Background: Telomerase synthesizes single-stranded telomeric repeats.
Results: The TERT N-terminal domain coordinates telomerase holoenzyme subunit association, DNA synthesis initiation, and rate of elongation.
Conclusion: TERT N-terminal domain recruitment of holoenzyme proteins increases overall activity and confers repeat addition processivity.
Significance: New understanding of DNA handling by a telomerase holoenzyme illuminates mechanisms for processive synthesis of single-stranded telomeric repeats.
Keywords: Protein-Nucleic Acid Interaction, Protein-Protein Interactions, Protozoan, Telomerase, Telomeres, Processivity, Single-stranded DNA Recognition, Telomerase Holoenzyme, Telomeric Repeat Synthesis
Abstract
Telomerase extends chromosome ends by the addition of single-stranded telomeric repeats. To support processive repeat synthesis, it has been proposed that coordination occurs between DNA interactions with the telomerase RNA template, the active site in the telomerase reverse transcriptase (TERT) core, a TERT N-terminal (TEN) domain, and additional subunits of the telomerase holoenzyme required for telomere elongation in vivo. The roles of TEN domain surface residues in primer binding and product elongation have been studied largely using assays of minimal recombinant telomerase enzymes, which lack holoenzyme subunits that properly fold and conformationally stabilize the ribonucleoprotein and/or control its association with telomere substrates in vivo. Here, we use Tetrahymena telomerase holoenzyme reconstitution in vitro to assess TEN domain sequence requirements in the physiological enzyme context. We find that TEN domain sequence substitutions in the Tetrahymena telomerase holoenzyme influence synthesis initiation and elongation rate but not processivity. Functional and direct physical interaction assays pinpoint a conserved TEN domain surface required for holoenzyme subunit association and for high repeat addition processivity. Our results add to the understanding of telomerase holoenzyme architecture and TERT domain functions with direct implications for the unique mechanism of single-stranded repeat synthesis.
Introduction
The eukaryotic reverse transcriptase (RT) telomerase elongates chromosome 3′ ends by the addition of single-stranded telomeric DNA repeats (1). This de novo DNA synthesis compensates for incomplete genome replication by DNA-templated DNA polymerases. Single-celled organisms and telomerase-positive cultured cells have constitutively active telomerase that is regulated to maintain telomere length homeostasis (2). In contrast, many cell types in multicellular organisms lack active telomerase. In telomerase-negative cell lineages, cumulative cell divisions are counted as a mitotic clock of progressive telomere shortening (3). Some somatic cell lineages gain renewal capacity by transiently activating telomerase, which can result in complete or only partial offset of telomere shortening (4). Mechanisms that limit the amount of telomere synthesis are still largely unclear, but recent studies have shown that the extent of elongation varies in part due to changes in the number of repeats added to a chromosome end before enzyme dissociation (5, 6).
The active site of telomerase reverse transcriptase (TERT)2 copies a template within the telomerase RNA component (TER). The template sequence typically encodes ∼1.5 units of the telomeric repeat, such that a product 3′ end released from the template after complete synthesis to the template 5′ end can reanneal at the template 3′ end for another round of repeat synthesis. As a prerequisite for this repeat addition processivity (RAP), product must be released from the template without release from enzyme per se. RAP is evident in vitro in the synthesis of a long product ladder even when substrate primer is present in excess (7). Telomerase capacity for RAP varies dramatically across species (8). RAP can also vary for telomerase complexes within the same cell assembled into different holoenzymes (9). To date, many molecular requirements for RAP have been characterized using minimal recombinant telomerase enzymes assembled from TERT and TER in rabbit reticulocyte lysate (RRL). However, only a few of these studies have aimed to resolve direct determinants of single-stranded DNA interaction from more global requirements for RNP conformation.
Telomerase from the ciliate Tetrahymena thermophila has been relatively well characterized due to its pioneering discovery and more recently its amenability to reconstitution (10, 11). Curiously, the physiologically assembled Tetrahymena telomerase holoenzyme has much higher RAP than the minimal recombinant RNP; holoenzyme RAP is also less dependent on dGTP concentration (12). Studies of holoenzyme architecture have shown that a multiprotein telomere adaptor subcomplex (TASC) bridges the physiological catalytic core RNP (p65-TER-TERT) to a telomeric repeat DNA binding subunit (Teb1), which is paralogous to the large subunit of replication protein A (13–15). Addition of TASC to the recombinant catalytic core RNP increases activity, and TASC-dependent Teb1 association converts the catalytic core RNP activity to holoenzyme high RAP activity (16). Surprisingly, the high affinity DNA binding domains of Teb1 are not essential for high RAP activity. Instead, high RAP requires the low affinity Teb1 C-terminal DNA binding domain (16, 17).
Here, we use Tetrahymena telomerase holoenzyme reconstitution in vitro to define TERT and TER requirements for RAP in the biologically functional enzyme complex. We find that holoenzyme RAP is not compromised by TERT N-terminal (TEN) domain sequence substitutions previously reported to interfere with sequence-specific DNA recognition or RAP. Instead, in holoenzyme context these substitutions reduce initiation efficiency or elongation rate. Using multiple reconstitution methods and direct binding assays, we define a surface of the TEN domain that is critical for holoenzyme assembly and high RAP. Findings described here provide new insights about TEN domain functional requirements and more broadly about telomerase holoenzyme interaction with single-stranded DNA.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Partial Proteolysis
Teb1BC and TEN domain polypeptides were expressed with an N-terminal His6 tag in Escherichia coli from pET28a using BL21(DE3) cells as described previously (15). Teb1BC was eluted in T2MG (20 mm Tris, pH 8.0, 1 mm MgCl2, 10% glycerol) with 300 mm NaCl, 0.1% IGEPAL, 5 mm β-mercaptoethanol, and 300 mm imidazole. TEN domains were eluted in T2MG with 50 mm NaCl, 5 mm β-mercaptoethanol, and 300 mm imidazole after washing with 250 mm NaCl. Proteins were supplemented with 5 mm DTT before freezing in liquid nitrogen and storage at −80 °C. TASC was purified from Tetrahymena expressing C-terminally tagged p45 as described previously, using a micrococcal nuclease digestion step to isolate TASC from the catalytic core RNP (16). Typical purifications gave yields of ∼0.2–0.5 ng/μl p75 subunit estimated by silver staining after SDS-PAGE. For limited proteolysis, 1 μg of purified TEN domain was incubated for 90 min at 37 °C in the absence or presence of 0.2 or 2 ng of trypsin in a total volume of 10 μl. Protein digestion products were analyzed using 12.5% SDS-PAGE and Coomassie Blue staining.
Telomerase Reconstitution and Activity Assays
Catalytic core RNP was assembled using RRL (Promega). Previous studies suggest that only about one quarter of the full-length Tetrahymena TERT expressed in RRL is competent for RNP assembly, corresponding to at most 0.5 ng/μl in the expression reaction (18). Therefore, TERT and p65 were expressed from a pCITE4a vector in separate reactions containing [35S]methionine (NEN/PerkinElmer Life Sciences) and subsequently combined at a 3:1 volume excess of TERT. The protein synthesis reactions were supplemented with 4 ng/μl purified in vitro transcribed TER and incubated for 10 min at 30 °C to allow RNP assembly. Radiolabeled methionine incorporation was monitored by SDS-PAGE and imaging with a Typhoon Trio (GE Healthcare). TEN domain trans-complementation assays included ¼ final volume of purified TEN domain in the RNP assembly reaction, with a variable final TEN domain concentration indicated in each figure.
To reconstitute holoenzyme, the catalytic core RNP assembled in RRL was diluted 5-fold with T2MG containing 5 mm DTT. A 10-μl volume of diluted catalytic core RNP was incubated for 10 min with 1 μl of TASC, 200 nm Teb1BC, and 200 nm (GT2G3)3 primer (unless indicated otherwise) in a 14-μl final volume at room temperature. Primer extension was initiated by addition of 6 μl of reaction buffer containing final assay concentrations of 50 mm Tris acetate, pH 8.0, 10 mm spermidine, 5 mm β-mercaptoethanol, 2 mm MgCl2, 0.2 mm dTTP, 0.3 μm dGTP, and trace [α-32P]dGTP (Easy Tide, 3000 Ci/mmol; NEN/PerkinElmer Life Sciences). Unless otherwise noted, reactions were incubated at room temperature for 10 min. After addition of a 32P-labeled oligonucleotide as a recovery control, telomerase reaction products were extracted, precipitated, and resolved on a 9% (19:1), 0.6 × TBE, 7 m urea gel (15). Dried gels were imaged using the Typhoon Trio. For chase experiments, reactions with radiolabeled dGTP and cold dGTP as above were incubated for 4 min before addition of excess unlabeled dGTP to 30 μm final concentration.
Physical Association Assays
TASC purification was performed on M2 FLAG antibody magnetic beads (Sigma-Aldrich). Binding and washes were performed in buffer A (T2MG with 50 mm NaCl, 0.1% IGEPAL, 1 mm DTT, and 1 mg/ml BSA). In parallel, [35S]methionine-labeled telomerase RNP was generated as described above. Telomerase holoenzyme was then assembled at room temperature by incubation of resin-bound TASC with recombinant RNP in the presence of 200 nm Teb1BC and 2 μm (GT2G3)3 for 30 min in buffer A without IGEPAL and with 0.1 mg/ml Saccharomyces cerevisiae tRNA (Sigma-Aldrich). After a wash of 45 min total including six changes of buffer A supplemented with 0.1 mg/ml tRNA, complexes were eluted at room temperature with 0.2 mg/ml 3×FLAG peptide (Sigma-Aldrich) in buffer A with 0.1 mg/ml tRNA, resolved by SDS-PAGE, and analyzed for radiolabeled proteins using the Typhoon Trio.
RESULTS
TER Motif Dependence of Reconstituted Holoenzyme Activity
To establish that assays of holoenzyme reconstituted in vitro are appropriately sensitive to subunit sequence changes, we first tested the impact of TER motif disruptions. Previous assays of Tetrahymena telomerase holoenzymes reconstituted with TER variants in vivo (19, 20) offer an opportunity for comparison with assays of holoenzymes reconstituted in vitro. To reconstitute holoenzyme in vitro we used a previously established system that combines the physiological p65-TER-TERT catalytic core RNP assembled in RRL, TASC isolated from Tetrahymena, and Teb1 purified from E. coli (Fig. 1A). For studies here we used an N-terminally truncated Teb1, termed Teb1BC, which is fully active for reconstituting holoenzyme RAP (16). Because Teb1BC lacks the highest affinity Teb1 DNA binding domain, the potential complication of primer sequestration from holoenzyme is avoided (16). To sensitize activity assays for the gain of RAP from holoenzyme assembly, we used a lower dGTP concentration than is optimal for RAP of the minimal recombinant RNP (12).
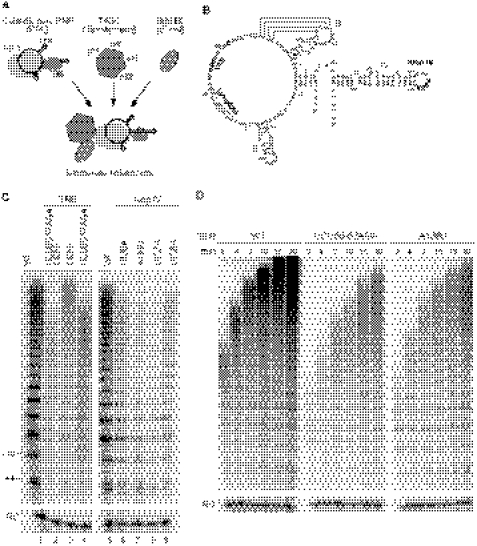
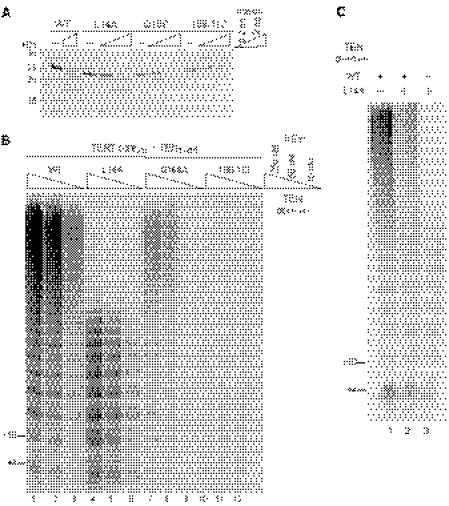
FIGURE 1.
TER sequence dependence of reconstituted holoenzyme activity. A, schematic of the holoenzyme reconstitution strategy. B, schematic of T. thermophila TER modeled after Ref. 34 with paired stems indicated by Roman numerals. The template region, template recognition element (TRE), and loop IV are labeled. Substituted nucleotides are indicated in boldface. C, activity assays of telomerase holoenzyme reconstituted with wild-type (WT) or sequence-variant TER. A 32P-labeled oligonucleotide was used as a recovery control (RC). Reactions were performed and analyzed in parallel with lanes cropped from the same exposure of the same gel. D, activity assay time course for holoenzymes reconstituted with wild type, U55A/C56G/U57A, or A136U TER.
The substitution U55A/C56G/U57A in the template recognition element (Fig. 1B) reduces the RAP of a Tetrahymena telomerase catalytic core RNP, but the same substitution has minimal if any impact on the RAP of holoenzyme purified after assembly in vivo (18, 19, 21). The shared phenotype of these two U55A/C56G/U57A TER enzymes is an increase in product intensity from incomplete repeat synthesis, which at least for in vivo reconstituted holoenzyme is caused by a slower rate of synthesis at the template 3′ end (19). Compared with U55A/C56G/U57A, the substitution C62G reduces activity overall in most assays of recombinant catalytic core RNPs (18, 19, 21, 22). A severe reduction in recombinant catalytic core RNP activity is also imposed by deletion or substitution of the evolutionarily conserved loop IV motif UAUU (Fig. 1B), but purified holoenzymes containing individual UAUU motif loop IV nucleotide substitutions have relatively normal specific activity and RAP (18–20, 23). One impediment to interpretations from holoenzyme reconstitution in vivo is that only a fraction of the variant holoenzyme was stable to affinity purification. Despite equal levels of steady-state TER accumulation in vivo, purifications of holoenzymes with TER variants recovered less RNP from extract (19, 20). Therefore, the lack of a RAP phenotype for a holoenzyme reconstituted in vivo could be due to degradation of the reconstituted holoenzyme population with altered catalytic activity during the interval of RNP purification from extract.
For each of the TER variants described above, we reconstituted holoenzyme in vitro and assayed for a change in catalytic activity compared with holoenzyme reconstituted with wild-type TER (Fig. 1C). TER sequence substitutions reduced the overall level of holoenzyme catalytic activity roughly in parallel with their impact on recombinant catalytic core RNP activity. In addition, TER sequence substitutions imposed changes in the product profile that paralleled their effect on the activity of holoenzyme reconstituted in vivo. At a 10-min reaction time point, the product ladder of the U55A/C56G/U57A TER holoenzyme was shorter than that of the wild-type TER holoenzyme, with an increase in product intensity from incomplete repeat synthesis (Fig. 1C, compare lanes 1 and 4). The shorter ladder does not result from premature product dissociation because products of both the wild-type and U55A/C56G/U57A TER holoenzymes continued to elongate through first 20 min of reaction time (Fig. 1D) and much longer (data not shown). Holoenzyme reconstitution of C62G TER gave a reduced level of high RAP activity additive with the phenotype of U55A/C56G/U57A substitution (Fig. 1C, lanes 1–4). Loop IV nucleotide substitutions also reduced overall activity, with some decrease in the length of the product ladder at the 10-min reaction time point (Fig. 1C, lanes 5–9). As with the template recognition element U55A/C56G/U57A substitution, the loop IV A136U substitution also slowed the rate of elongation but did not prevent high RAP product synthesis (Fig. 1D). We conclude that the product profile of holoenzyme reconstitution in vivo is recapitulated by holoenzyme reconstitution in vitro. Furthermore, holoenzyme reconstitution in vitro is sensitized to detection of reduced specific activity in the holoenzyme population overall. This reduction in specific activity was not detected following purification of in vivo reconstituted holoenzyme, presumably due to the low activity holoenzyme instability in cell extract.
Holoenzyme Reconstitution of TEN Domain Sequence Variants
Tetrahymena TEN domain requirements for RAP have not been studied in holoenzyme context. We began with an analysis of single amino acid substitutions in the TEN domain, including several previously characterized in Tetrahymena catalytic core RNP context (24–26). Reconstituted catalytic core RNP and holoenzyme activity were both strongly decreased by alanine substitution of the evolutionarily conserved Phe-158 and Gln-168 side chains (Fig. 2A, lanes 5 and 7; see supplemental Fig. S1A for assays of catalytic core RNPs alone). Consistent with a major DNA binding contribution of Tetrahymena telomerase holoenzyme subunits beyond the catalytic core RNP (15), the ∼10-fold change in primer Km reported for Q168A TERT in the catalytic core RNP context (25) was not evident comparing the primer concentration dependence of wild-type and Q168A TERT holoenzymes (Fig. 2B). Alanine substitution of Phe-178 and Trp-187 had, respectively, minor or no impact on holoenzyme activity (Fig. 2A, lanes 8 and 9), despite removing side chains from a proposed sequence-specific single-stranded DNA binding groove inferred from DNA cross-linking results (24). Alanine substitution of other nonconserved amino acids Asp-94, Gln-108, and Lys-160 affected the overall level of high RAP activity but not high RAP per se (Fig. 2A, lanes 3, 4, and 6).
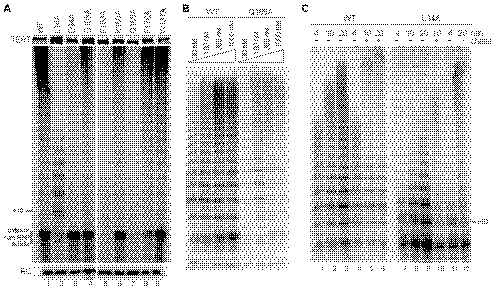
FIGURE 2.
Holoenzyme activity dependence on TEN domain single amino acid substitutions. A, activity assays of reconstituted holoenzymes with a single amino acid substitution of TERT, performed using 500 nm (GT2G3)3 primer. Reactions were performed and analyzed in parallel with lanes cropped from the same exposure of the same gel. An experiment with limiting TASC concentration is shown so that the substitution phenotype for catalytic core RNP activity can be compared with the substitution phenotype for holoenzyme activity within the same experiment (catalytic core RNP products are labeled). RC, recovery control. B, comparable reconstituted wild-type or Q168A TERT holoenzyme activity dependence on primer concentration. Reactions were performed and analyzed in parallel with lanes cropped from the same exposure of the same gel. C, pulse-chase analysis of RAP. After a 4-min pulse of radiolabeling (the 4-min time point is shown), reactions were supplemented or not with a 100-fold excess of unlabeled dGTP (the chase). Elongation was allowed to continue for a total of 10 or 20 min.
The Tetrahymena TEN domain substitution L14A was reported to eliminate RAP selectively without compromising primer binding, nucleotide addition processivity, or specific activity, leading to Leu-14-dependent models for RAP (26). Holoenzyme reconstituted with L14A TERT generated a product ladder shorter than that of the wild-type holoenzyme, with a notable increase in product intensity from incomplete repeat synthesis (Fig. 2A, lane 2). Product lengths increased with reaction time for the wild-type and L14A holoenzymes over a time course of 20 min (Fig. 2C, lanes 1–3 and 7-9) and much longer (data not shown). Although L14A holoenzyme products were shorter than wild-type holoenzyme products at any given time point, pulse radiolabeling of products for the initial 4 min of reaction time followed by a chase with excess unlabeled dGTP demonstrated conclusively that both holoenzymes retained high RAP: radiolabeled products increased in length during the chase interval for both the wild-type holoenzyme (Fig. 2C, lanes 5 and 6) and the L14A holoenzyme (lanes 10 and 12). The elevated dGTP concentration of the chase phase increased the rate of repeat synthesis for the wild-type holoenzyme (Fig. 2C, compare the longest products in lanes 3 and 6) and the L14A holoenzyme as well (compare lanes 9 and 12). Moreover, at least some of the incomplete repeat synthesis products of the L14A holoenzyme were chased to longer products (Fig. 2C, compare lanes 10 and 11), suggesting that synthesis by the L14A TERT holoenzyme is slowed rather than terminated at the template 3′ end in each cycle of template copying. Thus, similar to the other TEN domain single amino acid substitutions examined here, the L14A substitution does not eliminate any protein-protein, protein-DNA, or protein-RNA interaction critical for high RAP.
Block Substitutions of TEN Domain Sequence
To broaden the search for phenotypes from TEN domain sequence disruption, we assayed the impact of several block substitutions of consecutive six amino acid segments with the peptide sequence NAAIRS. This approach was successful in defining a central region of the TEN domain of human TERT that could be altered in sequence without precluding catalytic activity but abrogating telomere elongation in vivo (27, 28). We analyzed NAAIRS substitutions at six positions throughout the Tetrahymena TEN domain. TERT with the 184NAAIRS189 substitution reconstituted catalytic core RNP activity and holoenzyme activity comparable with wild-type when normalized to the product recovery control (Fig. 3A, lanes 6 and 7; see supplemental Fig. S1B for assays of catalytic core RNPs alone). As expected, some of the other NAAIRS substitutions eliminated enzyme activity (Fig. 3A, lanes 4 and 5) or reduced activity without changing the holoenzyme product profile (lane 1). Interestingly, two substitutions in the central region of the TEN domain (amino acids 102–107 or 108–113) in a β-hairpin surface region (see supplemental Fig. 1C) preserved catalytic core RNP activity but reduced or eliminated its conversion to holoenzyme high-RAP activity (Fig. 3A, lanes 2 and 3).
FIGURE 3.
Holoenzyme activity dependence on TEN domain block substitutions. A, activity assays of reconstituted holoenzymes with a six-amino acid block substitution of TERT, performed using 500 nm (GT2G3)3 primer. RC, recovery control. B, assays of catalytic core RNP activity stimulation by TASC or TASC and Teb1BC using RNPs with wild-type or 108NAAIRS113 TERT.
To investigate the dramatic inhibition of high RAP activity by the 108NAAIRS113 substitution, we compared enzyme activity across the individual steps of holoenzyme reconstitution in vitro. In assays with a catalytic core RNP containing wild-type TERT, TASC increased activity level, and the combination of TASC and Teb1BC converted the product synthesis profile to high RAP (Fig. 3B, lanes 1–3). In contrast, in assays with a catalytic core RNP containing 108NAAIRS113 TERT, activity was unchanged in amount by the addition of TASC and was also unchanged in product profile by the combined addition of TASC and Teb1BC (Fig. 3B, lanes 4–6). These observations suggest that disruption of TEN domain amino acids 108–113 in a protein region well separated from the proposed DNA interaction groove (24) interferes with communication between the catalytic core RNP and TASC.
Autonomous Folding and Function of TEN Domain
For human telomerase, the TERT core lacking the TEN domain supports single-repeat synthesis but not RAP (29). Complementation of the TEN domain and TERT core co-expressed as separate polypeptides demonstrated that the human TEN domain can fold and function autonomously (29), particularly when extended at its C terminus by the vertebrate-specific long linker between the TEN domain and the adjacent high affinity telomerase RNA binding domain (TRBD). TEN domain trans-complementation sensitized the detection of RAP for changes in TEN domain docking to the TERT core RNP (29). We therefore investigated whether TEN domain trans-complementation could be detected for Tetrahymena TERT, both to investigate the conservation of telomerase RNP architecture and as an approach to define further the role of the TEN domain in telomerase holoenzyme reconstitution.
A Tetrahymena TEN domain polypeptide including amino acids 1–195 was previously expressed in E. coli as a soluble protein and demonstrated to bind Tetrahymena TER (21). A slightly smaller region of the Tetrahymena TEN domain was crystallized and mostly solved in structure at high resolution (24). To investigate TEN domain trans-complementation, based on the previous results for human TERT, we assayed both our original TEN domain (amino acids 1–195) and a C-terminally extended TEN domain (amino acids 1–215), including a region that can be trimmed from the TRBD without any change in p65-stimulated binding of the TRBD to TER.3 We combined one of two versions of the bacterially expressed TEN domain and one of two matching versions of the RRL-expressed TERT core with the remaining holoenzyme subunits (Fig. 4, A and B).
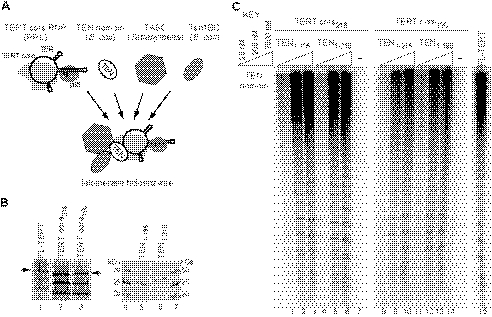
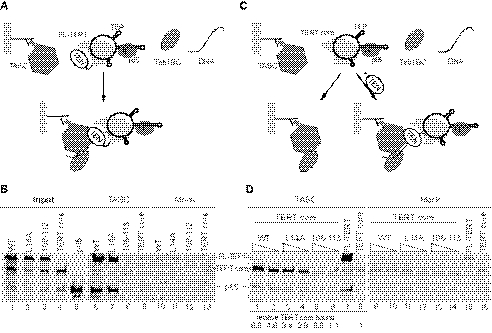
FIGURE 4.
Complementation of the TEN domain and TERT core for holoenzyme reconstitution. A, schematic of the TEN domain trans-complementation strategy for holoenzyme reconstitution. B, expression of the TERT domains for trans-complementation. At left, the intact full-length (FL) TERT or TERT core radiolabeled by expression in RRL is indicated with an arrow; the other translation products result from internal initiation. At right, the purified bacterially expressed TEN domains were detected by gel staining with Coomassie Blue. C, activity assays with or without the indicated concentration (20–2000 nm) of a TEN domain (TEN1–195 or TEN1–215) in combination with a TERT core polypeptide (TERT core196 or TERT core216). An assay with full-length TERT is shown for comparison (lane 15).
The Tetrahymena TERT core RNP without any added TEN domain showed extremely weak single-repeat synthesis even in the presence of other holoenzyme components (data not shown). Addition of either version of the purified TEN domain to either version of the TERT core RNP resulted in a dramatic gain of overall activity and RAP (Fig. 4C). Activity was marginally higher in reconstitutions using the TERT core starting at amino acid 216, so subsequent complementation assays used the bacterially expressed TEN domain and the TERT core with matching 215/216 breakpoints. Because the region spanning from amino acid 196 to amino acid 215 was not required on either the TEN domain or the TERT core for successful complementation of high RAP activity (Fig. 4C, lanes 1–3), this region is likely to be part of a linker between the TEN domain and TRBD (27). In assays of human TERT TEN domain trans-complementation, the atypically long vertebrate TERT linker was stimulatory but not required for activity reconstitution by holoenzyme assembly in vivo (29). Notably, a substantial region previously considered as part of the Tetrahymena TERT TEN domain could be deleted without precluding holoenzyme activity reconstitution by trans-complementation in vitro (supplemental Fig. S2). Our results suggest that the linker region can be entirely dispensable for catalytic activity reconstitution in nonvertebrate organisms.
TERT Core Complementation by Sequence-variant TEN Domains
We next expressed and purified isolated TEN domains with substitutions that imposed distinct activity phenotypes in full-length TERT holoenzyme context: the L14A substitution that reduced the rate of elongation, the Q168A substitution that reduced activity overall without altering the product profile, and the 108NAAIRS113 substitution that blocked conversion of catalytic core RNP activity to holoenzyme high RAP activity. To verify stable folding of the isolated TEN domains with these sequence substitutions, we performed limited proteolysis with trypsin. Trypsin cleavage produces a slightly smaller TEN domain (Fig. 5A), as observed in other partial proteolysis assays (24). Importantly, all of the sequence variant TEN domains showed a similar extent and specificity of proteolysis (Fig. 5A).
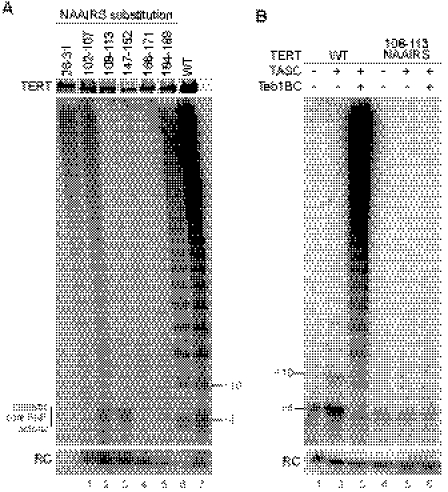
FIGURE 5.
Complementation of the TERT core with TEN domain sequence variants. A, limited proteolysis of the isolated wild-type (WT) or sequence-variant TEN domain. B and C, activity assays of TERT core complementation with different concentrations of wild-type or sequence-variant TEN domain (10–250 nm). C, 50 nm wild-type TEN domain and/or 200 nm L14A TEN domain used for reconstitution independently (lanes 1 and 3) or in combination (lane 2).
Variant TEN domains were then used for holoenzyme reconstitution with the TERT core, comparing activity across a titration of TEN domain concentration. The relative activity levels and product profiles obtained by TEN domain trans-complementation (Fig. 5B) closely matched the results for the full-length TERT proteins (Figs. 2A and 3A). For the wild-type TEN domain and the L14A and Q168A variants, activity increased with a similar dependence on TEN domain concentration (Fig. 5B, lanes 1–9). In contrast, the 108NAAIRS113 TEN domain failed to stimulate high RAP activity at any concentration (Fig. 5B, lanes 10–12; additional data not shown). We conclude that TEN domain sequence substitutions affect TEN domain function in an autonomous manner.
Curiously, although the TEN domain concentrations used in these assays represent a substantial molar excess over both the TERT core RNP and TASC (see “Experimental Procedures”), this excess stimulates rather than inhibits holoenzyme activity reconstitution. Given the role of the TEN domain in physically bridging the TERT core RNP and TASC (see below), it is possible that only some of the purified recombinant TEN domain adopts a conformation permissive for binding to these complexes. It is also possible that the isolated TEN domain binds to the TERT core RNP and TASC with low affinity, but after binding to one complex the TEN domain gains higher affinity for other complex thereby reducing free TEN domain competition for complete holoenzyme assembly. Mixing the wild-type and L14A TEN domains before holoenzyme assembly, with the excess of TEN domain still present in the activity assay, produced a mixed rather than hybrid product profile (Fig. 5C). This result suggests that there is no exchange of a holoenzyme-assembled TEN domain during high RAP elongation.
TEN Domain Requirements for Physical Association of TASC with TERT Core RNP
The activity assays above do not distinguish whether the 108NAAIRS113 substitution disrupts holoenzyme assembly or assembled holoenzyme activity. Therefore, we investigated the TEN domain sequence dependence of TASC physical interaction with the full-length TERT RNP or the trans-complemented TERT core RNP.
By omitting the final TASC purification step of elution from FLAG antibody resin, we immobilized TASC as the platform for holoenzyme assembly (see “Experimental Procedures”). RRL-assembled catalytic core RNP with radiolabeled TERT and p65 was added to immobilized TASC under conditions that parallel holoenzyme activity assays (Fig. 6A). As a negative control, binding reactions were done with FLAG antibody resin from a mock TASC preparation using cell extract lacking a FLAG-tagged protein. Some background binding of TERT was often detected (Fig. 6B, lanes 10–13). In comparison, TASC resin specifically co-purified full-length wild-type TERT and p65 (Fig. 6B, lane 6). Physical association of TASC with the catalytic core RNP was observed with or without Teb1BC and DNA (data not shown), consistent with previous findings (15). Physical association also did not require the TEN domain L14 side chain, but the 108NAAIRS113 substitution or deletion of the TEN domain reduced RNP association with TASC to a background level (Fig. 6B, lanes 7–9).
FIGURE 6.
TEN domain dependence of catalytic core RNP assembly with TASC. A and B, schematic and results of the interaction assay for holoenzyme reconstitution using full-length (FL) TERT. C and D, schematic and results of the interaction assay for holoenzyme reconstitution using an isolated TEN domain and TERT core RNP. A and C, arrows indicate steps of resin washing. B and D, TASC and Mock indicate the resin-bound complex. Radiolabeled, RRL-expressed FL TERT (WT, L14A, 108NAAIRS113) or TERT core216 combined with radiolabeled p65 and allowed to bind to TASC or mock resin. D, TEN domain omitted (lanes 8 and 16) or added at a final concentration of 50 or 250 nm. Bound TERT core signal intensity (given below the lane numbers) is quantified relative to nonspecific background in the absence of a TEN domain (lane 8).
We extended these results using TEN domain trans-complementation. RRL-assembled TERT core RNP was added to immobilized TASC in the presence or absence of a purified, bacterially expressed TEN domain (Fig. 6C). Like full-length TERT, the TERT core protein showed substantial background binding to resin without TASC (Fig. 6D, lanes 9–16). TASC resin specifically enriched the TERT core and p65 when bacterially expressed wild-type or L14A TEN domain was also present, but not in the presence of the 108NAAIRS113 TEN domain or with the TERT core RNP alone (Fig. 6D, lanes 1–6 and 8). Altogether, we conclude that physical association between the catalytic core RNP and TASC directly or indirectly requires the TEN domain surface disrupted by the 108NAAIRS113 substitution (see supplemental Fig. 1C). We note that the amount of TERT core RNP bound to TASC was always slightly lower in assays with L14A compared with wild-type TEN domain (Fig. 6D; quantification below the lane numbers has background binding normalized to 1). This could reflect a difference in L14A TEN domain conformation that is linked to slowed elongation or an unrelated property of the L14A TEN domain such as increased aggregation.
DISCUSSION
In this study we investigated how TEN domain sequence disruptions affect telomeric primer elongation by a telomerase holoenzyme. We found that the Tetrahymena telomerase holoenzyme retains high RAP even with sequence substitutions that were previously inferred to directly affect a TERT DNA binding site. TERT defects in DNA interaction might be compensated by other holoenzyme subunits, but previous disruptions of individual TEN domain surface residues could also have indirectly reduced catalytic core RNP activity and RAP by destabilizing productive enzyme conformation(s). In the catalytic core RNP alone even wild-type TERT does not adopt the active conformation in a stable or homogeneous manner, as shown by single-molecule FRET demonstrations of population heterogeneity (30).
Although the TEN domain sequence substitutions investigated here did not preclude high RAP, they did affect holoenzyme synthesis initiation and rate of elongation. The same changes in activity were observed whether the TEN domain functioned within full-length TERT or as a physically separate, autonomously folded domain. To explain these phenotypes, we propose that the primary function of the TEN domain is to favor placement or stabilization of a short primer-template hybrid in the active site of the TERT core RNP, which could occur with or without direct physical contact between the TEN domain and template hybrid. Also, anchored through a network of domain interactions (29), the TEN domain could reduce the release of single-stranded DNA product from the elongating enzyme. The topology of product DNA contact by the TEN domain merits some reconsideration because previous cross-linking approaches may have preferentially identified conformationally dynamic side chains of the linker between the TEN domain and TRBD instead of a potentially less reactive DNA contact surface within the stably folded TEN domain itself. Indeed, many amino acids previously considered to be part of the TEN domain can be truncated without loss of TEN domain function in trans-complementation of high RAP (supplemental Fig. S2). We also note that the most severe two of three substitutions (W187A and F178A) originally used to define a DNA binding groove (24) in our hands do not affect catalytic core RNP or holoenzyme activity in a manner that is more severe than other TEN domain single amino acid substitutions, and none of the three defining substitutions (including Q168A) affects holoenzyme RAP (Fig. 2A and supplemental Fig. 1A).
Of particular interest from our studies is the phenotype of the TERT 108NAAIRS113 substitution, which preserved the activity of the catalytic core RNP but precluded its conversion to high RAP holoenzyme activity. No holoenzyme activity reconstitution was observed when the 108NAAIRS113 substitution was assayed in full-length TERT or in an autonomously folded TEN domain used for trans-complementation of the TERT core. In human TERT this same surface could be disrupted by the G100V substitution that prevents stimulation of the RAP of the minimal recombinant enzyme by TPP1 and POT1 (31).
We suggest as a working model that one or more subunits of Tetrahymena TASC, the human protein TPP1, and the yeast protein Est3 are architecturally and functionally analogous in their respective telomerase holoenzymes: holoenzyme protein interaction(s) with the TEN domain contribute allosteric stabilization of the elongation-competent architecture of the catalytic core RNP and could also extend the direct surface of DNA contact (32, 33). Better understanding of the elongating telomerase holoenzyme architecture remains a critical goal for future studies. Our findings establish Tetrahymena telomerase holoenzyme reconstitution as an approach that should be broadly informative for this goal.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM54198.

This article contains supplemental Figs. S1 and S2.
C. M. O'Connor and K. Collins, unpublished data.
- TERT
- telomerase reverse transcriptase
- RAP
- repeat addition processivity
- RNP
- ribonucleoprotein
- RRL
- rabbit reticulocyte lysate
- TASC
- telomere adaptor subcomplex
- TEN
- TERT N-terminal
- TER
- telomerase RNA
- TRBD
- telomerase RNA binding domain.
REFERENCES
- 1. Blackburn E. H., Collins K. (2010) in RNA Worlds (Gesteland R. F., Atkins J. F., Cech T. R., eds) 4th Ed., pp. 205–213, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 2. Hug N., Lingner J. (2006) Telomere length homeostasis. Chromosoma 115, 413–425 [DOI] [PubMed] [Google Scholar]
- 3. Shay J. W., Wright W. E. (2010) Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 584, 3819–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins K., Mitchell J. R. (2002) Telomerase in the human organism. Oncogene 21, 564–579 [DOI] [PubMed] [Google Scholar]
- 5. Chang M., Arneric M., Lingner J. (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a T cel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21, 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Y., Abreu E., Kim J., Stadler G., Eskiocak U., Terns M. P., Terns R. M., Shay J. W., Wright W. E. (2011) Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol. Cell 42, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greider C. W. (1991) Telomerase is processive. Mol. Cell. Biol. 11, 4572–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins K. (2009) in Non-Protein Coding RNAs (Walter N. G., Woodson S. A., Batey R. T., eds) pp. 285–301, Springer-Verlag, Berlin [Google Scholar]
- 9. Greene E. C., Shippen D. E. (1998) Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev. 12, 2921–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greider C. W., Blackburn E. H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 11. Collins K. (2011) Single-stranded DNA repeat synthesis by telomerase. Curr. Opin. Chem. Biol. 15, 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hardy C. D., Schultz C. S., Collins K. (2001) Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276, 4863–4871 [DOI] [PubMed] [Google Scholar]
- 13. Witkin K. L., Collins K. (2004) Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18, 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Witkin K. L., Prathapam R., Collins K. (2007) Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 27, 2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Min B., Collins K. (2009) An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Min B., Collins K. (2010) Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 285, 16434–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng Z., Min B., Huang J., Hong K., Yang Y., Collins K., Lei M. (2011) Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20357–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Licht J. D., Collins K. (1999) Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 13, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cunningham D. D., Collins K. (2005) Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 25, 4442–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robart A. R., O'Connor C. M., Collins K. (2010) Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16, 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Connor C. M., Lai C. K., Collins K. (2005) Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280, 17533–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai C. K., Mitchell J. R., Collins K. (2001) RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21, 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sperger J. M., Cech T. R. (2001) A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40, 7005–7016 [DOI] [PubMed] [Google Scholar]
- 24. Jacobs S. A., Podell E. R., Cech T. R. (2006) Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13, 218–225 [DOI] [PubMed] [Google Scholar]
- 25. Romi E., Baran N., Gantman M., Shmoish M., Min B., Collins K., Manor H. (2007) High resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl. Acad. Sci. U.S.A. 104, 8791–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zaug A. J., Podell E. R., Cech T. R. (2008) Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 15, 870–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Armbruster B. N., Banik S. S., Guo C., Smith A. C., Counter C. M. (2001) N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21, 7775–7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. R., Wong J. M., Collins K. (2003) Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 278, 52531–52536 [DOI] [PubMed] [Google Scholar]
- 29. Robart A. R., Collins K. (2011) Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell 42, 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu J. Y., Stone M. D., Zhuang X. (2010) A single-molecule assay for telomerase structure-function analysis. Nucleic Acids Res. 38, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaug A. J., Podell E. R., Nandakumar J., Cech T. R. (2010) Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenfeld K. K., Ziv T., Goldin S., Glaser F., Manor H. (2011) Mapping of DNA binding sites in the Tetrahymena telomerase holoenzyme proteins by UV cross-linking and mass spectrometry. J. Mol. Biol. 410, 77–92 [DOI] [PubMed] [Google Scholar]
- 33. Yen W. F., Chico L., Lei M., Lue N. F. (2011) Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20370–20375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCormick-Graham M., Romero D. P. (1995) Ciliate telomerase RNA structural features. Nucleic Acids Res. 23, 1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.