Background: The mechanism involved in mitochondrial-nuclear translocation of AIF is unclear.
Results: SIP interacts with AIF in mitochondria and inhibits apoptosis. Apoptotic stimuli lead SIP degradation and AIF translocation to the nucleus, inducing apoptosis.
Conclusion: SIP is a novel regulator of apoptosis.
Significance: This work adds to our understanding of the biological function of SIP and the regulatory mechanism of programmed cell death.
Keywords: Apoptosis, Cancer Biology, Cell Proliferation, Mitochondria, Protein Degradation
Abstract
Apoptosis-inducing factor (AIF) is a caspase-independent death effector. Normally residing in the mitochondrial intermembrane space, AIF is released and translocated to the nucleus in response to proapoptotic stimuli. Nuclear AIF binds to DNA and induces chromatin condensation and DNA fragmentation, characteristics of apoptosis. Until now, it remained to be clarified how the mitochondrial-nuclear translocation of AIF is regulated. Here we report that steroid receptor coactivator-interacting protein (SIP) interacts directly with AIF in mitochondria and specifically inhibits caspase-independent and AIF-dependent apoptosis. Challenging cells with apoptotic stimuli leads to rapid degradation of SIP, and subsequently AIF is liberated from mitochondria and translocated to the nucleus to induce apoptosis. Together, our data demonstrate that SIP is a novel regulator in caspase-independent and AIF-mediated apoptosis.
Introduction
Apoptosis, or the programmed cell death, is an important physiological process that enables an organism to tightly control cell numbers and tissue size and to protect itself from rogue cells that threaten the well-being of the organism. Appropriate apoptosis is essential for animal development and homeostasis. Apoptosis is also triggered by various excitotoxins and functions as a safeguard mechanism to remove cells with deleterious mutations. Dysregulation of apoptosis leads to tumor initiation, progression, or metastasis. Apoptotic phenotypes include shrinkage of the cell, condensation of the chromatin, and disintegration of the cell into small fragments (1). In classical caspase-dependent apoptosis pathway, apoptosis insults induce the permeabilization of the outer mitochondrial membrane (MOMP),2 leading to cytochrome c release from mitochondria and subsequent activation of Apaf-1/caspase 9/caspase 3 (2). Alternatively, apoptosis-inducing factor (AIF), which normally resides in the mitochondrial intermembrane space, is translocated to the nucleus and causes chromatin condensation and large scale DNA fragmentation. The AIF-mediated cell death is mainly caspase-independent (3, 4).
In healthy cells, AIF is a mitochondrial FAD-dependent oxidoreductase that functions in oxidative phosphorylation and redox metabolism (5–8). Various apoptosis stimuli such as serum withdrawal (9), staurosporin (3, 10, 11), and N-methyl-N-nitroso-N′-nitroguanidine (MNNG) (12, 13) trigger AIF to be released from mitochondria and move to the nucleus. AIF exhibits an isoelectric point that is close to that of DNA-binding histones and binds DNA in a sequence-independent manner (4). AIF itself lacks nuclease activity; thus AIF-triggered DNA degradation depends on its associated proteins such as EndoG in Caenorhabditis elegans (14) and cyclophilin A in Homo sapiens (15).
The mechanism responsible for AIF release from mitochondria remains to be clarified. According to some reports, AIF is first synthesized as a 67-kDa precursor molecule (613 amino acid residues) in the cytosol and then is imported to the mitochondria by cleaved off the first 101 amino acid residues (3). This process gives rise to a 57-kDa molecule, which is also called the mature AIF (3). Mature AIF is soluble in the intermembrane space of mitochondria, and MOMP would be sufficient to release this form of AIF from the mitochondria (2, 3, 11). However, other reports suggested that mature AIF is a 62-kDa protein harboring a transmembrane region (residues 66–84), through which AIF is anchored in the inner membrane of mitochondria (16, 17). Calpain I, a calcium-dependent cysteine protease, has been suggested to be responsible for the cleavage and release of AIF from the mitochondrial inner membrane upon apoptosis induction (18, 19). However, it remains debatable whether calpain I is essential for the mitochondrial release of AIF (20). It has also been reported that overactivation of poly(ADP-ribose) polymerase 1 (PARP-1) and subsequent PAR polymer formation are associated with mitochondrial AIF release (13, 21, 22). On the other hand, heat shock protein 70 (HSP70) has been shown to antagonize AIF-mediated apoptosis by binding AIF in the cytosol and restraining the nuclear import of AIF (23–25).
In current study, we showed that steroid receptor coactivator (SRC)-interacting protein (SIP) (26) interacts with AIF in mitochondria and prevents its release under normal conditions. We demonstrated that SIP specifically inhibits caspase-independent and AIF-dependent cell apoptosis.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies used were: SIP, Tublin, FLAG (M2), and β-actin (Sigma); PARP-1 and pro-caspase 3 (Abcam); Tim23 (Active Motif); rabbit polyclonal antibodies against the AIF C-terminal were obtained from Upstate. Staurosporine and MG132 were from Sigma. Z-VAD-FMK was from Promega; FITC annexin V apoptosis detection kit I was from BD Pharminge. The mitochondrial isolation kit for mammalian cells was from Thermo Scientific. Control siRNA and siRNAs for SIP and AIF were synthesized by Shanghai GeneChem Inc. (Shanghai, China). The sequence for control siRNA was: 5′-UUCUCCGAACGUGUCACGU-3′. The sequences for SIP siRNA were: 5′-GCUACCAGCAACGUCCAUA-3′, 5′-GACAACGACAGCACAGAGA-3′, and 5′-GGAGAUCAGGAUGGAGCUA-3′. The sequences for AIF siRNA were 5′-GCGAUUCAAACAGUGGAAU-3′, 5′-CACAGUGGAAUUGGCAAAC-3′, and 5′-UGGUGGCUUCCGGGUAAAU-3′. The sequence for GAPDH was 5′-GUGGAUAUUGUUGCCAUCA-3′.
Plasmid Construction
The coding region of AIF was amplified in a human mammary gland library (Clontech) by PCR with primers 5′-CCGGAATTCATGTTCCGGTGTGGAGGC-3′ (forward) and 5′-ATAAGAATGCGGCCGCCAGTCTTCATGAATGTTGAA-3′ (reverse) and cloned into the pcDNA3.1 vector (Invitrogen) to generate a plasmid named pcDNA3.1-AIF. To generate GFP fusion construct, the entire coding region of AIF was amplified by PCR using primers 5′-CCGGAATTCATGTTCCGGTGTGGAGGCCTG-3′ (forward) and 5′-CGGGGTACCGTGTCTTCATGAATGTTGAA-3′ (reverse). The PCR product was digested and cloned into the pEGFP-N1 expression vector (Clontech). The resultant construct was named pEGFP-AIF. SIP and SIP deletion mutants were described previously in our paper.
Cell Culture and Cell Apoptosis Induction
U2OS and MCF-7 cells were from ATCC. To induce apoptosis, the cells were treated with staurosporin (STS; 1 μm), with MNNG (500 mm), or with serum-free medium or with UV irradiation. In MNNG experiments, the treating medium was replaced after 15 min of incubation by fresh medium devoid of MNNG, and the cells (80% confluence) were cultured for the appropriate times. In UV treatment, before UV irradiation, the cells were washed twice with prewarmed PBS and were exposed to UVC (254 nm) at a dose of 40 J/m2 with a Stratalinker 2400 UV cross-linker (Stratagene, La Jolla, CA) or were mock treated and incubated for the indicated periods of time. Z-VAD-FMK (40 mm) was added to the culture medium 30 min before induction of apoptosis.
Immunoprecipitation and Western Blotting
These experiments were performed as described previously (27–35). Briefly, U2OS cells lysates were prepared by incubating cells in the lysis buffer (20 mm Tris-HCl, pH 7.5, 1% Triton X-100, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 5% glycerol) containing protease inhibitor mixture for 20 min at 4 °C, followed by centrifugation at 14,000 × g for 15 min at 4 °C. The protein concentration of the lysates was determined using the BCA protein assay kit according to the manufacturer's protocol (Pierce). For immunoprecipitation, 500 μg of protein was incubated with a specific antibodies (1–2 μg) for 2 h at 4 °C with constant rotation; 50 μl of 50% protein A- and G-agarose bead mixtures were then added, and the incubation was continued for an additional 2 h. The beads were then washed five times using the lysis buffer. Between washes, the beads were collected by centrifugation at 3000 × g for 30s at 4 °C. The precipitated proteins were eluted from beads by resuspending the beads in 2× SDS-PAGE loading buffer and boiling for 5 min. The resulting materials from immunoprecipitation or cell lysates for detecting protein levels were separated using 10% or 12% SDS-PAGE and transferred onto nitrocellulose membranes. For Western blot analysis, the membranes were first blocked using 5% nonfat dry milk in TBS-T buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20) for 1 h at room temperature and then blotted using appropriate antibodies that were diluted in TBS-T buffer for 1 h at room temperature or overnight at 4 °C followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Immunoreactive bands were visualized using Western blotting Luminol reagent (Santa Cruz Biotechnology) according to the manufacturer's recommendation.
GST Pulldown Assays
These experiments were performed as described previously (36, 37). Briefly, GST fusion constructs were expressed in BL21 Escherichia coli cells, and crude bacterial lysates were prepared by sonication in TEDGN (50 mm Tris-HCl, pH 7.4, 1.5 mm EDTA, 1 mm dithiothreitol, 10% (v/v) glycerol, 0.4 m NaCl) in the presence of the protease inhibitor mixture. In vitro transcription and translation experiments were done with rabbit reticulocyte lysate (TnT system; Promega, Madison, WI), according to the manufacturer's recommendations. In GST pulldown assays, about 10 μg of the appropriate GST fusion proteins was mixed with 15 μl of the in vitro transcribed/translated products and incubated in binding buffer (75 mm NaCl, 50 mm HEPES, pH 7.9) at room temperature for 30 min in the presence of the protease inhibitor mixture. The binding reaction was then added to 40 μl of glutathione-Sepharose beads and mixed at 4 °C for 2 h. The beads were washed three times with binding buffer, separated on 10% SDS-PAGE, and analyzed by Western blotting.
Fluorescence Confocal Microscopy
HeLa cells were transfected with appropriate plasmids using Lipofectamine 2000. Twenty-four hours after transfection, the cells were washed with PBS, fixed in 4% paraformaldehyd, and permeablized with 1% Triton X-100. The cells were washed for four times, and a final concentration of 0.1 μg/ml 4,6-diamidino-2-phenylindole dihydrochloride (Sigma) was included in the final wash to stain the nuclei. The images were visualized with an Olympus inverted microscope equipped with a charge-coupled camera. The resulting images were deconvolved with Deltavision software. For immunostaining, 24 h after transfections, the cells were washed with PBS, fixed with 4% paraformaldehyde, and incubated with appropriate primary antibodies followed by the addition of rhodamine-conjugated secondary antibodies.
Flow Cytometry
Annexin V-FITC apoptosis detection kit I was used for the assessment of PS exposure; propidium iodide (PI, 0.5 mg/ml) was used for cell viability. Cell apoptosis was recorded in FACSCanto II (BD Biosciences) in total population (10,000 cells/nuclei).
RESULTS
SIP Inhibits Apoptosis through Caspase-independent Pathway
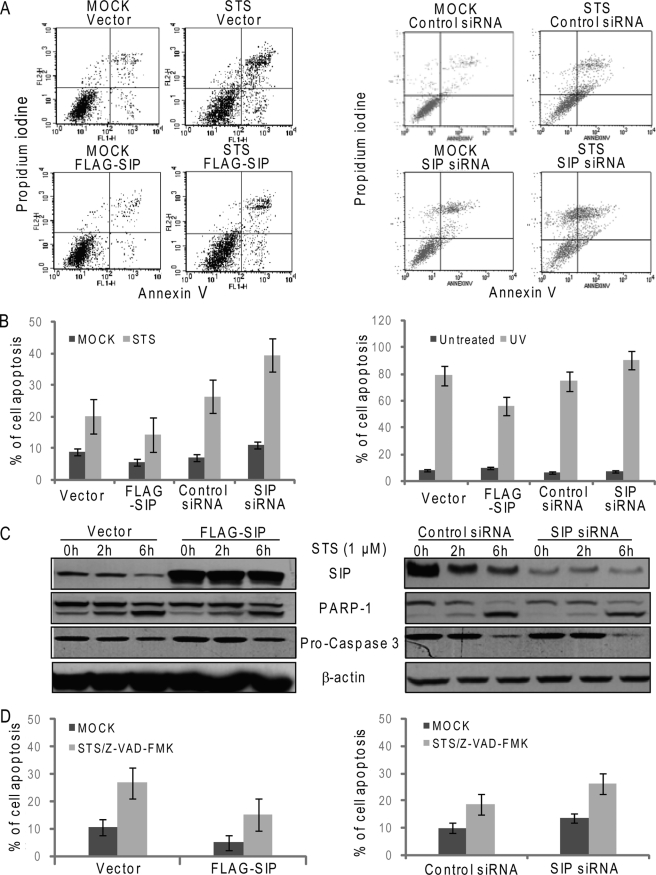
We have previously identified SIP, a SRC-interacting protein that is able to sequester SRC coactivators in the cytoplasm and inhibit estrogen-stimulated gene transcription and cell proliferation (26). To further explore the functional role of SIP in cell growth and proliferation, osteosarcoma U2OS cells were transfected with SIP expression construct or SIP-specific siRNAs, and the cells were then treated with apoptosis inducer STS and analyzed by flow cytometry. Compared with control cells, overexpression of SIP resulted in a decreased number of apoptotic cells, whereas silencing the expression of SIP led to an increase of apoptotic cells (Fig. 1A). The inhibitory effect of SIP on apoptosis treatment was also validated in MCF-7 cells (Fig. 1B, left panels) and U2OS cells exposed to UV irradiation (Fig. 1B, right panels). Because STS has been reported to induce both caspase-dependent and independent apoptosis (38), to investigate whether the anti-apoptotic effect of SIP is through regulation of the classical caspase-dependent pathway, U2OS cells were transfected with SIP expression construct or SIP-specific siRNAs and treated with 1 μm STS for various lengths of time. Cellular lysates were then collected for the measurement of the activation of caspase 3 and cleavage of PARP-1 by Western blotting. The results indicated that changes of SIP protein levels did not alter the levels of caspase 3 activation and PARP-1 cleavage (Fig. 1C), suggesting that SIP-regulated apoptosis did not involve caspase-mediated pathway. To examine whether SIP regulates apoptosis through a caspase-independent pathway, U2OS cells were transfected with SIP expression construct or SIP-specific siRNAs and treated with Z-VAD-FMK, a pan-caspase inhibitor, before apoptosis induction with STS. The experiments revealed that SIP inhibited STS-induced apoptosis to the same extent, despite the inhibition of caspase activity (Fig. 1D), indicating that SIP antagonizes apoptosis through a caspase-independent pathway.
FIGURE 1.
SIP inhibits apoptosis through caspase-independent mechanism. A, FACS analysis of apoptosis by double staining with annexin V and propidium iodide. U2OS cells were transfected with SIP expression construct or SIP siRNA. Forty-eight hours after transfection, the cells were untreated (control) or treated with STS (6 h), labeled with annexin V-FITC and PI, and analyzed by flow cytometry. Representative cytofluorometric plots are shown. B, FACS analysis of apoptosis in MCF-7 cells (left panel) and U2OS cells treated with UV irradiation (right panel). MCF-7 cells described in A or U2OS cells unexposed (control) or exposed to 40 J/m2 UV for 1 min, 7 h later, were labeled with annexin V-FITC and PI and analyzed by flow cytometry. The values are expressed as percentages of annexin V-positive versus total cells. The data are the means ± S.D. from triplicate experiments. C, Western analysis of apoptosis. Total cell lysates from U2OS described in A were prepared, and the expression levels of SIP and activated PARP-1 and caspase-3 were examined by immunoblotting. β-actin was used as a loading control. D, FACS analysis of apoptosis. U2OS described in A in the absence or presence of Z-VAD-FMK were untreated (control) or treated with STS (6 h), labeled with annexin V-FITC and PI, and analyzed by flow cytometry. The values are expressed as percentages of annexin V-positive versus total cells. The data are the means ± S.D. from triplicate experiments.
SIP Interacts with AIF in Vivo and in Vitro
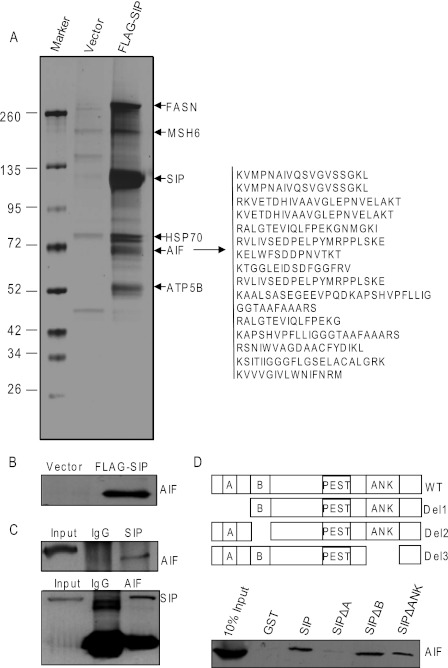
To gain mechanistic insights into the role of SIP in the inhibition of STS-induced apoptosis, we next employed affinity purification and mass spectrometry methods to identify SIP-associated proteins that may be implicated in the regulation of apoptosis. Lysates from HeLa cells that had been stably transfected with FLAG-tagged SIP (FLAG-SIP) were immunoprecipitated with anti-FLAG or control antibody, and bound proteins were eluted with FLAG peptide, resolved by SDS-PAGE, and visualized by silver staining (Fig. 2A). SIP-associated proteins were retrieved and analyzed by mass spectrometry. These experiments revealed the association of several proteins including FASN (fatty acid synthase), MSH6 (mutS homolog 6), ATP5B, and HSP70 with SIP. Interestingly, 15 peptides matching to AIF were also identified among the SIP-interacting proteins. Western blotting using AIF-specific antibody further confirmed the results (Fig. 2B).
FIGURE 2.
SIP associates with AIF in vivo and in vitro. A, immunoaffinity purification of SIP-interacting proteins. Cellular extracts from HeLa cells stably expressing FLAG-SIP were immunopurified with anti-FLAG affinity columns and eluted with FLAG peptide. The eluates were resolved by SDS-PAGE and silver-stained. The proteins bands were retrieved and analyzed by mass spectrometry. B, Western blotting analysis of the purified fractions using antibody against AIF. C, coimmunoprecipitation of SIP and AIF. Whole cell lysates from HeLa cells were prepared, and immunoprecipitation was performed with anti-SIP followed by immunoblotting with antibodies against AIF (upper panel) or immunoprecipitated with antibodies against AIF followed by immunoblotting with anti-SIP (lower panel). D, in vitro interaction between SIP and AIF. GST pulldown assays were performed with GST, GST-SIP, GST-SIP-Del 1, GST-SIP-Del 2, or GST-SIP-Del 3 and in vitro transcribed/translated AIF. Schematic diagrams of the deletion constructs are shown.
To further confirm the in vivo interaction between SIP and AIF, coimmunoprecipitation experiments were performed with HeLa cell extracts. Immunoprecipitation with antibodies against SIP followed by immunoblotting with antibodies against AIF demonstrated that AIF was indeed coimmunoprecipitated with SIP. Reciprocally, immunoprecipitation with antibodies against AIF and immunoblotting with antibodies against SIP also revealed that SIP interacted with AIF in vivo (Fig. 2C). Next, we performed GST pulldown assays to examine the molecular detail of the interaction between SIP and AIF. In these experiments, wild type SIP and its deletion mutants were expressed as GST fusion proteins, and AIF was transcribed/translated in vitro. The results of the GST pulldown experiments indicated that SIP and AIF interacted directly and that the coiled-coiled domain in SIP is responsible for the interaction with AIF (Fig. 2D).
Subcellular Localization of SIP and AIF
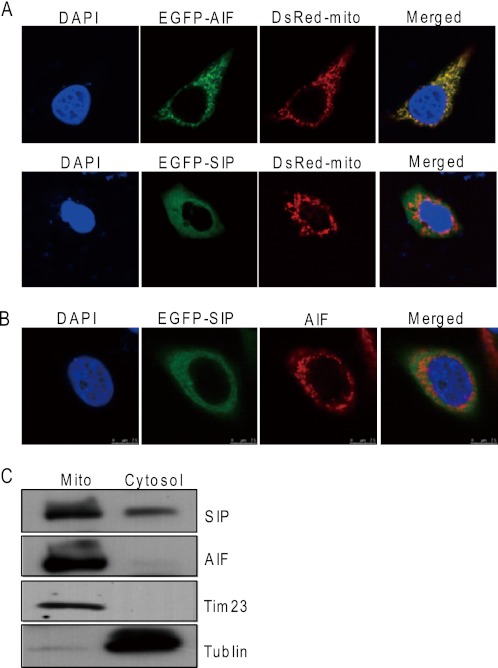
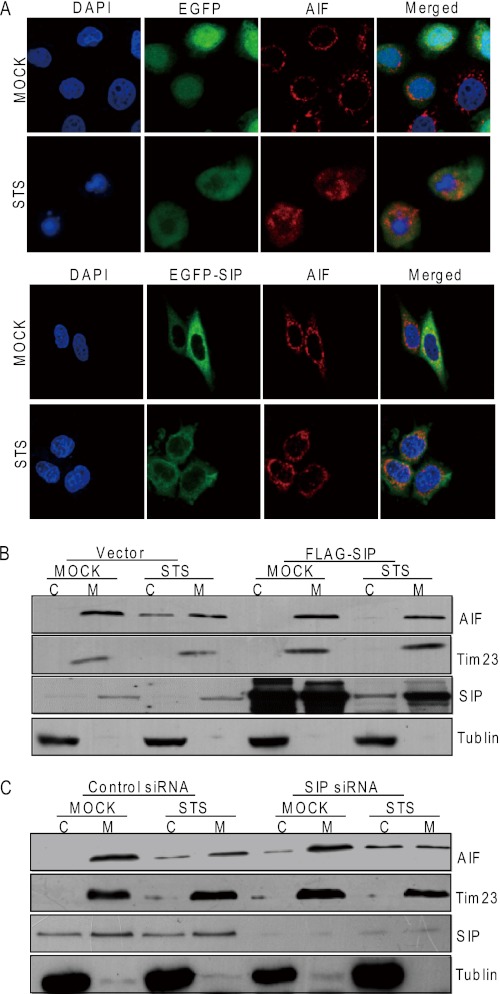
Our previous study indicated that that SIP is mainly a cytoplasmic protein (26). The association of SIP and AIF prompted us to investigate whether SIP could be also localized in mitochondria, because AIF is a mitochondrial protein. For this purpose, EGFP-AIF and DsRed-mito (a mitochondrial marker) or EGFP-SIP and DsRed-mito were cotransfected into HeLa cells. Confocal microscopy examination indicated that the intracellular localization of AIF was mitochondria-specific, whereas SIP showed diffused distribution in the cytoplasm, including in the mitochondria (Fig. 3A). Transfection of HeLa cells with GFP-AIF construct and immunostaining with anti-AIF revealed a partial overlap of the cellular distribution of SIP and AIF (Fig. 3B), suggesting that SIP and AIF could be colocalized in the mitochondria. We also employed subcellular fractionation assay to further examine the subcellular distribution of SIP. Western blotting using SIP-specific antibodies showed that a large amount of SIP appeared in the mitochondrial fraction (Fig. 3C).
FIGURE 3.
Subcellular localizations of SIP and AIF. A, subcellular localizations of SIP protein. pEGFP-AIF and DsRed-mito or pEGFP-SIP and DsRed-mito were cotransfected into HeLa cells. EGFP and DsRed fluorescence were visualized by fluorescence microscopy. 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI) staining was also included to visualize the cell nucleus. B, subcellular colocalization of SIP and AIF. HeLa cells were transfected with pEGFP-SIP. Twenty-four hours after transfections, EGFP fluorescence and rhodamine staining of AIF were visualized by fluorescence microscopy. C, analysis of subcellular localization of SIP by subcellular fractionation. HeLa cells were subjected to subcellular fractionation, and immunoblotting was performed with cytoplasm (Cytosol) and mitochondrial (Mito) fractions. Tublin and Tim23 were used as cytosolic and mitochondrial marker proteins, respectively.
SIP Inhibits Apoptosis in AIF-dependent Manner
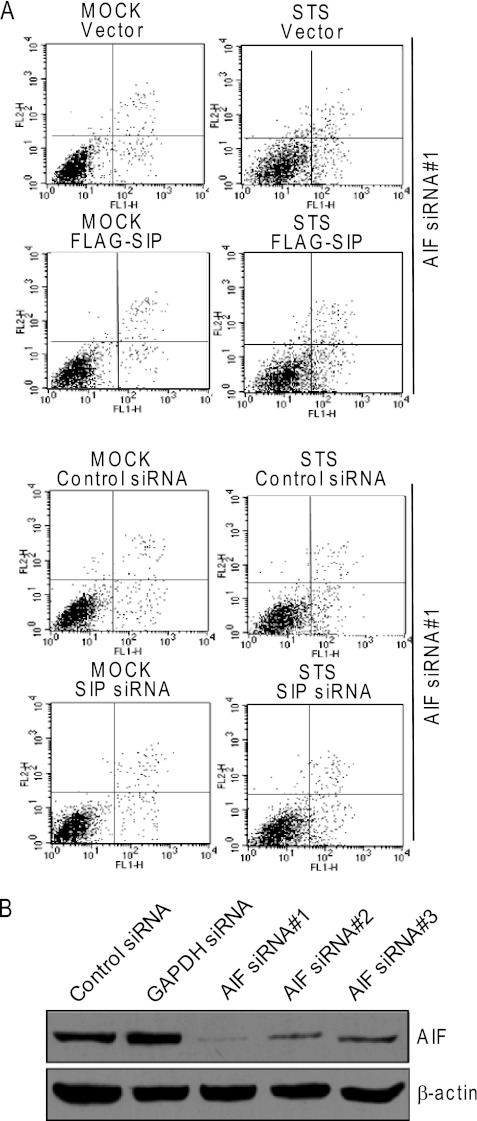
To investigate the biological significance of the physical interaction between SIP and AIF, SIP was overexpressed or knocked down in AIF-depleted U2OS cells, and the effect of gain-of-function and loss-of-function of SIP on STS-induced apoptosis in these cells was examined with annexin V and propidium iodide double staining and flow cytometry. In agreement with the experimental data described in Fig. 1A, overexpression of SIP was associated with a significant reduction of STS-induced apoptosis, and knockdown of SIP resulted in a significant elevation of STS-induced apoptosis of U2OS cells. However, in AIF-depleted cells, either overexpression or knockdown of SIP had only a limited effect in STS-induced apoptosis (Fig. 4A). These experiments clearly indicate that AIF is required for SIP-inhibited apoptosis. Notably, knockdown of the expression of AIF led to a significant reduction of apoptotic cell number under the treatment of STS (Fig. 4A). The knockdown efficiency was examined by Western blotting, and the most effective siRNAs were chosen for the experiments (Fig. 4B).
FIGURE 4.
SIP inhibits apoptosis in AIF-dependent manner. A, FACS analysis of apoptosis of AIF-depleted U2OS. AIF-depleted U2OS cells were transfected with SIP expression construct or SIP siRNA. Forty-eight hours after transfection, the cells were untreated (control) or treated with STS (6 h), labeled with annexin V-FITC and PI, and analyzed by flow cytometry. Representative cytofluorometric plots are shown. B, the knockdown efficiency of AIF. U2OS cells were transfected with three pairs of AIF siRNA. Forty-eight hours after transfection, the total cell lysates from U2OS cells were examined by Western blotting analysis of the protein expression of AIF. β-actin was used as a loading control.
SIP Prevents AIF from Being Released from Mitochondria
The mitochondrial-to-nuclear translocation of AIF is a key event in AIF-mediated caspase-independent cell apoptosis (3, 10, 11). Therefore, to gain insight into how SIP inhibits AIF-mediated apoptosis, we examined whether SIP could affect the subcellular localization of AIF. HeLa or U2OS cells transfected with SIP or an empty vector were treated with STS. The relative mitochondria/cytoplasm distribution of SIP and AIF was evaluated by both immunofluorescence microscopy (Fig. 5A) and cellular fractionation assays (Fig. 5B). In control cells, STS treatment resulted in the release of AIF from mitochondria to the cytoplasm, whereas overexpression of SIP inhibited AIF release from mitochondria. On the other hand, silencing SIP by specific siRNAs resulted in elevated release of AIF from mitochondria when cells were treated with STS (Fig. 5C). These experiments support the hypothesis that SIP functions to prevent AIF from being released from mitochondria.
FIGURE 5.
SIP prevents AIF from being translocated into the nucleus. A, subcellular localization of AIF under SIP overexpression. HeLa cells were transfected with pEGFP and pEGFP-SIP. Twenty-four hours after transfection, EGFP fluorescence and rhodamine staining of AIF were visualized by fluorescence microscopy in the presence or absence of STS treatment. B, analysis of AIF translocation by subcellular fractionation. U2OS cells were transfected with vector or SIP expression construct. Forty-eight hours after the transfections, the cells were untreated (control) or treated with STS (6 h) and then were subjected to subcellular fractionation into cytosolic and mitochondrial fractions. Tim23 and Tublin were probed as mitochondrial and cytosolic markers, respectively. C, U2OS cells were transfected with control siRNA or specific SIP siRNA. Forty-eight hours after the transfections, cells were untreated (control) or treated with STS (6 h) and then were subjected to subcellular fractionation into cytosolic (lanes C) and mitochondrial (lanes M) fractions. Tim23 and Tublin were probed as mitochondrial and cytosolic markers, respectively. DAPI, 4,6-diamidino-2-phenylindole dihydrochloride.
SIP Is Degraded in Response to Apoptosis Stimuli
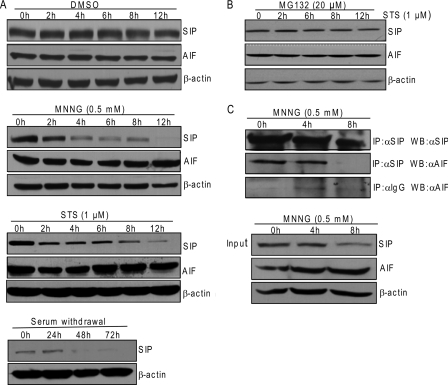
In our initial effort to investigate whether SIP plays a role in STS-induced apoptosis, we found that the protein levels of SIP decreased when cells were incubated with STS (Fig. 1C, right panels). This observation raised the possibility that SIP is degraded in response to apoptotic stimuli. To test this hypothesis, U2OS cells were challenged with different types of apoptotic stimuli, and the protein levels of SIP and AIF were examined under these conditions. As shown in Fig. 6A, various apoptotic stimuli such as MNNG (12, 13), STS, and serum withdrawal were associated with decreased protein levels of SIP in cells, whereas the protein levels of AIF remained unchanged. Cells treated with MG132, a potent proteasome inhibitor, stabilized SIP protein (Fig. 6B), indicating that proteasome-dependent protein degradation is responsible for the decrease of SIP upon induction of apoptosis. Degradation of SIP resulted in decreased association of SIP with AIF, as examined by coimmunoprecipitation experiments in MNNG-treated U2OS cells (Fig. 6C). These data support the notion that upon apoptotic stimuli, SIP is rapidly degraded and the association of SIP with AIF is attenuated, resulting in the liberation of AIF from mitochondria.
FIGURE 6.
SIP is degraded in response to apoptosis stimulus. A, Western blotting analysis of SIP expression during DMSO, STS, MNNG, or serum withdrawal-induced apoptosis. HeLa cells treated with DMSO, STS, MNNG, or serum withdrawal were collected at the indicated times. Cell lysates were prepared and analyzed by Western blotting for SIP, AIF, or β-actin. B, the degradation of SIP triggered by apoptosis stimulus can be blocked by MG132. HeLa cells were first treated with MG132 for 1 h and then treated with STS at the indicated times. Cell lysates were prepared and analyzed by Western blotting for SIP, AIF, or β-actin. C, interaction between SIP and AIF during apoptosis. U2OS cells were treated with MNNG at the indicated times. Whole cell lysates were first immunoprecipitated (IP) with anti-SIP or anti-IgG and then immunoblotted with anti-AIF or anti-SIP. Inputs from each sample were immunoblotted with anti-SIP, anti-AIF, or anti-β-actin.
DISCUSSION
Apoptosis-inducing factor AIF is normally confined to the intermembrane space of mitochondria. However, it can be liberated from mitochondria to participate in cell death-related processes in the nucleus (3, 11). Therefore, understanding the mechanisms controlling the subcellular localization of AIF becomes an issue of great importance. It was reported that the apoptotic release of AIF from mitochondria requires the inducible activity of an unknown protease and MOMP (3, 39). Indeed, it was demonstrated that BCL-2, a MOMP inhibitor, can prevent the translocation of AIF from mitochondria (3, 40, 41). Subsequently, it was shown that calpain I, a calcium-dependent cysteine protease of the calpain family, is responsible for cleavage and release of AIF from mitochondria (18, 19). However, it remains questionable whether or not calpain I is essential for mitochondrial release of AIF (20). It was also reported that releasing AIF from mitochondria could be triggered by activation of PARP-1, a nuclear enzyme that is activated by oxidative stress and DNA damage (13). However, the underlying mechanism remains unknown. After release from the mitochondria, AIF is translocated from the cytosol into the nucleus. This step is subject to another level of control: the inducible HSP70 binds to AIF in the cytosol, preventing it from being translocated to the nucleus (23, 24). Cyclophilin A, on the other hand, aids the lethal translocation of AIF to the nucleus (42).
Here we report that SIP, an ankyrin repeat-containing protein, interacts with AIF in the mitochondria and prevents its translocation to the nucleus. Significantly, we showed that SIP inhibits STS-induced apoptosis in an AIF-dependent fashion, unlike HSP70, which also reportedly interacts with Apaf-1, thereby preventing its interaction with procaspase-9, and leads to inhibition of activation of caspase (43, 44). SIP has no effect on the activation of caspase. Therefore, it appears that SIP inhibits apoptosis in a caspase-independent manner. In addition, our data indicated that under apoptotic stimulation, SIP is degraded by a proteasome-dependent pathway, leading to the liberation of AIF from mitochondria. If our interpretation is correct, the following scenario is in effect: SIP, through its physical interaction with AIF, functionally sequesters AIF in the mitochondrial space. Apoptotic stimulation signals the degradation of SIP protein, allowing the release of AIF from mitochondria and its translocation to the nucleus. In this sense, it will be interesting in future investigationsto explore the signaling events that lead to the degradation of SIP proteins, and in this context, it is worth noting that SIP protein harbors a PEST motif (rich in proline, glutamine, serine, and threonine) in its sequence that was shown to target the protein for degradation (45, 46).
SIP was also named Ankrd25 and was found to act as a growth promoter in U2OS osteosarcoma cells (47). Previously, our lab found that SIP is primarily a cytosolic protein and could inhibit estrogen receptor-regulated transcription and estrogen-stimulated proliferation of mammary carcinoma cells through sequestration of SRC coactivators in the cytoplasm (26). In our current study, we showed that SIP is not only localized in the cytoplasm but also distributes in mitochondria, where it reinforces the mitochondrial localization of AIF. It will be interesting to investigate in future studies the dynamic changes of SIP protein in cytoplasm versus mitochondria and to explore the mechanisms underlying these changes corresponding to the regulation of cell proliferation versus apoptosis.
AIF, originally discovered as a caspase-independent death effector (3), now emerges as an important factor participating in cellular redox metabolism and mitochondrial bioenergetics (6, 8, 48–50). AIF-deficient mice exhibit a reliable model of respiratory chain complex I deficiency and a defect in oxidative phosphorylation (5, 8, 50). Phenotypically, these mice show neurodegeneration (with ataxia because of cerebellar atrophia) and blindness because of retinal degeneration (6). Whether or not SIP could affect the function of AIF in these venues is currently unknown. In summary, we report here a novel regulator, SIP, in the AIF-mediated and caspase-independent apoptosis pathway, adding to our understanding of the biological function of SIP and the regulatory mechanism of programmed cell death.
This work was supported by Grants 81130048 and 30921062 from the National Natural Science Foundation of China (to Y. S.) and Grant 973 of Program 2011CB504204 from the Ministry of Science and Technology of China (to Y. S.).
- MOMP
- mitochondrial outer membrane permeabilization
- SRC
- steroid receptor coactivator
- SIP
- SRC-interacting protein
- AIF
- apoptosis-inducing factor
- MNNG
- N-methyl-N-nitroso-N′-nitroguanidine
- PARP
- poly(ADP-ribose) polymerase
- HSP
- heat shock protein
- STS
- staurosporin
- Z-VAD-FMK
- carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
- PI
- propidium iodide.
REFERENCES
- 1. Hengartner M. O. (2000) The biochemistry of apoptosis. Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 2. Green D. R., Kroemer G. (2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 3. Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., Brothers G. M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D. R., Aebersold R., Siderovski D. P., Penninger J. M., Kroemer G. (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446 [DOI] [PubMed] [Google Scholar]
- 4. Ye H., Cande C., Stephanou N. C., Jiang S., Gurbuxani S., Larochette N., Daugas E., Garrido C., Kroemer G., Wu H. (2002) DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat. Struct. Biol. 9, 680–684 [DOI] [PubMed] [Google Scholar]
- 5. Joza N., Oudit G. Y., Brown D., Bénit P., Kassiri Z., Vahsen N., Benoit L., Patel M. M., Nowikovsky K., Vassault A., Backx P. H., Wada T., Kroemer G., Rustin P., Penninger J. M. (2005) Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol. Cell. Biol. 25, 10261–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein J. A., Longo-Guess C. M., Rossmann M. P., Seburn K. L., Hurd R. E., Frankel W. N., Bronson R. T., Ackerman S. L. (2002) The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419, 367–374 [DOI] [PubMed] [Google Scholar]
- 7. Pospisilik J. A., Knauf C., Joza N., Benit P., Orthofer M., Cani P. D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C. R., Kroemer G., Rustin P., Burcelin R., Penninger J. M. (2007) Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131, 476–491 [DOI] [PubMed] [Google Scholar]
- 8. Vahsen N., Candé C., Brière J. J., Bénit P., Joza N., Larochette N., Mastroberardino P. G., Pequignot M. O., Casares N., Lazar V., Feraud O., Debili N., Wissing S., Engelhardt S., Madeo F., Piacentini M., Penninger J. M., Schägger H., Rustin P., Kroemer G. (2004) AIF deficiency compromises oxidative phosphorylation. EMBO J. 23, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joza N., Susin S. A., Daugas E., Stanford W. L., Cho S. K., Li C. Y., Sasaki T., Elia A. J., Cheng H. Y., Ravagnan L., Ferri K. F., Zamzami N., Wakeham A., Hakem R., Yoshida H., Kong Y. Y., Mak T. W., Zúñiga-Pflücker J. C., Kroemer G., Penninger J. M. (2001) Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410, 549–554 [DOI] [PubMed] [Google Scholar]
- 10. Loeffler M., Daugas E., Susin S. A., Zamzami N., Metivier D., Nieminen A. L., Brothers G., Penninger J. M., Kroemer G. (2001) Dominant cell death induction by extramitochondrially targeted apoptosis-inducing factor. FASEB J. 15, 758–767 [DOI] [PubMed] [Google Scholar]
- 11. Daugas E., Susin S. A., Zamzami N., Ferri K. F., Irinopoulou T., Larochette N., Prévost M. C., Leber B., Andrews D., Penninger J., Kroemer G. (2000) Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 14, 729–739 [PubMed] [Google Scholar]
- 12. Artus C., Boujrad H., Bouharrour A., Brunelle M. N., Hoos S., Yuste V. J., Lenormand P., Rousselle J. C., Namane A., England P., Lorenzo H. K., Susin S. A. (2010) AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J. 29, 1585–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 14. Wang X., Yang C., Chai J., Shi Y., Xue D. (2002) Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science 298, 1587–1592 [DOI] [PubMed] [Google Scholar]
- 15. Candé C., Vahsen N., Kouranti I., Schmitt E., Daugas E., Spahr C., Luban J., Kroemer R. T., Giordanetto F., Garrido C., Penninger J. M., Kroemer G. (2004) AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene 23, 1514–1521 [DOI] [PubMed] [Google Scholar]
- 16. Otera H., Ohsakaya S., Nagaura Z., Ishihara N., Mihara K. (2005) Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uren R. T., Dewson G., Bonzon C., Lithgow T., Newmeyer D. D., Kluck R. M. (2005) Mitochondrial release of pro-apoptotic proteins: electrostatic interactions can hold cytochrome c but not Smac/DIABLO to mitochondrial membranes. J. Biol. Chem. 280, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 18. Cao G., Xing J., Xiao X., Liou A. K., Gao Y., Yin X. M., Clark R. S., Graham S. H., Chen J. (2007) Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J. Neurosci. 27, 9278–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Polster B. M., Basañez G., Etxebarria A., Hardwick J. M., Nicholls D. G. (2005) Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 280, 6447–6454 [DOI] [PubMed] [Google Scholar]
- 20. Joshi A., Bondada V., Geddes J. W. (2009) Mitochondrial micro-calpain is not involved in the processing of apoptosis-inducing factor. Exp. Neurol. 218, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andrabi S. A., Kim N. S., Yu S. W., Wang H., Koh D. W., Sasaki M., Klaus J. A., Otsuka T., Zhang Z., Koehler R. C., Hurn P. D., Poirier G. G., Dawson V. L., Dawson T. M. (2006) Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. U.S.A. 103, 18308–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu S. W., Andrabi S. A., Wang H., Kim N. S., Poirier G. G., Dawson T. M., Dawson V. L. (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 103, 18314–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gurbuxani S., Schmitt E., Cande C., Parcellier A., Hammann A., Daugas E., Kouranti I., Spahr C., Pance A., Kroemer G., Garrido C. (2003) Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 22, 6669–6678 [DOI] [PubMed] [Google Scholar]
- 24. Matsumori Y., Hong S. M., Aoyama K., Fan Y., Kayama T., Sheldon R. A., Vexler Z. S., Ferriero D. M., Weinstein P. R., Liu J. (2005) Hsp70 overexpression sequesters AIF and reduces neonatal hypoxic/ischemic brain injury. J. Cereb. Blood Flow Metab. 25, 899–910 [DOI] [PubMed] [Google Scholar]
- 25. Ravagnan L., Gurbuxani S., Susin S. A., Maisse C., Daugas E., Zamzami N., Mak T., Jäättelä M., Penninger J. M., Garrido C., Kroemer G. (2001) Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 3, 839–843 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Zhang H., Liang J., Yu W., Shang Y. (2007) SIP, a novel ankyrin repeat containing protein, sequesters steroid receptor coactivators in the cytoplasm. EMBO J. 26, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Differential gene regulation by the SRC family of coactivators. Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Sun L., Liang J., Yu W., Zhang Y., Wang Y., Chen Y., Li R., Sun X., Shang Y. (2006) The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 25, 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi B., Liang J., Yang X., Wang Y., Zhao Y., Wu H., Sun L., Zhang Y., Chen Y., Li R., Zhang Y., Hong M., Shang Y. (2007) Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 27, 5105–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li R., Zhang H., Yu W., Chen Y., Gui B., Liang J., Wang Y., Sun L., Yang X., Zhang Y., Shi L., Li Y., Shang Y. (2009) ZIP. A novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 28, 2763–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun L., Shi L., Li W., Yu W., Liang J., Zhang H., Yang X., Wang Y., Li R., Yao X., Yi X., Shang Y. (2009) JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc. Natl. Acad. Sci. U.S.A. 106, 10195–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y., Sun L., Zhang Y., Wang D., Wang F., Liang J., Gui B., Shang Y. (2011) The histone modifications governing TFF1 transcription mediated by estrogen receptor. J. Biol. Chem. 286, 13925–13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi L., Sun L., Li Q., Liang J., Yu W., Yi X., Yang X., Li Y., Han X., Zhang Y., Xuan C., Yao Z., Shang Y. (2011) Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun L., Shi L., Wang F., Huangyang P., Si W., Yang J., Yao Z., Shang Y. (2011) Substrate phosphorylation and feedback regulation in JFK-promoted p53 destabilization. J. Biol. Chem. 286, 4226–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 37. Yang X., Yu W., Shi L., Sun L., Liang J., Yi X., Li Q., Zhang Y., Yang F., Han X., Zhang D., Yang J., Yao Z., Shang Y. (2011) HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44, 39–50 [DOI] [PubMed] [Google Scholar]
- 38. Belmokhtar C. A., Hillion J., Ségal-Bendirdjian E. (2001) Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 20, 3354–3362 [DOI] [PubMed] [Google Scholar]
- 39. Yuste V. J., Moubarak R. S., Delettre C., Bras M., Sancho P., Robert N., d'Alayer J., Susin S. A. (2005) Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 12, 1445–1448 [DOI] [PubMed] [Google Scholar]
- 40. Susin S. A., Zamzami N., Castedo M., Hirsch T., Marchetti P., Macho A., Daugas E., Geuskens M., Kroemer G. (1996) Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J. Exp. Med. 184, 1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kroemer G. (1997) The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat. Med. 3, 614–620 [DOI] [PubMed] [Google Scholar]
- 42. Zhu C., Wang X., Deinum J., Huang Z., Gao J., Modjtahedi N., Neagu M. R., Nilsson M., Eriksson P. S., Hagberg H., Luban J., Kroemer G., Blomgren K. (2007) Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J. Exp. Med. 204, 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beere H. M., Wolf B. B., Cain K., Mosser D. D., Mahboubi A., Kuwana T., Tailor P., Morimoto R. I., Cohen G. M., Green D. R. (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469–475 [DOI] [PubMed] [Google Scholar]
- 44. Saleh A., Srinivasula S. M., Balkir L., Robbins P. D., Alnemri E. S. (2000) Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2, 476–483 [DOI] [PubMed] [Google Scholar]
- 45. Rogers S., Wells R., Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins. The PEST hypothesis. Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 46. Spencer M. L., Theodosiou M., Noonan D. J. (2004) NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J. Biol. Chem. 279, 37069–37078 [DOI] [PubMed] [Google Scholar]
- 47. Harada J. N., Bower K. E., Orth A. P., Callaway S., Nelson C. G., Laris C., Hogenesch J. B., Vogt P. K., Chanda S. K. (2005) Identification of novel mammalian growth regulatory factors by genome-scale quantitative image analysis. Genome Res. 15, 1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Urbano A., Lakshmanan U., Choo P. H., Kwan J. C., Ng P. Y., Guo K., Dhakshinamoorthy S., Porter A. (2005) AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. EMBO J. 24, 2815–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miramar M. D., Costantini P., Ravagnan L., Saraiva L. M., Haouzi D., Brothers G., Penninger J. M., Peleato M. L., Kroemer G., Susin S. A. (2001) NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 276, 16391–16398 [DOI] [PubMed] [Google Scholar]
- 50. Bénit P., Goncalves S., Dassa E. P., Brière J. J., Rustin P. (2008) The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS One 3, e3208. [DOI] [PMC free article] [PubMed] [Google Scholar]