Background: MurA is critical for the biosynthesis of the bacterial cell wall.
Results: The covalent MurA-phosphoenolpyruvate adduct was captured in different reaction states.
Conclusion: The covalent adduct primes phosphoenolpyruvate for catalysis and enables feedback inhibition by UDP-N-acetylmuramic acid, the product of MurB.
Significance: Cellular MurA exists in a previously unrecognized and tightly locked complex, which presumably accounts for the failure of drug discovery efforts.
Keywords: Antibiotics, Covalent Regulation, Enzyme Mechanisms, Peptidoglycan, X-ray Crystallography, Murein
Abstract
The enzyme MurA has been an established antibiotic target since the discovery of fosfomycin, which specifically inhibits MurA by covalent modification of the active site residue Cys-115. Early biochemical studies established that Cys-115 also covalently reacts with substrate phosphoenolpyruvate (PEP) to yield a phospholactoyl adduct, but the structural and functional consequences of this reaction remained obscure. We captured and depicted the Cys-115-PEP adduct of Enterobacter cloacae MurA in various reaction states by X-ray crystallography. The data suggest that cellular MurA predominantly exists in a tightly locked complex with UDP-N-acetylmuramic acid (UNAM), the product of the MurB reaction, with PEP covalently attached to Cys-115. The uniqueness and rigidity of this “dormant” complex was previously not recognized and presumably accounts for the failure of drug discovery efforts toward the identification of novel and effective MurA inhibitors. We demonstrate that recently published crystal structures of MurA from various organisms determined by different laboratories were indeed misinterpreted and actually contain UNAM and covalently bound PEP. The Cys-115-PEP adduct was also captured in vitro during the reaction of free MurA and substrate UDP-N-acetylglucosamine or isomer UDP-N-acetylgalactosamine. The now available series of crystal structures allows a comprehensive view of the reaction cycle of MurA. It appears that the covalent reaction of MurA with PEP fulfills dual functions by tightening the complex with UNAM for the efficient feedback regulation of murein biosynthesis and by priming the PEP molecule for instantaneous reaction with substrate UDP-N-acetylglucosamine.
Introduction
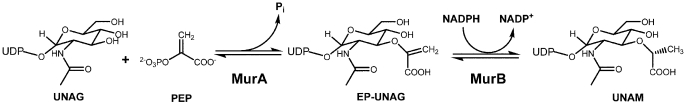
The enzyme MurA (UDP-N-acetylglucosamine 1-carboxyvinyltransferase, EC 2.5.1.7) catalyzes the first step in biosynthesis of the bacterial cell wall and is critical for bacterial survival (1, 2) (Fig. 1). Inhibition of MurA by the antibiotic fosfomycin (3) or the viral maturation protein A2 (4) results in bacterial cell lysis and death. The MurA reaction proceeds through a tetrahedral intermediate of substrates UDP-N-acetylglucosamine (UNAG)3 and phosphoenolpyruvate (PEP) followed by elimination of inorganic phosphate to yield the enolpyruvyl product (EP-UNAG) (5, 6). Enzyme mechanistic studies established that substrate PEP covalently reacts with the catalytically important residue Cys-115 to form a phospholactoyl adduct, suggesting that nucleophilic attack is a component of the reaction cycle (7–9). However, the discovery that Cys-115 could be replaced by an aspartate without loss of activity (a mutation found in some bacteria including Mycobacteria) led to the conclusion that Cys-115 functions as a general acid-base catalyst and not as a nucleophile in the reaction with PEP (10). Cys-115 is a member of a 10-residue loop in the N-terminal globular domain that undergoes drastic conformational changes upon binding of UNAG by approaching the interdomain cleft and positioning the Cys-115 side chain into the PEP binding site (11–15). Cys-115 has received considerable attention as the target of covalent modification by fosfomycin (12) and other electrophilic natural products such as terreic acid (16) and cnicin (17). Notably, no new drug candidates of MurA have been introduced since the discovery of fosfomycin 40 years ago, indicating a lack of critical information required to successfully target this enzyme by reversible inhibitors.
FIGURE 1.
Reactions catalyzed by MurA and MurB.
Despite the extensive knowledge collected by various laboratories over the years regarding the structure-function relationships of MurA, the PEP molecule bound covalently or non-covalently to the enzyme was never depicted in molecular detail. Hence, structural and functional consequences of the PEP interaction with MurA remained elusive. In an attempt to capture the PEP molecule in the course of reaction with MurA, we probed selected mutant enzymes and analogues of UNAG by x-ray crystallography and isothermal microcalorimetry (ITC). Through these studies we found that recombinant Enterobacter cloacae MurA isolated from E. coli expression strains exists in a tight complex with UDP-N-acetylmuramic acid (UNAM), the product of the MurB reaction, with PEP covalently bound to Cys-115. This dormant complex is extraordinarily resilient against dilution and salts commonly used in purification and crystallization but can be readily unlocked by incubation with UNAG (1 mm) or inorganic phosphate (50 mm). Without this treatment, however, MurA remains in the locked state. We demonstrate that previously published co-crystal structures of MurA from Escherichia coli, Haemophilus influenzae, and Aquifex aeolicus by three different laboratories were indeed misinterpreted and actually contain UNAM and covalently bound PEP. The rigidity of this complex presumably accounts for the failure of drug discovery efforts toward the development of reversible MurA inhibitors with antibacterial potential. Using ligand-free MurA, the Cys-115-PEP adduct was captured in two distinct reaction states with substrate or product. The now available series of crystal structures affords an unprecedented view of the individual steps of MurA catalysis in molecular detail. The findings suggest that formation of the Cys-115-PEP adduct fulfills dual functions by tightening the dormant complex with UNAM for the feedback regulation of murein synthesis and by activating the PEP molecule for catalysis.
EXPERIMENTAL PROCEDURES
PEP (potassium salt), UNAG, and UDP-N-acetylgalactosamine (UNAGal) were purchased from Sigma. Reagents for crystallization were purchased from Hampton Research (Aliso Viejo, CA). The concentration of crystallization-grade proteins was determined by A280 molar absorbance using a nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Cloning and Expression
Primers were designed and purchased from MWG Biotech (High Point, NC) to introduce C115D and R120A single mutations into wild-type MurA from E. cloacae in a pET-9d vector (Novagen, EMD4 Biosciences, San Diego, CA) using the QuikChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA). Wild-type and mutant proteins were separately overexpressed in E. coli BL21 (DE3) cells at 37 °C.
Protein Purification
Harvested cells were resuspended in 50 mm Tris buffer (pH 8.0) containing 100 mm NaCl, 1 mm EDTA, 2 mm DTT, 0.1 mg ml−1 lysozyme, and 0.01% Triton X-100 at 4 °C for 60 min. After sonication and centrifugation (60 min at 29,000 × g), solid ammonium sulfate ((NH4)2SO4) was added slowly to the supernatant over 60 min while stirring to achieve 25% saturation (1 m). The supernatant resulting from a second round of centrifugation (30 min at 29,000 × g) was subjected to hydrophobic interaction chromatography using a 70-ml phenyl-Sepharose high performance column (GE Healthcare). After washing the column with 1 liter of 50 mm Tris buffer (pH 8.0) containing 1 mm EDTA, 1 mm DTT, and 1 m (NH4)2SO4, MurA was eluted over a 1-liter linear gradient from 1 to 0 m (NH4)2SO4. Fractions containing MurA were pooled, concentrated, and exchanged into 50 mm Tris (pH 8.0), 1 mm EDTA, and 1 mm DTT using a Sephadex G-25 column (GE Healthcare) and were further purified using a 50-ml Q-Sepharose Fast Flow column (GE Healthcare) with a 1-liter linear gradient from 0 to 300 mm NaCl. MurA peak fractions were then concentrated and loaded onto a HiLoad 26/60 Superdex 200 column (GE Healthcare) equilibrated with 50 mm Tris (pH 8.0), 150 mm NaCl, 1 mm EDTA, and 1 mm DTT. The resulting eluate yielded crystallization-grade monomeric enzyme.
Protein Crystallography
After the last purification step, one third of the wild-type protein was loaded onto a PD-10 column (GE Healthcare) and eluted with 50 mm HEPES (pH 7.5). A second third of the protein was eluted into 50 mm sodium/potassium phosphate buffer (pH 6.8), and the final portion was first eluted into 50 mm sodium/potassium phosphate buffer (pH 6.8) followed by an additional PD-10 column to transfer the protein into 50 mm HEPES buffer (pH 7.5). The C115D and R120A mutant MurA proteins were directly transferred into 50 mm HEPES (pH 7.5) and 50 mm Tris (pH 8.0), respectively. All buffers used for PD-10 columns contained 2 mm DTT. Wild-type, C115D, and R120A MurA were concentrated to 72, 100, and 120 mg ml−1, respectively, using Amicon centrifugal filter devices (Millipore, Billerica, MA) at 4 °C. All crystals were grown using the hanging drop vapor diffusion method at 18 °C from a 1:1 (v/v) ratio of protein (40 mg ml−1 wild-type and C115D or 120 mg ml−1 R120A) and reservoir solutions (Table 1). For data collection, crystals were harvested in a cryoprotectant mixture consisting of the respective reservoir composition and including 20% (v/v) ethylene glycol and 0.5 mm concentrations each of the respective ligand when appropriate.
TABLE 1.
Reaction states of E. cloacae MurA determined by crystallography
| Complexa | MurA | Compounds provided | Ligands bound | Crystallization reservoir |
PDB ID | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Buffer | Salt | Precipitant | ||||||
| I | WTb | UNAM | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3350 | 3SU9 | This work | |
| +PEP-Cys | ||||||||
| II | R120A | 5 mm UNAG | UNAG | 0.1 m HEPES (pH 7.5) | 14% 2-Propanol, 20% PEG 4000 | 3SWA | This work | |
| + 5 mm PEP | +PEP-Cys | |||||||
| III | D305A | 5 mm UNAG | THI | 0.01 m MES (pH 6.4) | 10% PEG 20000 | 1Q3G | Ref. 5 | |
| + 5 mm PEP | ||||||||
| IV | C115S | 5 mm UNAG | EP-UNAG | 0.01 m MES (pH 6.4) | 10% PEG 20000 | 1RYW | Ref. 13 | |
| + 5 mm PEP | +Pi | |||||||
| V | WTc | 2 mm UNAGal | EP-UNAGal | 0.1 m HEPES (pH 7.5) | 0.05 m MgCl2 | 30% PEG MME 550 | 3SWI | This work |
| + 2 mm PEP | +PEP-Cys | |||||||
| VI | WTb | 2 mm UNAG | EP-UNAG | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3,350 | 3SWQ | This work |
| VII | WTc | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3350 | 1EJD, 1EJC, and 3SPB | Ref. 15 and this work | ||
| VIII | WTc | 2 mm UNAG | UNAG | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3350 | 3UPK | This work |
| D-VII | C115Db | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3350 | 3V5V | This work | ||
| D-VIII | C115Db | 2.5 mm UNAG | UNAG | 0.1 m Bis-Tris (pH 5.5) | 0.2 m NH4OAc | 25% PEG 3350 | 3V4T | This work |
X-ray diffraction data were recorded in the Moffitt Cancer Center Structural Biology Core at −180 °C using CuKα X-rays generated by a Rigaku Micro-Max 007-HF x-ray generator, focused by mirror optics and equipped with a Rigaku CCD Saturn 944 system, on single crystals frozen in liquid N2. Data were reduced with XDS (18). The structures were determined by molecular replacement using MOLREP (CCP4) (19) with PDB entry 1EJC or 3KQA as the search model. CNS (20) was employed for refinement (minimization and simulated annealing), and manual model building was performed using Coot (21). Initial models for the modified amino acid and small molecule ligands were generated using Chem3DPro (CambridgeSoft, Cambridge, MA) with parameters and topology files from the program xplo2d. Figures were drawn with PyMOL (Schrödinger, Cambridge, MA). The MurA structures 2RL1, 2YVW, and 3ISS were refined using the deposited structure factors converted to CNS format by sf-convert (RCSB Software Tools). Data collection and refinement statistics are shown in Tables 2 and 3. The atomic coordinates and structure factors for complexes I, II, V, VI, VII, VIII, D-VII, and D-VIII have been deposited under accession codes 3SU9, 3SWA, 3SWI, 3SWQ, 3SPB, 3UPK, 3V5V, and 3V4T, respectively. The re-refined atomic coordinates for the previously published structures of MurA from H. influenzae (PDB code 2RL1), A. aeolicus (PDB code 2YVW), and E. coli (PDB code 3ISS) have been deposited under accession numbers 3SWE, 3SWG, and 3SWD, respectively.
TABLE 2.
Summary of data collection and structure refinement
| Structure (PDB ID) | I (3SU9) | II (3SWA) | V (3SWI) | VI (3SWQ) | VII (3SPB) | VIII (3UPK) | D-VII (3V5V) | D-VIII (3V4T) |
|---|---|---|---|---|---|---|---|---|
| Data Collection | ||||||||
| Space group | I23 | P21 | I23 | I23 | P212121 | I23 | P212121 | P21 |
| Cell dimensions (a, b, c (Å); β, if >90°) | 138.6, 138.6, 138.6 | 60.1, 92.3, 71.5; 102.7 | 137.4, 137.4, 137.4 | 138.2, 138.2, 138.2 | 81.1, 101.2, 213.8 | 138.5, 138.5, 138.5 | 81.1, 101.7, 213.3 | 83.1, 153.1, 131.7; 102.4 |
| Resolution range (Å) | 20-2.20 (2.30-2.20) | 20-1.90 (1.95-1.90) | 20-2.80 (2.90-2.80) | 20-1.83 (1.90-1.83) | 20-2.30 (2.35-2.30) | 20-2.00 (2.10-2.00) | 20-2.70 (2.80-2.70) | 20-2.50 (2.60-2.50) |
| Unique reflections | 22,499 (2,771) | 59,238 (4,323) | 10,689 (1,095) | 38,692 (4,096) | 78,476 (4,864) | 29,840 (3,990) | 48,907 (4,852) | 110,457 (12,247) |
| Completeness (%) | 99.5 (99.8) | 95.5 (92.7) | 98.8 (99.3) | 99.7 (99.9) | 99.3 (99.9) | 99.7 (99.8) | 99.2 (97.0) | 99.5 (99.7) |
| I/σI | 33.1 (5.7) | 15.8 (3.9) | 18.5 (4.9) | 25.0 (5.8) | 19.7 (4.5) | 12.3 (4.5) | 18.5 (4.4) | 19.7 (4.5) |
| Rmerge (%)a | 7.9 (31.2) | 7.6 (30.6) | 12.0 (42.0) | 5.9 (29.4) | 9.4 (37.0) | 11.0 (34.9) | 12.1 (36.1) | 10.2 (42.5) |
| Structure refinement | ||||||||
| Protein atoms | 3157 | 6307 | 3153 | 3168 | 12568 | 3149 | 12576 | 25152 |
| Average B-factor (Å2) | 26 | 19 | 36 | 19 | 31 | 22 | 23 | 42 |
| Ligand atoms | 44 | 78 | 83 | 44 | 0 | 39 | 0 | 312 |
| Average B-factor (Å2) | 21 | 15 | 35 | 16 | 18 | 33 | ||
| Solvent molecules | 310 | 691 | 113 | 415 | 661 | 380 | 476 | 470 |
| Average B-factor (Å2) | 33 | 26 | 28 | 30 | 32 | 32 | 18 | 34 |
| r.m.s.d. bonds (Å)b | 0.011 | 0.010 | 0.010 | 0.011 | 0.010 | 0.010 | 0.009 | 0.010 |
| r.m.s.d. angles (°) | 1.8 | 1.7 | 1.7 | 1.7 | 1.8 | 1.7 | 1.9 | 1.8 |
| Rcryst (%)c | 16.2 | 17.5 | 18.8 | 16.0 | 20.0 | 16.8 | 19.1 | 20.8 |
| Rfree (%)d | 21.0 | 20.1 | 26.3 | 20.4 | 25.0 | 19.7 | 26.4 | 27.1 |
| Rfree reflection set size | 1103 (4.9%) | 1185 (2.0%) | 556 (5.2%) | 1121 (2.9%) | 1178 (1.5%) | 1105 (3.7%) | 1223 (2.5%) | 1216 (1.1%) |
| Ramachandran favored (%)e | 97.6 | 98.4 | 94.4 | 98.8 | 97.7 | 97.6 | 95.8 | 96.7 |
| Ramachandran outliers (%)e | 0.0 | 0.1 | 0.2 | 0.0 | 0.1 | 0.0 | 0.4 | 0.1 |
| Cross-validated coordinate errorf | ||||||||
| From Luzzati plot (Å) | 0.28 | 0.22 | 0.46 | 0.21 | 0.35 | 0.23 | 0.43 | 0.42 |
| From SigmaA (Å) | 0.22 | 0.19 | 0.55 | 0.12 | 0.30 | 0.16 | 0.52 | 0.39 |
a Rmerge = quality of amplitudes (F) in the scaled data set (29).
b r.m.s.d., root mean square deviation from ideal values.
c Rcryst = 100 × Σ |Fobs − Fmodel|/Fobs where Fobs and Fmodel are the observed and calculated structure factor amplitudes, respectively.
d Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement.
e Calculated by Molprobity (30).
f Cross-validated coordinate error was calculated by CNS (31).
TABLE 3.
Summary of structure refinement for previously deposited MurA structures
| Structure | 3SWE | 3SWG | 3SWD |
|---|---|---|---|
| Data collection | |||
| Source | H. influenzae | A. aeolicus | E. coli |
| Template | 2RL1 | 2YVW | 3ISS |
| Resolution range (Å) | 20-2.20 | 20-1.80 | 20-2.50 |
| Structure refinement | |||
| Protein atoms | 3,148 | 3,270 | 37,716 |
| Average B-factor (Å2) | 28 | 22 | 58 |
| Ligand atoms | 44 | 44 | 528 |
| Average B-factor (Å2) | 25 | 17 | 49 |
| Solvent molecules | 318 | 348 | 791 |
| Average B-factor (Å2) | 35 | 34 | 45 |
| r.m.s.d. bonds (Å)a | 0.011 | 0.010 | 0.013 |
| r.m.s.d. angles (°) | 1.6 | 1.4 | 1.9 |
| Rcryst (%)b | 18.6 | 16.5 | 22.6 |
| Rfree (%)c | 23.8 | 19.0 | 28.0 |
| Rfree reflection set sized | 2594 (10.2%) | 2516 (5.0%) | 8367 (5.0%) |
| Ramachandran favored (%)e | 97.1 | 98.3 | 96.7 |
| Ramachandran outliers (%)e | 0.0 | 0.0 | 0.0 |
| Cross-validated coordinate errorf | |||
| From Luzzati plot (Å) | 0.30 | 0.19 | 0.44 |
| From SigmaA (Å) | 0.30 | 0.17 | 0.60 |
a r.m.s.d., root mean square deviation from ideal values.
b Rcryst = 100 × Σ |Fobs − Fmodel|/Fobs where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
c Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement.
d Rfree reflections were inherited from original structure factor.
e Calculated by Molprobity (30).
f Cross-validated coordinate error was calculated by CNS (31).
ITC
The binding of ligands to wild-type (phosphate-treated) and C115D MurA in 50 mm HEPES (pH 7.5) and 2 mm DTT was analyzed using a MicroCal iTC200 titration calorimeter (GE Healthcare). After an initial 0.5-μl injection, a total of 18 or 24 aliquots (2.3 or 1.7 μl each, respectively) of the ligand solution was injected into 200 μl of the protein solution at 25 °C (wild type) or 35 °C (C115D). The solution was stirred constantly at 1000 rpm, and data were recorded for 120–150 s between injections. For binding of ligands to MurA only, wild-type (100 μm) or C115D (200 μm) MurA was titrated with UNAG (1.5 or 3.5 mm, respectively) or UNAGal (2.5 or 6 mm, respectively). To measure binding of fosfomycin or PEP to the MurA·UNAG-UNAGal binary complexes, wild-type or C115D MurA (50 μm) was preincubated with 1 mm UNAG or UNAGal before titration with fosfomycin (0.4 mm) or PEP (0.5 mm), respectively. Generation of heat due to dilution was determined in a separate experiment by diluting protein into buffer and subtracting these as blank values for each injection. Corrected heat values were fitted using a nonlinear least square curve-fitting algorithm (Microcal Origin 7.0) to obtain the stoichiometry (n), binding constants (Ka, Kd), and change in enthalpy for each enzyme-ligand interaction (ΔH).
RESULTS AND DISCUSSION
In an attempt to capture the intact PEP molecule in the reaction of MurA, we probed wild-type and single-site-directed mutant enzymes of recombinant E. cloacae MurA with analogues of UNAG for co-crystallization in the presence and absence of PEP. MurA generally crystallizes in different space groups and higher oligomeric states depending on the conformation of the 10-residue loop containing Cys-115 (5, 6, 11–13,15, 16, 22–25), which often negatively impacts the diffraction quality and feasibility of structure determination. We, therefore, broadened the spectrum of previously established crystallization conditions by subjecting the enzyme to new crystallization screening trials. The use of different buffers into which the enzyme was transferred before crystallization proved crucial to crystallization of the various enzyme-ligand complexes presented here (Table 1).
Recombinant MurA Expressed in E. coli Exists in Tight Complex with UDP-N-Acetylmuramic Acid and PEP
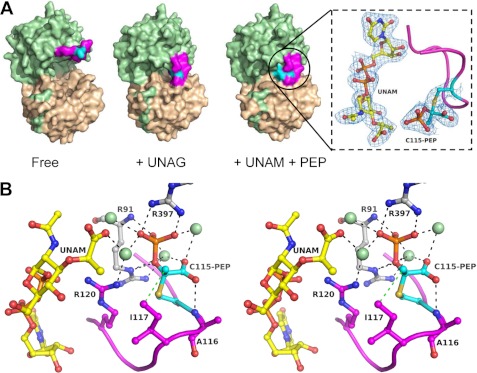
We found that E. cloacae MurA overexpressed in E. coli crystallized in the closed state with a fully occupied active site, although ligands were not present during purification or crystallization (Fig. 2A, Table 2). This result was unexpected, as we had previously crystallized the same enzyme in distinct open and ligand-free states (22). The only difference between this and our previous studies is that the enzyme was not treated with phosphate buffer before crystallization, as discussed below. The results indicate tight ligand binding within the complex, as evidenced by the extraordinary resilience of this complex against dilution during purification and in preparation for crystallization. Detailed inspection of the electron density revealed that the UNAG binding site is occupied with UNAM, the product of the MurB reaction, which was previously recognized by Berti and co-workers (26) as a potent inhibitor of MurA. Remarkably, the complex also contains a clearly defined PEP molecule covalently attached to Cys-115 and positioned next to the muramic acid moiety. This phospholactoyl adduct forms multiple electrostatic interactions with positively charged residues, thereby stabilizing the loop, tightening the complex, and effectively shielding the active site from solvent (Fig. 2B).
FIGURE 2.
MurA exists in a tight complex with UNAM and PEP covalently attached to Cys-115. A, surface representations of different states of MurA (ligand-free (VII), UNAG (VIII), and UNAM+PEP (I)) show the closure of the active site upon covalent reaction of Cys-115 with PEP. The 2Fo − Fc electron density at 2.2 Å resolution and contoured at 1σ is shown as blue mesh for UNAM and adduct of the dormant complex. The Fo − Fc omit map is shown in supplemental Fig. S2. The C-terminal domain of MurA is colored in beige, the N-terminal domain is in green, the loop 112–121 is in magenta, and Cys-115 or the Cys-115-PEP adduct is in cyan. B, shown is a stereo presentation of the PEP binding site in the dormant complex with UNAM. Black dotted lines indicate hydrogen bonding or electrostatic interactions, and the green spheres are water molecules.
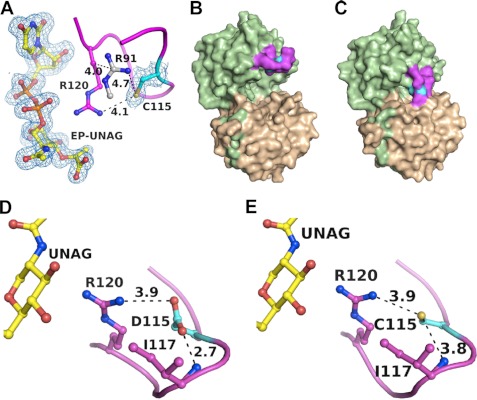
The Cys-115-PEP adduct of MurA was identified by biochemical methods almost two decades ago (8, 9), but the structure and physiological or mechanistic rationale behind this reaction remained elusive. Initially characterized as a covalent intermediate in the MurA reaction, the catalytic competence of the C115D mutant enzyme suggested that the covalent adduct is a mere side product of a general acid-base reaction cycle devoid of covalent intermediates (10). Our finding of a tight MurA-PEP·UNAM complex indicates that the covalent reaction with PEP is of functional significance. Co-crystallization with PEP rendered the complex unchanged, whereas co-crystallization with UNAG yielded a new complex with EP-UNAG and Cys-115 free of PEP (Fig. 3A). As PEP was not present during crystallization, it appears that UNAG displaced UNAM followed by transfer of PEP from Cys-115 to UNAG and yielding EP-UNAG in a single-turnover reaction. These findings corroborate earlier biochemical studies that the Cys-PEP adduct is catalytically competent and that incubation of purified MurA with UNAG yielded enzyme free of PEP (8, 9). Using the same expression system and purification procedures, the E. cloacae C115D MurA isolated from E. coli is open and free of ligands (Fig. 3B), emphasizing the importance of the nucleophilic cysteine side chain for the formation of the dormant complex. The crystal structure of the C115D MurA was also determined with UNAG bound (Fig. 3C), revealing for the first time that the cysteine and aspartate side chains adopt similar positions and conformations in the PEP binding site (Fig. 3, D–E).
FIGURE 3.
The dormant complex is catalytically competent but does not form with C115D MurA. A, upon incubation with UNAG, the dormant complex catalyzes a single turnover to yield EP-UNAG and free Cys-115 (VI). Shown as blue mesh is the 2 Fo − Fc electron density at 1.8 Å resolution, contoured at 1σ. Purified C115D mutant enzyme is free of ligands (D–VII) (B) and closes upon interaction with UNAG (D–VIII) (C). The conformation of the loop and positioning of the aspartate side chain in the closed state with UNAG (D) is similar to that of the wild-type enzyme (E).
The dormant complex opens and releases both UNAM and PEP after transfer into 50 mm sodium/potassium phosphate buffer (pH 6.8). Buffers such as Tris, MES, MOPS, and HEPES as well as salts such as ammonium sulfate (up to 1 m), KCl (150 mm), or NaCl (150 mm) did not affect the complex. Inorganic phosphate is a product of the forward reaction and was previously found to be present in the PEP binding site of the C115S mutant MurA (supplemental Fig. S1) (13). High phosphate concentrations may, therefore, disrupt the complex by forcing the PEP-Cys-115 adduct of the active site, thereby opening the enzyme and releasing bound UNAM. Upon exposure to solvent and reducing agents such as DTT in the buffer, PEP is liberated from Cys-115, and the enzyme adopts the familiar open, ligand-free state. Inorganic phosphate also binds to the open enzyme state in the space normally occupied by the diphospho bridge of the UNAG molecule (22). It is possible that additional low affinity phosphate binding sites exist that influence the stability of the dormant complex. However, the data suggest that cellular levels of inorganic phosphate are not sufficient to unlock the complex.
Previously Released Crystal Structures of MurA from Different Organisms Were Misinterpreted
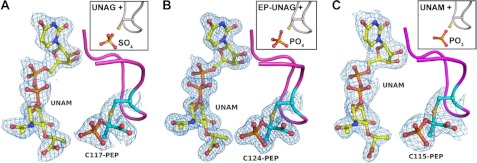
Previously we and others were unaware of the effect of inorganic phosphate on purified MurA to yield free enzyme. Our own studies (5, 11, 13–16, 22, 23, 27) were performed with enzyme routinely transferred into phosphate buffer in the preparation for structural (small angle x-ray scattering and crystallography) and biochemical (kinetics and fluorescence) experiments, enabling characterization of the distinct open and closed states of the enzyme. As structural studies of MurA by other laboratories did not involve treatment with phosphate before crystallization, we questioned whether crystal structures of MurA deposited in the PDB indeed contained the ligands as claimed or if the same MurA-PEP·UNAM complex had been crystallized unknowingly. We thoroughly reinvestigated the recently deposited crystal structures of E. coli MurA liganded with UNAM and phosphite (PDB code 3ISS (25)), H. influenzae MurA liganded with UNAG and sulfate (PDB code 2RL1 (24)), and A. aeolicus MurA liganded with EP-UNAG and phosphate (PDB code 2YVW, no associated publication). Using the deposited structure factors along with the respective coordinate sets, we performed refinement cycles in the presence and absence of ligands (Table 3). The results clearly demonstrate that the electron density of the respective active sites had previously been misinterpreted (Fig. 4). Each structure contains UNAM, and the originally assigned anions actually represent the phosphate group of covalently bound PEP. Combined, these findings indicate that MurA from various bacterial species exist predominantly in a dormant state under physiological conditions, supporting the previously proposed role of UNAM as a feedback inhibitor of murein biosynthesis and revealing a new function of PEP by locking the MurA·UNAM complex.
FIGURE 4.
MurA exists predominantly in a tightly locked state with UNAM and covalently attached PEP. Previous crystal structures of MurA were misinterpreted and actually contain UNAM and the Cys-115-PEP adduct. Structure factors deposited in the PDB were refined against MurA with and without ligands, and the resulting 2 Fo − Fc electron density maps contoured at 1σ are shown for MurA from H. influenzae (PDB entry 3SWE, original 2RL1), which was originally interpreted as UNAG and sulfate ion (inset) (24) (A), MurA from A. aeolicus (PDB entry 3SWG, original 2YVW), which was originally interpreted as EP-UNAG and phosphate ion (B), and MurA from E. coli (PDB entry 3SWD, original 3ISS), which was originally interpreted as UNAM and phosphite ion (25) (C). The Fo − Fc electron density maps omitting the ligands during the refinement are shown in supplemental Fig. S2.
Cys-115-PEP Adduct Also Forms with UNAG Analogues and Mutant Enzymes
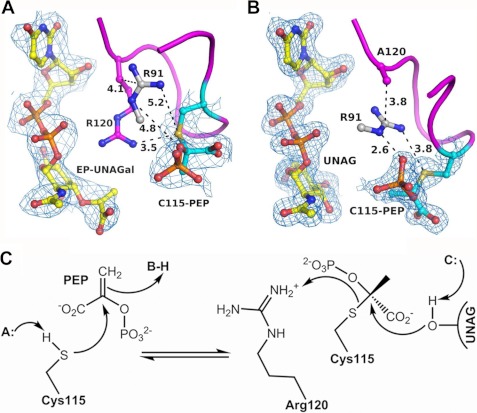
We next asked if the Cys-115-PEP adduct could also be generated in vitro by incubating ligand-free (phosphate-treated) MurA with PEP and UNAGal, an isomer of UNAG. The resulting crystal structure contained EP-UNAGal and PEP covalently bound to Cys-115 (Fig. 5A). The covalent adduct adopts the same conformation as in the dormant complex with UNAM, and the conformation of EP-UNAGal is identical to that of EP-UNAG, suggesting a single turnover or very slow reaction. The 4′-OH group of UNAGal interacts more favorably with the side chain of Asp-305, which may hinder product release. Interestingly, the crystal structure also shows that a single UNAGal molecule is bound outside the PEP binding site interacting with the Cys-115-PEP adduct through coordination with a Mg2+ ion (supplemental Fig. S3). Using the same conditions, MurA failed to crystallize in the presence of UNAG and PEP presumably due to the microheterogeneity arising from the various enzyme states interchanging rapidly reversible with substrates and products.
FIGURE 5.
The covalent adduct forms in vitro during the reaction of free MurA with UNAG and UNAGal. A, co-crystallization of free enzyme with UNAGal and PEP yielded EP-UNAGal and PEP covalently attached to Cys-115 (V), suggesting a single turnover and activation of PEP during the catalytic cycle. B, The Cys-115-PEP adduct also formed upon incubation of the R120A mutant enzyme with UNAG (II), suggesting that the guanidinium group of Arg-120 serves as proton donor in the transfer of PEP from Cys-115 to UNAG. C, in this mechanism the nucleophilic attack of the sulfanion on the C2 atom of PEP leads to formation of the thioketal, and the transfer of PEP to the target hydroxyl of UNAG is facilitated by protonation of Cys-115 by Arg-120. Shown as blue mesh is the 2 Fo − Fc electron density at 2.8 Å and 1.9 resolution, contoured at 1σ.
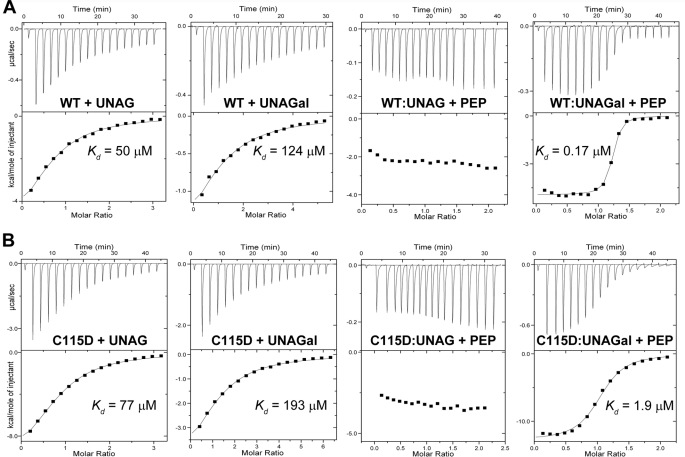
MurA did not display appreciable catalytic activity with UNAGal as a substrate, and direct binding studies using ITC support the crystallographic observations (Fig. 6). With a dissociation constant of 124 μm, UNAGal binds to free wild-type MurA ∼2-fold less efficiently than UNAG (Kd = 50 μm, as compared with Kd = 51 μm (28)). The heat generated during normal reaction with saturating UNAG (1 mm) and increasing PEP concentrations is not expected to change significantly between titrations, as PEP continues to convert immediately to product until equilibrium between forward and reverse reactions has been established. However, under single-turnover (very slow reaction) conditions with UNAGal as a substrate, PEP displayed very high affinity binding (Kd = 0.17 μm), presumably reflecting the covalent reaction with Cys-115 as shown in the crystal structure (complex V, Fig. 5A). PEP displayed significantly lower binding potential to the C115D mutant enzyme in complex with UNAGal (Kd = 1.9 μm) (Fig. 6B), indicating rapidly reversible binding. This 10-fold difference in binding potential suggests that the reaction of wild-type MurA proceeds through formation of the Cys-115-PEP adduct. Notably, PEP did not show binding potential to free MurA. Under steady-state conditions with both PEP and UNAG at saturation levels (1 mm each), the assay progress curves measured between 30 and 300 s for the dormant complex, free MurA, or the C115D enzyme did not show burst or lag phases. Under these conditions, bound UNAM is readily displaced by UNAG, and the reaction proceeds equally fast for the dormant complex and the free enzyme. However, it is likely that changes in the onset of the reaction are observable under presteady state conditions and varying substrate concentrations.
FIGURE 6.
Interaction of substrates with wild-type MurA (A) and C115D MurA (B) as determined by ITC.
The Cys-115-PEP adduct was also captured using the R120A mutant of MurA after incubation with UNAG and PEP (Fig. 5B). This mutant enzyme is catalytically incompetent and accumulates UNAG in the active site instead of EP-UNAG. PEP is covalently bound to Cys-115, and the loop adopts a slightly different conformation. The lack of the Arg-120 side chain allows Arg-91 to migrate into the active site and interact with the Cys-115-PEP adduct, possibly obstructing the transfer of PEP from Cys-115 to UNAG. The data suggest that in the wild-type enzyme, Arg-120 may donate a proton to the thiol group of the covalent adduct, such that Cys-115 becomes a good leaving group for the transfer of PEP to UNAG (Fig. 5C).
Reaction of Most MurA Enzymes Likely Proceeds through Cys-115-PEP Adduct
Taken together, the findings suggest that the reaction of PEP with Cys-115 holds significance for the function and regulation of MurA in the cell. The information gained from our crystal structures as well as those determined previously enable a comprehensive picture of the MurA reaction in atomic detail (Figs. 7 and 8). The existence of distinct complexes of MurA with PEP covalently attached to Cys-115 support the original proposal from the Wanke and Amrhein (9) and Anderson and co-workers (8) laboratories that the covalent adduct is a regular reaction intermediate.
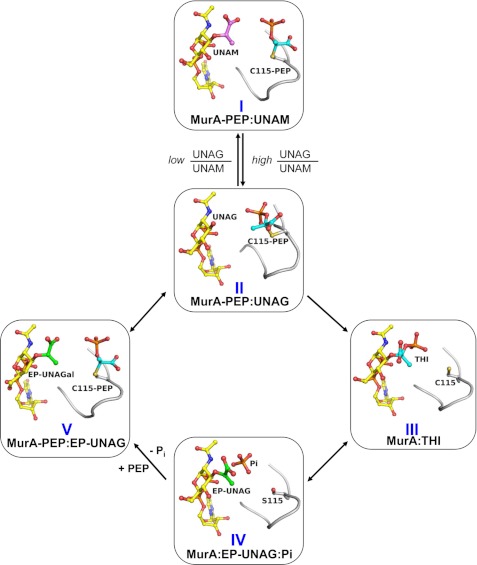
FIGURE 7.
Proposed reaction cycle of E. cloacae MurA involving the Cys-115-PEP adduct. The reaction cycle is derived from the different reaction states of MurA obtained by crystallography with wild-type or mutant enzyme liganded with UNAM, UNAG or UNAGal. The structure identifiers are the same as in Table 1. The cellular UNAG/UNAM ratio controls formation of the dormant complex I. Rapidly reversible exchange with UNAG yields complex II followed by transfer of PEP to UNAG to yield the tetrahedral intermediate, THI (III), which breaks down to the ternary products complex (IV). Covalent reaction of PEP with IV displaces Pi to yield complex V followed by rapidly reversible exchange of EP-UNAG and UNAG to complete the cycle. The PEP molecule is shown in cyan, the enolpyruvyl moiety is in green, and the muramic acid moiety is in magenta. The loop containing Cys-115 is shown in gray.
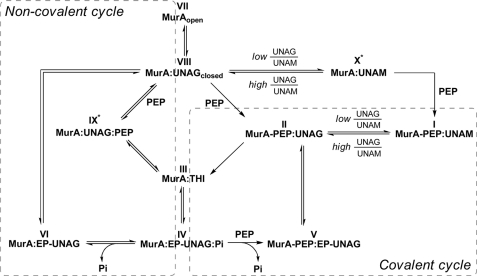
FIGURE 8.
Reaction mechanism of E. cloacae MurA according to the various enzyme states determined by crystallography. Shown are the covalent (right) and non-covalent (left) reaction cycles of MurA, which are interconnected via complexes III and IV. The structure identifiers are the same as in Table 1. Cellular MurA exists predominantly in the dormant complex (I), and the covalent reaction cycle may proceed as shown in Fig. 7. In vitro, the open unliganded enzyme (VII) interacts first with UNAG to form the binary complex (VIII). PEP may then react in a rapidly reversible manner to yield the ternary substrate complex (IX) or covalently with Cys-115 to yield complex II. Both complexes II and IX are catalytically competent and give rise to the THI (III). The product complex IV may release phosphate to yield complex VI followed by exchange with UNAG to revert to complex VIII, which completes the non-covalent cycle (VIII, IX, III, IV, and VI). It is possible that dormant complex I releases bound PEP in a single turnover reaction from II to III and that subsequent reactions proceed through the non-covalent cycle. With decreasing UNAG/UNAM ratios, a rapidly reversible complex with UNAM (X) may form to which PEP binds covalently and yields the dormant complex. IX* and X* denote that the crystal structures have not yet been determined.
The data suggest that cellular MurA exists predominantly in the dormant complex (I), which reacts in a rapidly reversible manner with UNAG (II) to yield the tetrahedral intermediate (THI) (III) (Fig. 7). The THI breaks down into reaction products EP-UNAG and inorganic phosphate (IV). PEP enters the active site and reacts with Cys-115, which displaces phosphate to yield complex V. Rapidly reversible exchange of EP-UNAG and UNAG (II) completes the cycle. This covalent reaction cycle (II, III, IV, and V) is driven by formation of the Cys-115-PEP adduct. Depletion of UNAG and an increase in UNAM by the MurB reaction favors formation of complex I, which likely plays a regulatory role in slowing flux through the pathway.
In vitro, the open unliganded enzyme (VII) interacts first with UNAG to form the binary complex (VIII) (Fig. 8). PEP may then react in a rapidly reversible manner to yield the ternary substrate complex IX or covalently with Cys-115 to yield complex II. Both complexes II and IX are catalytically competent and give rise to the THI (III). The product complex IV may release phosphate to yield complex VI followed by exchange with UNAG to revert to complex VIII, completing the non-covalent cycle (VIII, IX, III, IV, and VI). As both cycles are interconnected via complexes III and IV, it is possible that dormant complex I releases bound PEP in a single turnover reaction from II to III and that subsequent reactions proceed through the non-covalent cycle. With decreasing UNAG/UNAM ratios, a rapidly reversible complex with UNAM (X) may form to which PEP binds covalently and yields the dormant complex. The structure of complex X has not yet been determined, but it is likely that this complex forms readily under high UNAM concentrations. Intriguingly, although the formation of complex IX is central to the non-covalent cycle, it remains the only reaction state that was not able to be captured by crystallography despite numerous trials with different mutant enzymes and UNAG analogues. The presently available crystal structures support a covalent reaction cycle in MurA enzymes with an active site cysteine residue, whereas C115D enzymes exclusively catalyze the non-covalent reaction cycle.
Conclusions
Our findings indicate that cellular depletion of UNAG and concomitant accumulation of UNAM leads to formation of a tight MurA·UNAM complex with PEP covalently attached to Cys-115. This dormant complex displays extreme resilience against dilution and most salts and precipitants used in purification and crystallization. In this study only substrate UNAG, isomer UNAGal (at 1 mm), or product inorganic phosphate (at 50 mm) were able to unlock the complex. Using UNAG analogues and mutant enzymes we were able to generate the same covalent adduct with PEP in vitro, and the now available crystal structures support the original hypothesis that the reaction of MurA enzymes with an active site cysteine residue proceeds through a covalent intermediate.
Previous studies on MurA by other laboratories may have unknowingly been performed with the dormant complex, and we demonstrate using data deposited in the PDB by three different laboratories that those crystal structures were indeed misinterpreted. The tightness of this complex may explain the failure of drug discovery efforts toward the identification of novel reversible inhibitors of MurA with potential as antibiotic drugs. The natural product fosfomycin, discovered half a century ago, remains the only antibiotic drug specifically targeting MurA (12). Fosfomycin and other natural products such as cnicin (17) and terreic acid (16) are covalent MurA inhibitors that essentially compete with PEP for reaction with Cys-115. In contrast, reversible MurA inhibitors require the ability to unlock and open the dormant complex in competition with UNAM and UNAG. However, the mechanism by which UNAG or UNAM induces the open-closed transition is obscure, and the only drug-like molecules crystallized with MurA bind to or stabilize the open enzyme state (15, 23). It appears that identification of the molecular framework required to stabilize a closed enzyme-inhibitor complex with covalently bound PEP is an essential step in the future design of novel MurA inhibitors.
The C115D MurA variant found in Mycobacteria and other species catalyzes enolpyruvyl transfer through a general acid-base mechanism with rapidly reversible binding of PEP (Fig. 8) similar to that of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) (which has an active site glutamate) (10). As the C115D enzyme lacks the ability to react with PEP covalently, a tight complex with UNAM cannot be formed, and the enzyme is presumably vulnerable to inhibition by a wider range of small molecule inhibitors able to interfere with the open-closed transition. It is conceivable that the highly conserved active site cysteine residue provides an evolutionary advantage over aspartate, as the covalent reaction between Cys-115 and PEP enables tight regulation by UNAM, provides protection from potential inhibitors through tight complex formation, and primes PEP for instantaneous reaction with substrate UNAG.
Supplementary Material
Acknowledgments
We thank the Moffitt Chemical Biology Core for use of the protein crystallography and ITC instrumentation and Donna J. Ingles from the Schönbrunn laboratory for help with the manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01GM070633.

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3SU9, 3SWA, 3SWI, 3SWQ, 3SPB, 3UPK, 3V5V, 3V4T, 3SWE, 3SWG, and 3SWD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UNAG
- UDP-N-acetylglucosamine
- EP-UNAG
- enolpyruvyl-UNAG
- PEP
- phosphoenolpyruvate
- UNAGal
- UDP-N-acetylgalactosamine
- EP-UNAGal
- enolpyruvyl-UNAGal
- UNAM
- UDP-N-acetylmuramic acid
- ITC
- isothermal microcalorimetry
- THI
- tetrahedral intermediate.
REFERENCES
- 1. Brown E. D., Vivas E. I., Walsh C. T., Kolter R. (1995) MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177, 4194–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Heijenoort J. (2001) Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18, 503–519 [DOI] [PubMed] [Google Scholar]
- 3. Kahan F. M., Kahan J. S., Cassidy P. J., Kropp H. (1974) The mechanism of action of fosfomycin (phosphonomycin). Ann. N.Y. Acad. Sci. 235, 364–386 [DOI] [PubMed] [Google Scholar]
- 4. Bernhardt T. G., Wang I. N., Struck D. K., Young R. (2001) A protein antibiotic in the phage Qβ virion. Diversity in lysis targets. Science 292, 2326–2329 [DOI] [PubMed] [Google Scholar]
- 5. Eschenburg S., Kabsch W., Healy M. L., Schonbrunn E. (2003) A new view of the mechanisms of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and 5-enolpyruvylshikimate-3-phosphate synthase (AroA) derived from X-ray structures of their tetrahedral reaction intermediate states. J. Biol. Chem. 278, 49215–49222 [DOI] [PubMed] [Google Scholar]
- 6. Skarzynski T., Kim D. H., Lees W. J., Walsh C. T., Duncan K. (1998) Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA. Biochemistry 37, 2572–2577 [DOI] [PubMed] [Google Scholar]
- 7. Marquardt J. L., Brown E. D., Lane W. S., Haley T. M., Ichikawa Y., Wong C. H., Walsh C. T. (1994) Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry 33, 10646–10651 [DOI] [PubMed] [Google Scholar]
- 8. Brown E. D., Marquardt J. L., Lee J. P., Walsh C. T., Anderson K. S. (1994) Detection and characterization of a phospholactoyl-enzyme adduct in the reaction catalyzed by UDP-N-acetylglucosamine enolpyruvoyl transferase, MurZ. Biochemistry 33, 10638–10645 [DOI] [PubMed] [Google Scholar]
- 9. Wanke C., Amrhein N. (1993) Evidence that the reaction of the UDP-N-acetylglucosamine 1-carboxyvinyltransferase proceeds through the O-phosphothioketal of pyruvic acid bound to Cys-115 of the enzyme. Eur. J. Biochem. 218, 861–870 [DOI] [PubMed] [Google Scholar]
- 10. Kim D. H., Lees W. J., Kempsell K. E., Lane W. S., Duncan K., Walsh C. T. (1996) Characterization of a Cys-115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35, 4923–4928 [DOI] [PubMed] [Google Scholar]
- 11. Schönbrunn E., Sack S., Eschenburg S., Perrakis A., Krekel F., Amrhein N., Mandelkow E. (1996) Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin. Structure 4, 1065–1075 [DOI] [PubMed] [Google Scholar]
- 12. Skarzynski T., Mistry A., Wonacott A., Hutchinson S. E., Kelly V. A., Duncan K. (1996) Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure 4, 1465–1474 [DOI] [PubMed] [Google Scholar]
- 13. Eschenburg S., Priestman M., Schönbrunn E. (2005) Evidence that the fosfomycin target Cys-115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J. Biol. Chem. 280, 3757–3763 [DOI] [PubMed] [Google Scholar]
- 14. Schönbrunn E., Eschenburg S., Krekel F., Luger K., Amrhein N. (2000) Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA. Biochemistry 39, 2164–2173 [DOI] [PubMed] [Google Scholar]
- 15. Schonbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. (2000) Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc. Natl. Acad. Sci. U.S.A. 97, 6345–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han H., Yang Y., Olesen S. H., Becker A., Betzi S., Schönbrunn E. (2010) The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA). Biochemistry 49, 4276–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steinbach A., Scheidig A. J., Klein C. D. (2008) The unusual binding mode of cnicin to the antibacterial target enzyme MurA revealed by X-ray crystallography. J. Med. Chem. 51, 5143–5147 [DOI] [PubMed] [Google Scholar]
- 18. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 19. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 20. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Eschenburg S., Schönbrunn E. (2000) Comparative X-ray analysis of the un-liganded fosfomycin-target murA. Proteins 40, 290–298 [DOI] [PubMed] [Google Scholar]
- 23. Eschenburg S., Priestman M. A., Abdul-Latif F. A., Delachaume C., Fassy F., Schönbrunn E. (2005) A novel inhibitor that suspends the induced fit mechanism of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA). J. Biol. Chem. 280, 14070–14075 [DOI] [PubMed] [Google Scholar]
- 24. Yoon H. J., Lee S. J., Mikami B., Park H. J., Yoo J., Suh S. W. (2008) Crystal structure of UDP-N-acetylglucosamine enolpyruvyl transferase from Haemophilus influenzae in complex with UDP-N-acetylglucosamine and fosfomycin. Proteins 71, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 25. Jackson S. G., Zhang F., Chindemi P., Junop M. S., Berti P. J. (2009) Evidence of kinetic control of ligand binding and staged product release in MurA (enolpyruvyl UDP-GlcNAc synthase)-catalyzed reactions. Biochemistry 48, 11715–11723 [DOI] [PubMed] [Google Scholar]
- 26. Mizyed S., Oddone A., Byczynski B., Hughes D. W., Berti P. J. (2005) UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase). Biochemistry 44, 4011–4017 [DOI] [PubMed] [Google Scholar]
- 27. Schönbrunn E., Svergun D. I., Amrhein N., Koch M. H. (1998) Studies on the conformational changes in the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine enolpyruvyltransferase (MurA). Eur. J. Biochem. 253, 406–412 [DOI] [PubMed] [Google Scholar]
- 28. Samland A. K., Amrhein N., Macheroux P. (1999) Lysine 22 in UDP-N-acetylglucosamine enolpyruvyl transferase from Enterobacter cloacae is crucial for enzymatic activity and the formation of covalent adducts with the substrate phosphoenolpyruvate and the antibiotic fosfomycin. Biochemistry 38, 13162–13169 [DOI] [PubMed] [Google Scholar]
- 29. Diederichs K., Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.