Background: Y-family DNA polymerases may employ protein conformational changes to catalyze the bypass of various DNA lesions.
Results: The conformational dynamics of three structural domains of Dpo4 are altered by 8-oxo-7,8-dihydro-2′-deoxyguanine (8-oxoG).
Conclusion: Dpo4 undergoes different global conformational changes during 8-oxoG bypass than during the replication of undamaged DNA.
Significance: A DNA lesion alters the conformational dynamics of a DNA polymerase during catalysis.
Keywords: DNA Damage, DNA Polymerase, DNA Synthesis, Enzyme Catalysis, Enzyme Mechanisms, Fluorescence Resonance Energy Transfer (FRET), 8-oxoG, Conformational Dynamics, Lesion Bypass
Abstract
A major product of oxidative damage to DNA, 8-oxo-7,8-dihydro-2′-deoxyguanine (8-oxoG), can lead to genomic mutations if it is bypassed unfaithfully by DNA polymerases in vivo. However, our pre-steady-state kinetic studies show that DNA polymerase IV (Dpo4), a prototype Y-family enzyme from Sulfolobus solfataricus, can bypass 8-oxoG both efficiently and faithfully. For the first time, our stopped-flow FRET studies revealed that a DNA polymerase altered its synchronized global conformational dynamics in response to a DNA lesion. Relative to nucleotide incorporation into undamaged DNA, three of the four domains of Dpo4 undertook different conformational transitions during 8-oxoG bypass and the subsequent extension step. Moreover, the rapid translocation of Dpo4 along DNA induced by nucleotide binding was significantly hindered by the interactions between the embedded 8-oxoG and Dpo4 during the extension step. These results unprecedentedly demonstrate that a Y-family DNA polymerase employs different global conformational dynamics when replicating undamaged and damaged DNA.
Introduction
Unrepaired DNA lesions often alter DNA local structure and stall replicative DNA polymerases, leading to cell death. To counter this problem, cells employ specialized Y-family DNA polymerases to bypass lesions during DNA replication. In addition to the three polymerase core domains (finger, palm, and thumb) that are common to the members of all six DNA polymerase families (A, B, C, D, X, and Y), each Y-family DNA polymerase possesses a unique little finger (LF)2 domain, which plays an important role in defining the lesion bypass properties that are possibly inherent to each enzyme (1). DNA polymerases are hypothesized to undergo various conformational changes throughout the catalytic cycle, although the physical nature of these conformational changes and their relationship to enzymatic function remain poorly defined and constantly debated (2). For many DNA polymerases, crystal structure studies (3–7) and solution-phase FRET-based studies (8–10) have revealed a large domain-closing transition upon nucleotide binding to the enzyme·DNA binary complex characterized by the inward rotation of the finger domain, which helps position key residues in the active site for catalysis. In contrast, no large domain shift is observed when overlaying the binary (E·DNA) and ternary (E·DNA·dNTP) crystal structures of Y-family DNA polymerases (11). However, our stopped-flow FRET studies have revealed short-range pre- and post-catalytic conformational changes (0.5–3.7 Å) in each of the four domains of Dpo4, the lone Y-family enzyme from Sulfolobus solfataricus, as well as a rapid DNA translocation event previously hypothesized from the superposition of binary and ternary crystal structures of Dpo4 (11) during correct nucleotide incorporation into undamaged DNA (supplemental Fig. S1) (12). These newly discovered conformational change steps have led to an expanded minimal kinetic pathway for nucleotide incorporation catalyzed by Dpo4 (supplemental Fig. S1C). Interestingly, the LF domain and the polymerase core domains of Dpo4 move in opposite directions during catalysis (supplemental Fig. S1A). Therefore, we hypothesized that the global conformational dynamics of Dpo4, especially in the LF domain, have evolved to support translesion DNA synthesis (TLS), the in vivo role of Dpo4.
8-Oxo-7,8-dihydro-2′-deoxyguanine (8-oxoG) is the major DNA lesion formed by oxidative damage in vivo and has been identified as a biomarker for oxidative stress (13, 14). Crystallographic evidence suggests that Dpo4 makes several additional contacts with a DNA substrate containing an 8-oxoG lesion, which are not possible with an undamaged DNA substrate (15–17). Thus, although studies have shown that Dpo4 can bypass 8-oxoG and unmodified DNA bases with similar efficiency (17), it is possible that these additional interactions are able to perturb the conformational dynamics of Dpo4 during nucleotide incorporation in the vicinity of the damaged base. Here, this hypothesis was examined through transient kinetic and dynamic studies of the bypass of an 8-oxoG lesion and the subsequent extension catalyzed by Dpo4. Our results show conclusively that Dpo4 was able to bypass an 8-oxoG lesion with conformational dynamics that differed from those employed during the replication of undamaged DNA.
EXPERIMENTAL PROCEDURES
Preparation of Fluorescently Labeled Dpo4 and DNA Substrates
Unlabeled wild-type S. solfataricus Dpo4 was purified as described previously (18). The creation of the Dpo4 enzymes containing the C31S substitution and site-specific labeling with Alexa Fluor 594 (Invitrogen) at another engineered Cys substitution (i.e. N70C, E49C, S112C, S207C, or K329C) were described previously (12).
The 30-mer template in all DNA substrates (see Table 1) contains a site-specifically placed 8-oxoG and was purchased from Midland Certified Reagent Co. All other DNA oligomers in Table 1 were purchased from Integrated DNA Technologies, Inc. All oligonucleotides were purified by gel electrophoresis. Alexa Fluor 488 (Invitrogen) was attached to the ninth base from the 3′-end of the primer strand via 5-C6-amino-2′-deoxythymidine. Alexa Fluor 488-labeled DNA was purified according to the manufacturer's protocol. Each DNA substrate in Table 1 was annealed as described previously (19). For kinetic assays, the primer strand was 5′-radiolabeled with [γ-32P]ATP (MP Biomedicals) and OptiKinase (USB Corp.) before annealing (19).
TABLE 1.
Sequences of DNA substrates
T, C, and X denote Alexa Fluor 488 attached to a modified C6 linker on base dT, 2′,3′-dideoxycytidine, and 8-oxoG, respectively.
| Name | Sequence |
|---|---|
| O-1 | 5′-CGAGCCGTCGCATCCTACCGC-3′ |
| 3′-GCTCGGCAGCGTAGGATGGCGXCGTCGTAG-5′ | |
| O-2 | 5′-CGAGCCGTCGCATCCTACCGC-3′ |
| 3′-GCTCGGCAGCGTAGGATGGCGXCGTCGTAG-5′ | |
| O-3 | 5′-CGAGCCGTCGCATCCTACCGC-3′ |
| 3′-GCTCGGCAGCGTAGGATGGCGXCGTCGTAG-5′ | |
| O-4 | 5′-CGAGCCGTCGCATCCTACCGCC-3′ |
| 3′-GCTCGGCAGCGTAGGATGGCGXCGTCGTAG-5′ | |
| O-5 | 5′-CGAGCCGTCGCATCCTACCGCC-3′ |
| 3′-GCTCGGCAGCGTAGGATGGCGXCGTCGTAG-5′ |
Reaction Buffers
All stopped-flow, steady-state fluorescence, and rapid chemical quench kinetic assays were carried out in buffer R containing 50 mm HEPES (pH 7.5 at 20 °C), 50 mm NaCl, 6 mm MgCl2, 0.1 mm EDTA, and 10% glycerol. All reported concentrations indicate the final concentrations upon mixing all components. All assays were performed at 20 °C.
Steady-state Fluorescence Assays
Steady-state fluorescence spectra were recorded on a FluoroMax-4 system (HORIBA Jobin Yvon). The assay solution contained 100 nm Alexa Fluor 488-labeled DNA equilibrated at 20 °C. 600 nm Alexa Fluor 594-labeled Dpo4 and 1 mm dNTP were added sequentially to this solution. Fluorescence spectra were recorded at the excitation maximum of Alexa Fluor 488 (493 nm) with slits set at 3 nm for both excitation and emission. Sample temperature was controlled by a circulating water bath during the entire process. The distance between each Alexa Fluor 594-labeled Dpo4 residue (acceptor) and the Alexa Fluor 488-labeled primer base (donor) (see Table 1) was calculated using Equation 1,
where ET is the FRET efficiency; FAD and FD are the donor peak fluorescence intensity in the presence and absence of an acceptor, respectively; R0 is the Förster distance (60 Å) of the FRET pair of Alexa Fluor 488 and Alexa Fluor 594; and r is the calculated distance between the donor and acceptor.
Stopped-flow Assays
For all stopped-flow assays, a preincubated solution of Alexa Fluor 594-labeled Dpo4 (600 nm) and Alexa Fluor 488-labeled DNA (100 nm) was rapidly mixed with dNTP (1 mm), and the resulting fluorescence emission was recorded in real-time upon excitation of the donor at 493 nm on an SX20 stopped-flow apparatus (Applied Photophysics). Both emission and excitation slits were set to 5 nm. Donor and acceptor emission was measured separately using XF3084 (for Alexa Fluor 488, 510–570 nm) and XF3028 (for Alexa Fluor 594, 615–650 nm) band pass filters (Omega Optical). The rates for each fluorescent phase were determined by fitting stopped-flow fluorescence traces using the KinTek Explorer global fitting software (20). The software was set to fit each trace using a single-, double-, or triple-exponential equation depending on the number of observed FRET phases.
Pre-steady-state Kinetic Assays
To determine the observed incorporation rate (kobs) for Dpo4 enzymes with the single-Cys substitutions, a preincubated solution of Dpo4 (180 nm) and 5′-32P-labeled O-2 (30 nm) was rapidly mixed with dCTP (1 mm) and subsequently quenched at various time intervals with 0.37 m EDTA. Reaction products were resolved by denaturing PAGE, scanned using a Typhoon Trio system (GE Healthcare), and quantitated using ImageQuant (Molecular Dynamics). To obtain kobs, a plot of product concentration versus time was fit to Equation 2,
where A represents the reaction amplitude.
RESULTS
Design of FRET System to Monitor Conformational Changes during TLS
To investigate enzyme conformational dynamics during 8-oxoG bypass, we designed and performed steady-state and stopped-flow FRET assays similar to those used in our previous work with undamaged DNA (12). A FRET pair (supplemental Table S1) was formed by binding a Dpo4 enzyme containing a site-specific fluorescent label to a DNA substrate (Table 1) containing a donor fluorophore, Alexa Fluor 488, just outside of the DNA-binding cleft (12) (Fig. 1). An acceptor fluorophore, Alexa Fluor 594, was covalently attached to Dpo4 via a single engineered cysteine residue in a specific structure loop in one of the Dpo4 domains (supplemental Table S1). These single-residue substitutions did not affect polymerase structure or activity (12) or the single-turnover rate of dCTP incorporation opposite 8-oxoG (supplemental Table S2).
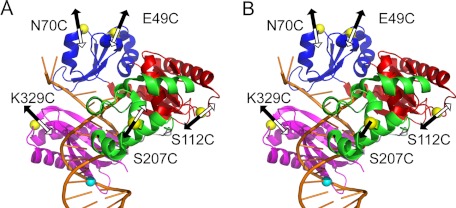
FIGURE 1.
Structure and dynamics of Dpo4 during 8-oxoG bypass and extension steps. The domains of Dpo4 are shown in blue (finger), red (palm), green (thumb), and purple (LF). The DNA is shown in gold. The positions of the Alexa Fluor 594 acceptor on Dpo4 and the Alexa Fluor 488 donor on DNA are shown as yellow and cyan spheres, respectively. The arrows indicate the directions of residue movement during dCTP incorporation opposite 8-oxoG (A) and the subsequent extension of the 8-oxoG bypass product (B) based on the FRET signal changes for phase P1 (black) and phase P2 (white).
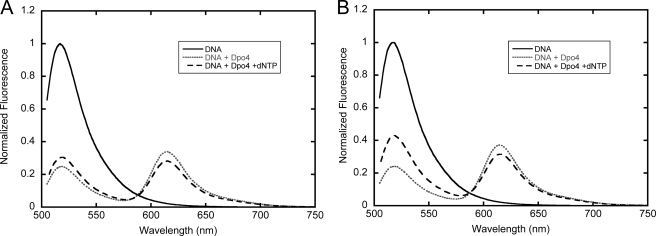
To verify that the conformational dynamics of lesion bypass would lead to an observable signal with our FRET system, we performed steady-state fluorescence assays to monitor conformational changes involved in both the bypass of the 8-oxoG lesion and the subsequent extension. As described previously (12), the addition of unlabeled enzyme to Alexa Fluor 488-labeled DNA or of Alexa Fluor 594-labeled enzyme to unlabeled DNA substrate resulted in no detectable change in fluorescence. In contrast, the addition of Dpo4 with an acceptor label at K329C in the LF domain (or each of the other labeled Dpo4 enzymes in supplemental Table S1) to donor-labeled DNA substrate O-2 with 8-oxoG in the templating position (Table 1) resulted in a large decrease in donor fluorescence and the appearance of an acceptor peak centered at 615 nm (Fig. 2A). Upon further addition of dCTP to the binary complex formed between the labeled Dpo4 and O-2, an increase in donor fluorescence and a corresponding decrease in acceptor fluorescence were observed (Fig. 2A). When the non-extendible O-3 DNA substrate (Table 1) was used, a larger change in FRET was observed (Fig. 2B), indicating that there was a greater distance between the two fluorophores in the nucleotide-bound ternary complex (E·O-3·dCTP) than in the subsequent binary complex resulting from the covalent incorporation of dCTP opposite 8-oxoG in O-2. This result was consistent with steady-state and stopped-flow data observed previously with undamaged DNA (12), which indicated a pre-catalytic outward motion of the LF domain upon nucleotide binding, followed by an inward motion after nucleotide incorporation. Similar steady-state fluorescence results were observed for the extension step after 8-oxoG bypass and with enzymes labeled at N70C, S112C, and S207C (supplemental Table S1).
FIGURE 2.
Steady-state fluorescence spectra of Dpo4 labeled at LF domain K329C at 20 °C. Alexa Fluor 488-labeled DNA (100 nm; solid black line) was excited at 493 nm. The sequential addition of Alexa Fluor 594-labeled Dpo4 (600 nm) and dTTP (1 mm) produced the dotted gray and dashed black lines, respectively. Spectra were normalized to one by using the donor in the absence of the acceptor as a reference. Emission spectra are shown for both O-2 (A) and O-3 (B) DNA substrates (Table 1).
Real-time Conformational Dynamics during Bypass of 8-oxoG
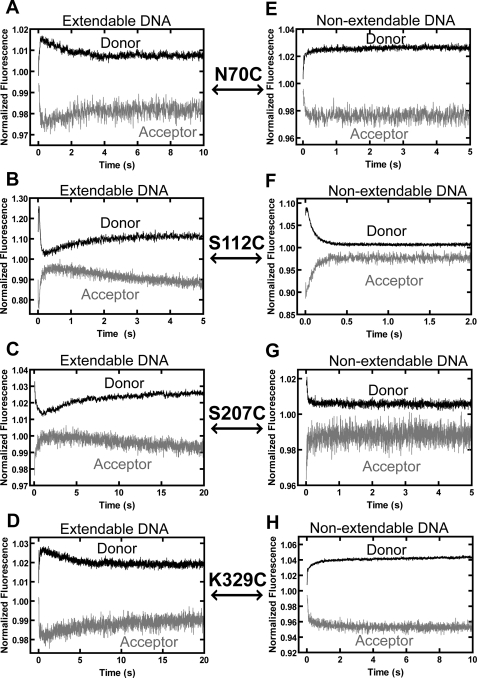
To investigate the real-time conformational dynamics involved in TLS, we preformed stopped-flow FRET studies to monitor the real-time conformational motions of each domain of Dpo4 during dCTP incorporation opposite 8-oxoG. Upon mixing a preincubated solution of Alexa Fluor 594-labeled Dpo4 and Alexa Fluor 488-labeled O-2 with dCTP, two to three phases of FRET signal changes were detected (Fig. 3). In all cases, donor and acceptor fluorescence intensity traces were anti-correlated, and hereafter, only the acceptor signal is discussed. As described previously, control experiments with either unlabeled DNA or unlabeled protein showed no change in fluorescence after mixing (12). Dpo4 labeled at S112C, S207C, and K329C exhibited time-dependent FRET changes similar to those observed for correct incorporation opposite undamaged DNA (12), albeit with slightly different rates (Table 2). For example, palm domain S112C (Fig. 3B) had a rapid decrease phase (P0, rate > 100 s−1) preceding a fast increase phase (P1, rate = 14 s−1) and a slow decrease phase (P2, rate = 0.17 s−1). Using non-extendable O-3, only P2 was not observed for S112C (Fig. 3F), thereby indicating that this phase represents a conformational change that occurs after phosphodiester bond formation, as has been rationalized with correct incorporation into undamaged DNA (12). Thus, phase rates and trends similar to those observed with undamaged DNA (12) suggest that P0, P1, and P2 likely represented the DNA translocation event, pre-catalytic closing motion of the palm domain, and post-catalytic reopening motion of the palm domain, respectively, during dCTP incorporation opposite 8-oxoG (Fig. 1A). As observed with undamaged DNA (12), only P1 and P2 were detected for S207C and K329C during 8-oxoG bypass, and P2 was absent with non-extendable O-3 (Fig. 3). The trends of the FRET signal changes indicate that the motions of the thumb and LF domains during 8-oxoG bypass were in opposite directions (Fig. 1A) and are similar to the corresponding motions observed with undamaged DNA (supplemental Fig. S1A) (12). The rate of P2 for each Dpo4 mutant was within 2-fold of the rate of dCTP incorporation opposite 8-oxoG measured by rapid chemical quench (Table 2 and supplemental Table S2), which lends further support to the conclusion that P2 represents a post-catalytic conformational change.
FIGURE 3.
Stopped-flow traces for dCTP incorporation and binding opposite 8-oxoG at 20 °C. A preincubated solution containing Alexa Fluor 594-labeled Dpo4 (600 nm) and either extendable DNA substrate O-2 (A–D) or non-extendable DNA substrate O-3 (E–H) (100 nm) (Table 1) was rapidly mixed with dCTP (1 mm), and the resulting donor and acceptor fluorescence signals upon excitation of the donor at 493 nm were recorded in real-time. Donor and acceptor traces are shown for the finger domain mutant N70C (A and E), the palm domain mutant S112C (B and F), the thumb domain mutant S207C (C and G), and the LF domain mutant K329C (D and H).
TABLE 2.
Rates of FRET phases
The phase rates (s−1) and errors at 20 °C were obtained by fitting stopped-flow fluorescence traces with the KinTek Explorer global fitting software (see “Experimental Procedures”) (20).
| Domain mutanta and DNA substrateb | Phase rate |
|||||
|---|---|---|---|---|---|---|
| Donor |
Acceptor |
|||||
| P0 | P1 | P2 | P0 | P1 | P2 | |
| s−1 | ||||||
| Finger N70C | ||||||
| O-2 | 15 ± 2 | 0.83 ± 0.04 | 14 ± 2 | 0.63 ± 0.06 | ||
| O-3 | 12 ± 1 | 18 ± 2 | ||||
| O-4 | 11.1 ± 0.8 | 0.38 ± 0.02 | 6 ± 2 | 0.41 ± 0.05 | ||
| O-5 | 9 ± 3 | 11 ± 2 | ||||
| Finger E49C | ||||||
| O-2 | 22 ± 4 | 0.41 ± 0.02 | 18 ± 5 | 0.49 ± 0.06 | ||
| O-3 | 22 ± 4 | 17 ± 3 | ||||
| Palm S112C | ||||||
| O-2 | >100 | 12 ± 0.8 | 0.4 ± 0.1 | >100 | 14 ± 1 | 0.17 ± 0.04 |
| O-3 | >100 | 12 ± 1 | >100 | 11 ± 2 | ||
| O-4 | 21 ± 2 | 1.33 ± 0.07 | 0.19 ± 0.01 | 13 ± 7 | 1.8 ± 0.5 | 0.19 ± 0.07 |
| O-5 | 21.8 ± 0.3 | 1.42 ± 0.02 | 21 ± 1 | 2.0 ± 0.1 | ||
| Thumb S207C | ||||||
| O-2 | 5.5 ± 0.8 | 0.24 ± 0.06 | 5.5 ± 0.7 | 0.12 ± 0.04 | ||
| O-3 | 17 ± 3 | 12 ± 2 | ||||
| O-4 | 6.4 ± 0.4 | 9 ± 1 | ||||
| O-5 | 8 ± 2 | 8.0 ± 0.5 | ||||
| LF K329C | ||||||
| O-2 | 9 ± 1 | 0.32 ± 0.03 | 5 ± 2 | 0.23 ± 0.07 | ||
| O-3 | 6.1 ± 0.5 | 5.4 ± 0.4 | ||||
| O-4 | 8.3 ± 0.3 | 0.51 ± 0.05 | 7.4 ± 0.6 | 0.30 ± 0.01 | ||
| O-5 | 5 ± 1 | 5.2 ± 0.4 | ||||
a Each of the Alexa Fluor 594-labeled Dpo4 mutants also contains the C31S mutation and is listed in supplemental Table S1.
b DNA substrates are listed in Table 1.
Compared with the three FRET phases observed with correct incorporation opposite undamaged DNA (supplemental Fig. S1B) (12), finger domain N70C exhibited very different conformational dynamics during 8-oxoG bypass: a fast initial decrease in FRET efficiency (P1, rate = 14 s−1), followed by a slow increase phase (P2, rate = 0.63 s−1) (Fig. 3A), and this P2 disappeared with non-extendable O-3 (Fig. 3E). It is likely that the DNA translocation step (P0) still led to a decrease in acceptor fluorescence, which could not be resolved from a slower pre-catalytic conformational change, which also caused a decrease in the fluorescence intensity of the acceptor, and therefore, P1 for N70C is due to a combination of these two events. The rate observed for the combined P1 with N70C (15 s−1) is similar to the P1 rates measured at the other positions (Table 2) and to those measured previously at all positions with undamaged DNA (12), suggesting that this combined phase is rate-limited by the same pre-catalytic conformational change step, rather than the DNA translocation step, which occurred at a much faster rate (>100 s−1). Based on the directions of the FRET signal changes in both P1 and P2, the finger domain moved away from DNA before catalysis and reversed its motion after 8-oxoG bypass (Fig. 1A), which is opposite what has been observed with undamaged DNA (supplemental Fig. S1B) (12). Notably, control experiments with dCTP incorporation opposite unmodified G produced FRET phases identical to those observed in the previous study with dTTP incorporation opposite A (data not shown), indicating that the difference in the conformational motions was due to the lesion and not the identity of the added nucleotide. In addition, we have previously proposed that the finger domain undergoes a rotational motion during nucleotide incorporation into undamaged DNA based on the observation that finger domain N70C and E49C move in opposite directions, indicating a rotational axis between the two locations (supplemental Fig. S1A) (12). To determine whether this rotational axis is maintained during 8-oxoG bypass, we monitored the FRET signal changes of E49C (supplemental Table S1) during dCTP incorporation opposite 8-oxoG. Interestingly, two FRET phases (P1 and P2) were observed with O-2 (supplemental Fig. S2A), and P2 disappeared with non-extendable O-3 (supplemental Fig. S2B). These observations are analogous to those observed with N70C (Fig. 3, A and E) and with undamaged DNA (12). Thus, the rotational axis between E49C and N70C did not exist during 8-oxoG bypass. Moreover, the motions of E49C confirmed that the finger domain indeed opened pre-catalytically and closed post-catalytically as derived from the motions of N70C (Fig. 1A).
Conformational Changes during Extension of 8-oxoG Bypass Product
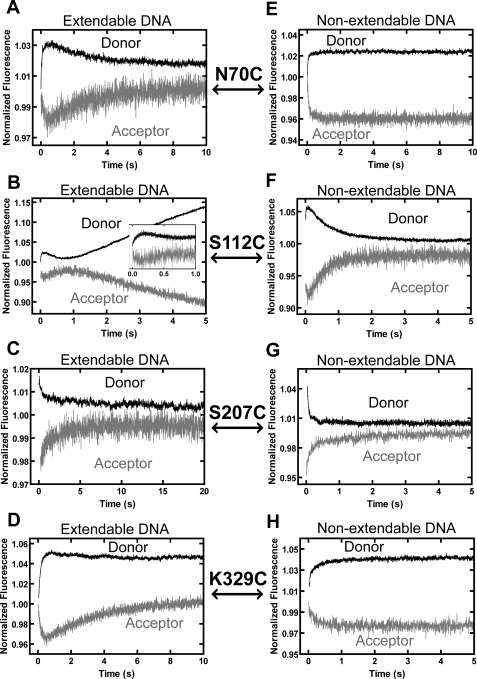
To explore the conformational dynamics of Dpo4 during the extension of the 8-oxoG bypass product, we also monitored the FRET changes in real-time by adding correct dGTP to a preincubated solution of Alexa Fluor 594-labeled Dpo4 (supplemental Table S1) and Alexa Fluor 488-labeled O-4 or non-extendable O-5 DNA substrates containing a terminal 8-oxoG:C base pair (Table 1). For finger domain N70C, the FRET signal changes with O-4 in Fig. 4A (or with non-extendable O-5 in Fig. 4E) were similar to those described above for O-2 and O-3 DNA substrates containing 8-oxoG at the templating position (Fig. 3, A and E), indicating that the finger domain underwent similar conformational dynamics during 8-oxoG bypass and the subsequent extension step (Fig. 1). In contrast, the FRET signal changes with palm domain S112C in Figs. 3B and 4B were similar in directions but differed significantly in rates between the two different steps of TLS. The rates of both P0 and P1 were at least 5-fold lower than the corresponding rates observed during 8-oxoG bypass, whereas the P2 rates were similar (Table 2). These differences suggest that both DNA translocation and the pre-catalytic closing motion of the palm domain were impeded by the embedded 8-oxoG during the extension step. Surprisingly, for thumb domain S207C, only P1 was observed for both O-4 (Fig. 4C) and non-extendable O-5 (Fig. 4G) during the extension step. In comparison, both P1 and P2 were detected during 8-oxoG bypass (Fig. 3C) or correct incorporation into undamaged DNA (12). It is possible that the opening motion of the thumb domain did occur but was too small to be detected by our FRET system. Finally, no differences in the rates (Table 2) or trends (Fig. 3, D and H versus Fig. 4, D and H) of the conformational dynamics of the LF domain were detected between 8-oxoG bypass and the extension step using LF domain K329C. Thus, among the four domains of Dpo4, the palm and thumb domains underwent different conformational dynamics during 8-oxoG bypass and the extension step (Fig. 1). For both steps, only the LF domain, a non-polymerase core domain, displayed motions (Fig. 1) similar to those observed with undamaged DNA (supplemental Fig. S1A) (12).
FIGURE 4.
Stopped-flow traces for dGTP incorporation during subsequent extension of 8-oxoG bypass product at 20 °C. A preincubated solution of Alexa Fluor 594-labeled Dpo4 (600 nm) and either extendable DNA substrate O-4 (A–D) or non-extendable DNA substrate O-5 (E–H) (100 nm) (Table 1) was rapidly mixed with dGTP (1 mm), and the resulting donor and acceptor fluorescence signals upon excitation of the donor at 493 nm were recorded over time. Donor and acceptor traces are shown for the finger domain mutant N70C (A and E), the palm domain mutant S112C (B and F), the thumb domain mutant S207C (C and G), and the LF domain mutant K329C (D and H).
Magnitudes of Conformational Changes
The magnitudes of the observed conformational changes were estimated by determining the distances between the donor and acceptor fluorophores in the pre- and post-insertion binary and ternary complexes for the bypass and extension steps as determined by steady-state fluorescence spectra (supplemental Table S3). Our results indicate that the translocation step (P0) leads to distance changes in the ranges of 0.4–1.4 Å during dCTP incorporation opposite 8-oxoG in O-2 (Table 1) and 0.3–3.7 Å during dGTP incorporation opposite dC in O-4 (Table 1) and that the changes in distances due to the subsequent domain motions (either P1 or P2) are in the ranges of 0.3–6.5 Å during 8-oxoG bypass and 0.2–4.3 Å during the subsequent extension reaction (supplemental Table S3). Consistently, predication based on the crystal structures of Dpo4 in various complexes with 8-oxoG-containing DNA substrates (15, 16) revealed changes in distance of similar magnitudes (supplemental Table S4). Interestingly, the motions of the LF and palm domains appear to be larger than those of the thumb and finger domains for both the bypass and extension steps based on both the steady-state FRET data (supplemental Table S3) and published crystal structures (supplemental Table S4).
DISCUSSION
Although our preliminary kinetic studies show that the overall kinetic mechanisms for both replication of undamaged DNA and 8-oxoG bypass are similar (data not shown), our results here indicate that the slight structural differences between 8-oxoG and an undamaged base dG are sufficient to significantly affect the conformational dynamics of Dpo4 during catalysis. However, the similarity in rates suggests that the P0, P1, and P2 phases observed during TLS in this work and those observed previously during the replication of undamaged DNA (12) correspond to the same mechanistic steps in the catalytic cycle (supplemental Fig. S1C). During 8-oxoG bypass, the four domains of Dpo4 moved in a synchronized manner, whereby the palm and thumb domains went through a “gripping-reopening” motion, whereas the finger and LF domains undertook an “opening-closing” transition during the P1 and P2 phases, respectively (Fig. 1A). In comparison, only the LF domain went through the opening-closing transition during correct incorporation into undamaged DNA (supplemental Fig. S1) (12). For both the bypass and extension steps, only the dynamics of the LF domain were unchanged compared with incorporation into undamaged DNA. This observation agrees with our earlier hypothesis that the conformational dynamics of the LF domain have evolved to support TLS. To allow for the stabilization of 8-oxoG in the anti-conformation and to promote correct dCTP incorporation opposite the lesion, the 5′-phosphate group must be shifted to avoid a steric clash with the O8 atom on the modified base. The outward motion of the LF domain therefore likely acts to pull the damaged template base and phosphate into this distorted configuration via interactions between the O8 and phosphate oxygen atoms on the DNA and the LF domain residues Arg-331 and Arg-332 (15, 17). Our steady-state fluorescence data indicate that this outward motion of the LF domain is larger than the changes observed in the other domains (supplemental Table S3), which underlines the importance of this conformational change. This mechanism involving the direct interactions between the LF domain residues and the O8 and phosphate oxygen atoms appears to be unique for Dpo4. For example, mammalian DNA polymerase β makes only minimal contact with the 8-oxoG phosphate backbone, which adopts a similarly kinked conformation without any stabilizing hydrogen bonds formed between the DNA backbone and enzyme residues (21). In contrast, in another Y-family polymerase, yeast polymerase η, no significant difference is observed between undamaged dG and 8-oxoG ternary complexes, as the DNA appears to bind to the enzyme with backbone torsion angles preset to accommodate the O8 atom without the need for a kink to avoid steric hindrance with the 5′-phosphate (22). Polymerase η only forms hydrogen bonding interactions with the O8 atom itself in the post-insertion complex after dCTP incorporation opposite the lesion (22).
Both stopped-flow (Fig. 3, A and E, and Fig. 4, A and E) and steady-state (supplemental Table S3) FRET data indicate that during the bypass and extension of the 8-oxoG lesion, finger domain N70C moves away from the Alexa Fluor 488-labeled DNA base in P1 and thus in an opposite direction than during nucleotide incorporation into undamaged DNA. The altered conformational dynamics of the finger domain as observed with N70C indicates there may be cross-talk between the LF and finger domains. Based on crystallographic evidence, there is a large interface of the LF domain that interacts with the DNA substrate. Moreover, the loop between β-sheets 2 and 3 of the finger domain interacts with β-sheet 9 of the LF domain, which is in direct contact with the DNA template strand (15, 17). Thus, the dynamics of the finger domain may be influenced by those of the LF domain. In support of this proposed role, a similar transmission of the LF domain's motions to the finger domain has been observed on a fast time scale through molecular dynamics simulation (23). When dCTP was incorporated opposite 8-oxoG, the simultaneous pre-catalytic opening motions of both the finger and LF domains created a wider and more solvent-accessible active site to better accommodate the damaged and distorted DNA substrates. The post-catalytic closing motions of these domains may help the polymerase to reform a strong binding interface with the DNA, thereby inhibiting DNA translocation by 1 bp during the next nucleotide incorporation (12). Indeed, the DNA translocation event observed with S112C was 5-fold slower during the extension step than during 8-oxoG bypass (Table 2). Notably, the slow DNA translocation during the extension step was likely caused by the breaking of some of the extra hydrogen bonds formed between Dpo4 and the embedded 8-oxoG in the binary complex (15). This was not a problem during the 8-oxoG bypass step based on the binary crystal structure of Dpo4·damaged DNA (15–17, 24), which displays no interactions between Dpo4 and the templating base 8-oxoG. The inhibition of DNA translocation after the first extension step may facilitate polymerase switching during TLS (25) by allowing the replicative DNA polymerase PolB1 more time to replace Dpo4 at the primer-template junction before Dpo4 moves into the next undamaged template position to incorporate another nucleotide. The closing motion of the palm domain was also slowed during extension from the 8-oxoG:C base pair. This result, coupled with the fact that the steady-state FRET measurements indicate that the palm domain moved a much larger distance than the finger and thumb domains (supplemental Table S3), suggests that the precise positioning of the catalytic residues in the palm domain requires a significant conformational shift throughout the domain to accommodate the damaged and distorted DNA substrate.
Our stopped-flow assays failed to reveal the post-catalytic closing of the thumb domain during the extension step, indicating that this motion was impeded by the embedded 8-oxoG lesion. Furthermore, the steady-state FRET data demonstrated that, relative to both the pre-insertion binary and ternary extension complexes, the distance between the Alexa Fluor 594-labeled S207C and the Alexa Fluor 488-labeled primer base decreased after incorporation of dGTP extending from the 8-oxoG:C base pair (supplemental Table S3), indicating a lack of domain reopening. In contrast, for dCTP incorporation opposite 8-oxoG, although there was an initial decrease in distance between the fluorophores upon dCTP binding, the subsequent dCTP incorporation led to an increase in distance due to the domain opening in P2 (supplemental Table S3).
Altogether, our FRET studies demonstrate that the rotational and translational conformational dynamics of Dpo4 were significantly affected by error-free TLS of 8-oxoG, perhaps more so when 8-oxoG was at the −1 position than at the templating position. It will be interesting to see how error-prone TLS affects global conformational dynamics of Dpo4 during catalysis.
Acknowledgment
The authors would like to thank Dr. Jessica A. Brown for critical reading of the manuscript.
This work was supported by National Science Foundation Career Award MCB-0447899 and Grant MCB-0960961 (to Z. S.).

This article contains supplemental Figs. S1 and S2, Tables S1–S4, and additional references.
- LF
- little finger
- TLS
- translesion DNA synthesis
- 8-oxoG
- 8-oxo-7,8-dihydro-2′-deoxyguanine.
REFERENCES
- 1. Boudsocq F., Kokoska R. J., Plosky B. S., Vaisman A., Ling H., Kunkel T. A., Yang W., Woodgate R. (2004) Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 279, 32932–32940 [DOI] [PubMed] [Google Scholar]
- 2. Joyce C. M., Benkovic S. J. (2004) DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324 [DOI] [PubMed] [Google Scholar]
- 3. Franklin M. C., Wang J., Steitz T. A. (2001) Structure of the replicating complex of a pol α family DNA polymerase. Cell 105, 657–667 [DOI] [PubMed] [Google Scholar]
- 4. Li Y., Korolev S., Waksman G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pelletier H., Sawaya M. R., Wolfle W., Wilson S. H., Kraut J. (1996) Crystal structures of human DNA polymerase β complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry 35, 12742–12761 [DOI] [PubMed] [Google Scholar]
- 6. Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 7. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 [DOI] [PubMed] [Google Scholar]
- 8. Santoso Y., Joyce C. M., Potapova O., Le Reste L., Hohlbein J., Torella J. P., Grindley N. D., Kapanidis A. N. (2010) Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. U. S. A. 107, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joyce C. M., Potapova O., Delucia A. M., Huang X., Basu V. P., Grindley N. D. (2008) Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry 47, 6103–6116 [DOI] [PubMed] [Google Scholar]
- 10. Rothwell P. J., Mitaksov V., Waksman G. (2005) Motions of the fingers subdomain of Klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell 19, 345–355 [DOI] [PubMed] [Google Scholar]
- 11. Wong J. H., Fiala K. A., Suo Z., Ling H. (2008) Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 379, 317–330 [DOI] [PubMed] [Google Scholar]
- 12. Xu C., Maxwell B. A., Brown J. A., Zhang L., Suo Z. (2009) Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 7, e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasai H. (1997) Analysis of a form of oxidative DNA damage, 8-hydroxy-2′-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res. 387, 147–163 [DOI] [PubMed] [Google Scholar]
- 14. Cadet J., Douki T., Gasparutto D., Ravanat J. L. (2003) Oxidative damage to DNA: formation, measurement, and biochemical features. Mutat. Res. 531, 5–23 [DOI] [PubMed] [Google Scholar]
- 15. Rechkoblit O., Malinina L., Cheng Y., Kuryavyi V., Broyde S., Geacintov N. E., Patel D. J. (2006) Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rechkoblit O., Malinina L., Cheng Y., Geacintov N. E., Broyde S., Patel D. J. (2009) Impact of conformational heterogeneity of oxoG lesions and their pairing partners on bypass fidelity by Y family polymerases. Structure 17, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zang H., Irimia A., Choi J. Y., Angel K. C., Loukachevitch L. V., Egli M., Guengerich F. P. (2006) Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 281, 2358–2372 [DOI] [PubMed] [Google Scholar]
- 18. Fiala K. A., Suo Z. (2004) Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43, 2106–2115 [DOI] [PubMed] [Google Scholar]
- 19. Zhang L., Brown J. A., Newmister S. A., Suo Z. (2009) Polymerization fidelity of a replicative DNA polymerase from the hyperthermophilic archaeon Sulfolobus solfataricus P2. Biochemistry 48, 7492–7501 [DOI] [PubMed] [Google Scholar]
- 20. Johnson K. A., Simpson Z. B., Blom T. (2009) Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 [DOI] [PubMed] [Google Scholar]
- 21. Krahn J. M., Beard W. A., Miller H., Grollman A. P., Wilson S. H. (2003) Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 22. Silverstein T. D., Jain R., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2010) Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase η. Structure 18, 1463–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Arora K., Schlick T. (2006) Subtle but variable conformational rearrangements in the replication cycle of Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) may accommodate lesion bypass. Protein Sci. 15, 135–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Irimia A., Loukachevitch L. V., Eoff R. L., Guengerich F. P., Egli M. (2010) Metal ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedberg E. C., Lehmann A. R., Fuchs R. P. (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell 18, 499–505 [DOI] [PubMed] [Google Scholar]