Background: Gephyrin is a postsynaptic scaffolding protein at inhibitory synapses and undergoes alternative splicing.
Results: Gephyrin splice variants expressed in insect cells were purified as stable hexamers and high-oligomers. Splice-specific folding and stability, glycine receptor β-loop binding, and phosphorylation of gephyrin were found.
Conclusion: Splicing and phosphorylation controls gephyrin clustering via conformational changes within the C domain.
Significance: Novel regulatory circuits controlling gephyrin clustering.
Keywords: Alternative Splicing, GABA Receptors, Glycine Receptors, Phosphorylation, Scaffold Proteins, Gephyrin
Abstract
The multimeric scaffolding protein gephyrin forms post-synaptic clusters at inhibitory sites, thereby anchoring inhibitory glycine (GlyR) and subsets of γ-aminobutyric acid type A (GABAA) receptors. Gephyrin is composed of three domains, the conserved N-terminal G- and C-terminal E-domain, connected by the central (C-) domain. In this study we investigated the oligomerization, folding and stability, GlyR β-loop binding, and phosphorylation of three gephyrin splice variants (Geph, Geph-C3, Geph-C4) after expression and purification from insect cells (Sf9). In contrast to Escherichia coli-derived trimeric gephyrin, we found that Sf9 gephyrins form hexamers as basic oligomeric form. In the case of Geph and Geph-C4, also high-oligomeric forms (∼900 kDa) were isolated. Partial proteolysis revealed a compact folding of the Gephyrin G and C domain in one complex, whereas a much lower stability for the E domain was found. After GlyR β-loop binding, the stability of the E domain increased in Geph and Geph-C4 significantly. In contrast, the E domain in Geph-C3 is less stable and binds the GlyR β-loop with one order of magnitude lower affinity. Finally, we identified 18 novel phosphorylation sites in gephyrin, of which all except one are located within the C domain. We propose two models for the domain arrangement in hexameric gephyrin based on the oligomerization of either the E or C domains, with the latter being crucial for the regulation of gephyrin clustering.
Introduction
Efficient synaptic transmission in the central nervous system requires high concentrations of ligand-gated ion channels in the postsynaptic membrane. Scaffolding proteins ensure accurate localization of neurotransmitter receptors at synaptic sites, thereby controlling both long-term stability as well as plasticity of synapses (1). At inhibitory synapses the scaffolding protein gephyrin provides docking sites for glycine receptors (GlyRs)2 and GABAA receptors (GABAAR) (2).
Gephyrin binds with high affinity to the large cytoplasmic loop of the GlyR β-subunit (GlyR β-loop) (3, 4) and a subset of GABAAR receptors via direct interactions (5–7) or other modes of binding (i.e. γ2) (8). Suppression of gephyrin expression either in cultured neurons by antisense oligonucleotides (9) or gene knock-out (10) illustrated the crucial requirement of gephyrin in maintaining inhibitory synapse formation. In contrast to the spastic mouse model with a lack of GlyR β-subunit, gephyrin-deficient mice are more severely affected, probably due to its additional role in GABAAR clustering as well as its metabolic function in catalyzing the terminal steps of molybdenum cofactor biosynthesis (11, 12).
Gephyrin is a 93-kDa multidomain protein consisting of three major domains: an N-terminal G domain, a central C domain, and a C-terminal E domain (13). Crystal structures of the conserved G and E domains were determined and show tightly folded trimers (14, 15) and dimers (16), respectively. This observed oligomerization behavior suggested a hexagonal lattice of gephyrin underneath the post-synaptic membrane (17). Consistently, insertion of charged residues within the oligomerization interfaces of either of the two conserved domains interfered with postsynaptic gephyrin clustering (18), suggesting the importance of the integrity of the entire gephyrin protein as also demonstrated by the use of a variety of gephyrin deletion variants (19).
The high resolution crystal structure of the E domain in complex with the GlyR β-loop unrevealed a key-and-lock mode of binding in the dimer interface (20). Recently, we identified a serine residue within the binding motif of GlyR β-loop that upon phosphorylation weakens gephyrin binding and thereby controls receptor scaffold interaction (21).
There is growing evidence that phosphorylation impacts gephyrin function, being a main determinant of gephyrin transport, interaction with receptors, and signaling molecules as well as clustering. The peptidyl-prolyl cis-trans isomerase 1 was found to interact with gephyrin in a phosphorylation-dependent manner. Poly proline-directed phosphorylation of serine residues 188, 194, and 200 was shown to be involved in peptidyl-prolyl cis-trans isomerase 1 recruitment (22), which in turn altered the overall conformation of gephyrin, thus enhancing its ability to bind the GlyR. This finding suggests a function of the C domain in gephyrin folding and thereby affects receptor binding via the E domain. Recently, another gephyrin phosphorylation site, Ser-270, was found to modulate GABAergic transmission (23). Blocking the phosphorylation at Ser-270 or interfering with glycogen synthase kinase 3β resulted in an increased density of gephyrin clusters, which is in line with the observed decrease in cluster size when protein phosphatase 1 was inhibited (24).
Beside phosphorylation, alternative splicing of the gephyrin gene is also believed to contribute to the functional complexity of gephyrin. To date more than 10 different variants have been reported (13, 25–27) affecting each of the three domains. The majority of modifications reside in the C domain, pointing toward its crucial contribution in controlling gephyrin folding and clustering. Among eight of the most frequently found gephyrin splice variants, we found no impact on gephyrin metabolic function for all C domain splice variants, whereas the insertion of the G2 cassette (previously named C5) (26), rendered dimer formation of the G-domain (28).
In this study we analyzed the oligomerization of different gephyrin splice variants. Using the baculovirus system three different gephyrin splice variants (Geph, Geph-C3, and Geph-C4) were expressed in Spodoptera frugiperda (Sf9) insect cells and purified to homogeneity. Size exclusion chromatography revealed stable and isolatable high molecular oligomers for Sf9-expressed gephyrin splice variants but not for Escherichia coli-derived gephyrin. Differential scanning calorimetry revealed two clearly distinguishable unfolding events, one for the G-C domain complex and one for the E domain, the later being significantly stabilized in the presence of the GlyR β-loop. Alterations between the splice variants were mirrored by the binding affinities observed by isothermal titration calorimetry revealing a major impact of the C3 splice cassette on E domain folding, stability, and receptor binding. Finally, we identified 18 novel phosphorylation sites, of which all except one were localized within the C domain. This study sets the ground for future functional and structural studies toward a better understanding of the molecular architecture of the major scaffolding protein at inhibitory synapses and identified numerous phosphorylation sites that link gephyrin-mediated receptor clustering to different signaling pathways.
EXPERIMENTAL PROCEDURES
Cloning and Expression of Gephyrin Splice Variants in Sf9 Insect Cells
For recombinant expression in Sf9 insect cells, rat gephyrin DNAs encoding the gephyrin splice variant (P1, residues 1–736), gephyrin C3 splice variant (C3 cassette encoding NHPFYTSPAVFMANHGQPIPGLISYSHHATGSADKR inserted after K243), and gephyrin C4 splice variant (C4 cassette encoding ARLPSCSSTYSVSE and inserted after K288) were cloned from pQE-30 as XmaI/NotI fragments with His6 tag (19) into the baculovirus transfer plasmid pVL1393 (Invitrogen) yielding pJHGeph, pJHGeph-C3, and pJHGeph-C4. To generate recombinant baculovirus-encoding gephyrin splice variants, pJHGeph, pJHGeph-C3, and pJHGeph-C4 plasmids and wild type viral-linearized AcPNV BaculoGold DNA (BD Biosciences) were co-transfected into insect Sf9 cells by calcium phosphate co-precipitation, and baculoviruses were amplified following standard protocols. Sf9 cells were grown at 27 ºC as suspension cultures in spinner flasks under constant agitation (60 rpm) in TNM-FH media (Applichem) supplemented with 50 μg/ml gentamicin, 2.5 μg/ml Fungizone, and 10% fetal bovine serum. Sf9 cells were infected with virus encoding gephyrin splice variants at a multiplicity of infection >5 and harvested 52–55 h post-infection.
Purification of Gephyrin Splice Variants
Harvested Sf9 insect cells were sedimented at 1000 × g for 10 min and washed 2 times with ice-cold phosphate-buffered saline buffer (PBS). Cells were resuspended in 25 mm Tris/HCl, 300 mm NaCl, 10 mm imidazole, 5 mm β-mercaptoethanol, pH 8.0, and lysed by sonication using a microtip (3 × 30 s, 45% power output). Protease inhibitor (Complete, Roche Applied Science) was added before resuspension. Disrupted cells were centrifuged at 50,000 × g for 15 min. Supernatants were first purified at 4 ºC using a nickel-nitrilotriacetic acid resin (Qiagen) according to the manufacturer's instructions. After the elution of protein-containing fractions, samples were buffer-exchanged to Su6 buffer (10 mm Tris/HCl, 250 mm NaCl, pH 8.0) using PD10 desalting columns (GE Healthcare), and proteins were concentrated to 5–20 mg/ml. Concentrated proteins were applied to a Superose 6 size exclusion column (GE Healthcare) equilibrated with Su6 buffer. To estimate the apparent molecular mass of the analyzed proteins, the Superose 6 column was calibrated with thyroglobulin (669 kDa), ferritine (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and chicken ovalbumin (44 kDa) as standard proteins. After the separation, fractions of different gephyrin oligomers were collected and applied again on an analytical Superose 6 column. Pure gephyrin oligomer fractions were pooled, concentrated to 25–100 μm, flash-frozen in 20-μl aliquots, and stored at −80 ºC.
Expression and Purification of Gephyrin Domain Constructs, EcGeph-C4, and GlyR β-loop in E. coli
For heterologous expression of gephyrin domains and Geph-C4 in E. coli, pQE80 constructs were generated. Gephyrin G-domain (residues 1–166), gephyrin G-C (residues 1–253), gephyrin E-C (including C4, residues 254–750), and gephyrin E (residues 333–750) were amplified via PCR and cloned in-frame as XmaI/NotI fragments into pQE80. Gephyrin domain constructs and Geph-C4 splice variant were expressed as His-tagged proteins in E. coli and purified using nickel-nitrilotriacetic acid, ion exchange, and size exclusion chromatography as described earlier (4). In brief, gephyrin domain and Geph-C4-containing E. coli cells were treated as described above except an additional step of cell lysis using a cell-disruptor (IUL Instruments) was introduced. The GlyR β-loop (residues 378–425) was expressed as intein fusion using the IMPACT-TWIN protein expression and purification system (New England Biolabs) and cleaved from the tag with 50 mm dithiothreitol for 20 h at 20 ºC as described (4). Eluted GlyR β-loop was concentrated and further purified by size exclusion chromatography using a preparative 26/60 Superdex 200 column (GE Healthcare). GlyR β-loop-containing fractions were pooled, concentrated to 4–8 mg/ml, and stored at −80 ºC in 20-μl aliquots after flash-freezing in liquid nitrogen.
Immunofluorescence Analysis
Sf9 cells infected with virus as described above were seeded onto glass coverslips coated with collagen. Coverslips were removed 48 h post-infection, and cells were fixed using 4% paraformaldehyde for 20 min. Thereafter, cells were washed once with PBS and incubated for 20 min in PBS containing 0.2% Triton X-100. Fixed cells were washed again with PBS and moistened with PBS containing 1% BSA for 1–2 min. Next, primary gephyrin antibodies (3B11) (19) were added and incubated at 37 ºC for 30–60 min. Cells were washed again with PBS and incubated with secondary antibodies (Alexa Fluor 488, goat anti-mouse IgG; Invitrogen) for 60 min and washed again with PBS and water. Finally, coverslips were embedded in Mowiol (Calbiochem) and DABCO (1,4-diazobicyclo-octane; Merck).
Partial Proteolysis
Gephyrin splice variants and trypsin (sequencing grade, Sigma Life Science; dissolved and diluted in 20 mm Tris/HCl, pH 8.0, 100 mm NaCl) were incubated in a 25-μl reaction mixture in an indicated protein/protease molar ratio (1 nmol/2 pmol) at room temperature. After different time points (0–2 h and overnight), trypsin treatment was inhibited by the addition of 5× SDS-PAGE sample buffer. Samples were analyzed using a 12% SDS-PAGE and either stained with Coomassie Blue or immunoblotted and stained either with the E domain-specific monoclonal antibody m3B11 (19) or G domain-specific polyclonal, “puszta serum” (29).
Differential Scanning Calorimetry (DSC)
DSC scans were performed with 20–50 μm gephyrin splice variants (domain- or holoproteins) with and without 200–500 μm GlyR β-loop using a VP-DSC microcalorimeter (Microcal Inc.) with a cell volume of 650 μl. All samples were buffer-exchanged to Su6 buffer and degassed for at least 10 min before loading into the calorimeter. A constant pressure of about 30 pounds/square inch over the liquids in the cells was applied to the calorimeter to prevent solvent evaporation during the thermal scan. A scanning rate of 25 ºC h −1 from 20 to 90 ºC was applied. The calorimetric data were corrected for the instrument base-line buffer-buffer scan and subtracted from the sample scan. MicroCal Origin Software (OriginLab) was used to analyze the obtained heat profiles. Curve-fitting of raw data was achieved by the non-linear Levenberg-Marquardt method. The peaks of the transition temperatures (Tm1) and ΔHdenaturation were calculated for each sample from several thermal scans.
Isothermal Titration Calorimetry (ITC)
For all ITC experiments, gephyrin and GlyR β-loop were extensively dialyzed against large volumes of 10 mm Tris/HCl, 250 mm NaCl, 1 mm β-mercaptoethanol, pH 8.0, at 4 ºC to ensure equal buffer conditions. As ligand, GlyR β-loop was titrated into the sample cell containing different gephyrin splice variants using a VP-ITC microcalorimeter (Microcal Inc.). Gephyrin concentrations varied from 20–50 μm, and GlyR β-loop was usually used with 10 times higher concentrations (0.2–0.5 mm) and titrated in 3–5-μl injections. The protein concentrations were determined spectrophotometrically at 280 nm in 0.1% SDS-containing buffer applying the calculated extinction coefficients for Geph, Geph-C3, Geph-C4, and GlyR β-loop. All experiments were performed at 25 ºC at least in triplicate. Using the ORIGIN software (OriginLab), the binding enthalpy for every injection was calculated by integration of the peak area. The association constant Ka and additional binding parameters (binding stoichiometry, enthalpy, and entropy) were obtained through curve-fitting with ORIGIN.
LC-MS/MS Analysis of Gephyrin Phosphopeptides
Different oligomers of gephyrin splice variants were analyzed using a 12% SDS-PAGE. Coomassie Blue-stained protein bands were excised from the gel and chopped into small cubes. For triple enzyme digests, gel cubes were dived into three aliquots and washed 3 times with acetonitrile-water (1:1). Gel pieces were shrunk with acetonitrile, rehydrated in 50 mm NH4HCO3, and dried in a SpeedVac. Next, 10 mm dithiothreitol in 50 mm NH4HCO3 was added to the dried gel pieces, and proteins were reduced for 45 min at 56 ºC. To alkylate reduced cysteine residues, the remaining liquid was removed, an equal volume of 50 mm iodoacetamide in 50 mm NH4HCO3 was added, and the reaction was allowed to proceed for 30 min in the dark. Before in-gel digestion, gel pieces were washed and dried as above. Depending on the desired protease, the gel pieces were rehydrated in an ice-cold solution of either 10 ng/μl trypsin (sequencing grade, Promega) or 10 ng/μl chymotrypsin in 10 mm NH4HCO3 or 20 ng/μl Gluc Protease (sequencing grade, New England Biolabs) in 50 mm Tris/HCl, 0.5 mm Glu-Glu, pH 8.0. After 45 min on ice, excessive enzyme solution was replaced by 20 μl of buffer without enzyme, and proteins were digested at 37 ºC over night. The digests were stopped by the addition of 20 μl 10% formic acid, and peptides were extracted for 30 min at 37 ºC. For each sample the extracts from multiple protease digests were combined before LC-MS analysis.
Liquid chromatography-MS data were acquired on a HCT ETD II iontrap mass spectrometer (Bruker Daltoniks) equipped with a nano-ESI source (Bruker Daltonics). Samples were introduced by an easy nano liquid chromatography system (Proxeon) using a vented column setup comprising a 0.1 × 20-mm trapping column and a 0.075 × 100-mm analytical column, both self-packed with ReproSil-Pur C18-AQ, 5 μm (Dr. Maisch, Ammerbuch, Germany). A 5–18-μl sample was aspirated into the sample loop, and a total of 25 μl was loaded onto the trap column using a flow rate of 6 μl/min. Loading pump buffer was 0.1% formic acid. Peptides were eluted with a gradient of 0% to 35% acetonitrile in 0.1% formic acid over 20 min and a column flow rate of 300 nl/min. Subsequently the acetonitrile content was raised to 100% over 2 min, and the column was regenerated in 100% acetonitrile for additional 8 min. Data-dependent acquisition of MS and tandem MS (MS/MS) spectra was controlled by the Compass 3.0 software. MS1 scans were acquired in standard enhanced mode. Five single scans in the mass range from m/z 400 to 1400 were combined for one survey scan. Up to three double- and triple-charged ions rising above a given threshold were selected for MS/MS experiments. Ultrascan mode was used for the acquisition of MS2 scans in the mass range from m/z 100 to 1600, and 3 single scans were added up. The ion charge control value was set to 250,000 for all scan types. Peaklists in MASCOT generic format (mgf) were generated from the raw data by using the Data Analysis software module (Bruker Daltoniks).
Data base searching of peptides and phosphopeptides was performed by searching expected protein sequences in a custom data base using a local installation of MASCOT 2.2 (Matrix Science Ltd, London, UK). Searches were submitted via PROTEINSCAPE 2.0 (Bruker Daltoniks) with the following parameter settings: enzyme, none; fixed modifications, carbamidomethyl; optional modifications, methionine oxidation and phosphorylation ST; missed cleavages, 1. The mass tolerance was set to 0.4 Da for peptide and fragment spectra.
Determination of Putative Kinase Sites
Using the open access NetPhos 2.0 server, phosphorylation status of gephyrin splice variants for serine, threonine, and tyrosine was predicted and compared with identified phosphorylated residues. Using a neural network prediction for phosphorylation sites in eukaryotic proteins, kinase recognition sites have been predicted with a threshold value of 0.45 (30).
RESULTS
Sf9 Insect Cell-expressed Gephyrins Do Not Form Typical Intracellular Aggregates
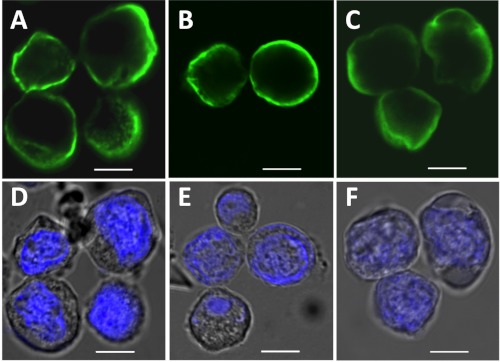
We expressed the three most common gephyrin splice variants (31, 32), gephyrin P1 (Geph), gephyrin C3 (Geph-C3), and gephyrin C4 (Geph-C4) in Sf9 insect cells using the baculovirus expression system as described under “Experimental Procedures.” We first investigated the subcellular localization of gephyrin splice variants 48 h after virus transfection. Cells were stained for gephyrin with the established 3B11 monoclonal antibody (19) and visualized using confocal laser scanning microscopy (Fig. 1). In contrast to the well known gephyrin blobs seen in HEK293 cells (26, 29) or COS7 (33), expression of all three gephyrin variants in Sf9 cells showed diffuse staining of gephyrin in the cytosol with clear enrichment in the vicinity of the plasma membrane for Geph (Fig. 1A) and Geph-C4 (Fig. 1B), whereas Geph-C3 (Fig. 1C) was more diffuse in the cytosol. Hoechst staining of the nucleus confirmed cell viability and a relatively narrow cytosolic compartment (Fig. 1, D–F).
FIGURE 1.
Subcellular localization of gephyrin splice variants in Sf9 cells. Sf9 cells were transfected with different gephyrin constructs and expressed for 48 h. Localization of Geph (A), Geph-C4 (B), and Geph-C3 (C) was visualized using mB311 antibodies (green) and laser scanning microscopy. D–F, nuclei were stained with Hoechst stain. Scale bars, 8 μm.
Gephyrin Splice Variants Form Hexamers and Large Oligomers
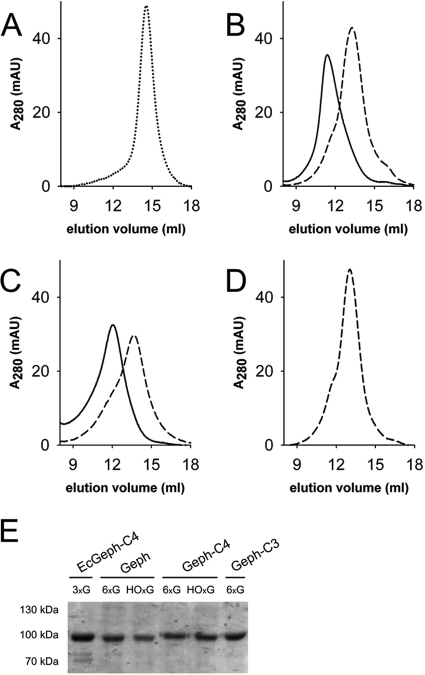
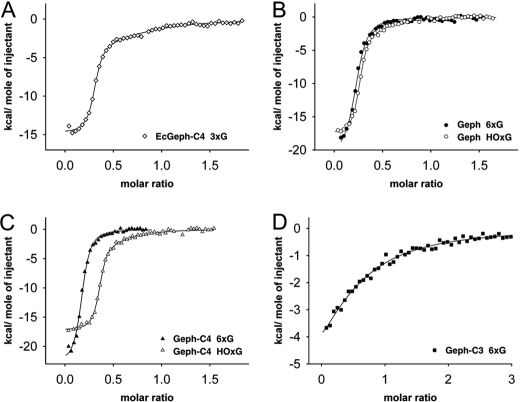
After showing a non-aggregated cellular distribution of Sf9-expressed gephyrins, we performed preparative expression cultures and purified all splice variants by nickel-nitrilotriacetic acid chromatography. Given the known sensitivity of gephyrin toward proteolytic degradation (4), purifications were performed rapidly at 4 ºC using small column sizes. Subsequently, nitrilotriacetic acid-enriched proteins were subjected to preparative size exclusion chromatography using a Superose-6 column. Except Geph-C3, for which only one major peak was observed, both other variants (Geph and Geph-C4) eluted in two peaks, one around 13.3 ml and the other at 11.5 ml (supplemental Fig. S1). Fractions under each peak were collected and rerun using the same column, which then resulted in the appearance of only one peak with a similar elution volume as the collected fraction from the first run (Fig. 2). In contrast to the trimeric gephyrin C4 variant expressed and purified from E. coli (EcGeph-C4, Fig. 2A), Geph (Fig. 2B) and Geph-C4 (Fig. 2C) were purified in at least two different oligomeric states, which remained stable during purification, whereas Geph-C3 (Fig. 2D) was mainly present in one oligomeric form. SDS-PAGE analysis confirmed the identity and homogeneity of purified gephyrins (Fig. 2E). Note, EcGeph-C4 shows some additional bands representing degradation products as proven by Western blot analysis (supplemental Fig. S1E).
FIGURE 2.
Molecular mass determination of purified gephyrin oligomers. A–D, size exclusion chromatography of EcGeph-C4 from E. coli (A), Geph (B), Geph-C4 (C), and Geph-C3 (D) derived from Sf9 cells using a 20-ml Superose 6 column. Samples were derived from a first size exclusion run (supplemental Fig. S1, A–D) where hexameric (6xG, dashed line), high oligomeric (HOxG, bold line), and in the case of EcGeph-C4, trimeric (dotted line, A) fractions were taken. Molecular mass of gephyrin oligomers was determined by comparison to marker proteins of known size. Different oligomerization states are indicated by symbols: HOxG, high oligomer; 6xG, hexamer; 3xG, trimer. Purity of gephyrin oligomers was probed by SDS-PAGE (E). AU, absorbance units.
Molecular masses of gephyrin oligomers were determined based on the elution volume of standard proteins under similar experimental conditions. EcGeph-C4 eluted as a single peak at 14.6 ± 0.1 ml corresponding to a mass of 314 ± 17 kDa (Fig. 2A). The two species of Geph (Fig. 2B) eluted at 13.5 ± 0.2 and 11.3 ± 0.4 ml with corresponding masses of 550 ± 20 and 976 ± 87 kDa (Fig. 2B), respectively. Also, Geph-C4 showed a similar elution pattern for both isolated peaks with masses of 579 ± 72 and 896 ± 101 kDa (Fig. 2C). Consistent with previous findings (4) the mass of EcGeph-C4 approximates to the theoretical mass of a trimer (243 kDa), and therefore, we conclude that the observed peak of Geph-C4 at 579 kDa represents an hexamer (6xG), given that the trimer is most likely the smallest building block of gephyrin (2, 4, 18). The molecular mass of the other Geph and Geph-C4 peaks are close to the exclusion volume of the Superose-6 column but eluted clearly in front of the largest marker protein (thyroglobulin, 669 kDa) used. Therefore, a mass assignment would be ambiguous, and we labeled this fraction as high oligomer (Fig. 2, HOxG). In contrast, Geph-C3 (Fig. 2D) eluted only as single peak at 12.9 ± 0.3 ml corresponding to 673 ± 61 kDa, which given the higher mass of the Geph-C3 monomer (86 kDa), approximates to a hexamer. Note that the presence of the C3 cassette strongly inhibits gephyrin ability to form larger oligomers as observed for Geph and Geph-C4.
As previously proposed, gephyrin oligomerization is believed to rely on E-domain-mediated interactions of gephyrin trimers (2, 17). Here, we found only hexamers as the smallest building block, which has been proposed previously (18) and suggests clear differences between gephyrin purified from E. coli and gephyrin derived from Sf9 cells, pointing to post-translational modifications affecting gephyrin structure and oligomerization.
Gephyrin Degrades into GC and E Domain Fragments
It is assumed that gephyrin oligomerization is a dynamic process essential for the assembly and disassembly of submembranous GlyR as well as GABAAR clusters. This process requires domain movement, which is most likely controlled and/or mediated by the C domain. The previously reported sensitivity of purified gephyrin toward proteolytic cleavage within the C domain (4) reflects the surface-exposed nature of this domain. Consequently, we asked the question to which the extent folding of the C domain is altered between the different splice variants as well as the E. coli-derived gephyrin EcGeph-C4. Therefore, we investigated gephyrin sensitivity against partial proteolysis (Fig. 3).
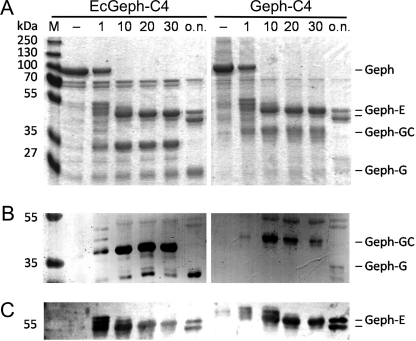
FIGURE 3.
Partial proteolysis of EcGeph-C4 and Geph-C4. E. coli-derived Geph-C4 (left) and Sf9 cell-derived Geph-C4 (right) were digested (6 μg each) with trypsin and separated by a 12% SDS-PAGE (A) together with equal amounts of untreated protein (−). Identification of gephyrin fragments was confirmed by Western blot using domain-specific antibodies detecting the G domain (B, puszta serum) and E-domain (C, m3B11).
As expected, a time-dependent degradation into semi-stable domain fragments as a function of trypsin exposure has been observed for both E. coli- as well as Sf9 cell-derived gephyrin. EcGeph-C4 converted within 10 min into an E and G domain-containing fragment, each of them further degraded slowly with time (Fig. 3A). The identity of both domain fragments has been confirmed by Western blot using domain-specific antibodies (Fig. 3, B and C). Although the E domain was truncated overnight to a 45-kDa fragment, the initial G domain-containing fragment reduced significantly in size from 35 to 25 kDa. This finding suggests that initial degradation targeted the C-terminal end of the C domain, probably due to a compact fold of both the G and C domains. Comparison between EcGeph-C4 and Geph-C4 revealed no alteration between the stability of the E domain; however, the size of the GC domain fragment derived from Geph-C4 was larger by ∼5 kDa (Fig. 3, A and B), suggesting a different fold of that C domain. Given the similar size of the released E domain fragments, it remains unclear if the initial proteolytic cut is similar to EcGeph-C4 or closer to the E domain boundary. However, the stability of the resulting domain fragments is similar between EcGeph-C4 and Geph-C4 (Fig. 3) as well as Geph and Geph-C3 (supplemental Fig. S2).
Alternative Splicing Affects Folding of Gephyrin
Our partial proteolysis results suggested differences in the folding of the C domain between E. coli- and Sf9-derived gephyrins. To study thermodynamic stability, tertiary structure, and possible alterations via splice cassette insertions, thermal unfolding of gephyrin splice variants was investigated using DSC (Fig. 4).
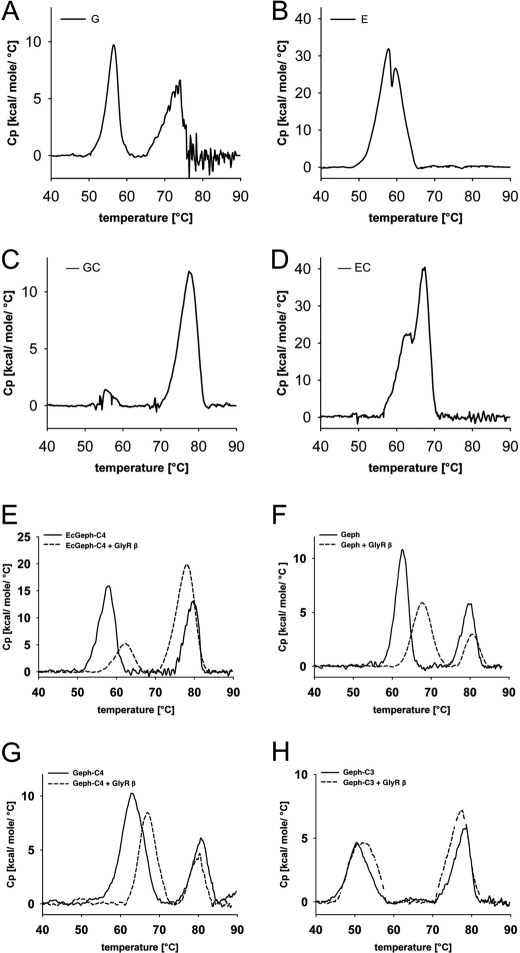
FIGURE 4.
Thermal stability of gephyrin splice variants. Shown are DSC heating thermograms of gephyrin domain fragments (A–D) and gephyrin splice variants (E–H). In each experiment 20–50 μm protein was used. A–D, shown are purified gephyrin domains G (A), E (B), G-C (C), and E-C (D) expressed in E. coli. E–H, shown are EcGeph-C4 (E) and Sf9 cell-derived Geph (F), Geph-C4 (G), and Geph-C3 (H) in the absence (bold lines) and presence of 200–500 μm GlyR β-loop (+GlyRβ, dotted line). The molar heat capacity profiles were recorded from 20–90 ºC at a scan rate of 25 ºC/h. Quantitative values of the corresponding peak transition temperatures of gephyrin E domains (Tm1), gephyrin G domains (Tm2), and with additional GlyR β-loop (ΔTm) are listed in Table 1.
First we investigated different domain variants to understand the unfolding profile of both domains (G and E domain) separately. Although the G domain showed two maxima, one at 56.2 ºC and one at 72.4 ºC, the E domain denatured with two closely associated peaks around 59 ºC (Fig. 4, A and B). As seen in the crystal structures of both domains (14, 20), the very compact and globular nature of the G domain suggests that its folding is more stable than the trimer interaction, whereas in the E domain both the domain folding as well as dimerization are coupled to each other, which is reflected by the two close unfolding maxima. However, the presence of the C domain remarkably changed the stability of the G domain (Fig. 4C). The signal for the proposed trimer dissociation became much weaker, and the majority of the unfolding heat was released at 77.3 ºC. Therefore, one can conclude that the C and G domain tightly pack into a compact structure, which unfolds together with the dissociation of the trimer. Also the E domain shows a gain in stability when fused to the C domain (62.7 and 67.2 ºC); however, the shift toward higher temperature is less pronounced, and there were still two unfolding maxima close to each other detected.
Next, all variants of holo-gephyrin were investigated and showed two peaks of heat release. In light of the unfolding profile of the domain variants, we conclude that the high temperature signal at 78–79 ºC derives from the G domain trimer together with the C domain. The fact that the low temperature signal (50–62 ºC) is below the signal of the E-C domain construct either suggests that the C domain does not express the same type of positive effect onto the E domain as seen for the isolated domain and/or that in holo-gephyrin the E domain adopts a different fold than in its isolated dimerized form.
The comparison of all three Sf9-derived variants and EcGeph-C4 shows a similar folding for their G domains (77.9–79.9 ± 0.1–0.8 ºC, Table 1). In contrast, E domains presented significant differences in their folding stability in different gephyrin splice variants. First, a direct comparison of Geph-C4 with EcGeph-C4 revealed a difference of 5.1 ºC, which in turn suggests that E domains in both proteins are folded and/or oriented differently. The fact that EcGeph-C4 is trimeric but Geph-C4 is a hexamer suggests additional E domain-mediated interaction (i.e. dimerization). Second, an even more pronounced difference is seen between Geph and Geph-C4 (62.1 ± 0.7 and 62.2 ± 0.8 ºC) on one hand and Geph-C3 (50.5 ± 0.5 ºC) on the other hand. Although the insertion of the C4 cassette did not alter E domain stability, the C3 cassette strongly impacted E domain stability, resulting in a reduction of the melting temperature by 11.9 ºC. This finding further supports an altered Geph-C3 oligomerization as seen already in our size exclusion studies.
TABLE 1.
Influence of GlyR β-loop binding on the unfolding temperatures of gephyrin G- and E-domains
| Sample | Tm1 | ΔTm1 + GlyR β loop | Tm2 | ΔTm2 + GlyR β loop |
|---|---|---|---|---|
| °C | °C | °C | °C | |
| EcGeph G | 56.2 | 72.4 | ||
| EcGeph GC | 56.1 | 77.3 | ||
| EcGeph E | 57.7 | 59.8 | ||
| EcGeph EC | 62.7 | 67.2 | ||
| Geph | 62.7 | +5.0 | 79.5 | +1.2 |
| Geph-C4 | 62.4 | +5.1 | 79.9 | +0.4 |
| Geph-C3 | 50.5 | +2.1 | 77.9 | −0.6 |
| EcGeph-C4 | 57.3 | +4.7 | 78.1 | −0.4 |
In summary, we conclude that the modular structure of gephyrin is well reflected by its unfolding characteristics with a compact and stably folded G domain, which is tightly associated to the C domain. Although previous studies suggested that the C domain is mostly unstructured, which is in line with a lack of any unfolding signal for the isolated C domain (data not shown), we propose a G domain-induced folding of the C domain. As expected from the crystal structure, folding of the E domain is less stable and is impacted by alternative splicing of the C domain, as seen for Geph-C3.
Binding of GlyRβ-loop Stabilizes Gephyrin E Domains
Given the previously seen differences in E domain stability and its known interaction with the GlyR β-subunit (4), we next performed DSC studies in the presence of excess of GlyR β-loop (residues 378–425). The addition of the GlyR β-loop (Fig. 4, E–H, dotted lines) showed no significant effects on the stability of gephyrin G-domains in all four investigated variants, as the change in melting temperature was no more than 1.2 ºC (Table 1). However, the stability of gephyrin E domain was increased by ∼5 ºC in all variants except Geph-C3 (Fig. 4, E–H), which was only moderately affected by an increase in stability of 2 ºC (Table 1). This finding demonstrates three aspects of gephyrin folding and GlyR binding. (i) Binding of GlyR β-loop increases E-domain stability. (ii) The binding site of the GlyR β-loop on the E-domain seems to be similar in Sf9 as well as E. coli-derived Geph-C4 given the comparable increase in stability upon GlyR β-loop binding. (iii) Insertion of the C3 cassette alters the folding of gephyrin in a way that GlyR β-loop binding cannot stabilize the E domain fold in the same way as in Geph or Geph-C4.
Alternative Splicing Alters GlyR Binding to Gephyrin
Our DSC studies suggested splice-specific differences impacting GlyR β-loop-mediated stabilization of gephyrin E domains. To quantify the binding of gephyrin splice variants to the GlyR, ITC was performed using the different oligomeric forms of gephyrin variants as well as isolated GlyR β-loop (Fig. 5, supplemental Fig. S3). As previously reported and recently confirmed (4, 21), EcGeph-C4 showed a bisphasic binding of GlyR β-loop with high affinity (KD = 51 ± 17 nm) and low affinity (KD = 6.25 ± 2.18 μm) binding sites (Fig. 5A, Table 2). Determined stoichiometries suggest one high and two low affinity sites per gephyrin trimer (Table 2). Similar biphasic binding curves were only observed for the high oligomeric forms of Geph and Geph-C4 with KD values for the high affinity (KD = 26–46 nm) and low affinity sites (2.66–3.00 μm, Table 2) being comparable to EcGeph-C4. However, binding stoichiometries are not clearly separated into one- and two- thirds of fractional occupation as seen in EcGeph-C4, and overall saturation was only 60–70%.
FIGURE 5.
Binding affinity of gephyrin-glycine receptor β-loop interaction. The binding affinity of gephyrin splice variants with GlyR β-loop was measured by ITC. Binding isotherms of trimeric EcGeph-C4 (A, white square), hexaoligomeric and high oligomeric Geph (B, black and white circle), Geph-C4 (C, black and white triangle), and hexameric Geph-C3 (D, black square) titrated with the GlyR β-loop are shown. All experiments were carried out under the same conditions. For data analysis, the heat release of the first injection in each experiment was omitted. Measured binding enthalpies (supplemental Fig. S3) were plotted as a function of the molar ratio of GlyR β-loop to gephyrin. Determined binding parameters are summarized in Table 2.
TABLE 2.
Binding parameters of gephyrin splice variants to GlyR β-loop
HOxG, high oligomer; 6xG, hexamer; 3xG, trimer.
| Geph variant | Oligomer | High affinity site |
Low affinity site |
||
|---|---|---|---|---|---|
| KD | n | KD | n | ||
| μm | μm | ||||
| EcGeph-C4 | 3xG | 0.05 ± 0.02 | 0.29 ± 0.01 | 6.25 ± 2.18 | 0.55 ± 0.10 |
| Geph | 6xG | 0.42 ± 0.04 | 0.22 ± 0.01 | ||
| HOxG | 0.05 ± 0.02 | 0.25 ± 0.01 | 3.00 ± 1.26 | 0.32 ± 0.08 | |
| Geph-C4 | 6xG | 0.54 ± 0.06 | 0.17 ± 0.01 | ||
| HOxG | 0.03 ± 0.01 | 0.35 ± 0.01 | 2.66 ± 1.28 | 0.36 ± 0.08 | |
| Geph-C3 | 6xG | 7.30 ± 0.91 | 0.21 ± 0.02 | ||
In contrast, the hexameric fractions of Sf9-expressed gephyrin variants (Geph and Geph-C4) showed a different binding behavior. Raw binding enthalpies could only be fitted with appropriate confidence intervals in the one-site binding model, suggesting a one-site model for the GlyR β-loop binding to Geph and Geph-C4. Surprisingly, the determined affinities were 10-fold lower (KD = 417–541 nm) than those for the high affinity sites in EcGeph-C4 as well as the respective high oligomers of Geph and Geph-C4. The fact that the binding stoichiometry was low (n = 0.172–0.220; Fig. 5, B and C, Table 2) suggests that the binding site observed in these hexameric gephyrins probably represents the previously identified high affinity binding site.
Finally, as already anticipated from our DSC studies, binding of GlyR β-loop to Geph-C3 was found to be one order of magnitude weaker (KD = 7.30 ± 0.91 μm) than for the corresponding hexameric gephyrins (Geph and Geph-C4), which clearly underlines a significant impact of the C3 splice cassette on GlyR β-loop binding. Again, binding to Geph-C3 followed a one-site model with a fractional occupation of n = 0.21 (Fig. 5D).
C Domain of Sf9-derived Gephyrins Is Highly Phosphorylated
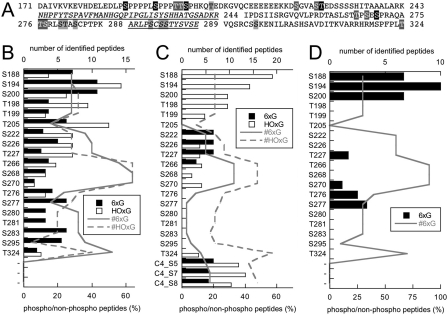
Phosphorylation of gephyrin has been described (22–24, 34, 35). Given the here-reported differences between E. coli- and Sf9-expressed gephyrins, we determined phosphorylation sites in both the hexameric and high oligomeric forms of each of the three splice variants using peptide mass fingerprinting. We found that all variants carry a high number of phosphorylated serine and some threonine residues, all of which, except one (Thr-324), are localized exclusively within the C domain (Fig. 6).
FIGURE 6.
Phosphorylation analysis of Sf9 cell-expressed gephyrin. Peptide mass fingerprinting of Geph, Geph-C4 and Geph-C3 revealed several phosphorylation sites, all localized within the C-domain of gephyrin, except Thr-324. A, residues that were identified in one, two, or three splice variants are shaded in gray and printed in black, shaded in gray and printed in white, and shaded in black and printed in white, respectively. B–D, shown is a plot of the percent ratio of phosphorylated over non-phosphorylated peptides containing the indicated residues found in Geph (B), Geph-C4 (C), and Geph-C3 (D). The total number of peptides identified for the hexameric (6xG) and high oligomeric (HOxG) pool of gephyrin are plotted with solid and dashed lines, respectively.
Residues Ser-188, Ser-194, and Ser-200, previously identified as the Pin1 binding motif (22), were found to be phosphorylated in all splice variants and oligomeric forms except in Geph-C4. However, the latter did not show any detectable peptides in the MS spectrum, and therefore, we are not in the position to conclude its phosphorylation status (Fig. 6, A-C). Among other identified phosphorylated residues, Ser-270 (23) was recently described, and we also found this residue phosphorylated in all variants, except in the hexameric form of Geph-C4 (Fig. 6C).
Furthermore, we found 18 additional phosphorylation sites in Geph, of which 10 were found in both the hexameric as well as the high oligomers. Only one of all the residues that were found in both Geph and Geph-C4 were also detected in Geph-C3 (Thr-227), suggesting significant differences in the phosphorylation pattern between the splice variants (Fig. 6). We also plotted the number of total peptides identified, showing that coverage over the C domain is not equal but relatively similar between the variants. The highest rate of phosphopeptide over non-phosphorylated peptides was found for the three residues in the Pin1 motif (67–100%, Fig. 6D), whereas most other peptides were in the range of 10–60%, suggesting substoichiometric phosphorylation. Finally, three novel serine phosphorylation sites were found in the C4 splice cassette with high coverage (Fig. 6C), suggesting a splice-specific regulation of gephyrin by phosphorylation.
DISCUSSION
For most biochemical studies on gephyrin using full-length protein as well as the individual domains, proteins were expressed in, and purified from E. coli. In the case of the G and E domain, highly purified preparations were obtained, enabling their structure determination (14–16, 20). Previous functional and biochemical studies demonstrated the full integrity of both domains (28) for which trimers were found for the G domains and dimers for the E domains. This finding, fostered the speculation that these two fundamentally different oligomerizations of both domains are key for the clustering function of gephyrin at inhibitory synapses, and consequently, the formation of a hexagonal lattice underneath the membrane has been proposed (2, 36–38).
In contrast to the individual domains, full-length gephyrin was sensitive to proteolytic degradation targeting the C domain of gephyrin, resulting in G and E domain-containing degradation products. Given the known domain oligomerizations, it remained a challenge for more than a decade to prepare highly pure holo-gephyrin, which would allow its crystallization and structure determination. Knowing that the concerted interaction of all three domains is required for proper neuronal function of gephyrin (18, 19, 28), it becomes critical to disclose the molecular interplay between all three domains on the atomic level.
Recently, several studies reported the importance of gephyrin phosphorylation for synaptic clustering (23, 24), and therefore, we developed and characterized a Sf9-based expression system for gephyrin, allowing post-translational modification and folding in a eukaryotic cell. In contrast to the expression of gephyrin in other non-neuronal cells (HEK or COS7 cells (26, 33)) where gephyrin forms large intracellular aggregates, upon expression in insect cells a diffuse cytosolic distribution of gephyrin was observed. In addition, two of the splice variants (Geph and Geph-C4) showed a clear enrichment near the plasma membrane, whereas Geph-C3 was more homogeneously distributed, indicating a different folding and/or interactions between the gephyrin subunits. It is generally assumed that intracellular gephyrin aggregates formed upon recombinant expression in HEK293 cells represent non-native clusters due to the lack of neuron-specific factors in those cells (2). After the co-expression with collybistin, gephyrin aggregates dissolve into microclusters that relocate to plasma membrane proximity (39). Recently, the expression and localization of gephyrin in liver cells has been reported (40) showing a diffuse cytosolic distribution. Consequently, we conclude that the diffuse cellular distribution of all gephyrin variants here found suggests that factors promoting gephyrin clustering/aggregation are missing in Sf9 cells. Therefore, Sf9 cells present an expression system that at least allows avoiding nonspecific gephyrin aggregation (as seen in HEK cells). Finally, Sf9 cells were successfully also used on other studies for the expression of neuronal proteins including functional GlyRs and GABAARs (41, 42).
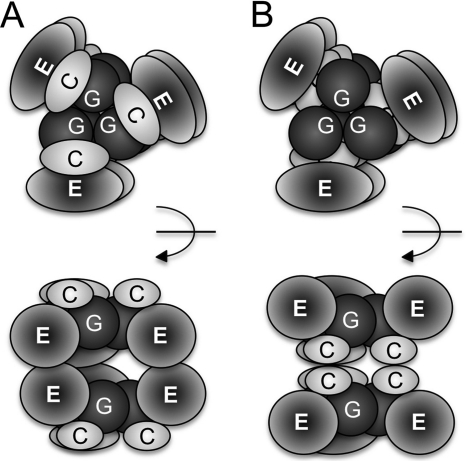
Sf9-expressed and purified gephyrin splice variants showed entirely different oligomerization behavior as compared with gephyrin expressed in E. coli, which forms trimers (4). All Sf9 gephyrins eluted from the gel filtration column with a molecular weight corresponding to a hexamer. Given the tight trimer interface seen in the G domain, one can assume that two trimers are linked to each other via the dimerization of either two adjacent E domains or C domains, thus resulting in a dimer of trimers (Fig. 7, A and B). To which extent the interaction between the E domains, if they contribute to hexamer formation, is similar to the known dimer interface in the isolated E domain remains open as previous studies found different motifs crucial for gephyrin clustering and oligomerization (18, 19).
FIGURE 7.
Models for the hexameric arrangement of gephyrin domains. G, C, and E domains are depicted in different shades. Shown are top and side views (90º rotated as indicated) of hexameric gephyrin either facing the E (A) or C domains (B) of each other. Note that based on our stability studies a interacting surface between G and C domain is depicted.
Besides hexamers, we also identified high oligomeric forms that we could not equivocally assign to a specific multimer, but given the 3-fold symmetry of gephyrin, we assume that these oligomers present either nonamers or dodecamers. When taking the apparent mass of the trimer as basis (317 kDa), the observed masses of 976 and 896 kDa for Geph and Geph-C4, respectively, would strongly support a nonamer. However, oligomerization could also trigger tighter packing of the subunits, and therefore, the calculated oligomer using a mass of 83–84 kDa would better fit with a dodecamer.
Interestingly, high oligomers were not found with Geph-C3, suggesting a major impact of the C3 splice cassette within the C domain, prohibiting the formation of high oligomers. This very different behavior of Geph-C3, which is highly expressed in Glia (28), liver, and kidney (27, 31), could link Geph-C3 to the non-neuronal metabolic function of gephyrin, the biosynthesis of the molybdenum cofactor, as seen already in previous studies (28). Finally, the observed membrane enrichment of Geph and Geph-C4, with Geph-C3 more homogeneously distributed in the cytosol, suggests that the high oligomeric forms prefer spatial proximity to the plasma membrane. Previous studies in Xenopus oocytes using Blue Native-PAGE analyses also reported gephyrin hexamers and higher oligomers (18), which add further strength to the proposal that post-translational modifications of gephyrin are crucial for native oligomerization.
Differences in oligomerization are accompanied by altered stability of gephyrin. In two different experimental setups we could show that the gephyrin domain structure is reflected by a modular stability profile. Both partial proteolysis and DSC revealed basically two semi-stable states, one G-C domain complex and one E domain fold. These findings suggest that the trimeric G domain tightly associates with the C domain, which on its own has no stable fold and shows no specific unfolding profile in the DSC (data not shown). Consequently, initial degradation by trypsin started at the C-terminal end of the C domain in both EcGeph-C4 and all Sf9-derived variants. Interestingly, the resulting fragment for Sf9 gephyrins appeared to be ∼5 kDa larger than for EcGeph-C4, suggesting a different fold of the C domain in those variants. Alternatively, the different migration behavior might also reflect the phosphorylation of the C domain, thereby causing a reduced mobility in the SDS-PAGE.
In addition to the partial proteolysis, DSC demonstrated that the folding stability of the G domain strongly increases in the presence of the C domain. As a result, first E domains unfold at lower temperature and the G-C domain complex remains stable up to 80 ºC. Although the stability of the G-C complex was relatively similar in both E. coli as well as Sf9-derived gephyrins, strong differences were seen for the E domains. In trimeric EcGeph-C4, E domains unfolded 5 ºC earlier than Sf9-derived Geph and Geph-C4. This finding would fit well with our conclusion that additional interdomain interactions mediated via the E domain could contribute to the increased stability of the E domains in Geph and Geph-C4. Consequently, we propose a dimerization of trimers in those variants (Fig. 7A). However, in Geph-C3, which is also hexameric, E domains are even less stable (12 ºC lower unfolding), suggesting an entirely different fold and/or oligomerization of those E domains triggered by the insertion of the C3 splice cassette. Therefore, an alternative mode of hexamer formation could be envisioned in which the C domains do interact, thereby linking two gephyrin trimers to each other (Fig. 7B). Furthermore, the lack of high oligomers in Geph-C3 further supports the view that E domains are key elements in controlling gephyrin oligomerization as well as clustering, which would further support the second model in which the C domains oligomerize, thus leaving the E domains free for higher order interactions as well as binding to the receptor. Finally, the addition of GlyR β-loop strongly increased the stability of E domains in all variants, except Geph-C3, suggesting (i) that the mode of binding and number of interactions is comparable between Geph, Geph-C4, and EcGeph-C4 and (ii) that the C3 splice cassette induced changes in gephyrin, thus severely impacting its ability to bind the GlyR β-loop.
The dramatically reduced binding of GlyR β-loop has been confirmed by ITC with a KD of 7 μm, which is nearly 100-fold higher than the KD for the previously reported high affinity site (4, 21). Interestingly, such a biphasic GlyR β-loop binding was only seen for the Geph and Geph-C4 high oligomeric forms, whereas both hexameric forms showed monovalent binding endotherms with KD values of 0.4–0.5 μm, respectively. This finding suggests that gephyrin high oligomers adopt E domain conformations that resemble those in EcGephE-C4 and present a high affinity GlyR β-loop binding site. Therefore, oligomerization of gephyrin seems to trigger increased receptor binding or vice versa. Alternatively, the observed high affinity site might additionally reflect GlyR β-loop-induced conformational changes within gephyrin that could contribute to receptor clustering.
Given the observed differences between E. coli- and Sf9-derived gephyrins in their oligomerization, stability, and receptor binding, we suggest post-translational modifications as major determinants. In early studies, gephyrin was identified as phosphoprotein (34), but only recently the importance of this modification for gephyrin clustering and receptor binding became recognized (22–24, 35). In our study we have confirmed the phosphorylation of all known and functionally investigated sites (Ser-188, Ser-194, Ser-200, Ser-270) in gephyrin and found 18 additional sites of phosphorylation. Except one, all phosphorylation sites localize to the C domain, again highlighting its importance in controlling gephyrin oligomerization, clustering, and receptor binding. In addition, the binding of various interacting proteins such as the peptidyl prolyl cis-trans-isomerase 1 (22), dynein light chain (43), and collybistin (44) is or could be regulated via phosphorylation.
Phosphorylation of Ser-188, Ser-194, Ser-200, and Ser-270 was found in all detected peptides derived from both oligomeric forms and splice variants, suggesting a high prevalence for the modification of these sites as described earlier (22, 23). Furthermore, additional proteomic studies in liver (45, 46) and brain (47–49) also revealed a robust phosphorylation of the Ser-188/Ser-194 cluster and spotted residues Thr-198, Thr-199, Ser-200, Thr-266, Ser-268, Ser-270, Ser-294, Ser-295, and Ser-305 as phospho residues.
A comparison between hexameric and high oligomeric forms revealed that Ser-226 and Ser-266 are phosphorylated in both variants but not in hexameric Geph-C3, and the cluster of Ser-280-Thr-281-Ser-283 was only phosphorylated in Geph hexamers. These residues have not been identified in previous studies and might reflect less abundant phosphorylation sites or Sf9 cell-specific modifications. Finally, we found three phosphorylated residues within the C4 splice cassette. Our data demonstrate significant sequence coverage, and therefore, we assume that given the high level expression of gephyrin in Sf9 cells and the large quantities of purified protein enabled us to selectively enrich gephyrin phosphopeptides for MS analysis.
In future studies the importance of each residue needs to be probed by structure-function studies and dissected to which extent the respective modification affects gephyrin neuronal or metabolic function. Nearly all identified phosphorylation sites were recognized in a bioinformatic screen for kinase motifs and should present the starting point for investigating regulatory circuits controlling gephyrin phosphorylation (Table 3).
TABLE 3.
Kinase motifs identified in gephyrin
| Kinasea | Phospho residue (prediction score)b |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ser-188 | Ser-194 | Thr-198 | Thr-199 | Ser-200 | Ser-222 | Ser 226 | Thr-227 | Thr-266 | Ser-268 | Ser-270 | Thr-276 | Ser-277 | Ser-280 | Thr-281 | Ser-283 | C4 Ser 5 | C4 Ser 7 | C4 Ser 8 | Ser-295 | Thr-324 | |
| PKC | 0.49 | 0.48 | 0.77 | 0.72 | 0.65 | 0.45 | 0.62 | ||||||||||||||
| PKA | 0.54 | 0.45 | 0.77 | 0.51 | |||||||||||||||||
| CaM-II | 0.45 | 0.44 | 0.45 | 0.47 | 0.45 | 0.50 | 0.47 | 0.46 | 0.47 | 0.48 | |||||||||||
| p38MAPK | 0.51 | 0.54 | 0.45 | 0.48 | 0.46 | 0.46 | |||||||||||||||
| GSK3 | 0.52 | 0.53 | 0.49 | 0.47 | 0.48 | 0.45 | 0.49 | 0.52 | 0.45 | 0.48 | 0.45 | 0.47 | 0.46 | 0.45 | 0.48 | 0.48 | 0.47 | ||||
| RSK | 0.49 | ||||||||||||||||||||
| CKII | 0.47 | 0.50 | 0.47 | 0.46 | |||||||||||||||||
| cdc2 | 0.45 | 0.47 | 0.50 | 0.51 | 0.49 | 0.49 | 0.51 | 0.47 | 0.49 | ||||||||||||
| cdk5 | 0.46 | 0.66 | 0.66 | 0.57 | 0.50 | ||||||||||||||||
a CaM-II, Ca2+/calmodulin-dependent protein kinases 2; p38MAPK, p38 mitogen-activated protein kinases; GSK3, glycogen synthase kinase 3; RSK, ribosomal s6 kinase; CKII, casein kinase 2; cdc2, cyclin-dependent kinase 2; cdk5, cyclin-dependent kinase 5.
b Prediction scores derived from Netphos 2.0.
In summary, we conclude that gephyrin phosphorylation is a tightly regulated process. Unrevealing the pattern of signal input will lead to a better understanding of the mosaic nature of different regulatory circuits controlling gephyrin oligomerization and receptor binding. Both gephyrin phosphorylation and splicing contribute to the complexity of gephyrin-mediated clustering of glycine and GABAA receptors, each of which underlie synaptic plasticity.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Müller and Dr. Franz-Georg Hanisch (Center for Molecular Medicine Cologne, University of Cologne, Germany) for performing peptide mass finger printing and phosphopeptide determination. Technical assistance by Simona Jansen, Joana Fischer, and Monika Gompert (University of Cologne, Germany) is gratefully acknowledged.
This work was supported by the German Science Foundation (DFG Schw 759/6-1, DFG-SFB635 TP-A11 (to G. S.) and Fonds der Chemischen Industrie (to G. S.).

This article contains supplemental Figs. S1–S3.
- GlyR
- glycine receptor
- GABAAR
- γ-aminobutyric acid type A receptor
- Sf9
- Spodoptera frugiperda
- GlyR β-loop
- glycine receptor β-subunit
- DSC
- differential scanning calorimetry
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Dumoulin A., Triller A., Kneussel M. (2009) Cellular transport and membrane dynamics of the glycine receptor. Front. Mol. Neurosci. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fritschy J. M., Harvey R. J., Schwarz G. (2008) Gephyrin. Where do we stand, where do we go? Trends Neurosci. 31, 257–264 [DOI] [PubMed] [Google Scholar]
- 3. Kneussel M., Hermann A., Kirsch J., Betz H. (1999) Hydrophobic interactions mediate binding of the glycine receptor β-subunit to gephyrin. J. Neurochem. 72, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 4. Schrader N., Kim E. Y., Winking J., Paulukat J., Schindelin H., Schwarz G. (2004) Biochemical characterization of the high affinity binding between the glycine receptor and gephyrin. J. Biol. Chem. 279, 18733–18741 [DOI] [PubMed] [Google Scholar]
- 5. Tretter V., Jacob T. C., Mukherjee J., Fritschy J. M., Pangalos M. N., Moss S. J. (2008) The clustering of GABA(A) receptor subtypes at inhibitory synapses is facilitated via the direct binding of receptor α2 subunits to gephyrin. J. Neurosci. 28, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tretter V., Kerschner B., Milenkovic I., Ramsden S. L., Ramerstorfer J., Saiepour L., Maric H. M., Moss S. J., Schindelin H., Harvey R. J., Sieghart W., Harvey K. (2011) Molecular basis of the γ-aminobutyric acid A receptor α3 subunit interaction with the clustering protein gephyrin. J. Biol. Chem. 286, 37702–37711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mukherjee J., Kretschmannova K., Gouzer G., Maric H. M., Ramsden S., Tretter V., Harvey K., Davies P. A., Triller A., Schindelin H., Moss S. J. (2011) The residence time of GABA(A)Rs at inhibitory synapses is determined by direct binding of the receptor α1 subunit to gephyrin. J. Neurosci. 31, 14677–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Essrich C., Lorez M., Benson J. A., Fritschy J. M., Lüscher B. (1998) Postsynaptic clustering of major GABAA receptor subtypes requires the γ2 subunit and gephyrin. Nat. Neurosci. 1, 563–571 [DOI] [PubMed] [Google Scholar]
- 9. Kirsch J., Wolters I., Triller A., Betz H. (1993) Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature 366, 745–748 [DOI] [PubMed] [Google Scholar]
- 10. Feng G., Tintrup H., Kirsch J., Nichol M. C., Kuhse J., Betz H., Sanes J. R. (1998) Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science 282, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 11. Stallmeyer B., Schwarz G., Schulze J., Nerlich A., Reiss J., Kirsch J., Mendel R. R. (1999) The neurotransmitter receptor-anchoring protein gephyrin reconstitutes molybdenum cofactor biosynthesis in bacteria, plants, and mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 96, 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Molybdenum cofactors, enzymes, and pathways. Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 13. Prior P., Schmitt B., Grenningloh G., Pribilla I., Multhaup G., Beyreuther K., Maulet Y., Werner P., Langosch D., Kirsch J. (1992) Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron 8, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 14. Schwarz G., Schrader N., Mendel R. R., Hecht H. J., Schindelin H. (2001) Crystal structures of human gephyrin and plant Cnx1 G domains. Comparative analysis and functional implications. J. Mol. Biol. 312, 405–418 [DOI] [PubMed] [Google Scholar]
- 15. Sola M., Kneussel M., Heck I. S., Betz H., Weissenhorn W. (2001) X-ray crystal structure of the trimeric N-terminal domain of gephyrin. J. Biol. Chem. 276, 25294–25301 [DOI] [PubMed] [Google Scholar]
- 16. Sola M., Bavro V. N., Timmins J., Franz T., Ricard-Blum S., Schoehn G., Ruigrok R. W., Paarmann I., Saiyed T., O'Sullivan G. A., Schmitt B., Betz H., Weissenhorn W. (2004) Structural basis of dynamic glycine receptor clustering by gephyrin. EMBO J. 23, 2510–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kneussel M., Betz H. (2000) Clustering of inhibitory neurotransmitter receptors at developing postsynaptic sites. The membrane activation model. Trends Neurosci. 23, 429–435 [DOI] [PubMed] [Google Scholar]
- 18. Saiyed T., Paarmann I., Schmitt B., Haeger S., Sola M., Schmalzing G., Weissenhorn W., Betz H. (2007) Molecular basis of gephyrin clustering at inhibitory synapses. Role of G- and E-domain interactions. J. Biol. Chem. 282, 5625–5632 [DOI] [PubMed] [Google Scholar]
- 19. Lardi-Studler B., Smolinsky B., Petitjean C. M., Koenig F., Sidler C., Meier J. C., Fritschy J. M., Schwarz G. (2007) Vertebrate-specific sequences in the gephyrin E-domain regulate cytosolic aggregation and postsynaptic clustering. J. Cell Sci. 120, 1371–1382 [DOI] [PubMed] [Google Scholar]
- 20. Kim E. Y., Schrader N., Smolinsky B., Bedet C., Vannier C., Schwarz G., Schindelin H. (2006) Deciphering the structural framework of glycine receptor anchoring by gephyrin. EMBO J. 25, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Specht C. G., Grünewald N., Pascual O., Rostgaard N., Schwarz G., Triller A. (2011) Regulation of glycine receptor diffusion properties and gephyrin interactions by protein kinase C. EMBO J. 30, 3842–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zita M. M., Marchionni I., Bottos E., Righi M., Del Sal G., Cherubini E., Zacchi P. (2007) Post-phosphorylation prolyl isomerization of gephyrin represents a mechanism to modulate glycine receptors function. EMBO J. 26, 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tyagarajan S. K., Ghosh H., Yévenes G. E., Nikonenko I., Ebeling C., Schwerdel C., Sidler C., Zeilhofer H. U., Gerrits B., Muller D., Fritschy J. M. (2011) Regulation of GABAergic synapse formation and plasticity by GSK3β-dependent phosphorylation of gephyrin. Proc. Natl. Acad. Sci. U.S.A. 108, 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bausen M., Weltzien F., Betz H., O'Sullivan G. A. (2010). Regulation of postsynaptic gephyrin cluster size by protein phosphatase 1. Mol. Cell Neurosci. 44, 201–209 [DOI] [PubMed] [Google Scholar]
- 25. Ramming M., Betz H., Kirsch J. (1997) Analysis of the promoter region of the murine gephyrin gene. FEBS Lett. 405, 137–140 [DOI] [PubMed] [Google Scholar]
- 26. Meier J., De Chaldée M., Triller A., Vannier C. (2000) Functional heterogeneity of gephyrins. Mol. Cell Neurosci. 16, 566–577 [DOI] [PubMed] [Google Scholar]
- 27. Rees M. I., Harvey K., Ward H., White J. H., Evans L., Duguid I. C., Hsu C. C., Coleman S. L., Miller J., Baer K., Waldvogel H. J., Gibbon F., Smart T. G., Owen M. J., Harvey R. J., Snell R. G. (2003) Isoform heterogeneity of the human gephyrin gene (GPHN), binding domains to the glycine receptor, and mutation analysis in hyperekplexia. J. Biol. Chem. 278, 24688–24696 [DOI] [PubMed] [Google Scholar]
- 28. Smolinsky B., Eichler S. A., Buchmeier S., Meier J. C., Schwarz G. (2008) Splice-specific functions of gephyrin in molybdenum cofactor biosynthesis. J. Biol. Chem. 283, 17370–17379 [DOI] [PubMed] [Google Scholar]
- 29. Giesemann T., Schwarz G., Nawrotzki R., Berhörster K., Rothkegel M., Schlüter K., Schrader N., Schindelin H., Mendel R. R., Kirsch J., Jockusch B. M. (2003) Profilin and Mena link the postsynaptic scaffold protein gephyrin to the microfilament system. J. Neurosci. 23, 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blom N., Gammeltoft S., Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 31. Ramming M., Kins S., Werner N., Hermann A., Betz H., Kirsch J. (2000) Diversity and phylogeny of gephyrin. Tissue-specific splice variants, gene structure, and sequence similarities to molybdenum cofactor-synthesizing and cytoskeleton-associated proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paarmann I., Schmitt B., Meyer B., Karas M., Betz H. (2006) Mass spectrometric analysis of glycine receptor-associated gephyrin splice variants. J. Biol. Chem. 281, 34918–34925 [DOI] [PubMed] [Google Scholar]
- 33. Kirsch J., Betz H. (1995) The postsynaptic localization of the glycine receptor-associated protein gephyrin is regulated by the cytoskeleton. J. Neurosci. 15, 4148–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langosch D., Hoch W., Betz H. (1992) The 93-kDa protein gephyrin and tubulin associated with the inhibitory glycine receptor are phosphorylated by an endogenous protein kinase. FEBS Lett. 298, 113–117 [DOI] [PubMed] [Google Scholar]
- 35. Charrier C., Machado P., Tweedie-Cullen R.Y., Rutishauser D., Mansuy I. M., Triller A. (2010) A cross-talk between β1 and β3 integrins controls glycine receptor and gephyrin trafficking at synapses. Nat. Neurosci. 13, 1388–1395 [DOI] [PubMed] [Google Scholar]
- 36. Kneussel M., Betz H. (2000). Receptors, gephyrin, and gephyrin-associated proteins. Novel insights into the assembly of inhibitory postsynaptic membrane specializations. J. Physiol. 525, Pt 1, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jockusch B. M., Rothkegel M., Schwarz G. (2004) Linking the synapse to the cytoskeleton. A breath-taking role for microfilaments. Neuroreport 15, 1535–1538 [DOI] [PubMed] [Google Scholar]
- 38. Luscher B., Fuchs T., Kilpatrick C. L. (2011) GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kins S., Betz H., Kirsch J. (2000) Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat. Neurosci. 3, 22–29 [DOI] [PubMed] [Google Scholar]
- 40. Nawrotzki R., Islinger M., Vogel I., Volkl A., Kirsch J. (2012) Expression and subcellular distribution of gephyrin in non-neuronal tissues and cells. Histochem. Cell. Biol. doi: 10.1007/s00418-012-0914-7 [DOI] [PubMed] [Google Scholar]
- 41. Srinivasan S., Nichols C. J., Lawless G. M., Olsen R. W., Tobin A. J. (1999) Two invariant tryptophans on the α1 subunit define domains necessary for GABA(A) receptor assembly. J. Biol. Chem. 274, 26633–26638 [DOI] [PubMed] [Google Scholar]
- 42. Cascio M., Shenkel S., Grodzicki R. L., Sigworth F. J., Fox R. O. (2001) Functional reconstitution and characterization of recombinant human alpha 1-glycine receptors. J. Biol. Chem. 276, 20981–20988 [DOI] [PubMed] [Google Scholar]
- 43. Fuhrmann J. C., Kins S., Rostaing P., El Far O., Kirsch J., Sheng M., Triller A., Betz H., Kneussel M. (2002) Gephyrin interacts with Dynein light chains 1 and 2, components of motor protein complexes. J. Neurosci. 22, 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiang S., Kim E. Y., Connelly J. J., Nassar N., Kirsch J., Winking J., Schwarz G., Schindelin H. (2006) The crystal structure of Cdc42 in complex with collybistin II, a gephyrin-interacting guanine nucleotide exchange factor. J. Mol. Biol. 359, 35–46 [DOI] [PubMed] [Google Scholar]
- 45. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han G., Ye M., Liu H., Song C., Sun D., Wu Y., Jiang X., Chen R., Wang C., Wang L., Zou H. (2010) Phosphoproteome analysis of human liver tissue by long-gradient nanoflow LC coupled with multiple stage MS analysis. Electrophoresis 31, 1080–1089 [DOI] [PubMed] [Google Scholar]
- 47. Xia Q., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., Peng J. (2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 49. Choudhary C., Olsen J. V., Brandts C., Cox J., Reddy P. N., Böhmer F. D., Gerke V., Schmidt-Arras D. E., Berdel W. E., Müller-Tidow C., Mann M., Serve H. (2009) Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell 36, 326–339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.