Background: The lipid-modified structures of bacterial lipoproteins in low-GC Gram-positive bacteria remains elusive.
Results: Three novel structures of bacterial lipoproteins were determined and functioned as TLR2 ligands.
Conclusion: Identified novel TLR2-stimulating lipoprotein structures are conserved in low-GC Gram-positive bacteria.
Significance: Results open further fields of research concerning functions and biosynthesis of bacterial lipoproteins.
Keywords: Host-Pathogen Interactions, Innate Immunity, Lipoprotein, Mass Spectrometry (MS), Toll-like Receptors (TLR), Low-GC Content, Gram-positive Bacteria
Abstract
Bacterial lipoproteins/lipopeptides inducing host innate immune responses are sensed by mammalian Toll-like receptor 2 (TLR2). These bacterial lipoproteins are structurally divided into two groups, diacylated or triacylated lipoproteins, by the absence or presence of an amide-linked fatty acid. The presence of diacylated lipoproteins has been predicted in low-GC content Gram-positive bacteria and mycoplasmas based on the absence of one modification enzyme in their genomes; however, we recently determined triacylated structures in low-GC Gram-positive Staphylococcus aureus, raising questions about the actual lipoprotein structure in other low-GC content Gram-positive bacteria. Here, through intensive MS analyses, we identified a novel and unique bacterial lipoprotein structure containing an N-acyl-S-monoacyl-glyceryl-cysteine (named the lyso structure) from low-GC Gram-positive Enterococcus faecalis, Bacillus cereus, Streptococcus sanguinis, and Lactobacillus bulgaricus. Two of the purified native lyso-form lipoproteins induced proinflammatory cytokine production from mice macrophages in a TLR2-dependent and TLR1-independent manner but with a different dependence on TLR6. Additionally, two other new lipoprotein structures were identified. One is the “N-acetyl” lipoprotein structure containing N-acetyl-S-diacyl-glyceryl-cysteine, which was found in five Gram-positive bacteria, including Bacillus subtilis. The N-acetyl lipoproteins induced the proinflammatory cytokines through the TLR2/6 heterodimer. The other was identified in a mycoplasma strain and is an unusual diacyl lipoprotein structure containing two amino acids before the lipid-modified cysteine residue. Taken together, our results suggest the existence of novel TLR2-stimulating lyso and N-acetyl forms of lipoproteins that are conserved in low-GC content Gram-positive bacteria and provide clear evidence for the presence of yet to be identified key enzymes involved in the bacterial lipoprotein biosynthesis.
Introduction
In innate immunity, mammalian Toll-like receptors (TLRs)3 play an important role in the recognition of invading microorganisms and in the induction of first-line host defense responses (1). To date, 11 human TLRs and 13 mouse TLRs have been identified, and each TLR appears to sense specific pathogen-associated molecular patterns derived from various microorganisms, including bacteria, viruses, protozoa, and fungi (2). Among these, TLR2 is unique in its cooperation with TLR1 or TLR6, and the TLR2/1 or TLR2/6 heterodimer recognizes bacterial lipoproteins and lipopeptides as a pathogen-associated molecular pattern. The bacterial lipoproteins/lipopeptides as well as a number of TLR ligand molecules are known to induce inflammatory cytokine secretion and to be involved in the establishment of adaptive immunity (3). Because bacterial lipoproteins/lipopeptides function as TLR2 ligands, they are also potential target molecules for the development of vaccines and adjuvants (4, 5). The identification and biochemical characterization of novel native bacterial lipoproteins as TLR2 ligands are important for elucidating the molecular basis of host-microbe interactions.
Bacterial lipoproteins are structurally divided into two groups: diacylated and triacylated forms (4). These two groups are distinguished by the absence or presence of a third enzyme involved in the maturation of bacterial lipoproteins. During the maturation process, the first enzyme Lgt, prolipoprotein diacylglycerol transferase, catalyzes the transfer of diacylglycerol to the sulfhydryl moiety of a cysteine residue conserved in the signal peptide of bacterial lipoprotein precursors (6). The second enzyme Lsp, prolipoprotein signal peptidase, subsequently cleaves the signal peptide from the lipoprotein precursors and leaves diacylated lipoproteins (7, 8), which contain S-diacyl-glyceryl-cysteine residues at their N termini. The third enzyme Lnt, apolipoprotein N-acyltransferase, transfers an additional acyl group to the amino group of the diacylated cysteine residue (9, 10), yielding a triacylated lipoprotein, which contains an N-acyl-S-diacylated cysteine residue. The conversion of the diacylated form to the triacylated form is generally considered to alter TLR binding specificity from the TLR2/6 to TLR2/1 heterodimer (11, 12).
Interestingly, diderm Gram-negative bacteria and high-GC content Gram-positive bacteria are reported to have Escherichia coli-type Lnt homolog(s) in their genomes, suggesting that these bacteria can produce triacylated lipoproteins (13). However, monoderm low-GC content Gram-positive bacteria, such as Bacillus, Enterococcus, Lactobacillus, Listeria, Staphylococcus, and Streptococcus and cell wall-less mycoplasmas, are assumed to produce diacylated lipoproteins because they do not have the E. coli-type Lnt in their genomes. However, our recent studies provided clear biochemical evidence that staphylococcal lipoproteins are triacylated (14, 15). In addition, Serebryakova et al. (16) reported that lipoproteins from Acholeplasma laidlawii, a mycoplasma strain, are in the triacyl form. These studies indicate that staphylococci and mycoplasma strains must have another type of Lnt whose structure is distinct from the E. coli-type Lnt protein. However, the exact N-terminal lipopeptide structures in other low-GC content Gram-positive bacteria have not yet been determined. TLR2-mediated bacterial lipoprotein recognition is essential for the activation of the innate immune response against infection by low-GC content Gram-positive pathogens as well as mycoplasmas (17–20). Therefore, determination of N-terminal lipid structures of lipoproteins from other low-GC content monoderm bacteria and characterization of their abilities to stimulate TLR2 for the induction of the innate immune responses are important and essential to understanding host-microbe interactions.
Here, through intensive MS analyses, we unexpectedly identified a novel lipoprotein structure containing N-acyl-S-monoacyl-glyceryl-cysteine, which we named the lyso structure, in Enterococcus faecalis, Bacillus cereus, Lactobacillus bulgaricus, and Streptococcus sanguinis. When the production of the proinflammatory cytokines was examined with purified native lyso-type lipoproteins using mice peritoneal macrophages, a B. cereus lyso-form lipoprotein induced tumor necrosis factor (TNF)-α and interleukin-6 (IL-6) by TLR2-dependent and TLR1- and TLR6-independent manners, similarly to the triacylated Streptococcus aureus SitC lipoprotein (14). By contrast, another E. faecalis lyso form lipoprotein induced cytokine secretion by TLR2- and TLR6-dependent and TLR1-independent pathways, similarly to the diacylated lipopeptide. Additionally, we identified two other new lipoprotein structures. One is an “N-acetyl” lipoprotein structure containing N-acetyl-S-diacyl-glyceryl-cysteine, which was identified from five bacterial species, including food-associated Bacillus subtilis and Bacillus licheniformis. The other is a “peptidyl” form, an unusual diacyl lipoprotein structure purified from Mycoplasma fermentans and containing two additional amino acids before the lipid-modified cysteine residue. Two lipoproteins with the N-acetyl structures induced the release of TNF-α and IL-6 in TLR2- and TLR6-dependent and TLR1-independent manners, supporting that lyso- and N-acetyl-form lipoproteins in low-GC content Gram-positive bacteria function as TLR2 ligand molecules. These results also implicate strong evidence for the presence of yet to be identified key enzymes involved in bacterial lipoprotein biosynthesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
E. faecalis strain ATCC19433, S. sanguinis strain NCTC10904, and L. bulgaricus strain ATCC11842 were grown in brain heart infusion broth, Todd Hewitt broth, and Lactobacilli Man-Rogosa-Sharpe broth, respectively. B. cereus strain ATCC14579 and B. licheniformis strain ATCC14580 were grown in nutrient broth. B. subtilis strain 168 was grown in Luria Bertani (LB) medium (1% tryptone, 0.5% yeast extract and 1% NaCl) containing 0.5% glucose. Listeria monocytogenes strain ATCC15313 was grown in tryptic soy broth containing 0.6% yeast extract or in brain heart infusion broth. Geobacillus kaustophilus strain HTA426 was grown in LB medium. Bacillus halodurans strain C125 was grown in Horikoshi II alkali medium (pH 9.5) (1% soluble starch, 0.5% polypeptone, 0.5% yeast extract, 0.1% K2HPO4, 0.02% MgSO4·7H2O, and 2% NaHCO3) (21). Oceanobacillus iheyensis strain HTE831 was grown in alkaline PY medium (pH 9.5) (1% polypeptone, 0.5% yeast extract, 0.1% K2HPO4, 0.02% MgSO4·7H2O, and 2% NaHCO3) (22). Bacterial cells were grown aerobically from an A600 of 0.05 until the late exponential growth phase at 37 °C, except for B. cereus (30 °C), O. iheyensis (30 °C), and G. kaustophilus (60 °C). The M. fermentans strain PG-18 and Mycoplasma pneumoniae strain M129 were grown in pleuro-pneumonia-like organism medium containing 10% horse serum. Mycoplasma genitalium strain G37 was grown in pleuro-pneumonia-like organism medium using 20% horse serum and fresh yeast extract.
Purification of Lipoproteins by Triton X-114 Phase Partitioning
Native bacterial lipoproteins were obtained using the Triton X-114 phase partitioning method (15). Briefly, the harvested bacterial cells were disrupted with glass beads and centrifuged at a low speed, and the supernatant was further centrifuged at high speed. The obtained supernatant was supplemented with Triton X-114 to a final concentration of 2% and was then incubated at 4 °C. The mixture was subsequently incubated at 37 °C and centrifuged for phase separation. The Triton X-114 phase was washed repeatedly and precipitated with ethanol. The precipitates were used for subsequent experiments as a Triton X-114 phase. Triton X-114 fractions of mycoplasmas were prepared as described previously (23, 24).
In-gel Digestion
Proteins in the Triton X-114 phase were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250. The protein bands excised from the gel were destained with 50% (v/v) methanol and dried by vacuum centrifugation. The dried gel pieces were rehydrated with 2 μl of a 10 ng/μl of trypsin solution (Promega) and then incubated in ∼20 μl of 50 mm Tris-HCl (pH 8.5) containing 0.1% (w/v) n-decyl-β-d-glucopyranoside (Sigma) at 37 °C for 18 h. The resulting digests were analyzed by MS directly or after chloroform/methanol extraction. The sample tubes used for in-gel digestion and subsequent analytical procedures were hydrophilic polypropylene tubes (Proteosave SS; Sumitomo Bakelite) to decrease the loss of lipoproteins or lipopeptides by nonspecific adsorption to the tubes.
Purification of Lipopeptides by Chloroform/Methanol Extraction
Lipopeptides from the purified native lipoproteins were extracted according to previous methods (15). Briefly, the peptide solution (10–20 μl) resulting from in-gel digestion was acidified with 1 μl of 70% formic acid, mixed with the same volume of chloroform/methanol (2/1, v/v) by vortexing, and then centrifuged. The organic phase contained the lipopeptides. The acidification step is necessary because the formation of a methyl ester at the C-terminal carboxyl group of lipopeptides is often observed in the organic phase in the presence of tryptic activity.
MALDI-TOF MS and MS/MS
MALDI-TOF MS was conducted using an Ultraflex (Bruker Daltonics) MALDI-TOF mass spectrometer in positive reflectron mode. Saturated α-cyano-4-hydroxycinnamic acid solution in chloroform/methanol (2/1, v/v) was used as the matrix. A thin layer of α-cyano-4-hydroxycinnamic acid matrix was prepared and the samples were deposited on the matrix. The resolution was about 10,000. The mass spectra were calibrated externally so that the typical mass error was about 50 ppm. The MS/MS spectra were acquired using a MALDI-TOF/TOF instrument (Ultraflex; Bruker Daltonics) using α-cyano-4-hydroxycinnamic acid as matrix. The MS/MS isolation width was ±2 Da. Fragments of tryptic lipopeptides observed using MALDI MS/MS are mainly C terminus-containing sequence ions (y-type) because the positive charge tends to localize at the C-terminal lysine or arginine of the peptides.
On-target Oxidation of Lipopeptides
To facilitate the neutral loss of the acylglyceride moiety from lipopeptides by MS/MS, the thioether sulfur at the N-terminal S-acyl-glyceryl-cysteine of a lipopeptide was oxidized to sulfoxide as described previously (15). One microliter of hydrogen peroxide (30% aqueous solution) was spotted onto the sample matrix co-crystal of a MALDI sample target, and the target was then completely dried at room temperature.
Lipoprotein Lipase Treatment
The organic phase containing the lipopeptides was dried under vacuum and redissolved in 10–20 μl of water by sonication for 5 min. The solution was heated at 100 °C for 1.5 h to inactivate trypsin and then incubated at 37 °C with 80 ng/μl of lipoprotein lipase from Pseudomonas sp. (Sigma). The resulting digests were directly analyzed by MALDI MS and MS/MS.
Protein Identification by LC-MS/MS
In-gel digests were analyzed by a nano-LC-MS/MS (1100 series; Agilent Technologies) coupled with a hybrid quadrupole-TOF instrument (Q-Tof2; Waters) as described previously (15). The mobile phases of a homemade capillary column packed with Inertsil ODS3 (GL Science) were: solvent A (0.075% (v/v) formic acid in water) and solvent B (0.075% (v/v) formic acid and 80% (v/v) acetonitrile in water). The resulting MS/MS data were applied to Mascot (Matrix Science) for protein identification.
Protein Sequence and Amino Acid Composition Analyses
After SDS-PAGE, the lipoproteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane and then stained with Coomassie Brilliant Blue R-250. The bands excised from the membrane were subjected to Edman degradation using a Procise 494HT protein sequencing system (Applied Biosystems). For identification of S-glycerylcysteine, the excised protein or the synthetic model peptide were individually dried in clean 6-mm × 32-mm glass tubes containing 50 pmol of norvaline as an internal standard. The tubes were placed in a glass vial that contained 200 μl of constant-boiling HCl and a piece of phenol crystal. The vial was sealed after evacuation for a few minutes using by a Mininert valve (Pierce). The samples were hydrolyzed for 20 h at 110 °C and then derivatized in situ by 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) for fluorophore detection. The AQC-amino acids were separated by ion-pair chromatography (25). The AQC-S-glyceryl-cysteine was eluted between AQC-glycine and AQC-threonine.
Mice Peritoneal Macrophages
Mice peritoneal macrophages were used as described previously (14) with some modifications. In brief, macrophages from parental C57BL/6, TLR1−/−, TLR2−/−, or TLR6−/− mice were collected 3 days after intraperitoneal injection of 2 ml of 4% thioglycollate and subsequently cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS, Invitrogen). The lipopolysaccharide (LPS) from Salmonella minesota RE595 and the synthetic Pam3CSK4 (N-palmitoyl-S-dipalmitoylglyceryl-Cys-Ser-(Lys)4) were purchased from Sigma. Macrophage-activating lipopeptide-2 was from Alexis Biochemicals. Each native lipoprotein was isolated through electroelution from an SDS-PAGE band from the Triton X-114 fraction. A synthetic lipopeptide in the N-acetyl form (N-acetyl-S-dipalmitoyl-glyceryl-Cys-Ser-Ser-Gly-Glu-Lys, which was derived from the N-terminal fragment of B. subtilis PrsA lipoprotein) was provided by Anygen (Gwang-ju, Korea). Macrophages were stimulated with purified lipoproteins for 24 h. The concentrations of both TNF-α and IL-6 from the culture supernatants were measured by ELISA (R&D Systems, Minneapolis, MN). To examine lyso form-mediated early response in immune cells, the mice peritoneal macrophages (2.5 × 105 cells) were stimulated with purified lipoproteins for 30 min, and cell lysates were determined for NF-κB activation or mitogen-activated protein kinase (MAPK) phosphorylation. Antibodies used in Western blotting were IκBα (C-21, Santa Cruz), phosphorylated extracellular signal-regulated kinase (ERK) (Thr202/Tyr294, Cell Signaling), ERK1 (K-23, Santa Cruz), phosphorylated p38 MAPK (Thr180/Thr182, Cell Signaling), and p38 MAPK (N-20, Santa Cruz).
RESULTS
Identification of a Novel Lyso Structure from Gram-positive Gut Microbes
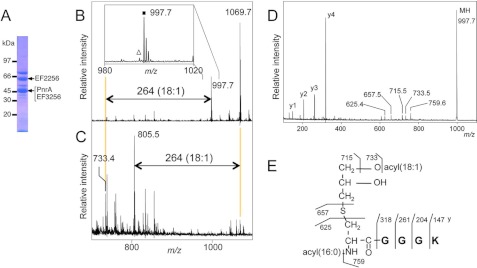
To determine the native lipoprotein structures of low-GC content Gram-positive bacteria, we first selected E. faecalis, a well known intestinal bacterium. Inappropriate activation of the innate immune system by gut microbes causes chronic inflammation, resulting in human inflammatory bowel diseases, such as Crohn disease and uncreative colitis (26). The lipoprotein-enriched Triton X-114 phase fraction was prepared and analyzed on SDS-PAGE (Fig. 1A). The 46-kDa purine nucleoside receptor PnrA (EF0177 in strain V583) (27) was in-gel digested with trypsin, extracted with chloroform/methanol, and analyzed using MALDI-TOF MS. The MS spectrum showed a peak at m/z 997.7 (Fig. 1B, inset), which corresponded to the N-terminal PnrA lipopeptide containing lipidated cysteine with two fatty acids of 34:1 (34 and 1 refer to the numbers of total carbons and double bond, respectively), but no peaks corresponding to conventional triacylated form. Although the MS spectra of triacylated lipoproteins from staphylococci contain 14-Da interval distinctive ions due to various numbers of methylene (-CH2-) groups in fatty acids (14, 15), the E. faecalis PnrA lipopeptide peak did not exhibit such accompanying ions (Fig. 1B, inset). When N-terminal PnrA lipopeptide was incubated with lipoprotein lipase, which can hydrolyze O-esterified fatty acids, we unexpectedly observed that only one 18:1 fatty acid was specifically removed (Fig. 1, B and C). Further MS/MS analysis of the peak at m/z 997.7 provided C terminus-containing y-series ions corresponding to the amino acid sequence (CGGK) of PnrA (Fig. 1D). Ions at m/z 715.5, 733.5, and 759.6 correspond to the product ions that have lost an 18:1 fatty acid (C17H33COOH), 18:1 ketene (C16H31CH = C = O), and 16:0 ketene (C14H29CH = C = O), respectively, and a characteristic fragment ion at m/z 625.4 corresponds to the N-acyl(16:0)-dehydroalanyl peptide, which is generated by the neutral loss of monoacyl(18:1)-thioglycerol. In addition, Edman sequencing of the N-terminal PnrA lipopeptide was blocked. Taken together, these results indicate that the N-terminal PnrA lipopeptide contains an N-acyl(16:0)-O-acyl(18:1)-S-glyceryl cysteine residue (Fig. 1C), and we named this lipoprotein structure the “lyso” form. Although we could not determine the exact O-acylation position, it is generally assumed that the sn-1 (R3 position)-acylated lyso form is the major species because the acyl group of lysophospholipids is known to transition between the sn-1 and sn-2 positions and the ratio of sn-1 to sn-2 acylation is about 9 (28). We also found the same lysolipidation in two other E. faecalis lipoproteins, EF2256 and EF3256 (Table 1). The N-terminal EF3256 lipopeptide peak and its deacylated peak were observed at m/z 1069.7 and 805.5, respectively (Fig. 1, B and C).
FIGURE 1.
The novel lyso form lipoprotein from E. faecalis. SDS-PAGE pattern of lipoproteins from E. faecalis cells (A), MALDI-TOF MS spectra of a fraction containing in-gel-digested E. faecalis PnrA and EF3256 incubated with lipoprotein lipase for 0 (B) or 120 h (C). Highlighted ions at m/z 733.4 or 805.5 correspond to the deacylated product from the N terminus lipopeptide of PnrA (m/z 997.7) or EF3256 (m/z 1069.7, diacylated(34:1)-CGADK), respectively. The inset in B is the magnified view. The closed square and open triangle indicate the monoisotopic lipopeptide ion containing two acyl chains with a total of one and two double bonds, respectively. The MS/MS spectrum (D) and elucidated structure of PnrA from E. faecalis (E) are shown. The C terminus-containing y-series ions and product ions that have lost a fatty acid, ketene, or monoacyl(18:1)-thioglycerol are highlighted. The position of O-acylation has not been determined (see text for details).
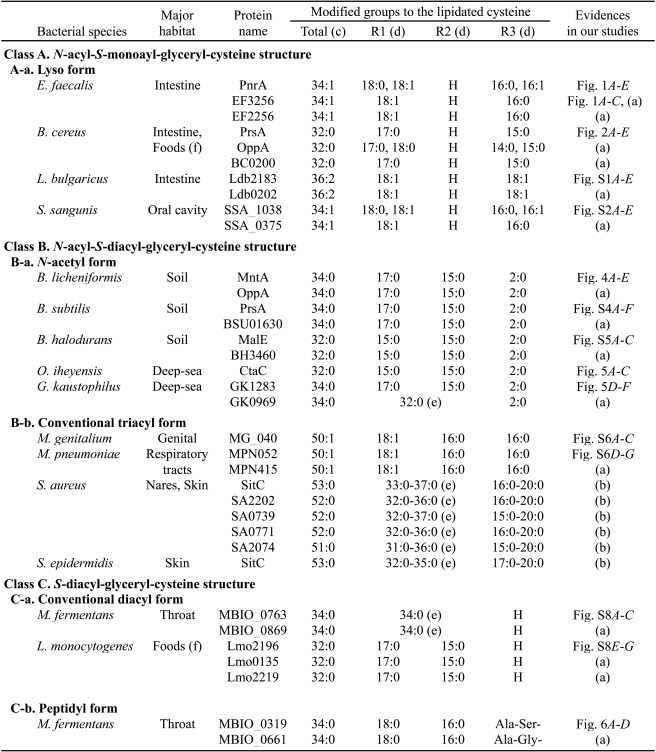
TABLE 1.
Newly determined lipoprotein structures by our group
(a) Data obtained in this study are not shown as figures.
(b) Data from our recent study (Asanuma et al. (15)), which used S. aureus strain RN4220.
(c) The total carbon number of modified acyl groups on the lipidated cysteine is caluculated from one MS/MS-analyzed lipopeptide peak. The most abundant N-terminal lipopeptide ion in MALDI-TOF MS was usually analyzed by MS/MS.
(d) R1 and R2 denote a hydrogen or an acyl group attached to the 3 and 2 position of the S-glyceryl group of the lipidated cysteine, respectively. R3 denotes a hydrogen, an acyl group, or a dipeptide attached to the α-amino group of the lipidated cysteine.
(e) The total carbon number of modified acyl groups onto R1 and R2 is shown.
(f) Foods are probably contaminated from human, animal, and/or soil.
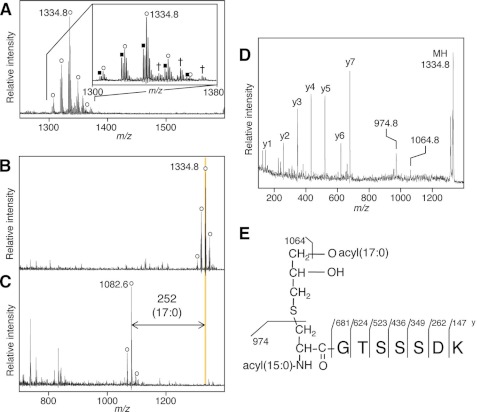
We also determined the structure of lyso form lipoproteins from B. cereus, one of components of the microbiota in the human gut and one of causative agents of food poisoning. A similar method of the E. faecalis PnrA lipoprotein as described above is applied. Lipoprotein-enriched Triton X-114 phase fraction was separated using SDS-PAGE, and the 33-kDa band containing peptidylprolyl isomerase PrsA (BC1043 of B. cereus strain ATCC14579) (29) was analyzed. The MS spectrum showed a main ion at m/z 1334.8 (Fig. 2A), which corresponds to the N-terminal CGTSSSDK lipopeptide of the PrsA containing lipid-modified cysteine with two saturated fatty acids, 32:0 in total. In contrast to other lyso form lipoproteins from E. faecalis, the B. cereus N-terminal lipopeptide ion consisted of 14-Da interval distinctive ions due to different lengths of saturated and monounsaturated fatty acids (Fig. 2A, inset). Ions corresponding to the triacyl form were not observed (Fig. 2A). To characterize an esterified fatty acid(s), the lipopeptide was treated with lipoprotein lipase. Lipoprotein lipase removed only one esterified fatty acid (17:0 as the major fatty acid) from the lipopeptide (Fig. 2, B and C). The MS/MS spectrum of the ion at m/z 1334.8 provided y-series ions that confirmed the GTSSSDK peptide sequence of B. cereus PrsA (Fig. 2, D and E). The m/z 1064.8 peak corresponded to a product fragment ion that lost a 17:0 fatty acid from the precursor ion, and a characteristic fragment ion at m/z 974.8 corresponded to N-acyl(15:0)-dehydroalanyl peptide generated by neutral loss of monoacyl(17:0)-thioglycerol. These results indicate that the lyso form of this B. cereus lipoprotein has an N-acyl(15:0)-O-acyl(17:0)-S-glyceryl-cysteine structure as the major form (Fig. 2E), in which we assumed that the sn-1 (R3 position) lyso form is the major component because of the same reason as described above. Additionally, the major forms of oligopeptide-binding OppA (BC3586) and BC0200 were also identified to be the same lyso form of N-acyl(15:0)-O-acyl(17:0)-S-glyceryl-cysteine (Table 1).
FIGURE 2.
Lyso structure of lipoprotein from B. cereus. MALDI-TOF MS spectra of a fraction containing in-gel-digested PrsA lipoprotein of B. cereus (A) or of the fraction incubated with lipoprotein lipase for 0 (B) or 116 h (C). A deacylated product ion is highlighted. The inset in A is the magnified view. The open circle and closed square indicate the monoisotopic lipopeptide ion containing two fatty acids with total number of double bonds of 0 and 1, respectively. The cross indicates a sodium-added form of the monoisotopic lipopeptide ion containing two saturated fatty acids. Shown at the right are MS/MS spectrum of the N-terminal lipopeptide ion at m/z 1334.8 (D) and elucidated lyso structure of B. cereus PrsA (E). The y-series ions and product ions that have lost a 17:0 fatty acid or monoacyl(17:0)-thioglycerol are highlighted.
In addition to E. faecalis and B. cereus, we also found lyso-form lipoproteins from Lactobacillus delbrueckii ssp. bulgaricus, a probiotic strain originating from Bulgarian yogurt, and S. sanguinis, a member of the human indigenous oral microflora (Table 1, supplemental text, and Figs. S1 and S2). These results suggest that the lyso form is a well distributed lipoprotein structure in low-GC content Gram-positive bacteria and that at least several Gram-positive bacterial species in gut microbes and a probiotic strain produce lyso-form lipoproteins.
TLR2 Stimulation by Lyso form Lipoproteins
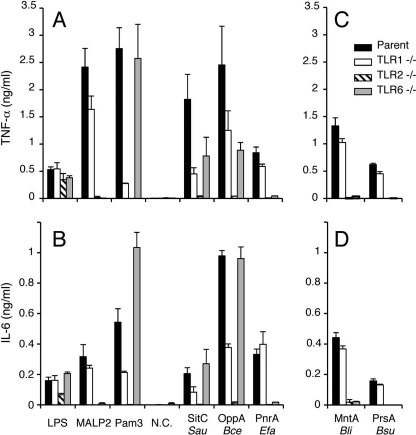
Recently published elegant crystal structures of TLR2/1 and TLR2/6 heterodimers complexed with a synthetic lipopeptide demonstrated that TLR2 holds both esterified fatty acids linked to the S-glyceryl group of lipopeptides (30, 31). Because lyso form lipoproteins lose one of two esterified fatty acids linked to the S-glyceryl group, we wondered whether lipoproteins harboring the novel lyso-type structure can stimulate host immune cells via the TLR2/1 or TLR2/6 heterodimer. Peritoneal macrophages from parental or TLR-deficient mice were stimulated with purified lyso form lipoproteins. Surprisingly, the lyso form B. cereus OppA lipoprotein induced proinflammatory cytokine production in TLR2-dependent and TLR1- and TLR6-independent manners (Fig. 3, A and B), similarly to the triacylated S. aureus SitC lipoprotein (14). Induction of the lyso form-mediated early cellular signaling response in mice macrophages was also examined. The lyso form OppA lipoprotein of B. cereus induced degradation of IκBα and phosphorylation of p38 and ERK1/2 MAPKs in TLR2-dependent manners (supplemental Fig. S3). TLR1 or TLR6 enhanced these early responses but was not essential. These results suggest that the lyso form OppA lipoprotein of B. cereus stimulates immune cells through both the TLR2/1 and TLR2/6 heterodimers. By contrast, another lyso-form E. faecalis PnrA lipoprotein induced cytokine secretion in TLR2- and TLR6-dependent and TLR1-independent manners (Fig. 3, A and B), indicating that E. faecalis PnrA mediates the immune signal through the TLR2/6 heterodimer. Therefore, the TLR2 heterodimer selectivity by lyso-form lipoproteins is not solely determined by the position of the acyl chains on the lipid-modified cysteine residue but also affected by the protein sequence. These results suggest that the newly identified lyso-form lipoproteins can function as the TLR2 ligand and that one of the two acyl chains of the S-diacylglyceryl group can be removable in the interaction between triacyl lipoproteins/lipopeptides and TLR2.
FIGURE 3.
TLR2-dependent cytokine induction by the lyso or N-acetyl forms of lipoproteins. Peritoneal macrophages (1 × 105 cells) prepared from the parental C57BL/6 (filled bar). TLR1−/− (open bar), TLR2−/− (hatched bar), or TLR6−/− (gray bar) mice were stimulated for 24 h with or without (N.C., negative control) 1 μg/ml of LPS, 100 ng/ml of macrophage-activating lipopeptide-2 (MALP-2), 10 ng/ml of synthetic Pam3CSK4, or 1 μg/ml of each purified lipoprotein/peptide. The lipoproteins tested were triacyl-SitC from S. aureus, the lyso form OppA from B. cereus, and lyso form PnrA from E. faecalis (A and B). The concentrations of tumor necrosis factor (TNF)-α (A and C) and interleukin (IL)-6 (B and D) in the culture supernatants were measured by ELISA. The data are shown as the mean ± S.D. (n = 4, each with duplicate or triplicate measurements). In addition, the N-acetyl-form MntA lipoprotein from B. licheniformis and synthetic N-acetyl form lipopeptide derived from B. subtilis PrsA were tested (C and D).
N-Acetyl Structure from Food-associated Gram-positive Bacillus
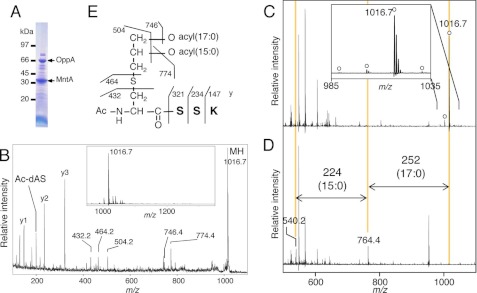
Next, we selected B. licheniformis, a soil bacterium capable of causing food poisoning. The MS analysis of the in-gel-digested 33-kDa band (Fig. 4A) showed a main peak at m/z 1016.7 (Fig. 4, B, inset, and C), which corresponds to the N-terminal lipopeptide of manganese-binding MntA (BLi03547 of strain ATCC14580) (32) possibly modified with two 35:0 fatty acids. The MS/MS spectrum showed the y-ions corresponding to the N-terminal amino acid sequence (SSK) of MntA (Fig. 4B). However, product ions at m/z 774.4, 746.4, and 504.2 suggest the loss of 15:0, 17:0, and both of the two fatty acids (32:0), respectively. These results indicate acyl modifications of 32:0 in total, which is smaller than the estimated 35:0. In addition, treatment with lipoprotein lipase also removed two O-esterified fatty acids of 15:0 and 17:0 (Fig. 4, C and D). Based on additional experimental data, an explanation for the apparent discrepancy was provided by acetylation of the α-amino group. A fragment ion at m/z 432.2 corresponds to the 42-Da-modified dehydroalanyl-SSK peptide, which is generated by the neutral loss of diacyl(17:0/15:0)-thioglycerol (Fig. 4B). As expected, Edman degradation did not provide an N-terminal sequence. Moreover, acid hydrolysis of B. licheniformis MntA generated S-glyceryl-cysteine, as did the same treatment of a synthetic model lipopeptide, N-acetyl-S-dipalmitoyl-glyceryl-cysteinyl. These results corroborate our predication of N-acetylation at the α-amino group and indicate that no acid-resistant modifications, such as trimethylation, occurred. Taken together, we conclude that the N terminus of B. licheniformis MntA has the N-acetyl-S-diacyl(15:0/17:0)-glyceryl-cysteine structure (Fig. 4E), which we named the N-acetyl form. Generally, modification on the sn-1 position of the glyceryl group of a phospholipid is more stable than the sn-2 modification (28). The peak at m/z 774.2 (17:0-containing) was higher than the peak at m/z 746.4 (15:0-containing) (Fig. 4B), suggesting that the sn-2 and sn-1 (2 and 3, respectively) positions of the S-glyceryl group were modified by 15:0 and 17:0 fatty acids, respectively (Fig. 4E). These fatty acid positions of the S-glyceryl group are consistent with literature on Bacillus phospholipids (33), the substrates of Lgt. We also identified the same N-acetyl form in another B. licheniformis lipoprotein OppA (BLi01232). Lipoproteins from neutrophilic B. subtilis, the best characterized low-GC content Gram-positive bacteria that is used in the production of Natto, a traditional Japanese dish of fermented soybeans, also contained the N-acetyl form (Table 1, supplemental text and Fig. S4, A–F).
FIGURE 4.
The novel N-acetyl form lipoprotein from B. licheniformis. SDS-PAGE image of lipoproteins from B. licheniformis cells (A), wide MS spectra of a fraction containing in-gel-digested B. licheniformis MntA (B, inset), and MS/MS spectrum of an ion obtained at m/z 1016.7 (B). The y-series ions and other major product ions are highlighted. Further shown are MS spectra of the fraction incubated with lipoprotein lipase for 0 (C) or 66 h (D). The deacylated product ions are highlighted in D. The inset in C is the magnified view. The open circle indicates the monoisotopic lipopeptide ion containing three saturated acyl chains. The elucidated structure of MntA from B. licheniformis is shown (E).
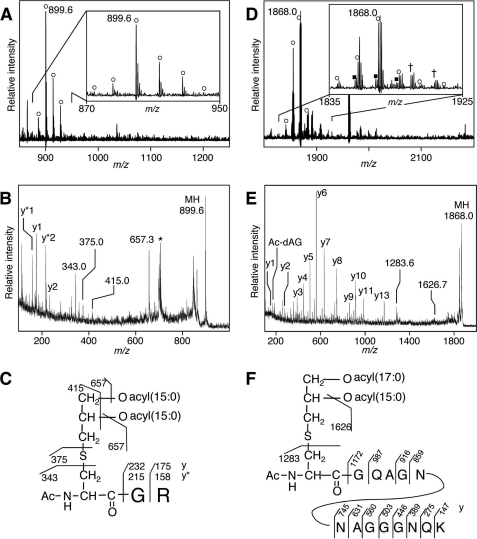
To examine whether the N-acetyl form is ubiquitous in Bacillus-related strains, we determined the N-terminal structures of lipoproteins from three other strains living in extremophilic conditions. One of the three strains was O. iheyensis strain HTE831, which was collected at a depth of 1,050 meters on the Iheya Ridge deep-sea sediment and is known to have extremely halotolerant and facultative alkaliphilic properties (34). O. iheyensis cells were grown aerobically at pH 9.5 until late-log phase. MS analysis revealed a main ion at m/z 899.6 and accompanying 14-Da interval ions (Fig. 5A). The main ion corresponded to the N-terminal CGR lipopeptide of cytochrome c oxidase subunit II CtaC (OB1437 of strain HTE831) (34) with two possible saturated fatty acids, 33:0 in total. The MS/MS spectrum of the ion at m/z 899.6 provided y- and y*-series ions describing the GR peptide sequence of O. iheyensis CtaC (Fig. 5B). The ion at m/z 657.3 corresponded to a product ion that had lost a 15:0 fatty acid. The MS/MS spectrum also provided a characteristic fragment ion at m/z 343.0 that corresponded to the N-acetyl-dehydroalanyl peptide generated by the neutral loss of diacyl(15:0/15:0)-thioglycerol. These results indicate that CtaC in O. iheyensis is the N-acetyl form: an N-acetyl-O-diacyl(15:0/15:0)-S-glyceryl-cysteine structure (Fig. 5C).
FIGURE 5.
N-Acetyl structure of lipoprotein from Bacillus-related species isolated from deep-sea. A–C, analyses of CtaC of O. iheyensis. Shown are MS spectrum of a fraction containing in-gel-digested CtaC of O. iheyensis cells grown in alkaline medium (A), MS/MS spectrum of an ion obtained at m/z 899.6 (B), and elucidated N-acetyl structure of O. iheyensis CtaC (C). The y-series ions, y* ions that have lost ammonium from y ions, and product ions that have lost one or two 15:0 fatty acid(s) or diacyl(15:0/15:0)-thioglycerol are highlighted. The asterisk indicates a contaminated ion derived from the matrix. D–F, analyses of GK1283 of G. kaustophilus. Shown are MS spectrum of a fraction containing in-gel-digested GK1283 of G. kaustophilus cells grown at 60 °C (D), MS/MS spectrum of an ion obtained at m/z 1868.0 (E), and elucidated N-acetyl structure of G. kaustophilus GK1283 (F). The y-series ions and product ions that have lost a 15:0 fatty acid or diacyl(17:0/15:0)-thioglycerol are highlighted. The inset in A or D is the magnified view. The open circle and closed square indicate the monoisotopic lipopeptide ion containing three acyl chains with total number of double bonds of 0 and 1, respectively. The cross indicates a sodium-added form of the monoisotopic lipopeptide ion containing three saturated acyl chains.
Next, we determined the N-terminal structures of lipoproteins of thermophilic G. kaustophilus strain HTA426, which was isolated at a depth of 10,897 meters in the Mariana Trench and can grow up to 74 °C (35). G. kaustophilus cells were grown aerobically at 60 °C until late-log phase. MALDI-TOF MS analysis revealed a main ion at m/z 1868.0 that accompanied 14-Da interval ions (Fig. 5D). The main ion corresponded to the N-terminal CGQAGNNAGGGNQK lipopeptide of an ABC transporter substrate-binding lipoprotein (GK1283 of strain HTA426) (35) with two possible saturated fatty acids, 35:0 in total. The lipopeptide ions having a monounsaturated fatty acid were also observed (Fig. 5D, inset, filled square). The MS/MS spectrum of the ion at m/z 1868.0 provided y-series ions describing the GQAGNNAGGGNQK peptide sequence of G. kaustophilus GK1283 (Fig. 5E). The ion at m/z 1626.7 corresponded to a product ion that had lost a 15:0 fatty acid. The MS/MS spectrum also provided a characteristic fragment ion at m/z 1283.6 that corresponded to the N-acetyl-dehydroalanyl peptide generated by the neutral loss of diacyl(17:0/15:0)-thioglycerol. These results indicate that GK1283 in G. kaustophilus is the N-acetyl form: an N-acetyl-O-diacyl(17:0/15:0)-S-glyceryl-cysteine structure (Fig. 5F). Regarding the fatty acid modification of the 3 and 2 positions of the S-glyceryl group, only the ion at m/z 1626.7 (17:0-containing), and not the 15:0-containing ion, was represented in the MS/MS spectrum (Fig. 5E), suggesting that the 2 and 3 positions of the S-glyceryl group could be modified by the 15:0 and 17:0 fatty acids, respectively (Fig. 5F). Additionally, GK0969 was identified to take the N-acetyl form, an N-acetyl-O-diacyl(32:0 in total)-S-glyceryl-cysteine, as its major form (Table 1). We also found N-acetyl form lipoproteins in other extremophilic Bacillus-related strains, namely alkaliphilic Bacillus halodurans grown at pH 9.5 (Table 1, supplemental text and Fig. S5). These results suggest that the N-acetyl form is also a well distributed structure of lipoproteins in low-GC content Gram-positive Bacillus-related strains. As described above, B. cereus lipoproteins are determined as the lyso form, indicating that the lipoprotein structure is not limited to the N-acetyl form in the Bacillaceae family. These N-acetyl form lipoprotein-producing bacteria grown in environments or food sources can be exposed to cutaneous immunity or to mucosal immune systems of the host gut after ingestion. So, we examined whether N-acetyl form lipoproteins can stimulate host immune cells through TLR2 and found that N-acetyl form lipoprotein/lipopeptides stimulate immune cells through TLR2/6 (Fig. 3, C and D).
Peptidyl Structure from Mycoplasma
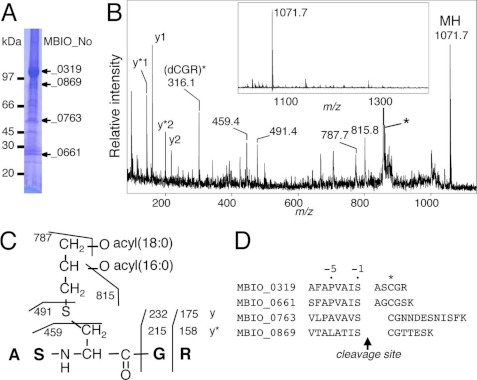
Next, we determined the structures of multiple mycoplasma lipoproteins. Consistent with our previous predictions (23, 24), lipoproteins from M. genitalium and M. pneumoniae adopted a conventional triacyl structure (Table 1, supplemental text and Fig. S6), suggesting the presence of a new type of Lnt enzyme in mycoplasmas, in accordance with a recent A. laidlawii study (16). In addition, macrophage-activating lipopeptide-2 precursor (MBIO_0763 of strain PG18) and MBIO_0869 from M. fermentans were determined to adopt the S-diacyl(34:0 in total)-glyceryl structure (Table 1, supplemental text and Fig. S7, A–C), consistent with a previous report (36). However, we identified a novel and unusual diacyl lipoprotein structure in M. fermentans. The N-terminal lipopeptide from a 104-kDa MBIO_0319 (Fig. 6A) produced a peak at m/z 1071.7 (Fig. 6B, inset). Intriguingly, the MS/MS analysis indicated the presence of additional alanyl-serine residues in front of the lipidated cysteine (Fig. 6C). Edman degradation confirmed the N-terminal amino acid sequence as ASXGR. Similarly, the N terminus of MBIO_0661 from M. fermentans had alanyl-glycine residues in front of the lipidated cysteine (Table 1). We named this new N-terminal lipoprotein structure the peptidyl form. We could not detect the conventional diacyl form in either MBIO_0319 or MBIO_0661 from MS analyses. Therefore, the lipoproteins in M. fermentans take only one of the two structures, either the conventional diacylated form or the peptidyl form, suggesting that lipoprotein structures are variable among mycoplasma species and within individual proteins of M. fermentans.
FIGURE 6.
Unusual peptidyl form lipoprotein from M. fermentans. SDS-PAGE pattern of lipoproteins from M. fermentans cells (A), MS spectrum (B, inset), and MS/MS spectrum (B) from a fraction containing in-gel-digested MBIO_0319 from M. fermentans. C, the elucidated structure of MBIO_0319. The y-series ions, y* ions that have lost ammonium from y ions, and other major product ions are highlighted. The asterisk indicates a contaminated ion derived from the matrix. D, putative cleavage sites of four M. fermentans lipoproteins. The lipidated cysteine residue is marked with an asterisk.
We examined the cleavage sites of the four M. fermentans lipoproteins listed above to be located at the C terminus of the serine residue in front of the lipidated cysteine (Fig. 6D). Twenty-seven predicted lipoproteins in the M. fermentans JER genome (37) have a serine residue(s) within three amino acids in front of the predicted lipid-modified cysteine (supplemental Fig. S7D). Therefore, we speculate that M. fermentans Lsp may have a unique specificity for a cleavage site compared with other bacteria. Unfortunately, because it was unable for us to obtain sufficient amounts of homogeneous M. fermentans native peptidyl form lipoproteins, we could not examine their TLR2 stimulation profiles.
Finally, we present the lipoprotein structures of L. monocytogenes, a food poisoning bacterium capable of living as an intracellular parasite. Studies using an lgt-deleted mutant showed critical roles of lipoproteins for virulence and TLR2-medated immune activation in L. monocytogenes (19). The MS and MS/MS spectra showed that N-terminal lipopeptides of Lmo2196 and two other lipoproteins (protein number of strain EGD-e) (38) have the S-diacyl(17:0/15:0)-glyceryl-cysteine structure (Table 1, supplemental text, and Fig. S7, E–G). These results suggest that the conventional diacyl form is the TLR2-stimulating lipoprotein structure of L. monocytogenes, that N-acylation is not essential for Listeria cell growth, and that the N-acyl state is not obligatory for all Firmicutes lipoproteins.
DISCUSSION
Determination of the exact lipoprotein structures from low-GC content Gram-positive bacteria is hampered due to difficulties in the purification of native bacterial lipoproteins and in their molecular structural analyses. Here, we first determined the native bacterial lipoprotein structures from 13 different bacterial species by MALDI-TOF-MS-based strategies. We found a novel and unique lyso form lipoprotein structure that functions as a TLR2 ligand. In addition, N-acetyl and peptidyl forms were also identified. Both the lyso and N-acetyl forms are conserved in low-GC content Gram-positive bacteria. Until this time, bacterial lipoproteins were mainly classified into two groups, diacylated and triacylated forms; this study identifies three additional new structures of bacterial lipoproteins and expands the possible structural variants of bacterial lipoproteins. These structural variants of lipoproteins are sensed by TLR2 heterodimers. The lyso form lipoproteins are specifically recognized by TLR2/1 and/or TLR2/6 (Fig. 3 and supplemental Fig. S3), whereas the N-acetyl form lipoproteins are recognized by TLR2/6 (Fig. 3), showing that the cooperation of TLR2 with TLR1 and TLR6 allowed the TLR2-mediated recognition of the structural variations of bacterial lipoproteins.
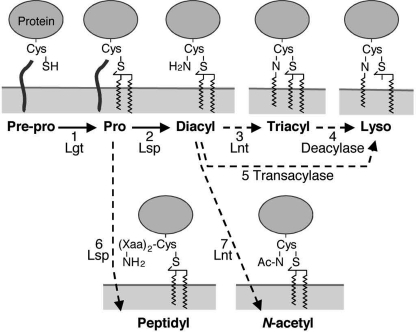
The lyso form is also an exceptionally unique structure from a bacterial lipoprotein biosynthesis viewpoint. To predict how newly identified bacterial lipoproteins are maturated, we summarized the N-terminal lipoprotein structures into three major classes (A to C) based on published and our current studies (Table 1) and made a schematic of possible bacterial lipoprotein biosynthetic pathways (Fig. 7). Class C has S-diacyl-glyceryl-cysteine structures, which are maturated by two enzymes, Lgt (Fig. 7, step 1) and Lsp (steps 2 or 6). Class B contains N-acyl-S-diacyl-glyceryl-cysteine structures, which are produced by a three-step modification by Lgt, Lsp, and Lnt that catalyzes N-acylation (steps 3 or 7). Because the low-GC content monoderm bacteria lack E. coli-type Lnt homologues, Lnt in these bacteria must have a new primary structure (14, 16) and may have distinct enzymatic properties. Finally, class A has an N-acyl-S-monoacyl-glyceryl-cysteine lyso structure whose maturation may require either a putative lipoprotein O-deacylase acting on the conventional triacyl form (step 4) or a putative lipoprotein transacylase acting on the conventional diacyl form and transferring a fatty acid from the S-glyceryl group to the α-amino group of the lipidated cysteine (step 5). Possible biochemical characteristics of the unidentified lipoprotein biosynthesis enzymes are discussed in the supplemental text and Table S1. The existence of the N-acetyl form lipoproteins indicates that, unlike E. coli-type Lnt, the yet to be discovered Lnt enzyme might not use membrane phospholipids as substrates. Taken together, the identification of novel lipoprotein structures supports the presence of novel modes of lipid modifications in bacterial lipoprotein biosynthesis.
FIGURE 7.
Bacterial lipoprotein biogenesis in low-GC content monoderm bacteria. The dotted lines indicate uncharacterized or putative modification steps.
Because the anchoring of lipoproteins onto bacterial membrane appears to be sufficiently achieved using two O-esterified fatty acids, the additional N-acylation may have a different role than just simple membrane association. N-Acylation in E. coli is required for sorting lipoproteins from the inner membrane to the outer membrane by the Lol system (39–41), as these monoderm bacteria do not have outer membranes. Possible roles of N-acylation may be analogous to the functions of N-myristoylation in eukaryotic proteins, such as protein localization and protein-protein interactions (42). Intriguingly, the N-acetyl form seems to be distributed among environmental bacteria isolated from soil and deep-sea environments, whereas normal inhabitants of humans tend to be N-long chain acylated (Table 1). Therefore, these structures may have been established during adaptation to environmental conditions including host immune systems.
In conclusion, we determined the molecular structures of novel TLR2-stimulating lyso, N-acetyl, and peptidyl form lipoproteins and also provide clear biochemical evidence for the presence of yet to be identified key enzymes in bacterial lipoprotein biosynthesis. Our discovery of the new lipid-modified lipoprotein structures will help to elucidate the biological functions of bacterial lipoproteins during microbe-host interactions.
Supplementary Material
Acknowledgments
We thank Dr. H. Takami (JAMSTEC, Japan) and Dr. Y. Choi (Seoul University, Korea) for providing bacterial strains. We also thank Dr. S. Rottem for information on the lipoproteins of the M. fermentans JER strain and Dr. Y-M. Kim (POSTEC, Korea) for critical reading of the manuscript.
This work was supported by Bio-Program Grant 2008-2004086 and the BK21 programs supported by the National Research Foundation of Korea (to B. L. L.).

This article contains supplemental text, Figs. S1–S7, and Table S1.
- TLR
- Toll-like receptor
- Pam3CSK4
- N-palmitoyl-S-[2,3-bis-(palmitoyloxy)-propyl]-(R)-cysteinyl-(lysyl)3-lysine
- Lgt
- prolipoprotein diacylglycerol transferase
- Lsp
- prolipoprotein signal peptidase
- Lnt
- apolipoprotein N-acyltransferase
- AQC
- 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate.
REFERENCES
- 1. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity. Update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 2. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 3. Iwasaki A., Medzhitov R. (2010) Regulation of adaptive immunity by the innate immune system. Science 327, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kovacs-Simon A., Titball R. W., Michell S. L. (2011) Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutchings M. I., Palmer T., Harrington D. J., Sutcliffe I. C. (2009) Lipoprotein biogenesis in Gram-positive bacteria. Knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 17, 13–21 [DOI] [PubMed] [Google Scholar]
- 6. Sankaran K., Wu H. C. (1994) Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 7. Hussain M., Ichihara S., Mizushima S. (1982) Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257, 5177–5182 [PubMed] [Google Scholar]
- 8. Tokunaga M., Loranger J. M., Chang S. Y., Regue M., Chang S., Wu H. C. (1985) Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J. Biol. Chem. 260, 5610–5615 [PubMed] [Google Scholar]
- 9. Tokunaga M., Tokunaga H., Wu H. C. (1982) Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. U.S.A. 79, 2255–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta S. D., Gan K., Schmid M. B., Wu H. C. (1993) Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J. Biol. Chem. 268, 16551–16556 [PubMed] [Google Scholar]
- 11. Takeuchi O., Kawai T., Mühlradt P. F., Morr M., Radolf J. D., Zychlinsky A., Takeda K., Akira S. (2001) Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13, 933–940 [DOI] [PubMed] [Google Scholar]
- 12. Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., Modlin R. L., Akira S. (2002) Cutting edge. Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169, 10–14 [DOI] [PubMed] [Google Scholar]
- 13. Tschumi A., Nai C., Auchli Y., Hunziker P., Gehrig P., Keller P., Grau T., Sander P. (2009) Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J. Biol. Chem. 284, 27146–27156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurokawa K., Lee H., Roh K. B., Asanuma M., Kim Y. S., Nakayama H., Shiratsuchi A., Choi Y., Takeuchi O., Kang H. J., Dohmae N., Nakanishi Y., Akira S., Sekimizu K., Lee B. L. (2009) The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for Toll-like receptor 2. J. Biol. Chem. 284, 8406–8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asanuma M., Kurokawa K., Ichikawa R., Ryu K. H., Chae J. H., Dohmae N., Lee B. L., Nakayama H. (2011) Structural evidence of α-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 278, 716–728 [DOI] [PubMed] [Google Scholar]
- 16. Serebryakova M. V., Demina I. A., Galyamina M. A., Kondratov I. G., Ladygina V. G., Govorun V. M. (2011) The acylation state of surface lipoproteins of mollicute Acholeplasma laidlawii. J. Biol. Chem. 286, 22769–22776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeuchi O., Hoshino K., Akira S. (2000) Cutting edge. TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165, 5392–5396 [DOI] [PubMed] [Google Scholar]
- 18. Henneke P., Dramsi S., Mancuso G., Chraibi K., Pellegrini E., Theilacker C., Hübner J., Santos-Sierra S., Teti G., Golenbock D. T., Poyart C., Trieu-Cuot P. (2008) Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J. Immunol. 180, 6149–6158 [DOI] [PubMed] [Google Scholar]
- 19. Machata S., Tchatalbachev S., Mohamed W., Jänsch L., Hain T., Chakraborty T. (2008) Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J. Immunol. 181, 2028–2035 [DOI] [PubMed] [Google Scholar]
- 20. Torres D., Barrier M., Bihl F., Quesniaux V. J., Maillet I., Akira S., Ryffel B., Erard F. (2004) Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect. Immun. 72, 2131–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takami H., Horikoshi K. (2000) Analysis of the genome of an alkaliphilic Bacillus strain from an industrial point of view. Extremophiles 4, 99–108 [DOI] [PubMed] [Google Scholar]
- 22. Lu J., Nogi Y., Takami H. (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol. Lett. 205, 291–297 [DOI] [PubMed] [Google Scholar]
- 23. Shimizu T., Kida Y., Kuwano K. (2008) A triacylated lipoprotein from Mycoplasma genitalium activates NF-κB through Toll-like receptor 1 (TLR1) and TLR2. Infect. Immun. 76, 3672–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimizu T., Kida Y., Kuwano K. (2007) Triacylated lipoproteins derived from Mycoplasma pneumoniae activate nuclear factor-κB through Toll-like receptors 1 and 2. Immunology 121, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masuda A., Dohmae N. (2010) Automated protein hydrolysis delivering sample to a solid acid catalyst for amino acid analysis. Anal. Chem. 82, 8939–8945 [DOI] [PubMed] [Google Scholar]
- 26. Podolsky D. K. (2002) Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429 [DOI] [PubMed] [Google Scholar]
- 27. Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R., Heidelberg J. F., Tettelin H., Dodson R. J., Umayam L., Brinkac L., Beanan M., Daugherty S., DeBoy R. T., Durkin S., Kolonay J., Madupu R., Nelson W., Vamathevan J., Tran B., Upton J., Hansen T., Shetty J., Khouri H., Utterback T., Radune D., Ketchum K. A., Dougherty B. A., Fraser C. M. (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 [DOI] [PubMed] [Google Scholar]
- 28. Nakanishi H., Iida Y., Shimizu T., Taguchi R. (2010) Separation and quantification of sn-1 and sn-2 fatty acid positional isomers in phosphatidylcholine by RPLC-ESIMS/MS. J. Biochem. 147, 245–256 [DOI] [PubMed] [Google Scholar]
- 29. Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., Kapatral V., Bhattacharyya A., Reznik G., Mikhailova N., Lapidus A., Chu L., Mazur M., Goltsman E., Larsen N., D'Souza M., Walunas T., Grechkin Y., Pusch G., Haselkorn R., Fonstein M., Ehrlich S. D., Overbeek R., Kyrpides N. (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91 [DOI] [PubMed] [Google Scholar]
- 30. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a triacylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 31. Kang J. Y., Nan X., Jin M. S., Youn S. J., Ryu Y. H., Mah S., Han S. H., Lee H., Paik S. G., Lee J. O. (2009) Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 [DOI] [PubMed] [Google Scholar]
- 32. Veith B., Herzberg C., Steckel S., Feesche J., Maurer K. H., Ehrenreich P., Bäumer S., Henne A., Liesegang H., Merkl R., Ehrenreich A., Gottschalk G. (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7, 204–211 [DOI] [PubMed] [Google Scholar]
- 33. Kaneda T. (1977) Fatty acids of the genus Bacillus. An example of branched-chain preference. Bacteriol. Rev. 41, 391–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takami H., Takaki Y., Uchiyama I. (2002) Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 30, 3927–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takami H., Takaki Y., Chee G. J., Nishi S., Shimamura S., Suzuki H., Matsui S., Uchiyama I. (2004) Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 32, 6292–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mühlradt P. F., Kiess M., Meyer H., Süssmuth R., Jung G. (1997) Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185, 1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rechnitzer H., Brzuszkiewicz E., Strittmatter A., Liesegang H., Lysnyansky I., Daniel R., Gottschalk G., Rottem S. (2011) Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 157, 760–773 [DOI] [PubMed] [Google Scholar]
- 38. Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Charbit A., Chetouani F., Couvé E., de Daruvar A., Dehoux P., Domann E., Domínguez-Bernal G., Duchaud E., Durant L., Dussurget O., Entian K. D., Fsihi H., García-del Portillo F., Garrido P., Gautier L., Goebel W., Gómez-López N., Hain T., Hauf J., Jackson D., Jones L. M., Kaerst U., Kreft J., Kuhn M., Kunst F., Kurapkat G., Madueno E., Maitournam A., Vicente J. M., Ng E., Nedjari H., Nordsiek G., Novella S., de Pablos B., Pérez-Diaz J. C., Purcell R., Remmel B., Rose M., Schlueter T., Simoes N., Tierrez A., Vázquez-Boland J. A., Voss H., Wehland J., Cossart P. (2001) Comparative genomics of Listeria species. Science 294, 849–852 [DOI] [PubMed] [Google Scholar]
- 39. Yakushi T., Masuda K., Narita S., Matsuyama S., Tokuda H. (2000) A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 [DOI] [PubMed] [Google Scholar]
- 40. Fukuda A., Matsuyama S., Hara T., Nakayama J., Nagasawa H., Tokuda H. (2002) Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 277, 43512–43518 [DOI] [PubMed] [Google Scholar]
- 41. Robichon C., Vidal-Ingigliardi D., Pugsley A. P. (2005) Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280, 974–983 [DOI] [PubMed] [Google Scholar]
- 42. Martin D. D., Beauchamp E., Berthiaume L. G. (2011) Post-translational myristoylation. Fat matters in cellular life and death. Biochimie 93, 18–31 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.