Background: TatA, TatB, and TatC are subunits of the Tat translocase allowing transport of folded pre-proteins across cellular membranes
Results: We identified TatC sites that interact with pre-proteins, TatA, TatB, and TatC
Conclusion: The cytosolic N terminus and first cytosolic TatC loop constitute part of a twin arginine recognition site
Significance: We developed a working model of how twin arginine pre-protein inserts into Tat translocase.
Keywords: Bacteria, Protein Cross-linking, Protein Export, Protein Secretion, Protein Targeting, Protein Translocation, Signal Sequence, Twin Arginine Translocation
Abstract
A number of secreted precursor proteins of bacteria, archaea, and plant chloroplasts stand out by a conserved twin arginine-containing sequence motif in their signal peptides. Many of these precursor proteins are secreted in a completely folded conformation by specific twin arginine translocation (Tat) machineries. Tat machineries are high molecular mass complexes consisting of two types of membrane proteins, a hexahelical TatC protein, and usually one or two single-spanning membrane proteins, called TatA and TatB. TatC has previously been shown to be involved in the recognition of twin arginine signal peptides. We have performed an extensive site-specific cross-linking analysis of the Escherichia coli TatC protein under resting and translocating conditions. This strategy allowed us to map the recognition site for twin arginine signal peptides to the cytosolic N-terminal region and first cytosolic loop of TatC. In addition, discrete contact sites between TatC, TatB, and TatA were revealed. We discuss a tentative model of how a twin arginine signal sequence might be accommodated in the Tat translocase.
Introduction
Twin arginine (Tat)-specific2 translocation of precursor proteins is known to occur at the plasma membranes of bacteria and archaea, as well as at the thylakoidal membrane of plant chloroplasts (recently reviewed in Refs. 1–3). The signal sequences of Tat-directed precursors possess the conserved SRRXFLK sequence motif with the almost invariant, name-giving RR pair. Many Tat substrates are fully folded when translocated across the respective membranes and therefore require protein conduits with variable pore sizes. These conduits are made in an unknown manner from large assemblies of two types of membrane proteins, a hexahelical TatC-type protein, and one to three representatives of the single-spanning TatA protein family, named TatA, TatB, and TatE.
The best studied Tat translocases are those of Escherichia coli and plant chloroplasts, both comprising TatA, TatB, and TatC subunits that associate to form high molecular mass hetero-oligomeric complexes. TatBC complexes have been shown to function as the first Tat-specific binding site for RR precursors (4–11), but an initial insertion of RR precursors into membrane lipids might also occur (12–14). Following interaction with the TatBC subunits, an advanced binding of RR signal sequences depends on the proton-motive force (PMF) (5, 15) and involves the recruitment of TatA (16–18). Oligomerization of TatA occurs in dependence on TatBC and RR precursors (17, 19). Pore-like oligomers of TatA (20, 21) or TatE (22) might be involved in the formation of a transmembrane protein conduit, although no direct evidence for a Tat substrate passing across an oligomeric TatA/TatE structure has thus far been provided (18, 23, 24). Translocation per se is energized by the PMF.
When RR precursors carry photo cross-linkers next to their RR consensus motif, they are found in close proximity to TatC (5, 7, 24). Cross-links between RR signal sequences and TatC are obtained even in the absence of TatB (5), suggesting that TatC might represent the primary Tat-specific docking site for RR precursor proteins. Single-alanine substitutions of residues spread over the entire N-terminal half of TatC were found to interfere with precursor binding (25), making it impossible to delineate a defined RR signal peptide-binding epitope on TatC. We have now performed an extensive site-specific cross-linking analysis of the E. coli TatC to narrow down the area of precursor recognition, as well as to identify contact sites with other subunits of the E. coli Tat translocase.
EXPERIMENTAL PROCEDURES
DNA Techniques
PCR were performed using Pfu polymerase (Stratagene). For mutagenizing PCR, template DNA was removed by digestion with DpnI (New England Biolabs). For further cloning steps, PCR products were extracted from agarose gels by use of a Qiagen gel extraction kit. The primers used are listed in supplemental Table S2.
Plasmids
Plasmids pPJ3 (pET22b+/TorA-mCherry) and pPJ5 (pET22b+/TorA(KK)-mCherry) (18), p8737 (pET22b+/TatABCD) (26), pKSMSufI-RR, and pKSMSufI-KK (5), as well as pSup-BpaRS-6TRN(D86R) (27) have been described elsewhere. Plasmid pPJ2 (pET22b+/TorA(KK)-MalE335) was constructed according to the QuikChange site-directed mutagenesis kit protocol (Stratagene) using as template DNA plasmid pPJ1 (pET22b+/TorA-MalE335) (18) and the primers 5-TorA KK for and 3-TorA KK rev.
Plasmid pPJ4 (pET22b+/TorA-SufI) was constructed from plasmid pPJ1 (18) digested with EcoRI and XhoI to excise the MalE335 fragment. The SufI-encoding DNA was amplified by PCR with EcoRI and XhoI restriction sites flanking each fragment. The template DNA used was pKSMSufI-RR (5), and the primers were 5-EcoRI oss SufI and 3-XhoI oss SufI. In the final construct, the TorA signal sequence was fused to SufI via a 10-amino acid long linker, which was derived from the early mature region of TorA and two amino acids encoded by nucleotides that had been introduced for cloning reasons.
Plasmid pPJ6 (pET22b+/TorA-SufIΔLinker) is a linker-less version of pPJ4. Plasmid pPJ6 was constructed by inverse PCR using pPJ4 as template and the primers 5-TorA-SufI ohne Linker and 3-TorA-SufI ohne Linker. The PCR mix was digested with DpnI for 1 h at 37 °C to remove the template DNA. The generated linear DNA fragment was phosphorylated with T4 polynucleotide kinase (New England Biolabs) for 1 h at 37 °C and blunt end-ligated overnight at room temperature using T4 ligase (Fermentas). The resulting plasmid was site-specifically mutagenized to change the signal peptide cleavage site from ATAAGQ to ASAAGQ using the primers TSdL SS SufI for and TSdL SS SufI rev.
To construct plasmid pPJ11 (pET22b+/TorA-MalE), TorA-MalE was amplified via PCR using pTorA-MalE (28) as template DNA and the oligonucleotide primers 5′-NdeI TorA-MalE and 3′-XhoI TorA-MalE. The PCR fragment and the recipient vector pET22b+ were digested with NdeI and XhoI and ligated. The KK mutation was introduced into pPJ11 as described above for plasmid pPJ2 yielding the new plasmid pPJ12 (pET22b+/TorA(KK)-MalE).
Plasmid pTF1 (pET22b+/AmiC) was constructed by PCR using genomic E. coli DNA as a template and the primers AmiC_nde_for and AmiC_xho_rev. The PCR product was blunt end-ligated into plasmid pSC-B of the StrataClone blunt PCR cloning kit (Stratagene). The plasmids pSC-B-AmiC and pET22b+ were digested with NdeI/XhoI, and the amiC fragment was ligated into the pET22b+ vector.
To construct plasmid pTF2 (pET22b+/TorA-Thioredoxin), DNA encoding the TorA signal sequence was amplified via PCR using plasmid pET28aTorA-PhoA (24) as template and the primers TorA_SS_for_nde and TorA_SS_rev. The thioredoxin-encoding DNA was amplified from genomic E. coli DNA using the primers TrxA_for2 and TrxA_rev_xho. The thioredoxin-encoding PCR product was used for a third PCR with primers TorA_TrxA_for and TrxA_rev_xho to extend the 5′-end of the thioredoxin DNA by the C-terminal codons of the TorA signal sequence. This PCR product was hybridized with the first one encoding the entire TorA signal peptide. The fused DNA was extended by DNA polymerase during ten PCR cycles. Following the addition of the primers TorA_SS_for_nde and TrxA_rev_xho, the reaction was continued to obtain a torA-thioredoxin construct flanked by NdeI and XhoI restriction sites. The PCR product was blunt end-ligated into plasmid pSC-A of the StrataClone blunt PCR cloning kit (Stratagene) and finally cloned into pET22b+ as described above for pTF1.
To construct plasmid pPS1 (pET22b+/AmiC-SufI), DNA encoding the signal sequence of AmiC was amplified and equipped with flanking restriction sites via PCR using pTF1 as template DNA and the primers AmiC 5-SS NdeI and AmiC 3-SS EcoRI. The PCR product thus obtained and plasmid pPJ4 (pET22b+/TorA-SufI) were digested with NdeI and EcoRI. The digested vector was dephosphorylated with antarctic phosphatase (New England Biolabs) and ligated with the AmiC signal sequence-encoding PCR product.
Plasmid pHCM1 encodes a His tag-free version of TorA-PhoA and was obtained from plasmid pET28aTorA-PhoA (24) by mutagenizing PCR using the primers TorA-PhoA(−His-tag)for and TorA-PhoA(−His-tag)rev.
In Vitro Reactions
Cell extracts used for the in vitro synthesis of RR precursor proteins were prepared from E. coli strain SL119 (29) according to Ref. 30. Coupled transcription/translation reactions were performed in 50-μl aliquots as described (30). INV were added 10 min after starting the synthesis reaction and incubated for 25 min at 37 °C. Protein translocation into INV was assayed as described (18). CCCP was added to a final concentration of 0.1 mm together with INV as detailed in Ref. 18. For cross-linking, samples were irradiated with UV light for 15 min as described (18). SDS electrophoresis using 8%, 10%, or 15% polyacrylamide gels was performed as described previously (30).
Membrane Vesicles
Tat+-INV were prepared as described (30) from E. coli strain BL21(DE3)* (Novagen) transformed with plasmids p8737 (26). Tat+-INVs containing Bpa variants of TatC were prepared from E. coli strain BL21(DE3)* transformed with plasmid pSup-BpaRS-6TRN(D86R) (27) and p8737 derivatives carrying the individual tatC amber mutants. Expression of tat genes was induced with 1 mm isopropyl β-d-thiogalactopyranoside when cell cultures had reached an optical density at 600 nm (A600) of 0.5 and growth was continued up to an A600 of 1.5–1.8. To suppress the amber stop codons, 1 mm Bpa was added at the same time as isopropyl β-d-thiogalactopyranoside from a 1 m Bpa stock solution prepared in 1 m NaOH. To detect contacts between the Tat subunits, 4 μl each of INV (∼40 A280 units/ml) carrying an individual Bpa variant of TatC were diluted with 96 μl of INV Puffer (30), UV-irradiated on ice for 20 min, and further processed for Western blotting using polyclonal antibodies raised against TatA, TatB, and TatC as described (18).
RESULTS
Clustering of Precursor Contacts in N Terminus and First Cytosolic Loop of TatC
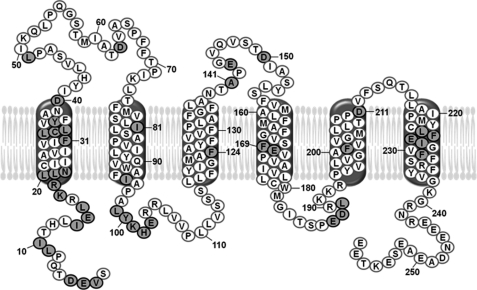
To identify molecular contacts of TatC, we replaced the amino acids of TatC highlighted in Fig. 1 and listed in Table 1 by the photo cross-linker p-benzoyl-phenylalanine (Bpa). This was achieved by exchanging the corresponding codons in the tatC DNA to amber stop codons using mutagenizing PCR. As template DNA, we used the tatABCD operon of plasmid p8737, because it allows T7 promoter-dependent high expression of the Tat proteins. This has proven to be a prerequisite for obtaining inner membrane vesicles with sufficient Tat-specific translocation activity (26, 31). An E. coli BL21(DE3)* derivative expressing an amber suppressor tRNA and a cognate Bpa-specific tRNA synthetase was individually transformed with the plasmids carrying the amber stop codon variants of tatC. Growth of these cells in the presence of Bpa leads to the suppression of the amber stop codons and concomitant incorporation of Bpa resulting in full-length TatC variants. This was verified for each Bpa variant by immunoblot analysis using anti-TatC antibodies (examples are given below in Figs. 5 and 6, bands labeled TatC).
FIGURE 1.
Amino acids of E. coli TatC that were exchanged against the photo cross-linker p-benzoyl-phenylalanine. Depicted is the amino acid sequence of E. coli TatC with the modified residues highlighted in gray. Numbering starts with the N-terminal start methionine, which has been omitted. The six predicted transmembrane α-helices (46–48) are drawn according to Ref. 49.
TABLE 1.
Cross-linking of Bpa variants of TatC to different RR precursors
TatC variants that gave rise to the most prominent cross-links are underlined. Indicated are cross-linking intensities (+) and failure to cross-link (−). Blank spaces indicate that cross-linking was not determined.
| TorA-mCherry | TorA-MalEa | TorA-PhoA | TorA-Thioredoxin | TorA-SufIb | SufI | AmiC-SufI | AmiC | |
|---|---|---|---|---|---|---|---|---|
| Val-3 | +++ | +++ | +++ | +++ | + | ++ | + | |
| Glu-4 | + | + | + | − | + | |||
| Asp-5 | + | + | + | − | + | |||
| Leu-9 | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Ile-10 | ++ | + | + | + | − | +++ | ++ | |
| Ile-14 | − | + | − | − | + | − | ||
| Glu-15 | ++ | ++ | ++ | ++ | − | + | − | |
| Leu-16 | + | + | + | − | − | − | ||
| Lys-18 | + | + | − | − | − | − | ||
| Arg-19 | − | − | − | − | − | − | ||
| Leu-20 | − | − | − | − | − | ++ | − | − |
| Leu-21 | − | − | − | − | − | − | ||
| Asn-22 | − | − | − | − | − | − | ||
| Phe-31 | − | − | − | − | − | − | ||
| Leu-32 | − | − | − | − | − | |||
| Cys-33 | − | − | − | − | − | |||
| Leu-34 | − | − | − | − | − | |||
| Tyr-36 | − | − | − | − | − | |||
| Asp-40 | − | − | − | − | − | |||
| Leu-49 | − | − | − | − | − | |||
| Asp-63 | − | − | − | − | ||||
| Ile-81 | − | − | − | − | − | |||
| Ile-95 | − | − | − | − | − | |||
| Leu-99 | − | − | − | − | − | |||
| Tyr-100 | + | + | + | − | + | − | ||
| Lys-101 | ++ | ++ | ++ | − | + | − | ||
| His-102 | − | − | − | − | − | |||
| Glu-103 | − | − | − | − | − | |||
| Phe-124 | − | − | − | − | − | |||
| Ala-141 | − | − | − | − | − | |||
| Glu-143 | − | − | − | − | ||||
| Asp-150 | − | − | − | − | ||||
| Phe-169 | − | − | − | − | − | |||
| Glu-170 | − | − | − | |||||
| Glu-187 | + | + | + | − | − | |||
| Asp-188 | − | − | − | − | ||||
| Leu-189 | − | − | − | − | ||||
| Phe-201 | − | − | − | |||||
| Asp-211 | − | − | − | − | ||||
| Leu-225 | − | − | − | |||||
| Phe-226 | − | − | − | |||||
| Glu-227 | + | − | − | + | ||||
| Ile-228 | − | − | − | |||||
| Phe-231 | − | − | − | − | − |
a Comparable results were obtained for TorA-MalE and the truncated version TorA-MalE335.
b Different from the experiments shown in Fig. 4, the results listed here were obtained with the variant TorA-SufIΔLinker (see “Experimental Procedures”).
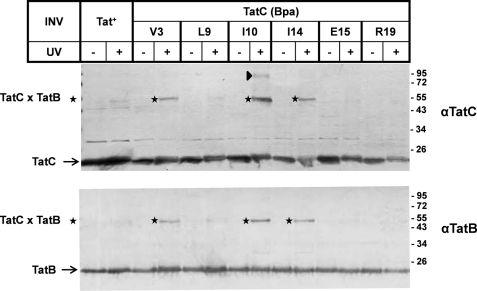
FIGURE 5.
Molecular proximity between the N terminus of TatC and TatB. Membrane vesicles carrying wild-type TatC (Tat+) or the indicated Bpa variants of TatC were irradiated with UV light to initiate cross-linking to TatC in the absence of added precursor proteins. Vesicle proteins were resolved by SDS-PAGE and immunoblotted against anti-TatC and anti-TatB antibodies (αTatC and αTatB). The adducts of ∼55 kDa (stars) obtained with TatC(V3Bpa), TatC(I10Bpa), and TatC(I14Bpa) were recognized by both antibodies, indicating 1:1 complexes between TatC and TatB. An ∼95-kDa adduct to TatC(I10Bpa) (triangle) consistently cross-reacted with αTatC but usually not clearly with αTatB.
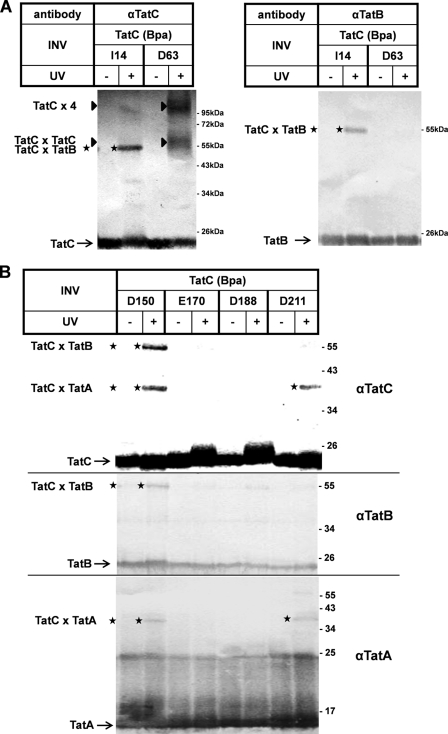
FIGURE 6.
Site-specific contacts of TatC with TatA, TatB, and TatC. UV-induced cross-linking of INV carrying Bpa variants of TatC. Experimental details are as described in the legend for Fig. 5 using the indicated antibodies. A, the adduct (stars) to TatC(I14Bpa) is recognized by αTatB and αTatC, and those to TatC(D63Bpa) are recognized only by αTatC (triangle). B, adducts to TatC(D150Bpa) and TatC(D211Bpa) (top panel) are also recognized by αTatB (middle panel) and αTatA (bottom panel), respectively. TatC(E170Bpa) and TatC(D188Bpa) serve as negative controls that did not give rise to any αTat-reactive cross-link.
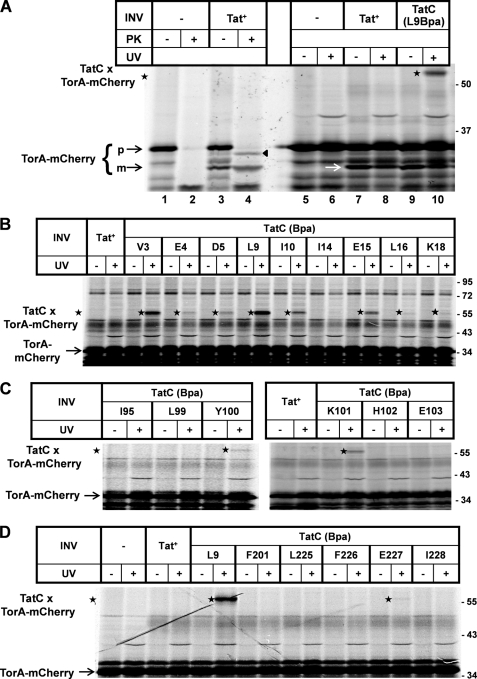
The Bpa variants of TatC thus obtained were then probed for contacts to a variety of twin arginine (RR)-containing precursors. To this end, RR precursors were synthesized by in vitro transcription/translation in the presence of INV that had been prepared from the various tatC mutant strains. In this way, RR precursors could bind to and translocate into the lumen of the INV. These experiments are depicted in Fig. 2 for the model Tat substrate TorA-mCherry, which had been constructed by fusing the RR signal sequence of E. coli TMAO reductase (TorA) to mCherry. When TorA-mCherry DNA was transcribed and translated in vitro in the presence of [35S]methionine/cysteine, a major translation product of the expected molecular mass of this precursor (35 kDa) became visible by SDS-PAGE and phosphorimaging (Fig. 2A, lane 1, p). When during synthesis of TorA-mCherry, INV were present that had been prepared from a strain overproducing wild-type TatABC (Tat+), an additional, smaller translation product of ∼30 kDa (lane 3, m) appeared. This species turned out to be resistant toward PK (lane 4), indicating that it represents the mature form of mCherry that was translocated into the protease-inaccessible lumen of the vesicles and simultaneously cleaved by the vesicle-borne signal peptidase. As routinely observed for those INV of E. coli (30), a certain fraction of RR precursor was translocated but failed to be processed by the signal peptidase of the vesicles. This is inferred from the finding that the precursor TorA-mCherry, when synthesized in the absence of INV, was virtually completely digested by PK (lane 2) but was partially protected in the presence of INV (triangle). The protected species was, however, slightly smaller than the non-PK-treated precursor (compare lanes 3 and 4). We have repeatedly observed this shift in molecular mass for many TorA signal sequence-containing precursors when treated with PK in the presence of INV. Most likely it is due to a PK-mediated removal of a few N-terminal amino acids from the otherwise membrane-protected TorA signal peptide. A prominent PK-resistant band running at the bottom of the gel appeared independently of the addition of INV (lanes 2 and 4), probably representing a stably folded part of the mCherry domain.
FIGURE 2.
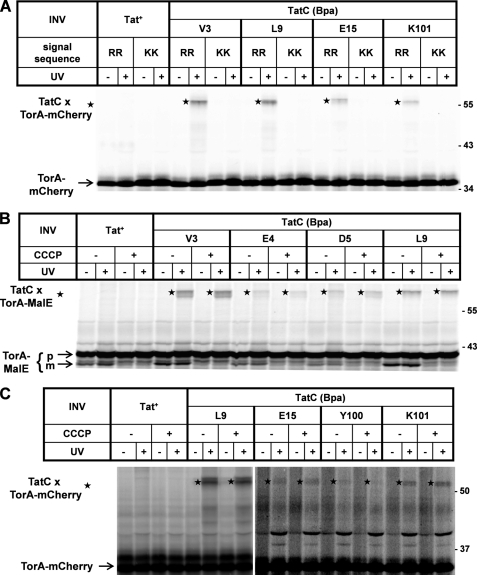
Cross-links between TatC and the model RR precursor TorA-mCherry. A, TorA-mCherry was synthesized by in vitro transcription/translation in the presence of [35S]methionine/cysteine. Radiolabeled translation products were separated by SDS-PAGE and are visualized by phosphorimaging. Numbers to the right indicate the molecular masses in kDa of marker proteins. Where indicated, INV were present during synthesis. INV were prepared from an E. coli wild-type strain overexpressing TatABC (Tat+) or a strain expressing in the same genetic background a TatC variant, in which the leucine at position 9 had been replaced by Bpa (L9Bpa). Indicated are the precursor of TorA-mCherry (p) and the mature form (m) obtained by INV-mediated signal sequence cleavage. Besides processing, translocation of TorA-mCherry into the lumen of the INV is also indicated by the acquirement of PK resistance of the precursor (black triangle) and the mature form. Only in the presence of TatC(L9Bpa)-INV did irradiation of the samples with UV-light (UV) give rise to an adduct (star) that by size corresponds to a 1:1 complex between TatC and TorA-mCherry. B, as in A except that cross-linking was performed with different INV each one carrying a Bpa substitution at the indicated positions within the N terminus of TatC. C, as in B using Bpa variants of the first cytosolic loop of TatC. D, as before comparing the L9Bpa and E227Bpa variants of TatC, the latter being located in the sixth TM of TatC.
Fig. 2A (lanes 9 and 10) depicts the results obtained when Tat+-INV containing wild-type TatC were replaced by vesicles carrying a TatC variant having leucine at position 9 exchanged against Bpa (TatC(L9Bpa)). Whereas these vesicles also allowed transport as indicated by the appearance of processed mCherry (white arrow), UV irradiation gave rise to a strong radiolabeled adduct of ∼55 kDa (lanes 9 and 10, star). This adduct was neither obtained with the Bpa-less control INV (Tat+, lane 8) nor in the absence of any INV (lane 6). Because the cross-linker had been incorporated into TatC, the 55-kDa adduct most likely results from a 1:1 cross-link between TatC and the radiolabeled TorA-mCherry.
Fig. 2 (B–D) depicts essentially all cross-links obtained between TorA-mCherry and TatC variants. Cross-linking sites on TatC localize to its N terminus (compare Fig. 2B with Fig. 1), its first cytosolic loop (compare Fig. 2C with Fig. 1), and its sixth transmembrane domain (TM) (compare Fig. 2D with Fig. 1). In addition, TorA-mCherry cross-linked to position Glu-187 (Table 1) that is located in the second cytosolic loop of TatC (cf. Fig. 1). In total, 44 positions of TatC had been replaced by Bpa. Most of them were chosen on the ground that by mutational analysis they themselves or nearby residues had previously been shown to be critical for the activity of TatC. As summarized in Table 1, of the 44 positions only the 12 named above yielded clearly discernable cross-links to TorA-mCherry. By far the strongest adducts were obtained for positions in the extreme N terminus, in particular Val-3, Leu-9, Ile-10, and Glu-15, followed by Lys-101 in the first cytosolic loop of TatC (Table 1; see also Fig. 2B). It should be noted that although most Bpa variants of TatC were less active in translocation than wild-type TatC (supplemental Table S1), there was no direct correlation between the extent of cross-linking of an individual variant and the degree by which the mutation affected the translocation activity. This was actually not to be expected because targeting to TatC has been characterized as a pretranslocational event (5, 7).
Substrate Binding by TatC Occurs via Signal Peptide
By incorporating a photo cross-linker site-specifically into the signal peptides of Tat substrates, it was previously shown that RR precursors interact with TatC via their signal sequences (5, 7, 24). This implies that the precursor docking site on TatC should be the same for RR precursors differing only in their mature moieties. To examine this, we performed cross-linking experiments using the Bpa variants of TatC and four additional fusion proteins constructed from the TorA signal sequence and maltose-binding protein (TorA-MalE), alkaline phosphatase (TorA-PhoA), thioredoxin (TorA-Thioredoxin), and SufI (TorA-SufI). The results are listed in Table 1 and are compared with those obtained for TorA-mCherry. As far as determined, the residues of TatC that cross-linked to the RR precursors were basically the same for all TorA fusions. The most prominent adducts were obtained for the TatC variants having Bpa incorporated at Val-3, Leu-9, Glu-15, and Lys-101. Thus the protein moieties that had been fused to the TorA signal peptide were in fact without influence on the contact sites with TatC, strongly suggesting that it is the signal sequence of TorA that was interacting with TatC.
N Terminus and First Cytosolic Loop of TatC Recognize Twin Arginine Consensus Motif
Cross-linking to TatC required an intact RR motif in the TorA signal peptide. This is shown in Fig. 3A for the strongly cross-linking TatC variants TatC(V3Bpa), TatC(L9Bpa), TatC(E15Bpa), and TatC(K101Bpa). All four of them failed to form adducts to a mutant TorA-mCherry precursor, in which the consensus RR pair had been replaced by two consecutive lysines. This is completely consistent with the results of previous studies probing the cross-linking behavior of RR signal sequences that had the photo cross-linker incorporated at the Phe position in the consensus box SRRXFLK. Whereas in this sequence context the cross-linker formed adducts to TatC, no interaction was observed when the preceding RR pair had been replaced by two lysines (5, 7). Thus the N terminus and the first cytosolic loop of TatC not only represent a global signal sequence-binding site, but they also seem to harbor the recognition center for the twin arginine consensus motif.
FIGURE 3.
Cross-linking of RR precursors to TatC requires an intact RR motif and occurs independently of the proton-motive force. Experimental details are as in the legend for Fig. 2. A, cross-linking to four prominent contact sites of TatC is compared between wild-type TorA-mCherry (RR) and a mutant derivative, in which the RR pair had been exchanged against two lysines (KK). Tat+-INV were included as Bpa-free control vesicles. B, in vitro synthesis of TorA-MalE in the presence of the indicated INV. Where indicated, CCCP was added to dissipate the proton-motive force of the INV. Control samples received an equal amount of the solvent Me2SO. The addition of CCCP resulted in a block in translocation, i.e. production of the mature form (m) of TorA-MalE but did not influence cross-linking to TatC (stars). C, as in B except for using TorA-mCherry as RR precursor. For unknown reasons, the TatC adducts to various precursors often appeared as double bands (B and C). The size difference is too small to result from cross-linking to precursors and the signal sequence-less forms.
Cross-links between TatC and RR Precursors Are Not Dependent on Proton-motive Force
Consistent with the binding of RR precursors to cytosolically exposed sites of TatC reflecting only a superficial contact, the cross-links to TatC were not negatively affected by dissipating the proton-motive force (PMF) (Fig. 3, B and C). How exactly the proton gradient is transduced into Tat-dependent translocation is not well understood. Although the electrical potential was shown to be required for an undefined early transport event (32), many studies substantiate an involvement of the PMF directly in the translocation step. This can be seen in Fig. 3B, in which the mature form of TorA-MalE (m) indicating successful transport into INV is reduced to a background band in the presence of the uncoupler CCCP for wild-type (Tat+) and all TatC(Bpa) mutant vesicles. In contrast to transport, CCCP did not diminish cross-linking of TorA-MalE to the Val-3, Glu-4, Asp-5, and Leu-9 positions of TatC (Fig. 3B, stars). Likewise, cross-linking of the TorA-mCherry precursor to TatC was unimpaired in the presence of CCCP (Fig. 3C, stars). These results demonstrate that the interaction of TorA precursors with TatC as visualized here by photo cross-linking was in fact independent of the PMF.
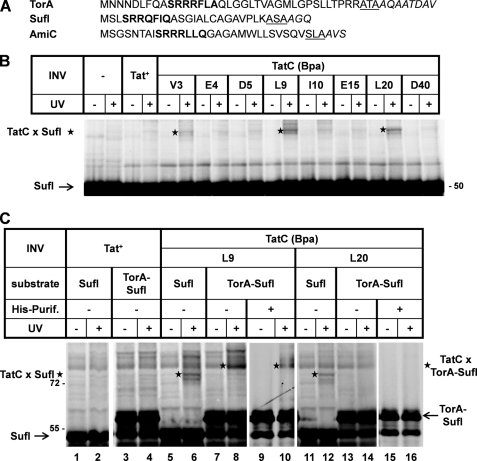
Signal Sequence of Natural Tat Substrate SufI Uniquely Cross-links to Residue Leu-20 of TatC
The results presented so far identified two major cytosolically exposed areas on TatC, by which the TorA signal sequence seems to be recognized. To see whether these contact points were generally involved in the recognition of RR precursors, we next probed for contacts between TatC and the natural Tat substrates SufI and AmiC. Both periplasmic proteins harbor RR signal sequences that are significantly shorter than that of TorA (Fig. 4A). The obtained cross-links are listed in Table 1. Whereas in general fewer Bpa variants of TatC were found to cross-link to SufI and AmiC than to the TorA derivatives, the major TorA-interacting residues Val-3 and Leu-9 yielded also the most prominent adducts with the precursors of SufI (Fig. 4B) and AmiC. The similarity in the cross-linking pattern of the TorA signal sequence with those of SufI and AmiC became even more evident for the fusion protein AmiC-SufI, in which the signal sequence of SufI had been replaced by that of AmiC. With this fusion protein, adducts to Tyr-100 and Lys-101 in the first cytosolic loop of TatC also became manifest, much like for the TorA fusions (Table 1). These findings suggest that the differences in TatC cross-linking observed for TorA and SufI/AmiC precursors are rather quantitative than qualitative in nature. Nevertheless we cannot entirely rule out that the length of an RR signal peptide and possibly also subsequent areas of the mature protein might have some influence on the binding area of the TatC receptor. In any case, the bottom line of the comparison between the various RR precursors is that the presumably cytosolic N terminus of TatC and the area around Tyr-100 and Lys-101 constitute a recognition site for RR signal peptides.
FIGURE 4.
The natural Tat substrate of E. coli, pSufI, uniquely cross-links to the position leucine 20 of TatC. A, signal sequences of the Tat substrates TorA, SufI, and AmiC. The consensus motifs are printed in bold, and amino acids following the signal peptides are in italics. The three residues preceding each signal sequence cleavage site are underlined. B, experimental details are as described for previous figures, now showing cross-links (stars) between TatC and pSufI. C, comparing cross-linking of SufI and TorA-SufI to TatC(L9Bpa) and TatC(L20Bpa). To separate TatC-TorA-SufI cross-links from the background bands, TorA-SufI species were purified via nickel affinity purification (His-Purif.). An adduct to TatC(L20Bpa) (star) is obtained only with pSufI.
There was, however, one notable exception to the uniformity, with which the tested RR precursors cross-linked to TatC. SufI was the only precursor that strongly cross-linked to Leu-20 located at the predicted beginning of the first TM of TatC (Table 1 and Fig. 4B). Because it was one of the strongest SufI-TatC adducts obtained, we examined whether it in fact reflected a specific interaction of the SufI signal sequence with TatC. To this end, we compared cross-linking to TatC(L9Bpa) and TatC(L20Bpa) between SufI and TorA-SufI, in which the signal sequence of SufI had been replaced by that of TorA. As illustrated in Fig. 4C, TatC(L9Bpa) yielded the clear UV-dependent cross-link to SufI (lane 6, star) as seen before (Fig. 4B), whereas a presumed adduct to TorA-SufI was hidden in the UV-independent background noise (lane 8, star). We therefore purified TorA-SufI products via a C-terminal His tag and could then clearly visualize the TorA-SufI-TatC(L9Bpa) adduct (lane 10, star). The same experimental approach was performed with TatC(L20Bpa) vesicles (lanes 11–16). Those vesicles, however, cross-linked only to the SufI signal sequence but not to that of TorA. It is not clear why the authentic SufI signal sequence yielded this additional contact site, which incidentally lies close to the recognition area of the N terminus of TatC (further discussed below). Irrespective of the reason why SufI showed this peculiar behavior, the different cross-linking pattern of pSufI and TorA-SufI lends further support to the notion that the cross-links observed here specifically involve the signal sequence of Tat substrates.
Signal Sequence Recognition Site of TatC Is Juxtaposed to TatB
Next we UV-irradiated all of the membrane vesicles carrying one of the Bpa variants of TatC (Fig. 1), to probe for adducts to TatC in the absence of added RR precursors. These adducts were visualized by immunoblotting using anti-TatC antibodies. As summarized in Figs. 5 and 6, the positions of TatC that yielded clearly visible adducts were Val-3, Ile-10, Ile-14, Asp-63, Asp-150, and Asp-211. The V3Bpa, I10Bpa, and I14Bpa variants of the cytosolic N terminus of TatC gave rise to cross-linking products of ∼55 kDa (Fig. 5, stars). Because these adducts were recognized by antibodies directed both against TatC and TatB (Fig. 5), they must represent 1:1 complexes between TatC and TatB. Ile-10 yielded an additional weaker adduct of ∼95 kDa that cross-reacted with anti-TatC antibodies (triangle). The origin of this adduct is not clear. By size it could represent a tetramer of TatC, although it does not exhibit the fuzzy appearance of other TatC oligomers (Fig. 6A, below), and we never observed a TatC(I10Bpa) adduct of the size expected for a TatC dimer. On the other hand, it theoretically could result from a dimer of a 1:1 TatBC complex, but it was never clearly recognized by anti-TatB antibodies (Fig. 5, lower panel). Notably, the three positions of TatC that cross-linked to TatB largely overlap with the precursor recognition site of TatC.
Trans-sided Residues of TatC Serve Homo-oligomerization and Interaction with TatA and TatB
Asp-63 is located in a predicted periplasmic loop of TatC (cf. Fig. 1). TatC(D63Bpa) gave rise to two TatC-cross-reacting adducts that in contrast to the adduct of TatC(I14Bpa) were not recognized by anti-TatB antibodies (Fig. 6A, left and right panels). They are therefore likely dimers and tetramers of TatC, indicating that TatC can homo-oligomerize via a periplasmic domain.
The two other interacting residues were Asp-150 located in the predicted second periplasmic loop of TatC and Asp-211 at the periplasmic end of TM 5. Both Bpa variants of TatC each yielded an adduct that was only ∼40 kDa in size (Fig. 6B, top panel). Consistent with the small size, these adducts were recognized by anti-TatA antibodies (bottom panel). In addition, TatC(D150Bpa) yielded another cross-link of ∼55 kDa that was decorated with anti-TatB antibodies (middle panel). Thus Asp-211 seems to be an intramembrane contact site for TatA, and Asp-150 seems to be a potentially nearby residue that can interact with both TatA and TatB.
DISCUSSION
RR Precursor Recognition Site of TatC
The strongest cross-links with all RR precursors tested were obtained when Bpa was located in Val-3 and Leu-9 of TatC, but the interacting area extended over almost the entire cytosolic N terminus of TatC up to Glu-15 and for some precursors even up to Lys-18 and Leu-20 (cf. Fig. 1). In addition, clear precursor adducts were obtained for Tyr-100 and Lys-101 located within the first predicted cytosolic loop of TatC. Consistent with these cytosolically exposed domains of TatC interacting with RR precursors, the signal sequences of membrane-bound Tat substrates are still accessible to proteases and can be detached by disrupting electrostatic interactions (15, 33).
In two recent reports, single amino acid exchanges in the extreme N terminus and the first cytosolic loop of TatC were described that suppress the translocation defect of KQ and KK mutant precursors (9, 34). Suppressors mapped to residues Leu-9, Lys-18, and Gln-22 each in the N terminus (9), as well as to Leu-99 and Phe-94 in the first cytosolic loop of TatC (34). One such suppressor simultaneously showed mutations of the nearby residues Trp-92 and Pro-97 (34). If these TatC suppressors function by restoring the impaired recognition of the KQ and KK signal sequences, the mutated amino acids might well be part of a signal peptide-binding epitope. The fact that this genetic approach mapped RR signal peptide-interacting sites to the same domains of TatC as our cross-linking strategy virtually rules out the possibility that by exchanging individual amino acids against Bpa we could have created artificial contact sites for RR precursors.
The data presented here provide only indirect evidence that it is the signal sequence of an RR precursor to which the N terminus and the first cytosolic loop of TatC cross-link. Thus independently of the nature of their mature domains, all RR precursors tested cross-linked to the same sites of TatC. Furthermore complementary studies with the cross-linker Bpa incorporated into surface-exposed residues of the mature parts of Tat substrates yielded adducts to TatA and predominantly to TatB but totally failed to show contacts with TatC (11). On the other hand, precursors carrying photo cross-linkers in the immediate vicinity of the RR pair do give rise to TatC adducts (5, 7, 24). Moreover, the above discussed tatC suppressors most likely function by allowing a relaxed recognition of the RR mutant signal peptides. These results collectively suggest that TatC recognizes the RR motif through its extreme N terminus and its first cytosolic loop. This is illustrated in the model shown in Fig. 7.
FIGURE 7.
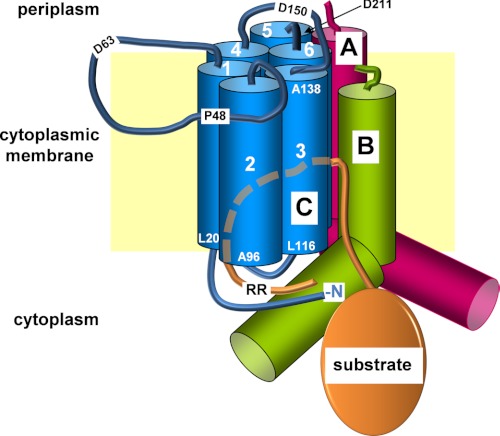
Model of the TatBC binding site for RR precursors. Shown are TatA, TatB, and TatC (labeled A, B, and C, respectively) embedded in the cytoplasmic membrane (yellow rectangle). The cylinders represent the six TMs (numbers) of TatC, as well as the TMs and cytosolic amphipathic helices of TatA and TatB. The blue lines symbolize the periplasmic loops of TatC and two of its cytosolic domains including the N terminus (N) of the protein. A folded Tat substrate (orange ellipse) with its RR-containing signal peptide (orange line) is depicted. The dashed line represents part of the RR signal peptide inserting possibly between the TMs of TatC. The approximate locations of distinct amino acids of TatC either flanking TMs or residing in the periplasmic loops are indicated using one-letter codes and the position numbers. This model focuses on the findings described here, whereas a functional Tat translocase very likely comprises multiple such Tat(A)BC units.
Contacts between TatC and TatB Reflecting Concerted Precursor Recognition by TatBC
Our cross-linking analysis further revealed that the RR signal sequence-recognizing N terminus of TatC is also in close contact to TatB. It is reasonable to assume that the cytosolic N terminus of TatC contacts cytosolic parts of TatB such as its amphipathic helix (Fig. 7). Because we obtained these TatB-TatC cross-links with isolated membrane vesicles in the bona fide absence of any RR precursor, a conceivable scenario would be that TatB blocks the RR recognition site of TatC, until Tat substrates displace TatB from TatC. We have, however, not been able to interfere with the TatB-TatC cross-linking by adding chemical amounts of purified pSufI. Therefore, it is more likely that the cross-links between TatC and TatB merely reflect their molecular neighborhood. A close association between TatC and TatB is not only indicated by the complex formation between both Tat subunits (35–38). Numerous data suggest that TatB also functionally cooperates with TatC by constituting one part of a joint binding pocket for RR signal peptides (Fig. 7). Thus TatB has repeatedly been demonstrated to directly interact with RR signal peptides (5, 7, 9, 24, 25), including residues adjacent to the RR pair (5, 7).
Nevertheless, several findings suggest that the RR pair is preferentially recognized by TatC. As shown here, cross-linking between TatC and an RR precursor is prevented by the KK mutation. Analogously, KK precursors having the photo cross-linker incorporated adjacent to the KK-pair fail to cross-link to TatC (5, 7), yet they still cross-link to TatB at sites outside of the consensus motif (5). KK precursors were shown also by other experimental approaches to still associate with the Tat translocase (8, 24, 39). These data collectively suggest that although RR signal peptides are bound by a probably spacious TatBC binding pocket, it is TatC that directly recognizes the RR motif via its cytosolic domains.
In a previous study, single alanine substitutions of Pro-48, Phe-94, Tyr-100, Glu-103, and Tyr-126 of TatC were reported to impair precursor binding (25). Whereas Phe-94, Tyr-100, and Glu-103 are located again in the first cytosolic loop of TatC, Tyr-126 is part of the third TM, and Pro-48 resides in the first periplasmic loop of TatC (Fig. 7). Therefore the Tyr-126 and Pro-48 mutations likely exert indirect effects on the precursor binding site. In accordance with such an assumption, the P48A mutation was shown to destabilize the TatBC receptor complex (40), which could lead to a disruption of the concerted TatBC binding site. A similar explanation might apply to residue Pro-142 of TatC, which when mutated to serine was found to suppress the defective transport of a KK precursor protein (34). According to the model shown in Fig. 7, Pro-142 is predicted to be part of the second periplasmic loop of TatC. Interestingly, we found the nearby Asp-150 to cross-link to TatB when exchanged to Bpa, suggesting that this periplasmic loop of TatC might be involved in maintaining the stability and functional interaction between TatB and TatC.
Additional Interactions between TatC and RR Precursors
Glu-187 and Glu-227 are two residues of TatC that showed weaker cross-links with some of the RR precursors tested (Table 1). Glu-187 is situated in the second predicted cytosolic loop of TatC connecting TMs four and five (not visible in the model of Fig. 7) and might well be in close proximity to the RR recognition epitope formed by the extreme N terminus and first cytosolic loop of TatC.
On the contrary, Glu-227 resides in the middle of the sixth transmembrane helix. If the Glu-227 adducts of TatC reflected contacts with the signal sequence, it could indicate that the RR signal peptide forms a hairpin loop on the outside of, or even in between, the TMs of TatC, similar to what is depicted in the model of Fig. 7. Such a hairpin-like insertion of an RR signal sequence into a concerted TatBC binding pocket is actually suggested by several findings. First, complete translocation can occur with the N terminus of an RR signal covalently attached to TatC (7). Second, tatB suppressors of KQ variant precursors (see above) were described that map to residue Glu-8 near the trans-sided N terminus of TatB (9), and third, the periplasmic end of the TM of TatB was found to contact RR precursors prior to translocation (11). The embedment of an RR signal peptide within TatBC as suggested by the model of Fig. 7 complies with these described interactions between RR signal peptides and TatB and TatC, respectively.
Contacts between TatC and TatA
Our cross-linking analysis of TatC also revealed contacts to TatA, when Bpa was incorporated into TatC at the two positions Asp-150 and Asp-211. Asp-150 is located in a periplasmic loop, and Asp-211 is at the periplasmic end of TM 5 (Fig. 1). Both residues might therefore lie in the vicinity of the periplasmic N terminus of TatA (Fig. 7). Recent data actually support such an association of monomeric TatA with the TatBC signal sequence receptor complex. Thus intramembrane proximity between TatA and TatC was demonstrated by bimolecular fluorescence complementation (41). The TM of TatA was found by site-specific cross-linking to contact not only TatC but also RR precursors (18). Conversely, earlier studies had revealed that the signal peptide of an RR precursor cross-links to TatA (5).
Oligomeric Nature of the Tat Translocase
TatC by itself and in complex with TatB has the propensity to oligomerize (4, 35, 36, 42, 43), and recent data obtained with fused TatC oligomers suggest that TatC forms at least a functional dimer (44). The cooperation of several TatC monomers during Tat-specific translocation is also invoked by the finding that covalently coupled precursor dimers and tetramers are simultaneously transported by a single oligomeric TatBC complex (45). Disulfide cross-linking experiments using single cysteine mutants throughout all six TMs of E. coli TatC revealed that each helix of TatC is in close proximity with the same helix of a neighboring TatC molecule (46). Our data now add to these findings by showing that homo-oligomerization of TatC also occurs via the presumably extended and flexible first periplasmic loop of TatC (Asp-63 in Fig. 7). Because of the numerous reports on TatC-TatC contacts and the oligomeric nature of isolated TatBC complexes (10), it is very likely that the model shown in Fig. 7 needs to be extended by including several more Tat(A)BC protomers in a speculative representation of what might be a functional Tat translocase.
Supplementary Material
Acknowledgment
We gratefully acknowledge Anne-Sophie Blümmel for technical support.
This work was supported by Sonderforschungsbereich 746 and Forschergruppe 929 of the Deutsche Forschungsgemeinschaft and an F. F. Nord grant by the University of Freiburg (to S. Z.).

This article contains supplemental Tables S1 and S2.
- Tat
- twin arginine translocation
- Bpa
- p-benzoyl-phenylalanine
- CCCP
- carbonyl cyanide m-chlorophenyl-hydrazone
- INV
- inside-out inner membrane vesicles
- KK
- twin-lysine
- RR
- twin-arginine
- PMF
- proton-motive force
- TM
- transmembrane domain or helix
- PK
- proteinase K.
REFERENCES
- 1. Palmer T., Sargent F., Berks B. C. (2010) In EcoSal-Escherichia coli and Salmonella: Cellular and Molecular Biology (Böck A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D., eds) ASM Press, Washington, D. C [Google Scholar]
- 2. Robinson C., Matos C. F., Beck D., Ren C., Lawrence J., Vasisht N., Mendel S. (2011) Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta 1808, 876–884 [DOI] [PubMed] [Google Scholar]
- 3. Fröbel J., Rose P., Müller M. (2012) Phil. Trans. R. Soc. B 367, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cline K., Mori H. (2001) Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 6. Richter S., Brüser T. (2005) Targeting of unfolded PhoA to the TAT translocon of Escherichia coli. J. Biol. Chem. 280, 42723–42730 [DOI] [PubMed] [Google Scholar]
- 7. Gérard F., Cline K. (2006) Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 8. McDevitt C. A., Buchanan G., Sargent F., Palmer T., Berks B. C. (2006) Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels. FEBS J. 273, 5656–5668 [DOI] [PubMed] [Google Scholar]
- 9. Kreutzenbeck P., Kröger C., Lausberg F., Blaudeck N., Sprenger G. A., Freudl R. (2007) Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J. Biol. Chem. 282, 7903–7911 [DOI] [PubMed] [Google Scholar]
- 10. Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. (2009) Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. U.S.A. 106, 13284–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maurer C., Panahandeh S., Jungkamp A. C., Moser M., Müller M. (2010) TatB functions as an oligomeric binding site for folded Tat precursor proteins. Mol. Biol. Cell 21, 4151–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou B., Frielingsdorf S., Klösgen R. B. (2006) Unassisted membrane insertion as the initial step in ΔpH/Tat-dependent protein transport. J. Mol. Biol. 355, 957–967 [DOI] [PubMed] [Google Scholar]
- 13. Bageshwar U. K., Whitaker N., Liang F. C., Musser S. M. (2009) Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol. Microbiol. 74, 209–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlesier R., Klösgen R. B. (2010) Twin arginine translocation (Tat)-dependent protein transport. The passenger protein participates in the initial membrane binding step. Biol. Chem. 391, 1411–1417 [DOI] [PubMed] [Google Scholar]
- 15. Gérard F., Cline K. (2007) The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J. Biol. Chem. 282, 5263–5272 [DOI] [PubMed] [Google Scholar]
- 16. Mori H., Cline K. (2002) A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 157, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dabney-Smith C., Mori H., Cline K. (2006) Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281, 5476–5483 [DOI] [PubMed] [Google Scholar]
- 18. Fröbel J., Rose P., Müller M. (2011) Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43679–43689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. (2008) Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. (2005) The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White G. F., Schermann S. M., Bradley J., Roberts A., Greene N. P., Berks B. C., Thomson A. J. (2010) Subunit organization in the TatA complex of the twin arginine protein translocase. A site-directed EPR spin labeling study. J. Biol. Chem. 285, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baglieri J., Beck D., Vasisht N., Smith C., Robinson C. (2012) Structure of TatA paralog, TatE, suggests a structurally homogeneous form of Tat protein translocase that transports folded proteins of differing diameter. J. Biol. Chem. 287, 7335–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cline K., McCaffery M. (2007) Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 26, 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Panahandeh S., Maurer C., Moser M., DeLisa M. P., Müller M. (2008) Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 283, 33267–33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holzapfel E., Eisner G., Alami M., Barrett C. M., Buchanan G., Lüke I., Betton J. M., Robinson C., Palmer T., Moser M., Müller M. (2007) The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46, 2892–2898 [DOI] [PubMed] [Google Scholar]
- 26. Alami M., Trescher D., Wu L. F., Müller M. (2002) Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277, 20499–20503 [DOI] [PubMed] [Google Scholar]
- 27. Ryu Y., Schultz P. G. (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 28. Blaudeck N., Kreutzenbeck P., Freudl R., Sprenger G. A. (2003) Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J. Bacteriol. 185, 2811–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lesley S. A., Brow M. A., Burgess R. R. (1991) Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 266, 2632–2638 [PubMed] [Google Scholar]
- 30. Moser M., Panahandeh S., Holzapfel E., Müller M. (2007) In vitro analysis of the bacterial twin-arginine-dependent protein export. Methods Mol. Biol. 390, 63–79 [DOI] [PubMed] [Google Scholar]
- 31. Yahr T. L., Wickner W. T. (2001) Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20, 2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bageshwar U. K., Musser S. M. (2007) Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 179, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fincher V., McCaffery M., Cline K. (1998) Evidence for a loop mechanism of protein transport by the thylakoid ΔpH pathway. FEBS Lett. 423, 66–70 [DOI] [PubMed] [Google Scholar]
- 34. Strauch E. M., Georgiou G. (2007) Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 374, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Behrendt J., Lindenstrauss U., Brüser T. (2007) The TatBC complex formation suppresses a modular TatB-multimerization in Escherichia coli. FEBS Lett. 581, 4085–4090 [DOI] [PubMed] [Google Scholar]
- 36. Orriss G. L., Tarry M. J., Ize B., Sargent F., Lea S. M., Palmer T., Berks B. C. (2007) TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 581, 4091–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDevitt C. A., Hicks M. G., Palmer T., Berks B. C. (2005) Characterisation of Tat protein transport complexes carrying inactivating mutations. Biochem. Biophys. Res. Commun. 329, 693–698 [DOI] [PubMed] [Google Scholar]
- 38. Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. (2001) TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276, 20213–20219 [DOI] [PubMed] [Google Scholar]
- 39. Alder N. N., Theg S. M. (2003) Protein transport via the cpTat pathway displays cooperativity and is stimulated by transport-incompetent substrate. FEBS Lett. 540, 96–100 [DOI] [PubMed] [Google Scholar]
- 40. Barrett C. M., Mangels D., Robinson C. (2005) Mutations in subunits of the Escherichia coli twin-arginine translocase block function via differing effects on translocation activity or tat complex structure. J. Mol. Biol. 347, 453–463 [DOI] [PubMed] [Google Scholar]
- 41. Kostecki J. S., Li H., Turner R. J., DeLisa M. P. (2010) Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation. PLoS One 5, e9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Leeuw E., Granjon T., Porcelli I., Alami M., Carr S. B., Müller M., Sargent F., Palmer T., Berks B. C. (2002) Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes. J. Mol. Biol. 322, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 43. Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. (2005) The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J. Mol. Biol. 346, 295–305 [DOI] [PubMed] [Google Scholar]
- 44. Maldonado B., Buchanan G., Müller M., Berks B. C., Palmer T. (2011) Genetic evidence for a TatC dimer at the core of the Escherichia coli twin arginine (Tat) protein translocase. J. Mol. Microbiol. Biotechnol. 20, 168–175 [DOI] [PubMed] [Google Scholar]
- 45. Ma X., Cline K. (2010) Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J. 29, 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Punginelli C., Maldonado B., Grahl S., Jack R., Alami M., Schröder J., Berks B. C., Palmer T. (2007) Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC Component. J. Bacteriol. 189, 5482–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ki J. J., Kawarasaki Y., Gam J., Harvey B. R., Iverson B. L., Georgiou G. (2004) A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J. Mol. Biol. 341, 901–909 [DOI] [PubMed] [Google Scholar]
- 48. Behrendt J., Standar K., Lindenstrauss U., Brüser T. (2004) Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol. Lett. 234, 303–308 [DOI] [PubMed] [Google Scholar]
- 49. Buchanan G., de Leeuw E., Stanley N. R., Wexler M., Berks B. C., Sargent F., Palmer T. (2002) Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43, 1457–1470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.