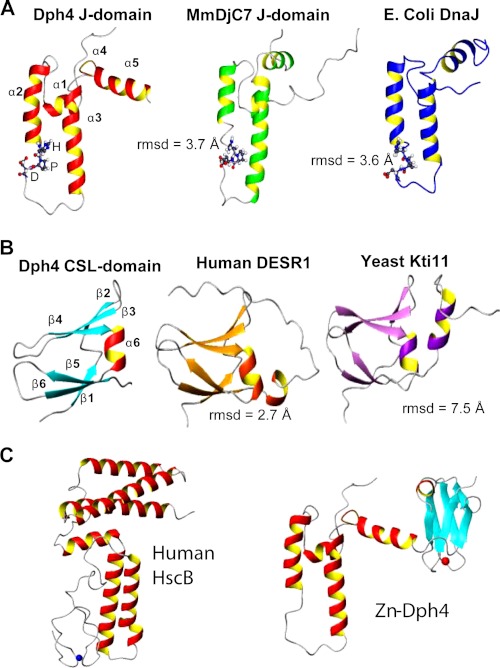
The structure of human Dph4 was compared with previously known three-dimensional structures of J-domains and CSL-domains (Fig. 5). A notable difference is in the length of helices; α1 and α2 are longer and α4 is shorter in J-domain of human Dph4 compared with that of a mouse Dph4 (sequence identity 88%) and E. coli DnaJ (sequence identity 31%) (24). However, the helix α3 of Dph4 is longer than the corresponding helix of E. coli DnaJ and shorter with respect to mouse Dph4 (MmDjC7). The additional linker helix, α5, in Dph4 acquires the position similar to helix α4 of canonical J-domains but arguably does not form a part of the J-domain. The CSL-domain was compared with human DESR1 (sequence identity 28%) and yeast Kti11 (sequence identity 25%) (16, 25). A salient feature is the absence of a helix at the C terminus end of the CSL-domain. We also compared the recently solved crystal structure of human HscB with human Dph4 (28). Notably, unlike Dph4 the metal binding domain in HscB is structurally disordered (28). These structural differences may have implications for the function of the protein and presumably lead to its functional divergence.
FIGURE 5.
Comparison of Dph4 subdomains with representative structures of similar folds. A, J-domain of Dph4, MmDjC7 (PDB code 1WJZ), and E. coli DnaJ (PDB code 1XBL) (top). B, CSL-domain of Dph4, human DESR1 (PDB code 2JR7) and yeast Kti11 (PDB code 1YOP) (middle). The r.m.s. deviations among the domains are indicated. The signature HPD motif of J-domains is represented in a ball and stick model. C, comparison of full-length human Dph4 and human HscB.
Interdomain Mobility in Dph4
The orientation of two domains in full-length Dph4 could not be fixed due to lack of interdomain NOEs. The high flexibility at the N and C termini of the central linker region Dph4(77–92) presumably gives rise to spatial heterogeneity of the two domains with respect to each other (supplemental Fig. S2A).
The absence of interdomain contacts does not imply the loss of interdomain orientation. Hence, chemical shift perturbation and a NOESY-based approach are generally coupled with either NMR-based relaxation studies or Residual Dipolar Couplings (RDC) data analysis (29). However, in Dph4, signal overlap and the absence of a significant amount of backbone amide resonances in the J-domain precluded obtaining accurate NMR relaxation data and RDC measurements for individual resonances. Therefore, we performed MD simulations on NMR-derived structures using the program GROMACS to probe interdomain dynamics.
Two structures from the NMR ensemble were subjected to 15 ns of unrestrained MD simulation after equilibration. The backbone r.m.s. deviation relative to the initial structure as a function of time is shown in supplemental Fig. S3A. The individual domains are essentially stable throughout the simulation, whereas the trajectory of the full-length protein is not stabilized even at the end of 10 ns. This behavior corresponds to conformational movements that alter the relative orientation of the domains, indicating pronounced interdomain mobility. This conformational flexibility between two domains may account for the fact that only a handful of crystal structures for full-length J-proteins have been reported. Interestingly, the trajectory of linker-helix (supplemental Fig. S3B) shows a transition in r.m.s. deviation at ∼4 ns, indicating a change in the conformation. A similar trend was also observed in radius of gyration, which starts decreasing around 4 ns (supplemental Fig. S3C). This suggests that the two domains of Dph4 adopt “open” and “closed” conformations in solution mediated by linker, which are interconvertible. A conformational exchange on a slower time scale is also observed in NMR for residues 91–95 in the two-dimensional 15N-1H HSQC spectrum (Fig. 4A). The average rotational correlation time for J- and CSL-domain in full-length protein was measured as 9.6 ± 0.8 and 10 ± 0.9 ns, respectively, which is similar to that of the full-length protein (10 ± 0.5 ns). On the whole, this suggests that while the two domains have ample conformational freedom in solution, they probably communicate through the central linker-helix and influence the mobility of each other.
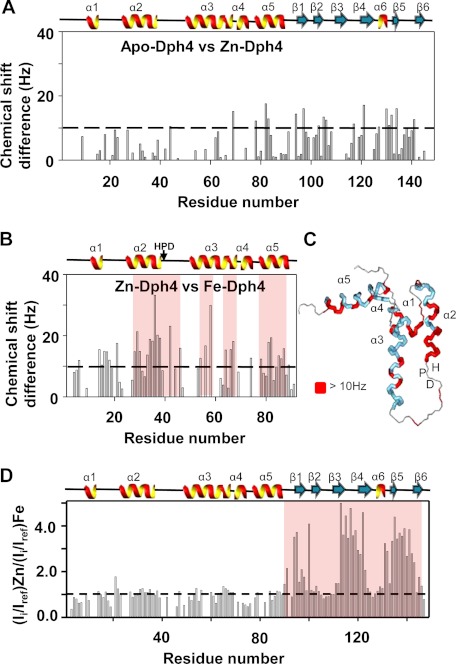
To probe conformational differences between Fe- and Zn-Dph4, leading to their differential stimulation activity, we analyzed the chemical shift differences in the apo- and metal-bound state. Fig. 6A shows the chemical shift perturbation in apo- and Zn-bound state. The perturbations are mostly localized on to the metal binding domain, whereas some also spread to linker and J-domain, clearly indicating the interdomain cross-talk. As shown in Fig. 6D, in Fe-Dph4, the paramagnetic effect of iron is highly localized on the CSL-domain; hence, we mapped the perturbation only on the J-domain and linker. While comparing Zn- and Fe-Dph4, we observed differences and mapped them on the J-domain structure (Fig. 6, B and C). The most notable ones were the continuous stretch on helix II near the HPD motif of J-domain and the interdomain linker (Fig. 6C). This region in J-proteins was reported to be crucial for both interaction and stimulation of Hsp70s (30–34). Interestingly, Fe-Dph4 and Zn-Dph4 exhibit similar overall mobility/flexibility, as revealed by 15N-1H heterogeneous NOE data (supplemental Fig. 4, A and B).
FIGURE 6.
Chemical shift perturbation analysis of apo-, Zn-, and Fe-Dph4. Chemical shift differences between apo- and Zn-Dph4 (A) and Zn- and Fe-Dph4 (B) as a function of residues. Bars crossing 10 Hz (as shown by the dashed line) indicate significantly perturbed amino acid residues. C, chemical shift differences of >10 Hz (red) between Zn- and Fe-Dph4 mapped onto the protein structure. Helices and loops are indicated in blue and gray, respectively. D, plot showing the ratio of peak intensities in 15N-1H HSQC spectra of Zn-Dph4 and Fe-Dph4. Bars crossing the threshold (shown as a dotted line) indicate broadening due to paramagnetic effect of Fe3+. The absence of a bar indicates a missing assignment. The secondary structure of the protein is plotted at the top of each panel, and the HPD-motif is indicated. The perturbed regions are highlighted in red.
Fe-Dph4 Is Redox-active and Functions as Electron Carrier Protein
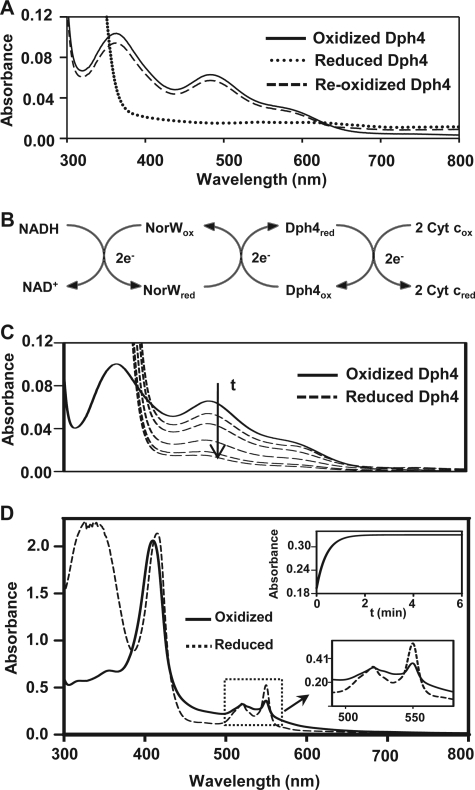
The iron-binding property of Dph4 prompted us to investigate the redox nature of the protein in vitro. As seen in Fig. 7A, a complete reduction of Fe-Dph4 was observed under anaerobic conditions upon the addition of 1.5 mm sodium dithionite at 25 °C, as monitored by loss of absorption in the UV-visible region. Importantly, overnight air oxidation at 25 °C led to a complete recovery of the characteristic absorption signals in the visible region, indicating the redox nature of Fe-Dph4 (Fig. 7A). A similar pattern has been reported for the rubredoxins (35).
FIGURE 7.
Redox and electron carrier properties of Fe-Dph4. A, UV-visible absorption spectra of 20 μm Fe-Dph4 reduced by 1.5 mm sodium dithionite in anaerobic conditions followed by reoxidation in air. B, scheme of in vitro electron carrier assay showing the transfer of electrons from NADH to cytochrome c via E. coli NorW and Fe-Dph4. C, anaerobic reduction of 18 μm Fe-Dph4 by 1 μm NorW at different time intervals after the addition of 400 μm NADH at 37 °C. D, demonstration of electron transfer from Fe-Dph4 (20 μm) to cytochrome c (300 μg/ml) in the presence of NorW (2 μm) and NADH (300 μm) under anaerobic conditions at 37 °C. Upper inset, Absorbance traces at 550 nm following the time course for reduction of 200 μg/ml cytochrome c by 5 μm Fe-Dph4 in the presence 1 μm NorW and 300 μm NADH. Lower inset, an expanded view of reduction of cytochrome c observed at 550 nm.
To characterize the electron carrier activity of Fe-Dph4, we adopted the scheme illustrated in Fig. 7B as reported previously (15). The assay utilized NADH and nonspecific NADH-dependent rubredoxin reductase NorW from E. coli as an electron donor and cytochrome c as a terminal electron acceptor. The reduction of Fe-Dph4 was monitored in the presence of fixed concentrations of NorW and NADH under anaerobic conditions at 37 °C. As shown in Fig. 7C, a time-dependent reduction of peak intensity at 485 nm is an indication of changes in the oxidation state of the Fe3+ center of Fe-Dph4.
The redox nature of Fe-Dph4 was further substantiated by transfer of electrons from reduced Fe-Dph4 to cytochrome c, as depicted schematically in Fig. 7B. A time-dependent reduction of cytochrome c was observed by monitoring the changes in absorbance at 550 nm at 37 °C under anaerobic conditions (Fig. 7D). The reduction of cytochrome c followed first order kinetics, and the observed rate constant (k = 1.6 ± 0.04 min−1) at 37 °C was relatively lower than rubredoxins (36) (Fig. 7D). Importantly, when the reaction was carried out by doubling the concentration of NorW or Fe-Dph4, a linear increase in the rate constants to 2.9 ± 0.01 and 2.6 ± 0.01 min−1, respectively, was obtained. This direct dependence of rate constants in presteady state conditions as a function of NorW and Fe-Dph4 concentrations indicates that the electron transfer between NorW and Fe-Dph4 is the rate-limiting step, implying that Fe-Dph4 can function as an electron transfer protein in a manner similar to rubredoxins (18).
DPH4 Is an Ortholog of Yeast JJJ3 and Functionally Complements Diphtheria Toxin Sensitivity in Vivo
The DPH genes have been identified in S. cerevisiae, presumably having a similar function (8). Of these, JJJ3 encodes a J-protein implicated in the biosynthesis of diphthamide. Yeast cells lacking JJJ3 or other DPH genes are resistant to DT activity (8, 10). Based on sequence and domain structural organization, Dph4 has been predicted to be an ortholog of yeast Jjj3, having similar functions. To test the function of Dph4 in vivo, the overexpression constructs were transformed into Δjjj3 cells harboring a plasmid encoding DT catalytic A-chain under the control of a galactose-inducible promoter. As expected, the Δjjj3 strain was resistant and able to grow in galactose media under conditional expression of DT A-chain. On the other hand, complementation of wild type Jjj3 imparts sensitivity toward DT, reconfirming its role in DPH biosynthesis. Furthermore, a similar sensitivity toward DT was observed upon Dph4 overexpression as compared with controls grown in glucose media, signifying its in vivo functional overlap in DPH biosynthesis (Fig. 8A). Thus, our result provides the first evidence to indicate that human Dph4 is an ortholog of yeast Jjj3 having a similar in vivo function.
FIGURE 8.
In vitro and in vivo functional analysis of Dph4. A, in vivo rescue of Δjjj3 phenotype by DPH4. All strains indicated contain DT plasmid under galactose regulation. Serial dilutions of wild type strain with empty vector (W303), Δjjj3 with empty vector (−), or 2μ-GPD vector expressing full-length JJJ3 (Jjj3) or full-length DPH4 (Dph4) were spotted on dropout media containing 2% glucose or galactose and incubated for 72 h at 30 °C. B, UV-visible absorption spectra of 200 μm Fe-Dph4 and iron-bound Jjj3 (Fe-Jjj3) purified from E. coli grown in iron-supplemented M9 medium recorded at 25 °C. Inset, nickel bead-bound Fe-Dph4 and iron-bound Jjj3 proteins. C, iron-bound Jjj3 separated on native-PAGE and stained for iron. D, immunodetection of iron-bound Jjj3 by anti-His antibody. Oligomers are marked with arrows.
To investigate whether Jjj3 possesses an iron-binding property similar to Dph4, we purified Jjj3 recombinantly from E. coli grown in iron-supplemented M9 minimal medium. The nickel column purified Jjj3 protein was dark brown-colored, similar to Dph4 (Fig. 8B, inset). Interestingly, the UV-visible spectra of Jjj3 was found to be similar to that of Dph4 and also stained specifically for iron (Fig. 8, B and C), thus highlighting its iron binding ability similar to that of Dph4. However, its iron binding ability was weaker compared with Dph4, and the bound iron was lost upon dialysis. Additionally, the protein also showed oligomers on native PAGE confirmed by immunodetection using anti-His antibodies (Fig. 8D). Therefore, we conclude that Jjj3 and Dph4 are true orthologs having similar in vivo functions.
DISCUSSION
Structurally, type III J-proteins are a poorly investigated class of heat shock proteins. Only a handful of J-protein structures have been elucidated for individual J-domains or C-terminal fragments using NMR and x-ray crystallography (1). At the same time, the molecular basis of conferring functional specificity through regions outside of the J-domain of type III J-proteins, including Dph4, is very elusive across the kingdoms (7). Our report uncovers the first structure of a full-length human type III J-protein, Dph4, and reveals multiple insights into its moonlighting cellular functions. These functional roles are broadly divided into J-dependent and J-independent activities.
J-domain-dependent Function; Enhanced Stimulation of Hsp70 by Fe-Dph4
Dph4 performs a classical J-protein function by catalytically stimulating the ATPase activity of Hsp70, thus regulating the chaperone cycle. Remarkably, Fe-Dph4 is very stable and exhibits a unique enhanced ability to stimulate Hsp70 ATPase activity as compared with any other J-proteins reported so far. This is due to the difference in the conformation between the iron- and zinc-bound forms, which is supported by several lines of experimental evidence. The trypsin digestion analysis suggests that the linker-helix is better exposed for limited proteolysis in Fe-Dph4 as compared with apo- and Zn-Dph4. At the same time, NMR chemical shift perturbation between zinc- and iron-bound forms of Dph4 indicates significant differences for residues in the linker-helix and around the HPD motif, which is known to be crucial for Hsp70 stimulation. Besides, the linker region sequence is critically required to maintain the ATPase stimulation function by enhancing the stability of J-domain alone. Based on these findings, it is reasonable to believe that linker α5-helix, which connects both of the domains, generates significant conformational changes in the iron-bound state, thus altering the exposure of the J-domain to enhance the stimulation of Hsp70 ATPase activity. For in vivo J-domain function, additional amino acid linker sequence outside of the J-fragment is required (10, 21). Based on the Dph4 results, we envisioned that the linker sequence plays a profound role in modulating the conformational flexibility associated with J-domain alone, thus enabling it to perform better J-dependent functions.
J-domain-independent Functions of Dph4
Based on the three-dimensional structure, it is evident that due to orientational freedom, both the J-domain and the CSL-domain can function independently under physiological conditions. The structure of the C-terminal CSL-domain closely resembles the full-length yeast and human Kti11, which are involved in multiple cellular functions, including diphthamide biosynthesis (16, 25, 37). Similar to Kti11, our finding uncovers novel functions for the CSL-domain of Dph4, which are independent of J-domain function. These results provide the first evidence to demonstrate a robust iron-binding property associated with human Dph4 protein. The specificity of iron binding to human Dph4 in the eukaryotic physiological context is further illustrated by overexpressing and purifying the protein from yeast. Dph4 could sequester iron in vivo, as revealed by its ability to show reactivity toward iron-specific 3,5-diaminobenzoic acid staining (12). Notably, yeast Jjj3, an ortholog of human Dph4 having similar function, also showed an ability to bind iron, suggesting that the iron-binding property is conserved across phylogenetic boundaries.
Iron Binding and Oligomerization
Typically, J-proteins are known to bind the zinc-metal ion via their C-terminal zinc-binding motifs (3, 6). Interestingly, DpH4 is the first J-protein that displayed an ability to bind iron preferentially over zinc at the C-terminal domain. The property of iron binding to Dph4 highlights that potentially it can function as iron donor. In the mitochondrial compartment, mitochondrial Hsp70 (Ssq1 in yeast), together with the J-protein HscB (Jac1 in yeast), plays an essential role in the formation of iron-sulfur cluster assembly (38, 39). The iron-binding protein frataxin (Yfh1 in yeast) functions as a putative iron donor in the mitochondrial iron-sulfur cluster assembly process (39). A similar Hsp70/J-protein pair and a putative iron donor have not been uncovered in the cytosolic compartment, whereas a related iron-sulfur cluster machinery has been recently identified in humans (40–42). Thus, we propose that human Dph4 can dually serve as a J-protein and an iron donor together with Hsc70/HspA1A in the process of specialized function, such as iron-sulfur cluster generation in the cytosolic compartment. The other noteworthy aspect of iron binding is that it promotes oligomerization of Dph4 similar to frataxin (43). As observed for ferritin and frataxin, the intermediate binding affinity of iron to Dph4 also suggests that J-protein Dph4 can function as a transient intracellular “iron store” (39, 43–46). The iron-binding property of Dph4 may therefore also serve as a “buffer” to reduce the iron toxicity, thereby tightly regulating the intracellular iron homeostasis.
Role in Dipthamide Biosynthesis
Our findings reveal that Fe-Dph4 exhibits redox and electron carrier activity similar to rubredoxins (35, 36). Fe-Dph4 can be reduced under anaerobic conditions using a reducing agent like sodium dithionite and slowly reoxidized in the presence of oxygen. In addition to that, Fe-Dph4 possesses a unique ability to oxidize “rubredoxin reductases” by direct transfer of electrons. An efficient transfer of electrons was observed from Fe-Dph4 in a chain reaction involving NorW and cytochrome c as a terminal acceptor. Fe-Dph4 can thus perform vital functions as an “electron reservoir” required for several important metabolic pathways, which involve oxidation and reduction reactions. Hence, it is conceivable that this is a novel co-evolved function attributed to the CSL-domain of Dph4, which is independent of the J-domain.
The electron carrier biological activity described above has implications for involvement of Dph4 in a specialized function for DPH biosynthesis in eukaryotic cells. In yeast Jjj3, despite having a functional J-domain, mutations within the CSL-domain distinctly showed a tolerance to the diphtheria toxin, thus highlighting the critical role played by the CSL-domain in DPH biosynthesis (10). Furthermore, both CSL-domains of Jjj3 and human Dph4 showed an ability to bind iron. As a result, iron-dependent activity of these proteins might play a crucial role in maintaining the redox nature of cytosolic iron-sulfur cluster-containing proteins, including DPH biosynthesis proteins such as Dph2 (47). In a recent report, it has been proposed that Dph4 and Dph3 proteins may be directly involved in maintaining the redox nature of Dph2 protein, which is the key component of DPH biosynthesis machinery (47). Our results provide first direct experimental evidence in favor of this hypothesis. In such a case, an iron-bound CSL-domain can selectively interact with Dph2 client protein as a multiprotein complex, thus defining its functional specificity.
In summary, we have elucidated the solution structure of the type III J-protein, Dph4, and uncovered its iron-mediated functional diversity. It has been reported that the splice site dph4 mutant identified in a mouse model has associated defects like prenatal lethality and limb phenotype (9). Interestingly, these developmental defects are specific to Dph4 and were not reported to be restored by J- or CSL-domain-containing proteins. The mutant phenotype of Dph4 might be a cumulative effect of the loss of multiple functions attributed to Dph4 protein. The biological functions of J-proteins have largely been known to be Hsp70-dependent. However, they have been co-evolved with additional specialized functions for regions outside of the J-domain, which predominantly determines the functional specificity. In some organisms, the J-domain has been found to be dispensable for their imperative function. For instance, the J-protein Cwc23 of S. cerevisiae does not require its J-domain for the essential disassembling process of spliceosome machinery (48). Similarly, the J-domain of Rsp16 is primarily expendable for regulating the flagellar stroke movement in Chlamydomonas reinhardtii (49). This is, therefore, a beginning for unraveling additional physiologically relevant functions of J-proteins other than folding of client proteins. Our findings provide invaluable evidence in this direction.
Supplementary Material
Acknowledgments
We thank R.M. Bhaskara for MD simulations, Rivula Tirupathi for fluorescence measurements, Prof. S.V. Bhat for EPR studies, Alexander F. Yakunin (University of Toronto) for NorW expression constructs, and Prof. E.A. Craig (University of Wisconsin, Madison, WI) for the Δjjj3 strain.

This article contains supplemental Experimental Procedures, References, and Figs. S1–S4.
The atomic coordinates and structure factors (code 2L6L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Hsp70
- heat shock protein 70
- Hsc70
- heat shock cognate protein 70
- DPH
- diphthamide
- DT
- diphtheria toxin
- Fe-Dph4
- iron-bound Dph4
- Zn-Dph4
- zinc-bound Dph4
- MD
- molecular dynamics
- PDB
- Protein Data Bank.
REFERENCES
- 1. Craig E. A., Huang P., Aron R., Andrew A. (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156, 1–21 [DOI] [PubMed] [Google Scholar]
- 2. Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 3. Walsh P., Bursać D., Law Y. C., Cyr D., Lithgow T. (2004) The J-protein family. Modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayer M. P., Brehmer D., Gässler C. S., Bukau B. (2001) Hsp70 chaperone machines. Adv. Protein Chem. 59, 1–44 [DOI] [PubMed] [Google Scholar]
- 5. Mayer M. P., Bukau B. (2005) Hsp70 chaperones. Cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheetham M. E., Caplan A. J. (1998) Structure, function and evolution of DnaJ. Conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery. J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu S., Milne G. T., Kuremsky J. G., Fink G. R., Leppla S. H. (2004) Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell Biol. 24, 9487–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Webb T. R., Cross S. H., McKie L., Edgar R., Vizor L., Harrison J., Peters J., Jackson I. J. (2008) Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J. Cell Sci. 121, 3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahi C., Craig E. A. (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U.S.A. 104, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mattheakis L. C., Shen W. H., Collier R. J. (1992) DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol. Cell Biol. 12, 4026–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuo C. F., Fridovich I. (1988) A stain for iron-containing proteins sensitive to nanogram levels of iron. Anal. Biochem. 170, 183–185 [DOI] [PubMed] [Google Scholar]
- 13. D'Silva P. D., Schilke B., Walter W., Andrew A., Craig E. A. (2003) J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U.S.A. 100, 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pareek G., Samaddar M., D'Silva P. (2011) Primary sequence that determines the functional overlap between mitochondrial heat shock protein 70 Ssc1 and Ssc3 of Saccharomyces cerevisiae. J. Biol. Chem. 286, 19001–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Proudfoot M. (2008) Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a zinc ribbon-like domain. J. Mol. Biol. 375, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun J., Zhang J., Wu F., Xu C., Li S., Zhao W., Wu Z., Wu J., Zhou C. Z., Shi Y. (2005) Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module. Biochemistry 44, 8801–8809 [DOI] [PubMed] [Google Scholar]
- 17. Rao K. K., Evans M. C., Cammack R., Hall D. O., Thompson C. L., Jackson P. J., Johnson C. E. (1972) Mössbauer effect in rubredoxin. Determination of the hyperfine field of the iron in a simple iron-sulfur protein. Biochem. J. 129, 1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon K. S., Hille R., Hemann C., Tabita F. R. (1999) Rubredoxin from the green sulfur bacterium Chlorobium tepidum functions as an electron acceptor for pyruvate ferredoxin oxidoreductase. J. Biol. Chem. 274, 29772–29778 [DOI] [PubMed] [Google Scholar]
- 19. Gething M. J., Sambrook J. (1992) Protein folding in the cell. Nature 355, 33–45 [DOI] [PubMed] [Google Scholar]
- 20. Sadis S., Hightower L. E. (1992) Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry 31, 9406–9412 [DOI] [PubMed] [Google Scholar]
- 21. Karzai A. W., McMacken R. (1996) A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236–11246 [DOI] [PubMed] [Google Scholar]
- 22. Bertini I., Turano P., Vila A. J. (1993) Nuclear magnetic resonance of paramagnetic metalloproteins. Chem. Rev. 93, 2833–2932 [Google Scholar]
- 23. Szyperski T., Luginbühl P., Otting G., Güntert P., Wüthrich K. (1993) Protein dynamics studied by rotating frame 15N spin relaxation times. J. Biomol. NMR 3, 151–164 [DOI] [PubMed] [Google Scholar]
- 24. Szyperski T., Pellecchia M., Wall D., Georgopoulos C., Wüthrich K. (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone. Secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc. Natl. Acad. Sci. U.S.A. 91, 11343–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu F., Zhang J., Sun J., Huang H., Ji P., Chu W., Yu M., Yang F., Wu Z., Wu J., Shi Y. (2008) Solution structure of human DESR1, a CSL zinc-binding protein. Proteins 71, 514–518 [DOI] [PubMed] [Google Scholar]
- 26. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 27. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bitto E., Bingman C. A., Bittova L., Kondrashov D. A., Bannen R. M., Fox B. G., Markley J. L., Phillips G. N., Jr. (2008) Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J. Biol. Chem. 283, 30184–30192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clore G. M., Gronenborn A. M. (1998) New methods of structure refinement for macromolecular structure determination by NMR. Proc. Natl. Acad. Sci. U.S.A. 95, 5891–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greene M. K., Maskos K., Landry S. J. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. U.S.A. 95, 6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fewell S. W., Pipas J. M., Brodsky J. L. (2002) Mutagenesis of a functional chimeric gene in yeast identifies mutations in the simian virus 40 large T antigen J domain. Proc. Natl. Acad. Sci. U.S.A. 99, 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garimella R., Liu X., Qiao W., Liang X., Zuiderweg E. R., Riley M. I., Van Doren S. R. (2006) Hsc70 contacts helix III of the J domain from polyomavirus T antigens. Addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb. Biochemistry 45, 6917–6929 [DOI] [PubMed] [Google Scholar]
- 33. Whalen K. A., de Jesus R., Kean J. A., Schaffhausen B. S. (2005) Genetic analysis of the polyomavirus DnaJ domain. J. Virology 79, 9982–9990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P., Lafer E. M., Sousa R. (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang S. S., Ljungdahl L. G., Dervartanian D. V., Watt G. D. (1980) Isolation and characterization of two rubredoxins from Clostridium thermoaceticum. Biochim. Biophys. Acta 590, 24–33 [DOI] [PubMed] [Google Scholar]
- 36. Petitdemange H., Marczak R., Blusson H., Gay R. (1979) Isolation and properties of reduced nicotinamide adenine dinucleotiderubredoxin oxidoreductase of Clostridium acetobutylicum. Biochem. Biophys. Res. Commun. 91, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 37. Bär C., Zabel R., Liu S., Stark M. J., Schaffrath R. (2008) A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes. Kti11/Dph3. Mol. Microbiol. 69, 1221–1233 [DOI] [PubMed] [Google Scholar]
- 38. Craig E. A., Marszalek J. (2002) A specialized mitochondrial molecular chaperone system. A role in formation of Fe/S centers. Cell Mol. Life Sci. 59, 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lill R., Mühlenhoff U. (2008) Maturation of iron-sulfur proteins in eukaryotes. Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 40. Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. (2010) Cytosolic iron-sulfur cluster assembly (CIA) system. Factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong W. H., Rouault T. (2000) Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19, 5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Broderick J. B. (2007) Assembling iron-sulfur clusters in the cytosol. Nat. Chem. Biol. 3, 243–244 [DOI] [PubMed] [Google Scholar]
- 43. Mühlenhoff U., Richhardt N., Ristow M., Kispal G., Lill R. (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet. 11, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 44. Liu X., Theil E. C. (2005) Ferritins. Dynamic management of biological iron and oxygen chemistry. Acc. Chem. Res. 38, 167–175 [DOI] [PubMed] [Google Scholar]
- 45. Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoon T., Cowan J. A. (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., Zhu X., Torelli A. T., Lee M., Dzikovski B., Koralewski R. M., Wang E., Freed J., Krebs C., Ealick S. E., Lin H. (2010) Diphthamide biosynthesis requires an organic radical generated by an iron-sulfur enzyme. Nature 465, 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sahi C., Lee T., Inada M., Pleiss J. A., Craig E. A. (2010) Cwc23, an essential J protein critical for pre-mRNA splicing with a dispensable J domain. Mol. Cell Biol. 30, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang C., Owen H. A., Yang P. (2008) Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J. Cell Biol. 180, 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.