Background: PINK1 loss of function induces mitochondrial dysfunction and dopaminergic neuron loss in Drosophila.
Results: Sir2 shows a specific genetic interaction with PINK1 and rescues PINK1 null mutant phenotypes via FOXO.
Conclusion: The strong genetic and functional interactions suggest that Sir2 and FOXO protect mitochondria and dopaminergic neuron downstream of PINK1.
Significance: Understanding the molecular roles of PINK1 will be helpful for deciphering the molecular pathogenesis of Parkinson disease.
Keywords: Drosophila, Genetics, Mitochondria, Neurobiology, Signal Transduction
Abstract
PTEN-induced kinase 1 (PINK1), which is associated with early onset Parkinson disease, encodes a serine-threonine kinase that is critical for maintaining mitochondrial function. Moreover, another Parkinson disease-linked gene, parkin, functions downstream of PINK1 in protecting mitochondria and dopaminergic (DA) neuron. In our fly genetic screening, knockdown of Sir2 blocked PINK1 overexpression-induced phenotypes. Consistently, ectopic expression of Sir2 successfully rescued mitochondrial defects in PINK1 null mutants, but unexpectedly, failed in parkin mutants. In further genetic analyses, deletion of FOXO nullified the Sir2-induced mitochondrial restoration in PINK1 null mutants. Moreover, overexpression of FOXO or its downstream target gene such as SOD2 or Thor markedly ameliorated PINK1 loss-of-function defects, suggesting that FOXO mediates the mitochondrial protecting signal induced by Sir2. Consistent with its mitochondria-protecting role, Sir2 expression prevented the DA neuron loss of PINK1 null mutants in a FOXO-dependent manner. Loss of Sir2 or FOXO induced DA neuron degeneration, which is very similar to that of PINK1 null mutants. Furthermore, PINK1 deletion had no deleterious effect on the DA neuron loss in Sir2 or FOXO mutants, supporting the idea that Sir2, FOXO, and PINK1 protect DA neuron in a common pathway. Overall, these results strongly support the role of Sir2 and FOXO in preventing mitochondrial dysfunction and DA neuron loss, further suggesting that Sir2 and FOXO function downstream of PINK1 and independently of Parkin.
Introduction
Parkinson disease (PD)3 is one of the most common neurodegenerative diseases characterized by movement disorders such as rigidity, tremor, bradykinesia of the limbs, and postural instability. In addition, selective loss of dopaminergic (DA) neurons in the substantia nigra is the neuropathological hallmark of the disease (1). PD probably occurs sporadically as the result of many different environmental factors, but it could also occur genetically by mutations in a number of genes such as α-synuclein (PARK1), parkin (PARK2), PTEN-induced kinase 1 (PINK1 (PARK6)), DJ-1 (PARK7), and leucine-rich repeat kinase 2 (LRRK2 (PARK8)) (2–6). Among them, PINK1, parkin, and DJ-1 were found to be associated with early-onset autosomal recessive parkinsonism (3–5).
PINK1 is a serine-threonine kinase mainly localized to the mitochondrial membrane via an N-terminal mitochondrial targeting sequence (4). Cells from patients with a PINK1 mutation demonstrated reduced mitochondrial function when compared with controls (7). Moreover, a PINK1 deficiency in Drosophila resulted in obvious phenotypes that resemble human PD symptoms, such as locomotive defects and loss of DA neuron cells (8–10). Further examination of PINK1 mutants showed indirect flight muscle degeneration accompanied with severe reduction in ATP levels and mitochondrial mass. In addition, mitochondrial swelling occurred in indirect flight muscles and DA neurons. Further genetic analysis with the mitochondrial protein Bcl-2 demonstrated that the mitochondrial defect is the main cause of the defective phenotypes in PINK1 mutants (8).
Because the fly mutants of parkin, the most commonly affected PD gene, which encodes an E3 ubiquitin ligase (11–13), showed phenotypes remarkably similar to PINK1 mutants (14, 15), the phenotypic analysis of these mutants led to the prediction that PINK1 and Parkin act in a common pathway. Indeed, transgenic expression of Parkin dramatically suppressed all PINK1 loss-of-function phenotypes, but not vice versa, establishing that PINK1 and Parkin are linked in a linear pathway in maintaining mitochondrial integrity and function with Parkin acting downstream of PINK1 (8–10). This result initiated further investigation of the roles of PINK1 and Parkin in mitochondria. Recent fly genetics studies clearly demonstrated that these two genes regulate the mitochondrial remodeling process including mitochondrial fusion and fission (16–19). These findings strongly suggested that mitochondrial dysfunction is the key cause of PINK1-parkin-related PD pathogenesis and implied that proper maintenance and reestablishment of mitochondrial function are critical to prevent and cure Parkinson disease.
Recently, proteomics analyses found stable complex formation between PINK1 and TNF receptor-associated protein 1 (TRAP1) (20). In in vitro kinase assays, PINK1 could directly phosphorylate TRAP1. Subsequent cell biological studies suggested that TRAP1 is a key signaling molecule mediating the cell-protective action of PINK1 under oxidative stress (20). A mitochondrial protease, high temperature requirement A2 (HtrA2/Omi), was also shown to be phosphorylated in a PINK1-dependent manner (21). Further genetic analysis supported that HtrA2 acts downstream of PINK1 independently from Parkin (22). In addition, loss of Drosophila phosphoglycerate mutase 5 (PGAM5) gene successfully suppressed mitochondrial degeneration in PINK1 mutants, but failed to modulate the phenotypes induced by loss of parkin (23). Furthermore, up-regulation of DJ-1 can ameliorate PINK1, but not parkin, Drosophila mutants (24). These results suggest that PINK1 can protect mitochondria and cells using other signaling molecules in addition to Parkin.
In the present study, we found that PINK1 genetically interacts with Sir2, the nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylase, which has a neuroprotective action in animal models (25–28). Transgenic expression of Sir2 successfully rescued mitochondrial defects and ameliorated the PD-related phenotypes in PINK1 mutants. These effects of Sir2 transgene were markedly suppressed by the mutations of FOXO, the transcription factor downstream of Sirt1, a mammalian homolog of Sir2 (29–31). Further genetic analysis confirmed that the activation of Sir2-FOXO pathway is sufficient to rescue DA neuron loss and indirect flight muscle degeneration induced by PINK1 deletion, suggesting that Sir2 and FOXO act downstream of PINK1.
EXPERIMENTAL PROCEDURES
Drosophila Strains
The generation of PINK1B9, parkin1, UAS-parkin, and UAS-PINK1 was described previously (8, 15). FOXO cDNA was subcloned into the pUAST vector and microinjected into w1118 embryos. The FOXO21 and FOXO25 lines were from E. Hafen. The UAS-mitoGFP line was from H. J. Bellen. The UAS-Sir2 line was from K. T. Min. The tyrosine hydroxylase (TH)-GAL4 fly line was a gift from S. Birman. The armadillo (arm)-GAL4, eyeless (ey)-GAL4, heat shock (hs)-GAL4, UAS-SOD2, UAS-Thor, Sir2EP2300, and Sir22A-7–11 were obtained from the Bloomington Stock Center. whiteGD30033, Sir2GD23201, and Sir2KK105502 RNAi lines were purchased from the Vienna Drosophila RNAi Center (VDRC).
Climbing Assays
Climbing assays were performed as described with some modifications (15). Groups of 15 3-day-old males were transferred into climbing ability test vials and incubated for 1 h at room temperature for environmental acclimatization. After the flies were tapped down to the bottom, the numbers of the climbing flies in 10 s were counted. For each group, 10 trials were performed, and the climbing score (percentage ratio of the number of climbed flies against the total number) was obtained. The average climbing score (± S.D.) was calculated for four independent tests.
Muscle Section and TUNEL Assay
The thoraces from 3-day-old flies were embedded in Spurr's resin and sectioned as described previously (8). The serial sections were then stained with toluidine blue dye and observed with BX-50 microscope (Olympus). For TUNEL assay, apoptosis in the thoraces of 3-day-old flies was detected using the in situ cell death detection kit (Roche Applied Science). DAPI (Sigma) was used to visualize the nucleus of muscle. Fluorescence images were obtained by BX-50 microscope (Olympus).
mtDNA PCR and ATP Assay
For mitochondrial DNA (mtDNA) PCR, total DNA from five thoraces of 3-day-old flies was extracted. Then, quantitative real-time PCR was performed as described previously on a Prism 7000 real-time PCR system (Applied Biosystems) (8). Genomic DNA levels of rp49 were measured for an internal control. Results were expressed as -fold changes when compared with the control. For ATP assay, five thoraces from 3-day-old flies were dissected, and ATP measurement was performed as described previously (8). The relative ATP level was calculated by dividing the measured ATP concentration by the total protein concentration, which was determined by the bicinchoninic acid (BCA) protein assay (Sigma). In mtDNA PCR and ATP assay, average ± S.D. is from three experiments.
Immunostaining
Adult brain was fixed with 4% paraformaldehyde and stained with anti-TH rabbit antibody (1:50, Pel-Freez) as described previously (8). Brains were observed and imaged by LSM 510 confocal microscope (Zeiss) and BX-50 microscope (Olympus). mitoGFP-tagged mitochondria (over 2 μm in diameter) and TH-positive neurons were counted under blinded conditions.
Quantification and Statistical Analyses
For quantification of wing and thorax phenotypes, the percentage of defective thorax and wing phenotypes of 3-day-old males was measured (n > 200). For quantification of DA neurons, dorsolateral region 1 (DL1) clusters from 20 brains of each genotype were observed in a blind fashion to eliminate bias (n = 40). To quantify DA cells with enlarged mitochondria, we calculated the percentage of the number of DA cells with mitochondria larger than 2 μm in diameter over the total number of DA cells in DL1 clusters from 10 brains of each genotype. To obtain the average percentage of DA cells with enlarged mitochondria, we performed three replicate experiments (n = 3). To compare three or more groups, we used a one-way ANOVA with Bonferroni's correction. For two-group comparison, we used two-tailed Student's t test. All statistical significance was calculated at p = 0.05, using GraphPad Prism 5.
RESULTS
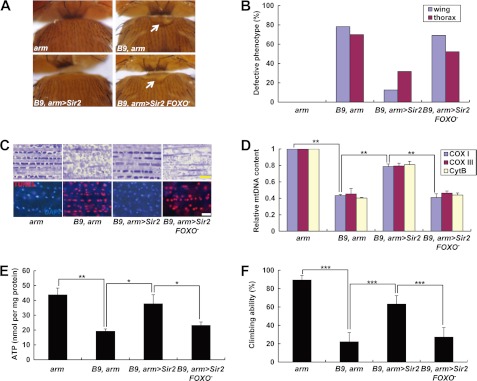
Deletion of PINK1 induced severe defects in Drosophila (8–10). Thoraces were crushed, particularly in the mid-anterior regions (Fig. 1, A and B). In the indirect flight muscle, mitochondria immensely swelled (Fig. 1C), and the levels of mtDNA and ATP were markedly reduced (Fig. 1, D and E). Locomotor activity was also severely decreased (Fig. 1F). In a genetic modifier screening to discover a novel signaling component of the PINK1 pathway, down-regulation of Sir2 highly suppressed eye size reduction and disrupted ommatidial patterns induced by PINK1 expression using an eye-specific GAL4 driver, eyeless (ey)-GAL4 (supplemental Fig. 1, A and C). Moreover, although PINK1 was deleted, overexpression of Sir2 successfully recapitulated the phenotypes induced by overexpression of PINK1 (supplemental Fig. 1, B and C). Based on the genetic interactions between these two genes, we tested whether Sir2 expression can ameliorate the various defects caused by loss of PINK1. Under the control of heat shock (hs)-GAL4, Sir2 expression induced lethality. Thus, we chose armadillo (arm)-GAL4, which induces weaker gene expression in whole body than hs-GAL4, to express Sir2 in PINK1 null mutants. The crushed thorax and downturned wing phenotypes of PINK1 null mutants were markedly rescued by Sir2 expression (Fig. 1, A and B). Muscle sections showed the intact structure of mitochondria in Sir2-expressed PINK1 null mutants (Fig. 1C and supplemental Fig. 2). Moreover, mtDNA content and ATP level in the indirect flight muscle were rescued by Sir2 expression (Fig. 1, D and E). Sir2-expressing PINK1 null mutants also showed increased climbing ability (Fig. 1F) and no TUNEL signal in the indirect flight muscle (Fig. 1C), confirming that Sir2 expression successfully abrogates the muscle degeneration and mitochondrial impairment in PINK1 null mutants. Collectively, these data demonstrated that Sir2 has an important role in regulating mitochondrial function and integrity downstream of PINK1. In contrast, Sir2 expression could not rescue the defective mitochondrial function and indirect flight muscle structure in parkin mutants (supplemental Figs. 2 and 3, A–E). Moreover, Sir2 and parkin double mutants failed to develop into adult (data not shown), suggesting that two genes downstream of PINK1, Sir2 and Parkin, are involved in different pathways to protect mitochondria.
FIGURE 1.
Expression of Sir2 rescues PINK1 null mutant phenotypes in a FOXO-dependent manner. A, light stereo micrographs of the thoraces of PINK1 null mutants (B9, arm), Sir2-expressing PINK1 null mutants (B9, arm>Sir2), and Sir2-expressing PINK1 and FOXO double mutants (B9, arm>Sir2, FOXO−). arm-GAL4/+ (arm) flies were used as wild type controls. White arrows indicate collapsed-thorax phenotypes. B, percentage of defective thorax and wing phenotypes. C, toluidine blue-stained longitudinal sections (top panels) and merged images of TUNEL (red) and DAPI (blue) staining (bottom panels) of indirect flight muscle in the thoraces. D, quantification of the mtDNA of thoraces (n = 3). Cox I, cytochrome c oxidase subunit I; Cox III, cytochrome c oxidase subunit III; Cyt B, cytochrome b. E, comparison of the ATP content of thoraces (n = 3). F, comparison of climbing ability (n = 4). Significance was determined by one-way ANOVA with Bonferroni's correction (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate mean ± S.D. Scale bars: yellow, 5 μm; white, 10 μm. Details of all the indicated genotypes in this and other figures are described in the supplemental Experimental Procedures.
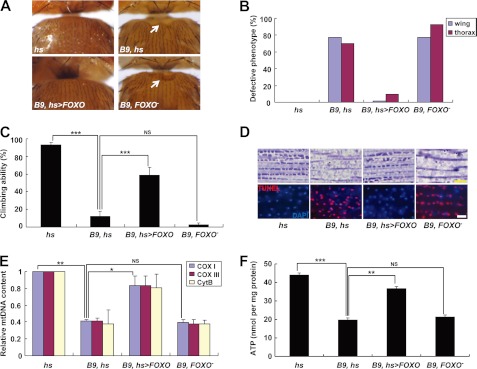
Recently, intensive genetic analyses and cell biological studies demonstrated that transcription factor FOXO mediates various physiological functions of Sir2. In Caenorhabditis elegans, DAF-16, the FOXO homolog in C. elegans, is critical in Sir2-induced neuroprotection (26). Sirt1, the mammalian Sir2 homolog, regulates cell survival and stress response signals by deacetylating FOXO (29–31). This close interaction between two genes was also found in genetic analysis using Drosophila (32). Therefore, we hypothesized that FOXO mediates the rescue of mitochondrial integrity and function by Sir2 in PINK1 null mutants. Excitingly, the null mutation of FOXO almost nullified the Sir2-mediated rescue of the defects in PINK1 null mutants (Fig. 1). Moreover, the phenotypes of PINK1 null mutants were successfully rescued by expression of FOXO. An almost complete recovery of thorax morphology and wing posture was observed after expression of FOXO in PINK1 null mutants (Fig. 2, A and B). In climbing assays, the locomotor activity of PINK1 null mutants was also rescued by FOXO expression (Fig. 2C). Muscle sections showed that FOXO expression ameliorates mitochondria disruption and apoptotic cell death in the indirect flight muscle of PINK1 null mutants (Fig. 2D and supplemental Fig. 2). Further biochemical analysis of the indirect flight muscle showed a marked rescue of the levels of mtDNA and ATP in FOXO-expressed PINK1 null mutants (Fig. 2, E and F). Overall, these results clearly demonstrated that FOXO is a critical downstream signaling molecule of Sir2 in regulating mitochondrial integrity and function downstream of PINK1. In addition, we generated and observed PINK1 and FOXO double mutants and found that FOXO mutation has no significant detrimental effect on the phenotypes of PINK1 null mutants (Fig. 2), excluding the possibility that FOXO may play mitochondrial protective roles in PINK1-independent pathways.
FIGURE 2.
FOXO expression rescues PINK1 null mutant phenotypes. A, light stereo micrographs of the thoraces of PINK1 null mutants (B9, hs), FOXO-expressing PINK1 null mutants (B9, hs>FOXO), and PINK1 and FOXO double mutants (B9, FOXO−). hs-GAL4/+ (hs) flies were used as wild type controls. White arrows indicate collapsed-thorax phenotypes. B, percentage of defective thorax and wing phenotypes. C, comparison of climbing ability (n = 4). D, toluidine blue-stained longitudinal sections (top panels) and merged images of TUNEL (red) and DAPI (blue) staining (bottom panels) of indirect flight muscle in the thoraces. E, quantification of the mtDNA of thoraces (n = 3). Cox I, cytochrome c oxidase subunit I; Cox III, cytochrome c oxidase subunit III; Cyt B, cytochrome b. F, comparison of the ATP content of thoraces (n = 3). Significance was determined by one-way ANOVA with Bonferroni's correction (*, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant). Error bars indicate mean ± S.D. Scale bars: yellow, 5 μm; white, 10 μm.
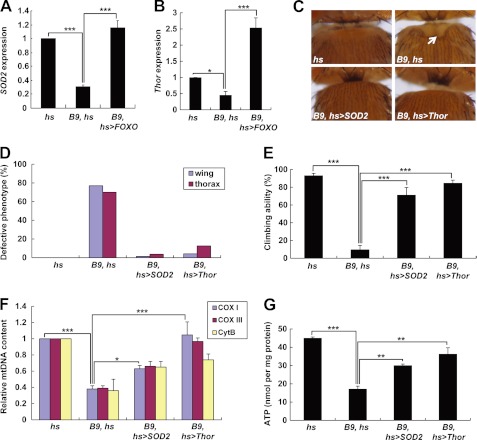
FOXO transcription factors regulate crucial cellular processes by inducing expression of various target genes (33). To find a key FOXO target gene in rescuing the mitochondrial defects in PINK1 null mutants, we checked mRNA expression of FOXO target genes in our Drosophila models using quantitative real-time RT-PCR (Fig. 3, A and B). When compared with the controls, PINK1 null mutants showed about a 3-fold reduction in expression of the mitochondrial superoxide dismutase SOD2, a FOXO target gene involved in stress resistance (Fig. 3A) (34). In addition, expression of Thor, another FOXO target gene encoding the Drosophila 4E-binding protein (4EBP) (35, 36), was reduced 2-fold in PINK1 null mutants (Fig. 3B). This reduction was completely rescued by expression of FOXO (Fig. 3, A and B). In FOXO-expressing PINK1 null mutants, gene expression of these two genes increases even more than that of controls (Fig. 3, A and B). From these results, we hypothesized that SOD2 and Thor are important mediators of the FOXO-induced mitochondrial protection and tested whether expression of them rescues PINK1 null mutant phenotypes. Excitingly, the downturned wing position and crushed thorax in PINK1 null mutants were almost completely rescued by ectopic expression of SOD2 or Thor (Fig. 3, C and D). We also observed markedly increased locomotor activities in PINK1 null mutants expressing SOD2 or Thor (Fig. 3E). mtDNA content and ATP level in the indirect flight muscle were also successfully rescued by SOD2 or Thor (Fig. 3, F and G). These data supported our hypothesis that SOD2 and Thor are critical FOXO target genes in rescuing mitochondrial dysfunction of PINK1 null mutants.
FIGURE 3.
FOXO target genes SOD2 and Thor rescue PINK1 null mutant phenotypes. A, comparison of SOD2 mRNA level in the thoraces from wild type controls (hs), PINK1 null mutants (B9, hs), and FOXO-expressing PINK1 null mutants (B9, hs>FOXO) (n = 3). B, comparison of Thor mRNA level in the thoraces (n = 3). C, light stereo micrographs of the thoraces of wild type controls (hs), PINK1 null mutants (B9, hs), SOD2-expressing PINK1 null mutants (B9, hs>SOD2), and Thor-expressing PINK1 null mutants (B9, hs>Thor). A white arrow indicates collapsed-thorax phenotypes. D, percentage of defective thorax and wing phenotypes. E, comparison of climbing ability (n = 4). F, quantification of the mtDNA of thoraces (n = 3). Cox I, cytochrome c oxidase subunit I; Cox III, cytochrome c oxidase subunit III; Cyt B, cytochrome b. G, comparison of the ATP content of thoraces (n = 3). Significance was determined by one-way ANOVA with Bonferroni's correction (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate mean ± S.D.
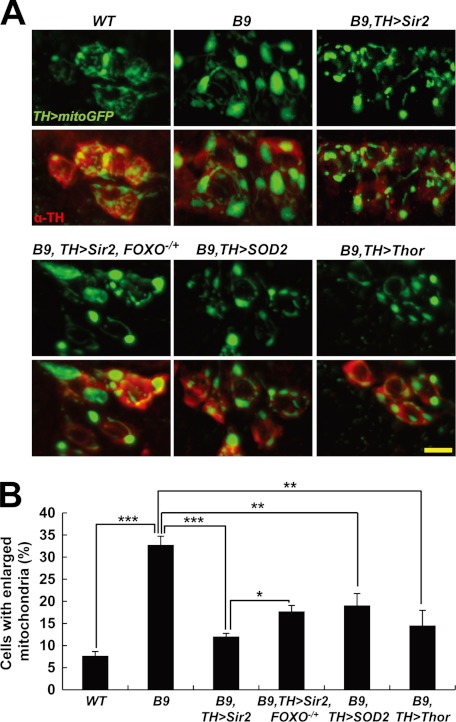
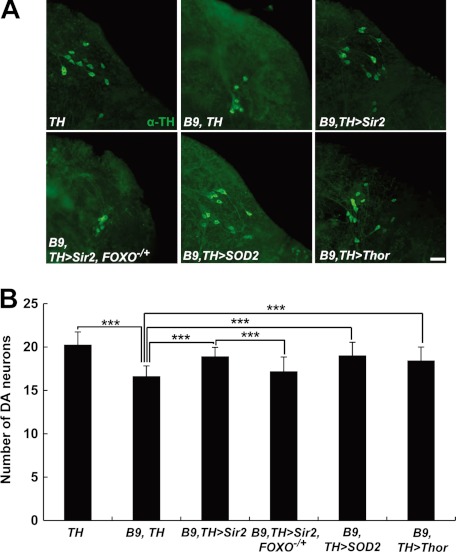
We previously reported that dopaminergic neurodegeneration, one of the major characteristics of early-onset autosomal recessive parkinsonism patients, is also observed in the brains of Drosophila PINK1 null mutants (8). Mitochondria-targeted green fluorescent protein showed enlarged mitochondria in the DA neurons of 3-day-old PINK1 null mutants (Fig. 4A). In 30-day-old flies, PINK1 null mutants exhibited a significant decrease in the number of DA neurons (Fig. 5A). In particular, the DA neurons in DL1 showed the most severe mitochondrial enlargement (8). To test the protective role of Sir2 in DA neurons, we expressed Sir2 using DA neuron-specific TH GAL4 driver (TH-GAL4) (Figs. 4 and 5). In PINK1 null mutants, about 30% of the DA neuron cells in DL1 cluster contained enlarged mitochondria similar to our previous data (Fig. 4B). After Sir2 expression, a 3-fold reduction was observed in the percentage of the DA neurons containing enlarged mitochondria (Fig. 4B). These results demonstrated that Sir2 rescues damaged mitochondria in DA neurons as well as the indirect fight muscle as shown in Fig. 1C. Moreover, reduction of FOXO gene dosage significantly suppressed the rescue activity of Sir2 in DA neuron, and expression of FOXO target genes also substantially rescued the enlarged mitochondria in DA neurons (Fig. 4). These data indicated that FOXO transcription factor is a critical downstream target of Sir2 in rescuing the damaged mitochondria of the DA neurons in PINK1 null mutants. In addition, overexpression of Sir2 also rescued the DA neuron loss in a FOXO-dependent manner (Fig. 5). Moreover, SOD2 or Thor transgene successfully prevented DA neuron loss similar to Sir2 (Fig. 5). These results demonstrated that Sir2 and FOXO are critical signaling molecules in rescuing the DA neuron degeneration and mitochondrial defects caused by PINK1 deficiency.
FIGURE 4.
Sir2 and FOXO suppress mitochondrial enlargement in DA neurons of PINK1 null mutants. A, examination of the mitochondria in DA neurons within the DL1 cluster of adult brain from wild type control (WT), PINK1 null mutants (B9), Sir2-expressing PINK1 null mutants (B9, TH>Sir2), Sir2-expressing PINK1 null mutants with a heterozygous FOXO mutation (B9, TH>Sir2, FOXO−/+), SOD2-expressing PINK1 null mutants (B9, TH>SOD2), and Thor-expressing PINK1 null mutants (B9, TH>Thor). TH-GAL4-drived expression of mitochondria-targeted green fluorescent protein (TH>mitoGFP, green) showed mitochondrial shape and size in the DA neurons stained with anti-TH antibody (red). B, graph showing the percentage of the number of DA cells with mitochondria larger than 2 μm in diameter over the total number of DA cells in DL1 clusters (n = 3). Significance was determined by one-way ANOVA with Bonferroni's correction (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate mean ± S.D. Scale bars: yellow, 5 μm.
FIGURE 5.
Sir2 and FOXO ameliorate DA neuronal degeneration in PINK1 null mutants. A, images of the DA neurons within the DL1 cluster of adult brain from wild type control (TH), PINK1 mutants (B9, TH), Sir2-expressing PINK1 null mutants (B9, TH>Sir2), Sir2-expressing PINK1 null mutants with a heterozygous FOXO mutation (B9, TH>Sir2, FOXO−/+), SOD2-expressing PINK1 null mutants (B9, TH>SOD2), and Thor-expressing PINK1 null mutants (B9, TH>Thor). DA neurons were stained with anti-TH antibody (green) B, graph showing the average number of DA neurons in DL1 clusters (n = 40). Significance was determined by one-way ANOVA with Bonferroni's correction (***, p < 0.001). Error bars indicate mean ± S.D. Scale bars: white, 20 μm.
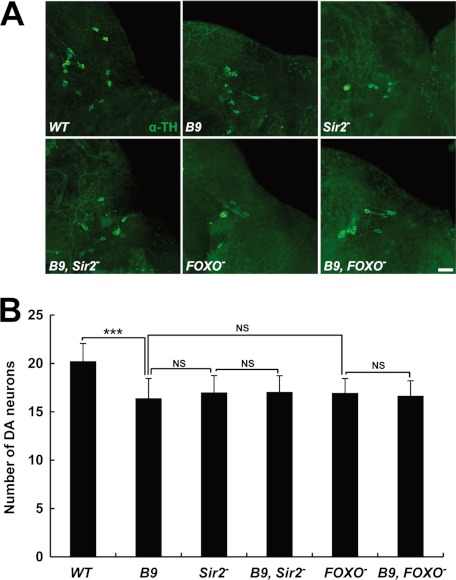
To further confirm the role of Sir2 and FOXO in DA neuron protection, we examined the number of DA neurons in the DL1 cluster of their Drosophila mutants. Interestingly, loss of Sir2 or FOXO induced DA neuron loss similar to that of PINK1 null mutants (Fig. 6), demonstrating their critical roles in protecting DA neuron. Because these three mutants were very similar in terms of the loss of DA neuron, we performed genetic analysis between them. Notably, deletion of PINK1 had no detrimental effect on the loss of DA neuron in Sir2 or FOXO mutants (Fig. 6), further supporting the idea that Sir2, FOXO, and PINK1 act in the same pathway in preventing DA neuronal degeneration.
FIGURE 6.
PINK1, Sir2, and FOXO act in same pathway in preventing DA neuron loss. A, images of the DA neurons within the DL1 cluster of adult brain from wild type control (WT), PINK1 null mutants (B9), Sir2 mutants (Sir2−), PINK1 and Sir2 double mutants (B9, Sir2−), FOXO mutants (FOXO−), and PINK1 and FOXO double mutants (B9, FOXO−). DA neurons were stained with anti-TH antibody (green). B, graph showing the average number of DA neurons in DL1 clusters (n = 40). Significance was determined by one-way ANOVA with Bonferroni's correction (***, p < 0.001; NS, not significant). Error bars indicate mean ± S.D. Scale bar: white, 20 μm.
DISCUSSION
In the genetic modifier screening and following genetic analyses, we found that Sir2 is a critical mediator of Drosophila eye phenotypes induced by PINK1 overexpression (supplemental Fig. 1). Interestingly, in contrast to PINK1, overexpression of Parkin showed normal eye phenotypes (supplemental Fig. 1). Moreover, deletion of Sir2 had no deleterious effect on PINK1 null mutants (Fig. 6), but Sir2 and parkin double mutants could not develop into adults (data not shown). These genetic interaction data suggested that Sir2 acts downstream of PINK1 in a Parkin-independent manner. Furthermore, expression of Sir2 successfully rescued the defects in the indirect flight muscle of PINK1 null mutants (Fig. 1), but failed to rescue defects in parkin mutants (supplemental Fig. 3, A–E), demonstrating specific interactions between Sir2 and PINK1.
Although Sir2 and Sirt1 shuttle between the nucleus and the cytosol, they have profound effects on mitochondrial functions (37). The specific genetic interaction between Sir2 and PINK1 in this study may provide a clue to resolve the cross-talk between mitochondria and Sir2. Also, it raises a question; how does PINK1 signal to Sir2? Because PINK1 localizes in the outer membrane of mitochondria facing the cytoplasm or in the cytoplasm, PINK1 can directly access its cytosolic targets (38). A recent proteomics study reported 13 in vivo phosphorylation sites on Sirt1 (39). For example, c-Jun N-terminal kinase 1 (JNK1), casein kinase 2 (CK2), and dual-specificity tyrosine phosphorylation-regulated kinase (DYRK) can phosphorylate Sirt1 and regulate its activity (40–42). Therefore, we suspected that PINK1 may directly phosphorylate and regulate Sir2 in the cytoplasm. However, we have not observed direct evidence for that, such as the protein-protein interaction between two molecules (data not shown). Otherwise, PINK1 may also indirectly signal to Sir2 through its upstream regulators including active regulator of Sirt1 (AROS) or deleted in breast cancer 1 (DBC1) (43–45).
Further genetic analysis identified that FOXO is a critical mediator of Sir2 in rescuing the PINK1 loss-of-function phenotypes. Deletion of FOXO almost completely abrogated the Sir2-induced rescue of mitochondrial defects in PINK1 null mutants (Fig. 1), and expression of FOXO markedly ameliorated the PINK1 null mutant phenotypes previously rescued by Sir2 (Fig. 2). These strong interactions between Sir2 and FOXO were also observed in recent studies using various animal models and cell lines (26, 29–32). Moreover, PINK1 null mutants showed a 2- or 3-fold reduction in expression of SOD2 or Thor, key FOXO target genes in mitochondrial protection (Fig. 3). Consistently, Kops et al. (34) showed that FOXO3a protects mitochondria through SOD2 during glucose deprivation. In addition, Thor extends Drosophila life span by enhancing mitochondrial activity (46), and rapamycin can suppress mitochondrial defects through stimulation of Thor, supporting our finding (47). In contrast, deletion of parkin failed to reduce the expression of these two genes (supplemental Fig. 3, F and G). Furthermore, the decreased expression of SOD2 or Thor was completely recovered by FOXO expression in PINK1 null mutants (Fig. 3, A and B), suggesting that FOXO mediates the mitochondria-protective roles of Sir2 in the PINK1 signaling pathway.
Consistent with its role in the indirect flight muscle, Sir2 also can rescue the defective mitochondria in DA neuron and the DA neuron loss in Drosophila PINK1 null mutants (Figs. 4 and 5). Further genetic analysis revealed that FOXO mediates this DA neuronal role of Sir2 in the mutants (Figs. 4 and 5). Remarkably, loss of Sir2 or FOXO induced DA neuron degeneration very similar to that of Drosophila PINK1 mutants (Fig. 6). Moreover, deletion of these genes also induced a substantial decrease in climbing ability and ATP level of the indirect flight muscle, especially in 15-day-old flies (supplemental Fig. 4). These data clearly showed that Sir2 and FOXO have protective roles in DA neuron and the indirect flight muscle, tissues with high demand for ATP generated by mitochondria (48). Because these defects were observed in 15- or 30-day-old flies, some may argue that these defects in Sir2 or FOXO mutants result from premature aging induced by loss of two genes. However, Sir2 and FOXO could rescue mitochondrial defects in only 3-day-old flies (Figs. 1 and 2). Moreover, loss of PINK1 had no additional deleterious effect on the DA neuron loss in Sir2 or FOXO mutants (Fig. 6), strongly suggesting that Sir2 and FOXO act in the same pathway at the downstream PINK1 in preventing mitochondrial dysfunction and DA neuron degeneration.
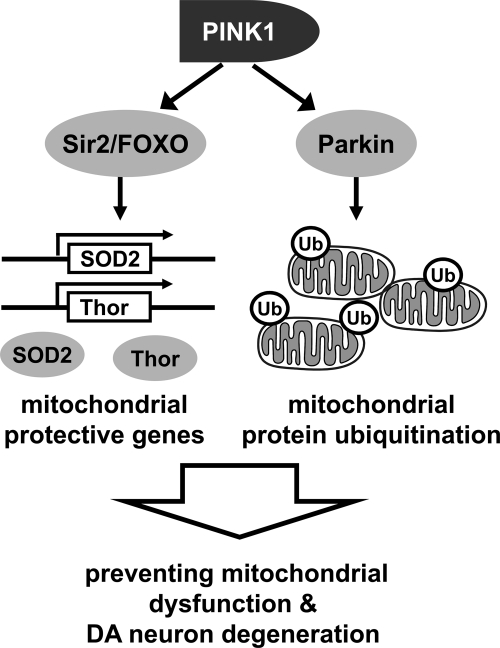
From these findings, we propose the following model for PINK1-mediated mitochondrial protection (Fig. 7). To protect mitochondria, PINK1 translocates Parkin to mitochondria and activates its E3 ubiquitin ligase activity (49–52). In mitochondria, Parkin ubiquitinates mitochondrial proteins such as voltage-dependent anion channel 1 (VDAC1) and mitofusin (Mfn) to regulate the mitochondrial remodeling process (53–55). In addition to the direct action in mitochondria, PINK1 transduces signals to the cytosol and activates Sir2. Sir2 deacetylates FOXO and induces the FOXO-dependent transcription of mitochondrial protective genes including SOD2 and Thor in the nucleus. The expressed proteins locate to the cytosol or mitochondria and play their roles such as scavenging harmful reactive oxygen species and enhancing production of mitochondrial proteins (34, 46, 47). Through the direct regulation of mitochondrial protein turnover and the induction of mitochondrial protective gene expression, PINK1 can efficiently protect cells from mitochondrial damages.
FIGURE 7.
Dual roles of PINK1 in mitochondria protection. Ub, ubiquitin.
In summary, we found novel genetic interactions between PINK1 and Sir2-FOXO pathway. In further genetic analyses, Sir2 and FOXO markedly complemented the mitochondrial defects, indirect flight muscle degeneration, and DA neuron loss in PINK1 null mutants, suggesting that Sir2 and FOXO play novel mitochondrial protective roles downstream of PINK1. Our findings provide a new perspective to the diverse molecular function of PINK1, which may help the development of more effective treatment strategies for early-onset autosomal recessive parkinsonism and possibly other forms of Parkinson disease.
Supplementary Material
Acknowledgments
We are grateful to Drs. H. J. Bellen, S. Birman, E. Hafen, and K. T. Min for flies. We also thank the members of Chung laboratory and MHRC for discussions and encouragement.
This work was supported by the National Research Foundation of Korea (NRF) (Grants 2008-0059520 and 2009-0093188) funded by the Korean government (Ministry Of Education, Science And Technology (MEST)) (to H. K.) and the National Creative Research Initiatives Program (Grant 2010-0018291) from MEST (to J. C.).

This article contains supplemental Figs. 1–4 and supplemental Experimental Procedures.
- PD
- Parkinson disease
- Sir2
- silent information regulator 2
- FOXO
- Forkhead box O
- PINK1
- PTEN-induced kinase 1
- PTEN
- phosphatase and tensin homolog
- DA
- dopaminergic
- TRAP1
- TNF receptor-associated protein 1
- TH
- tyrosine hydroxylase
- SOD
- superoxide dismutase
- DL1
- dorsolateral region 1
- ANOVA
- analysis of variance
- hs
- heat shock
- arm
- armadillo.
REFERENCES
- 1. Lang A. E., Lozano A. M. (1998) Parkinson disease: first of two parts. N. Engl. J. Med. 339, 1044–1053 [DOI] [PubMed] [Google Scholar]
- 2. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Mutation in the α-synuclein gene identified in families with Parkinson disease. Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 3. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 4. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Hereditary early-onset Parkinson disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 5. Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., van Dongen J. W., Vanacore N., van Swieten J. C., Brice A., Meco G., van Duijn C. M., Oostra B. A., Heutink P. (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259 [DOI] [PubMed] [Google Scholar]
- 6. Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson disease. Neuron 44, 595–600 [DOI] [PubMed] [Google Scholar]
- 7. Hoepken H. H., Gispert S., Morales B., Wingerter O., Del Turco D., Mülsch A., Nussbaum R. L., Müller K., Dröse S., Brandt U., Deller T., Wirth B., Kudin A. P., Kunz W. S., Auburger G. (2007) Mitochondrial dysfunction, peroxidation damage, and changes in glutathione metabolism in PARK6. Neurobiol. Dis. 25, 401–411 [DOI] [PubMed] [Google Scholar]
- 8. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 9. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 10. Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. (2006) Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imai Y., Soda M., Takahashi R. (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 [DOI] [PubMed] [Google Scholar]
- 12. Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. (2000) Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cha G. H., Kim S., Park J., Lee E., Kim M., Lee S. B., Kim J. M., Chung J., Cho K. S. (2005) Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 10345–10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng H., Dodson M. W., Huang H., Guo M. (2008) The Parkinson disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., Lu B. (2008) Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U.S.A. 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park J., Lee G., Chung J. (2009) The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem. Biophys. Res. Commun. 378, 518–523 [DOI] [PubMed] [Google Scholar]
- 20. Pridgeon J. W., Olzmann J. A., Chin L. S., Li L. (2007) PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. Plos Biol. 5, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R. J., McDonald N., Wood N. W., Martins L. M., Downward J. (2007) The mitochondrial protease HtrA2 is regulated by Parkinson disease-associated kinase PINK1. Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 22. Tain L. S., Chowdhury R. B., Tao R. N., Plun-Favreau H., Moisoi N., Martins L. M., Downward J., Whitworth A. J., Tapon N. (2009) Drosophila HtrA2 is dispensable for apoptosis but acts downstream of PINK1 independently from Parkin. Cell Death Differ. 16, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imai Y., Kanao T., Sawada T., Kobayashi Y., Moriwaki Y., Ishida Y., Takeda K., Ichijo H., Lu B., Takahashi R. (2010) The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. Plos Genet. 6, e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hao L. Y., Giasson B. I., Bonini N. M. (2010) DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc. Natl. Acad. Sci. U.S.A. 107, 9747–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Araki T., Sasaki Y., Milbrandt J. (2004) Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 26. Parker J. A., Arango M., Abderrahmane S., Lambert E., Tourette C., Catoire H., Néri C. (2005) Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet. 37, 349–350 [DOI] [PubMed] [Google Scholar]
- 27. Kim D., Nguyen M. D., Dobbin M. M., Fischer A., Sananbenesi F., Rodgers J. T., Delalle I., Baur J. A., Sui G., Armour S. M., Puigserver P., Sinclair D. A., Tsai L. H. (2007) SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvári M., Piper M. D., Hoddinott M., Sutphin G. L., Leko V., McElwee J. J., Vazquez-Manrique R. P., Orfila A. M., Ackerman D., Au C., Vinti G., Riesen M., Howard K., Neri C., Bedalov A., Kaeberlein M., Soti C., Partridge L., Gems D. (2011) Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 30. Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A. (2004) Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 10042–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van der Horst A., Tertoolen L. G., de Vries-Smits L. M., Frye R. A., Medema R. H., Burgering B. M. (2004) FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2SIRT1. J. Biol. Chem. 279, 28873–28879 [DOI] [PubMed] [Google Scholar]
- 32. Griswold A. J., Chang K. T., Runko A. P., Knight M. A., Min K. T. (2008) Sir2 mediates apoptosis through JNK-dependent pathways in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 8673–8678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Accili D., Arden K. C. (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 34. Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 35. Puig O., Marr M. T., Ruhf M. L., Tjian R. (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jünger M. A., Rintelen F., Stocker H., Wasserman J. D., Végh M., Radimerski T., Greenberg M. E., Hafen E. (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finley L. W., Haigis M. C. (2009) The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 8, 173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. (2008) The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 105, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) Phosphorylation regulates SIRT1 function. Plos One 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. Plos One 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kang H., Jung J. W., Kim M. K., Chung J. H. (2009) CK2 is the regulator of SIRT1 substrate binding affinity, deacetylase activity, and cellular response to DNA damage. Plos One 4, e6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo X., Williams J. G., Schug T. T., Li X. (2010) DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim E. J., Kho J. H., Kang M. R., Um S. J. (2007) Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell 28, 277–290 [DOI] [PubMed] [Google Scholar]
- 44. Kim J. E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 45. Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., Benzer S., Kapahi P. (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tain L. S., Mortiboys H., Tao R. N., Ziviani E., Bandmann O., Whitworth A. J. (2009) Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 12, 1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park J., Kim Y., Chung J. (2009) Mitochondrial dysfunction and Parkinson disease genes: insights from Drosophila. Dis. Model. Mech. 2, 336–340 [DOI] [PubMed] [Google Scholar]
- 49. Kim Y., Park J., Kim S., Song S., Kwon S. K., Lee S. H., Kitada T., Kim J. M., Chung J. (2008) PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 377, 975–980 [DOI] [PubMed] [Google Scholar]
- 50. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. Plos Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 54. Ziviani E., Tao R. N., Whitworth A. J. (2010) Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poole A. C., Thomas R. E., Yu S., Vincow E. S., Pallanck L. (2010) The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. Plos One 5, e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.