Background: Polη implements translesion DNA synthesis but has low fidelity in replication.
Results: Acetylation-stabilized IRF1 transactivates the POLH gene in response to the chemical carcinogen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG).
Conclusion: Abnormal up-regulation of Polη through IRF1 transactivation is responsible for mutation frequency increases in cells exposed to MNNG.
Significance: IRF1-induced Polη activity is a new mechanism leading to mutation accumulation and carcinogenesis in cells exposed to an environmental chemical carcinogen.
Keywords: Carcinogenesis, DNA Damage, DNA Polymerase, Mutagenesis, Transcription Regulation, IRF1, MNNG, Polη, Acetylation
Abstract
DNA polymerase η (Polη) implements translesion DNA synthesis but has low fidelity in replication. We have previously shown that Polη plays an important role in the genesis of nontargeted mutations at undamaged DNA sites in cells exposed to the carcinogen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). Here, we report that MNNG-induced Polη expression in an interferon regulatory factor 1 (IRF1)-dependent manner in human cells. Mutagenesis analysis showed that four critical residues (Arg-82, Cys-83, Asn-86, and Ser-87) located in the IRF family conserved DNA binding domain-helix α3 were involved in DNA binding and POLH transactivation by IRF1. Furthermore, Polη up-regulation induced by IRF1 was responsible for the increase of mutation frequency in a SupF shuttle plasmid replicated in the MNNG-exposed cells. Interestingly, IRF1 was acetylated by the histone acetyltransferase CBP in these cells. Lys → Arg substitution revealed that Lys-78 of helix α3 was the major acetylation site, and the IRF1-K78R mutation partially inhibited DNA binding and its transcriptional activity. Thus, we propose that IRF1 activation is responsible for MNNG-induced Polη up-regulation, which contributes to mutagenesis and ultimately carcinogenesis in cells.
Introduction
DNA damage blocks the progression of the replication fork, which switches highly stringent replicative DNA polymerases to low-fidelity DNA polymerases that perform translesion synthesis (TLS).4 TLS DNA polymerases contain relatively nonrestrictive active sites for the accommodation of altered bases, and lack 3′-5′ exonuclease activity for proofreading. Therefore, TLS balances the gain in survival with a tolerable mutational cost; disturbing this balance increases mutations, which may play a role in carcinogenesis (1, 2). The human POLH gene encodes an important member of the Y family of TLS DNA polymerases, Polη. Polη is specialized for the error-free bypass of pyrimidine dimers introduced by UV radiation (3–5). Defects in the gene result in xeroderma pigmentosum variant syndrome with an increased incidence of skin cancer (6, 7). Polη has also been shown to replicate across a wide spectrum of DNA lesions induced by environmental or chemotherapeutic agents (8). In addition to TLS, Polη is also involved in gene conversion and strand invasion during homologous recombination (9, 10). However, Polη generates replication errors at an average frequency of about 3 × 10−2 mutations per base pair, and copies undamaged DNA with much lower fidelity than any other template-dependent DNA polymerase studied (11). It can bypass some DNA lesions through an error-prone pathway, which is a mutation-generating molecular mechanism (12). Deficient or dysregulated Polη has severely harmful consequences for cells.
The monofunctional alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) is a widely accepted model chemical carcinogen for studying the mechanisms of mutagenesis and carcinogenesis induced by N-nitroso compounds. It generates adducts with DNA and protein, such as O6-methylguanine, leading to point mutations, chromosomal aberrations, or even cell death. We reported that MNNG also induces mutations in undamaged DNA sequences (nontargeted mutations) in mammalian cells, and this is caused by a decrease of DNA replication fidelity resulting from aberrant expression of TLS DNA polymerases upon MNNG exposure (13, 14). We demonstrated that the spontaneous mutation frequency in human FL amnion epithelial cells (as model normal cells) increases when Polη is depleted, indicating an important role of Polη in maintaining genetic stability. However, when the POLH gene is knocked down, the MNNG-induced mutation frequency of the SupF tRNA gene replicated in the cells is significantly reduced, suggesting that Polη induces error-prone replication and is involved in the genesis of nontargeted mutations in cells exposed to MNNG (15, 16). More recently, we found that MNNG not only causes DNA damage but also disturbs other cellular processes by activating signaling pathways and inducing endoplasmic reticulum stress (17–19). Using genomic and proteomic methodologies, we further revealed that many genes and proteins are involved in the response to MNNG exposure, including those participating in the regulation of transcription, metabolism, cytoskeletal organization, the cell cycle, cell proliferation, signal transduction, and transport (20–22). Although these findings help our understanding of the molecular mechanisms of MNNG-caused damage, how chemical carcinogens regulate the low-fidelity TLS DNA polymerases that contribute to mutagenesis and carcinogenesis remains unclear.
Interferon regulatory factors (IRFs) were originally identified as transcriptional regulators of interferon (IFN) and IFN-stimulated genes. IRF1 is a member of the IRF family, and functions in antiviral response, regulation of the cell cycle and apoptosis, and oncogenesis (23–25). IRF1 also plays an important role in DNA damage responses and DNA repair. Mouse embryonic fibroblasts lacking IRF1 are deficient in the ability to undergo the DNA damage-induced cell cycle arrest that normally occurs in wild-type cells under genotoxic stress (26). IRF1-deficient hepatocytes show reduced DNA repair activity compared with the wild-type after UV irradiation (27). A recent study using the ChIP-chip approach revealed that IRF1 is involved in regulating many genes that encode components of the DNA damage response and DNA repair pathways after IFN stimulation (28). The spectrum of IRF-1-responsive genes is dependent on a number of factors including stimulus, cell type, and stage of development. However, the exact role of IRF1 and the process by which it acts during DNA damage have not been fully characterized.
In the present study, we demonstrated that MNNG increased the expression of POLH by up-regulating the IRF1 protein level via lysine acetylation. IRF1 directly bound to the POLH promoter and transactivated gene expression. Consequently, the abnormally stimulated Polη led to an increase in DNA mutation frequency in cells exposed to MNNG. Thus, we propose that IRF1-dependent transcriptional regulation of TLS DNA polymerases is probably a new mutagenic and carcinogenic pathway stimulated by environmental chemical carcinogens.
EXPERIMENTAL PROCEDURES
Chemicals
The monofunctional alkylating agent MNNG, the protein synthesis inhibitor cycloheximide, the protein deacetylase inhibitors trichostatin A (TSA) and nicotinamide, the DNA-damaging agent camptothecin, and the solvent DMSO were from Sigma. Stock solutions were prepared in DMSO and used immediately.
Cells and Culture
Human amnion epithelial FL cells (ATCC CCL-62, Manassas, VA) were cultured in Eagle's minimal essential medium supplemented with 10% newborn bovine serum, and HEK293T, RKO, and A549 cells were maintained in Dulbecco's modified Eagle's medium or RPMI 1640 supplemented with 10% fetal bovine serum in a 5% CO2 humidified atmosphere at 37 °C.
Plasmids and Constructs
The putative promoter of the human POLH gene was amplified from the extracted genomic DNA of human FL cells by PCR. The PCR products were purified and inserted into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). This recombinant plasmid p1687(−984/+703) was used as the template for PCR to generate a series of deletion constructs with different primers. The mutations were introduced into p1687(−984/+703) with different primers containing mutated IRF1-binding sites using the QuikChange® XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The expression plasmid pcDNA3.1 c-myc-IRF1 was constructed by inserting IRF1 cDNA and 6 × c-myc sequences into the vector pcDNA3.1(+)-basic (Invitrogen). The mutations were introduced into pcDNA3.1 c-myc-IRF1 using primers with mutation of specific sites. The primers for the above constructs are listed in supplemental Tables S1 and S2. The sequences of all constructs were confirmed by DNA sequencing (Invitrogen).
Quantitative Real Time RT-PCR
FL cells were exposed to different doses of MNNG or the solvent control DMSO. Total RNA was isolated with RNAsio Plus reagents (TaKaRa Bio Inc., Shiga, Japan). qRT-PCR was performed using SYBR® Premix Ex TaqTM (TaKaRa Bio Inc.) on an ABI Prism 7500 real-time PCR system (Applied Biosystems, Foster City, CA). β-Actin was used as loading control.
Immunoblotting and Immunoprecipitation
The treated cells were lysed in 20 mm HEPES (pH 8), 0.1 mm EDTA, 5 mm MgCl2, 0.5 m NaCl, 20% glycerol, 1% Nonidet P-40, and proteinase inhibitors. A 30-μg aliquot of each sample was used for Western blotting, probed with primary antibodies against Polη (Abcam, MA), IRF1, c-myc, p53, or CBP (Santa Cruz Biotechnology, Santa Cruz, CA), stained with IRDye® 800CW- or IRDye® 680-conjugated secondary antibodies (LI-COR, Lincoln, NE), and then detected by an Odyssey® infrared imaging system (LI-COR). GAPDH was used as loading control.
For immunoprecipitation, 0.5–1 mg of protein was incubated with 1 μg of different antibodies for 4–16 h at 4 °C. Immunoprecipitates were collected on protein G-agarose beads, washed, and then denatured by boiling in Laemmli sample buffer. The supernatant was analyzed by Western blot with different primary antibodies against IRF1, c-Myc, or pan-acetylation-lysine (Santa Cruz or Cell Signaling Technology, Inc., Danvers, MA). ImageJ (National Institutes of Health) was used for measuring the intensity of bands.
Immunofluorescence Assay
The treated cell layers were fixed in ice-cold 4% paraformaldehyde for 15 min, washed with PBS, blocked with 1% BSA in PBS for 1 h at room temperature, and then incubated with primary antibodies against Polη, IRF1, or γH2AX (Santa Cruz) at 4 °C overnight. After rinsing in PBS, the cells were incubated with FITC- and TRITC-conjugated secondary antibodies for 1 h at room temperature. After a final wash, the cells were stained with DAPI and viewed using a fluorescence microscope (AX70, Olympus, Japan).
Luciferase Reporter Assays
Cells were plated onto 24-well plates at 1 × 105 cells per well the day before transfection. The cells were co-transfected with 0.5 μg of firefly luciferase reporter constructs, 0.02 μg of pRL-SV40 Renilla luciferase reporter plasmids (Promega, Madison, WI), and c-myc-IRF1 wild-type or mutant constructs using the SuperFect® transfection reagent (Qiagen). The luciferase activity was examined by a dual-luciferase reporter assay system (Promega).
DNA Pull-down Assay
The DNA pull-down assay was performed as described (29). The DNA sequences from −2066 or −984 to +703 of the human POLH promoter were amplified by PCR with the primers (one of which was labeled with biotin). Ten micrograms of the purified biotin-labeled PCR product was attached to streptavidin-agarose beads according to the manufacturer's guidelines (Invitrogen). The beads were washed 3 times with binding buffer (10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1.0 mm dithiothreitol, 1.0 mm EDTA, and 5% glycerol (v/v)), and then incubated with 100 μg of nuclear extract proteins in 350 μl of binding buffer at 4 °C for 45 min. The precipitates were washed 3 times with washing buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.0 mm dithiothreitol, 1.0 mm EDTA, and 5% glycerol (v/v)). The bound proteins were analyzed by SDS-PAGE and Western blotting. Ku80 (Santa Cruz) was used as loading control.
RNA Interference (RNAi)
FL cells were transfected with siRNAs targeting the IRF1, POLH, or CBP mRNA sequence, or with a negative control siRNA (Santa Cruz Biotechnology) to a final concentration of 100 nm using Lipofectamine 2000 (Invitrogen).
EdU Incorporation Assay
The EdU (5-ethynyl-2′-deoxyuridine) incorporation assay was used to represent TLS in cells after DNA damage (30). FL cells were cultured on 96-well plates and treated with MNNG for 12 h, or transfected with IRF1 wild-type or mutant constructs. Next, cells were washed 3 times with PBS, and then incubated in serum-free DMEM supplemented with 10 μm EdU for 2 h. After extensive washing with PBS, cells were blocked with 10% FBS in PBS for 30 min. Incorporated EdU was detected by the fluorescent azide coupling reaction (Invitrogen). Images of the cells were captured with a fluorescence microscope (Nikon, Tokyo, Japan). ImageJ was used to count the fluorescent points.
SupF-pZ189 Shuttle Plasmid Mutation Assay
DNA nontargeted mutation was measured using the shuttle plasmid pZ189 mutation assay as described (16). FL cells were transfected with IRF1 wild-type or mutant plasmid together with or without siRNA-Polη. After 24 h, the pZ189 plasmid containing the SupF tRNA gene was introduced into the cells. Forty-eight hours later, the replicated plasmid DNA was extracted from the cells and separated from cellular DNA as described by Hirt (31). Before the plasmid was used to transform the indicator bacterium Escherichia coli MBM7070, it was treated with DpnI to digest any DNA that still had the bacterial methylation pattern generated when the plasmid was first prepared (32). E. coli MBM7070 were transformed with the pZ189 plasmid and selected for ampicillin resistance on LB plates containing 40 mg/liter of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), 20 mg/liter of isopropyl 1-thio-β-d-galactopyranoside, and 100 mg/liter of ampicillin. White and light-blue colonies were picked and each colony was streaked on a fresh plate to confirm the phenotype. The mutation frequency of the SupF tRNA gene was equal to the number of mutants isolated/number of transformants.
Software
The software for promoter prediction and the prediction of transcription factor binding sites are shown in supplemental Table S3.
Statistical Analysis
All data are representative of at least three independent experiments and expressed as mean ± S.D. Statistical data analysis was performed using the unpaired Student's t test. Pearson χ2 analysis was used to evaluate the mutation frequency of the SupF tRNA gene in the treated FL cells.
RESULTS
MNNG Up-regulated Human Polη Expression
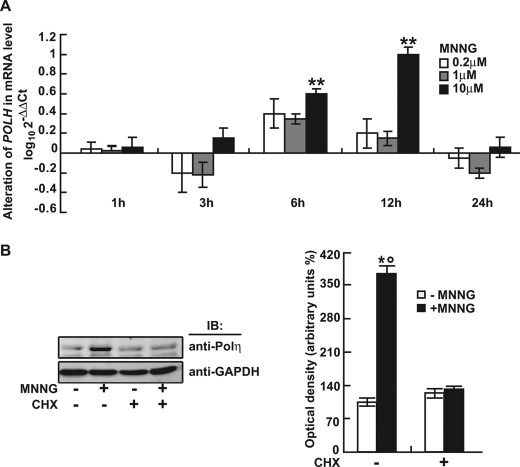
The effect of MNNG on Polη expression was evaluated by qRT-PCR and Western blot analysis. MNNG (10 μm) significantly up-regulated POLH mRNA expression at 6 and 12 h after treatment (Fig. 1A). The protein level of Polη increased more than 3-fold at 12 h (Fig. 1B). Pretreatment with 20 μg of cycloheximide (a specific inhibitor of protein synthesis) for 1 h completely abrogated this effect (Fig. 1B), suggesting that MNNG up-regulated the Polη protein level by transcriptional activation of the POLH gene.
FIGURE 1.
MNNG induced Polη expression in human FL cells. A, transcription of human POLH gene in response to 0.2, 1, and 10 μm MNNG for different times determined by qRT-PCR. **, p < 0.01 relative to control (DMSO). B, Left panel shows that protein levels of human Polη at 12 h following 10 μm MNNG treatment in the presence or absence of cycloheximide (CHX) measured by Western blot. GAPDH was used as loading control. Right panel shows quantitative data (mean ± S.D.) for three independent experiments analyzed by Student's t test. *, p < 0.05, MNNG-treated versus DMSO-treated cells; °, p < 0.05, non-pretreated versus cycloheximide-pretreated cells.
MNNG Transactivated the POLH Gene by Targeting Two IRF1-binding Motifs of Promoter
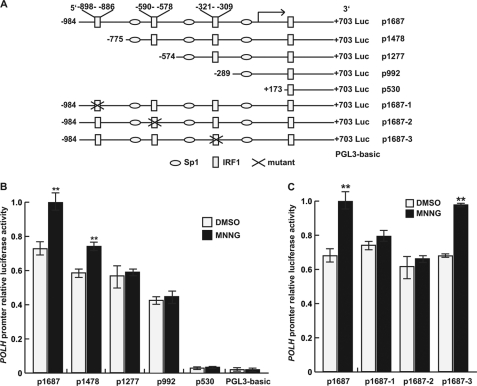
The mechanism of transcriptional activation of the human POLH gene in response to MNNG was first investigated by both computer-assisted DNA sequence analysis and luciferase reporter assay. We identified a conserved region located from −1100 to +600 relative to the transcriptional start position of the human POLH gene (data not shown). Several algorithms were used for the prediction of promoter and transcriptional factor binding sites (supplemental Table S3). The sequence from −984 to +703 (p1687) was chosen as the putative promoter for further functional analysis. Prediction analysis for transcription factor binding sites revealed that the POLH promoter sequence contained four IRF1-binding motifs (Fig. 2A).
FIGURE 2.
MNNG transactivated the human POLH gene dependent on the IRF1 binding motif of the promoter. A, schematic maps of the POLH promoter-luciferase reporter constructs used in this study. B and C, the successive 5′-deletion constructs and IRF1-binding motif mutants were transfected into FL cells and treated with 10 μm MNNG in serum-free medium for 2 h, and after 12 h, luciferase activity was determined. The data are from at least three independent experiments analyzed by Student's t test relative to control DMSO (**, p < 0.01).
To determine MNNG-responsive elements within the human POLH promoter, we cloned the promoter sequence of POLH p1687 (−984 to +703) from human FL cells, and a series of luciferase reporters for the POLH promoter and its 5′-deletion sequences were constructed: p1478 (−775 to +703), p1277 (−574 to +703), p992 (−289 to +703), and p530 (+173 to +703) (Fig. 2A). These luciferase reporter constructs were each then transfected into FL cells, followed by MNNG treatment. The transcriptional activity responsive to MNNG was optimal with p1687 but gradually attenuated with the reduction in promoter length (Fig. 2A). The construct p530 completely lost responsiveness to MNNG, suggesting that the IRF1-binding motif residing between +173 and +703 was not necessary for the POLH promoter activation by MNNG. To test whether three other upstream IRF1-binding motifs were critical for MNNG-dependent activation of the promoter, we constructed p1687-1, P1687-2, and p1687-3 luciferase reporters with the individual motifs mutated (Fig. 2B). Luciferase reporter assays showed that p1687-1 and p1687-2 but not p1687-3 significantly abolished the stimulatory effect of MNNG (Fig. 2C). These results indicated that the two IRF1 binding motifs located in −898 to −886 and −590 to −578 were responsible for MNNG-induced activation of the human POLH promoter.
IRF1-induced POLH Expression Was Stimulated by MNNG through Direct Binding of Its DNA-binding Domain (DBD)-Helix α3 to Promoter
To further evaluate the involvement of IRF1 in transactivation of the POLH gene, the c-myc-IRF1 expression plasmid and the POLH promoter luciferase reporter were co-transfected into FL cells, and luciferase activity was analyzed. The results showed that IRF1 overexpression increased promoter transcriptional activity of both p1687 and p1687-3 luciferase reporters (Fig. 3A). In contrast, neither p1687-1 nor p1687-2 luciferase reporters responded to IRF1 by activation. These results further supported the finding that the IRF1 binding motifs located in −898 to −886 and −590 to −578 were essential for the POLH promoter to respond to MNNG.
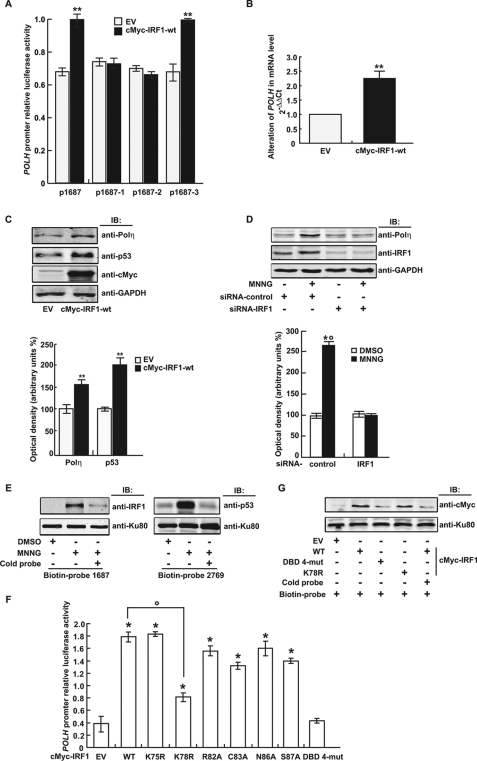
FIGURE 3.
IRF1 transactivated the human POLH gene in response to MNNG by direct binding of its DBD to the promoter. A, the expression plasmid pcDNA3.1 c-myc-IRF1 wild-type (WT) or the control plasmid pcDNA3.1 c-myc (EV) and the human POLH promoter reporter constructs with or without mutated IRF1 binding sites were co-transfected into FL cells. The luciferase activity was measured 48 h later. **, p < 0.01 compared with control transfected with EV. B, mRNA expression of Polη in FL cells induced by IRF1-wt overexpression. C, Upper panel shows the protein level of Polη induced by IRF1-wt overexpression. GAPDH was used as loading control. Bottom panel shows the data (mean ± S.D.) of three independent experiments. **, p < 0.01 compared with EV control. D, Upper panel shows that FL cells were transfected with siRNA-IRF1 or siRNA-control and treated with 10 μm MNNG. After 12 h, Western blot analysis was performed with anti-IRF1 and anti-Polη, respectively. GAPDH was used as loading control. Bottom panel shows the data (mean ± S.D.) of three independent experiments. *, p < 0.05, MNNG-treated versus DMSO-treated cells; °, p < 0.05, siRNA-control versus IRF1 knockdown transfected cells. E, FL cells were treated with 10 μm MNNG or DMSO for 2 h. Twelve hours later, nuclear proteins were extracted, and the binding ability of IRF1 to the biotin-labeled POLH promoter probe −984 to +703 (biotin-probe 1687) or p53 to the biotin-labeled POLH promoter probe −2066 to +703 (biotin-probe 2769), was analyzed by DNA pulldown assay. Nonlabeled POLH promoter probes (cold-probe) were used for competitive inhibition. Ku80 was used as control. F, IRF1 expression plasmids with or without various site mutations in the DBD were transfected together with the human POLH promoter reporter into 293T cells. The luciferase activity was measured 48 h later. *, p < 0.05 compared with EV control; °, p < 0.05, IRF1-K78R versus IRF1-wt-expressing plasmid-transfected cells. The data are from at least three independent experiments analyzed by Student's t test. DBD 4-mut, IRF1 expression plasmid with co-mutations of R82A, C83A, N86A, and S87A in DBD. G, expression plasmids of IRF1-wt or its mutants were transfected into 293T cells, and DNA pulldown assays were performed with the biotin-labeled or unlabeled human POLH promoter probe as described above.
We then examined POLH gene regulation by manipulating the IRF1 expression level in 293T cells (a cell line expressing little IRF1). Both mRNA and protein analysis revealed a basal level of POLH expression in these cells. Ectopic expression of IRF1 up-regulated the Polη level as well as that of p53, a biomarker of DNA damage (Fig. 3, B and C). In contrast, IRF1 depletion with siRNA-IRF1 decreased Polη induction by MNNG in FL cells (Fig. 3D). Furthermore, DNA binding assays with a biotin-labeled DNA probe covering the POLH promoter sequence −984 to +703 (biotin-probe 1687) demonstrated that IRF1-DNA binding was increased in the MNNG-treated cells (Fig. 3E). POLH has been reported as a p53 target gene (33). By DNA pull-down assay, we found that p53 bound directly to the POLH promoter −2066 to +703 (biotin-probe 2769), used as a positive control (Fig. 3E).
DBD-helix α3 is highly conserved among all IRF family members (34). To determine key residues in the DBD-helix α3 of IRF1 for POLH promoter binding, we performed site-directed mutagenesis by substituting the most conserved residues in this domain to alanine (R82A, C83A, N86A, and S87A) separately or together (DBD 4-mutant). These DBD-helix α3 mutants of IRF1 were overexpressed in 293T cells and tested for their transactivation ability with luciferase reporter assays. Although single-site mutants showed a fairly weak effect on POLH promoter activation, the DBD 4-mutant completely abolished its transactivation ability (Fig. 3F). We also constructed IRF1 expression plasmids with K75R or K78R mutations (nonacetylatable mutation) in DBD-helix α3, and tested their transactivation activity with luciferase reporter assays. The K78R mutant but not the K75R mutant, dramatically reversed the POLH promoter-driven luciferase activation (Fig. 3F). DNA binding analysis revealed that the IRF1 DBD 4-mutant, and to a lesser extent, the IRF1-K78R mutant, lost their DNA binding activity (Fig. 3G). Together, these data demonstrated that IRF1-mediated MNNG induced POLH transactivation by directly binding to the promoter, and IRF1 DBD-helix α3 was critical for POLH promoter activation.
MNNG-induced IRF1 Lys-78 Acetylation for IRF1 Protein Accumulation
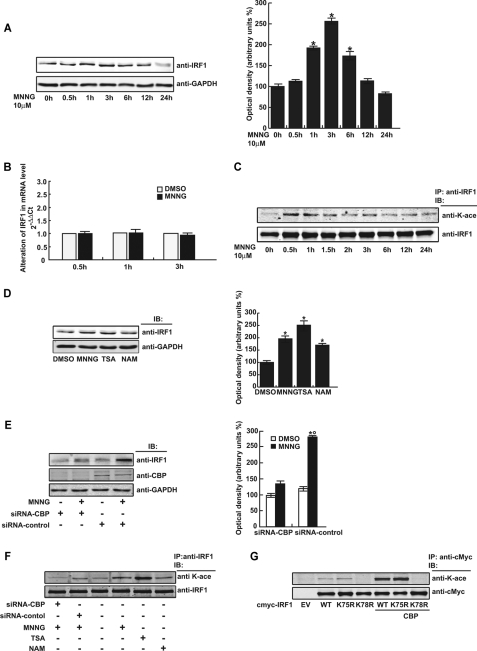
We next investigated the mechanism by which MNNG affected IRF1. Time course analysis revealed that the IRF1 protein level was significantly increased at 1 and 3 h in FL cells after exposure to MNNG, but IRF1 mRNA expression was not changed at these time points, indicating that up-regulation of the IRF1 protein did not result from mRNA transcription (Fig. 4, A and B). Interestingly, IRF1 acetylation was induced within 30 min after MNNG treatment but gradually decayed with time (Fig. 4C). TSA and nicotinamide are specific inhibitors of the histone deacetylase and Sirt deacetylase families, respectively. Treating FL cells with TSA and to a lesser extent with nicotinamide increased the IRF1 protein level (Fig. 4D). On the other hand, knockdown of the histone acetyltransferase CREB-binding protein (CBP) with siRNA-CBP inhibited the increase of IRF1 protein content induced by MNNG (Fig. 4E). Furthermore, siRNA-CBP and TSA did regulate IRF1 acetylation (Fig. 4F). Our data demonstrated that the IRF1 protein level was not regulated by an increase in gene expression but by post-translational acetylation that contributed to stabilizing the protein in the MNNG-exposed cells. Lys-78 within IRF1-DBD-helix α3 was the major site for the IRF1 acetylation mediated by CBP (Fig. 4G).
FIGURE 4.
MNNG up-regulated IRF1 protein level by lysine acetylation. A and B, the protein and mRNA levels of IRF1 in FL cells were measured by Western blot and qRT-PCR, respectively, at the indicated time points after 10 μm MNNG treatment. GAPDH was used as protein loading control. C, FL cells were treated with 10 μm MNNG and the protein lysates were collected at the indicated time points. Immunoprecipitation was performed with anti-IRF1 antibody, and lysine acetylation was detected with anti-pan-K-acetylation antibody. IRF1 immunoprecipitation efficiency was controlled by anti-IRF1 antibody. D and E, FL cells were treated or transfected as indicated, and then the protein lysates were analyzed by Western blot using the specific antibodies indicated. GAPDH was used as loading control. F, FL cells were treated or transfected as indicated; the protein lysates were immunoprecipitated (IP) and measured by Western blot. G, various expression plasmids of IRF1 were transfected with or without CBP expression plasmid into FL cells. Forty-eight hours later, the protein lysates were immunoprecipitated with anti-c-Myc antibody and then analyzed by Western blot using anti-pan-K-acetylation. Anti-c-Myc was also used for immunoprecipitation efficiency control.
IRF1 Increased Mutation Frequency by Up-regulating Polη in Response to MNNG
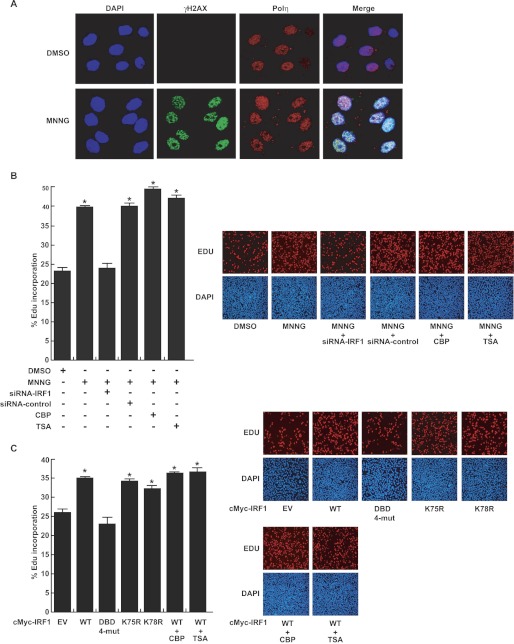
It is known that human Polη plays a key role in DNA translesion synthesis, and the activity of the enzyme is precisely regulated because of its low fidelity for DNA replication. Abnormally excessive activation of human Polη is mutagenic and carcinogenic (35). We showed above that MNNG transactivated the expression of human POLH in an IRF1-dependent manner in FL cells. Immunofluorescence assays revealed that in the nuclei of FL cells exposed to 10 μm MNNG, Polη was increased and co-localized with γH2AX at DNA-damaged sites over time (supplemental Fig. S1) and mostly at 12 h (Fig. 5A). The EdU incorporation assay demonstrated that MNNG exposure increased DNA synthesis for repair in FL cells, whereas siRNA-IRF1 abolished the MNNG-induced effect (Fig. 5B). Overexpression of CBP or pretreatment with TSA slightly enhanced the MNNG effect (Fig. 5B). Furthermore, overexpression of wild-type IRF1 increased the EdU incorporation, but the DBD 4-mutant of IRF1 totally lost this ability. Modulating the IRF1 acetylation level using the K78R mutation, CBP overexpression, or TSA pretreatment had a trivial influence on the EdU incorporation (Fig. 5C). These data indicated that IRF1 transactivated the POLH gene by binding to the promoter after exposure to MNNG, then the up-regulated Polη implemented TLS in the exposed cells.
FIGURE 5.
IRF1 mediated the biological consequences of Polη up-regulation in response to MNNG. A, co-localization of Polη with γH2AX at the DNA damaged sites in the FL cell nuclei at 12 h following MNNG treatment measured by immunofluorescent staining and confocal microscopy. Red, Polη; green, γH2AX; and blue, DAPI. B, right panel shows that FL cells were treated with 10 μm MNNG, and after 12 h, DNA synthesis was measured by EdU incorporation assays in the presence of siRNA-IRF1 knockdown, CBP overexpression, or TSA pretreatment. C, right panel shows that various expression plasmids of IRF1 were transfected into FL cells, with or without CBP overexpression or TSA pretreatment. DNA synthesis was measured by EdU incorporation assays. The percentage of Edu incorporation was calculated as the number of Edu-positive nuclei (red) divided by the total nuclei (blue). We selected 10 visual fields under the microscope for each well of cells and examined more than 10,000 cells for each well. The counting was performed with ImageJ software. Left panel of B and C shows quantitative data (mean ± S.D.) for three independent experiments analyzed by Student's t test. *, p < 0.05, treated versus control cells.
We previously showed that deregulation of Polη induces nontargeted DNA mutations in mammalian cells after exposure to MNNG (15, 16). The shuttle-plasmid pZ189 mutation assay was performed to investigate the involvement of IRF1 in nontargeted DNA mutations after MNNG treatment. We found that overexpressed wild-type IRF1 significantly increased the mutation frequency of the supF tRNA gene in the plasmid replicated in FL cells, the K78R mutant induced fewer mutations, whereas the DBD 4-mutant did not increase the mutation frequency over control (Table 1). Furthermore, we co-transfected siRNA-Polη and IRF1 expression plasmid in FL cells and repeated the mutation assay. The result showed that knockdown of Polη reversed the IRF1-enhanced mutation frequency, suggesting that IRF1 increased the mutation frequency in a Polη-dependent manner (Table 2). The efficiency of siRNA-Polη was evaluated by Western blotting (supplemental Fig. S2E). These data indicated that the IRF1-transactivated POLH was responsible for the increase in the nontargeted mutation frequency in FL cells.
TABLE 1.
Increased mutation frequency of SupF tRNA gene in the plasmid pZ189 replicated in the FL cells with overexpressed IRF1
| EV | IRF1 | IRF1-DBD 4-mut | IRF1-K78R | |
|---|---|---|---|---|
| Transformants | 15,745 | 33,710 | 17,751 | 25,738 |
| Mutants | 1 | 13 | 1 | 9 |
| Mutation frequencya (×10−5) | 6.3 | 38.5b | 5.6 | 34.9b |
| Induced mutation frequencyc (×10−5) | 32.2 | −0.7 | 28.5 |
a Mutation frequency: No. of mutants/No. of transformants.
b p < 0.01 versus EV-transfected FL cells (χ2-test).
c Induecd mutation frequency: mutation frequency of IRF1 or IRF1-DBD or IRF1-K78R minus mutation frequency of EV.
TABLE 2.
Decreased mutation frequency of SupF tRNA gene in the plasmid pZ189 replicated in the FL cells with Polη knocked down
| siRNA control |
siRNA-Polη |
||
|---|---|---|---|
| EV | IRF1 | IRF1 | |
| Transformants | 11,012 | 11,284 | 10,844 |
| Mutants | 3 | 16 | 4 |
| Mutation frequencya (×10−4) | 2.7 | 14.2b | 3.7 |
| Induced mutation frequencyc (×10−4) | 32.2 | 1.0 | |
a Mutation frequency: No. of mutants/No. of transformants.
b p < 0.01 versus EV-transfected FL cells (χ2-test).
c Induecd mutation frequency: mutation frequency of IRF1 minus mutaion frequency of EV.
DISCUSSION
In this study, we demonstrated that exposure to the carcinogen MNNG increased human POLH gene expression through IRF1 transactivation in human FL cells. MNNG caused an accumulation of IRF1 by Lys-78 acetylation associated with CBP. IRF1 directly bound to the POLH promoter by key residues (Arg-82, Cys-83, Asn-86, Ser-87, and Lys-78) in the conserved DBD-helix α3 and transactivated POLH gene expression. MNNG-stimulated Polη implemented TLS and caused an increase in mutation frequency in the exposed cells. In addition, we investigated whether this phenomenon is cell line or chemical specific. The results demonstrated that MNNG exposure up-regulated the IRF1 protein level that in turn transactivated POLH in A549 cells (supplemental Fig. S2, A and B), and camptothecin also induced a similar phenomenon in RKO cells (supplemental Fig. S2, C and D), which probably suggests a common transactivational regulation mechanism in the cellular response to DNA damage.
MNNG-induced POLH expression (Fig. 1). Two IRF1-binding sites in the POLH promoter were responsible for MNNG-induced transactivation of the gene (Figs. 2 and 3A). Although overexpression of IRF1 up-regulated Polη and transactivated the POLH gene, IRF1 knockdown abrogated these effects in the MNNG-treated cells, and DNA pulldown assays revealed that the binding ability of IRF1 to the POLH promoter was enhanced in MNNG-treated cells, confirming that MNNG activates POLH expression in an IRF1-dependent manner (Fig. 3, B–F).
All IRF family proteins carry a conserved DBD consisting of ∼110 residues in their N-terminal region, through which they bind to the IFN-stimulated response element in IFN-inducible gene promoters. Co-crystallization of the helix turn helix-containing DBD of IRF1 with the IFN-β promoter element PRD I showed that four residues (Arg-82, Cys-83, Asn-86, and Ser-87) project from the DBD-helix α3 of IRF1, and interact with a GAAA core sequence (existing in almost all IRF response elements) within the major groove of the DNA (34). Furthermore, acetylation of transcription factors often affects DNA binding and transcriptional activity, so we speculated that the neighboring Lys-75 (conserved in IRF1 and -2) and Lys-78 (conserved throughout IRF family members), which are also located in DBD-helix α3, may be implicated in the transcriptional function of IRF1 as well. Therefore, we performed site-mutation analysis of these 6 important amino acids. The results demonstrated that the 4-site co-mutant (R82A, C83A, N86A, and S87A) of IRF1 had no binding and transactivation ability with the POLH promoter, and the K78R mutant partially but definitely lost this activity (Fig. 3, F and G), suggesting that these sites in the DBD-helix α3 were critical for IRF1 transcriptional activity.
IRF1 is an immediate-early gene, whose transcription increases from 15 min to 1 h, is down-regulated by 4 h, and is then up-regulated a second time at 8–10 h after prolactin stimulation (36). In response to IR and etoposide, IRF1 protein levels peak 2–6 h post-treatment (37, 38). We found that the IRF1 protein content was enhanced between 1 and 6 h following MNNG exposure, whereas the mRNA level of IRF1 was not changed (Fig. 4, A and B). Correlated with this, IRF1 lysine acetylation increased beginning at 30 min in the MNNG-exposed cells (Fig. 4C). Protein lysine acetylation is involved in various cellular functions. The most apparent effect is the inhibition of ubiquitination-mediated protein degradation by the proteasome (39–42). It has been reported that recombinant IRF1 is acetylated by p300 in vitro (43). We showed that treatment with the deacetylase inhibitor TSA, and to a lesser degree with nicotinamide, increased both the protein content and acetylation level of IRF1, and siRNA-CBP inhibited the MNNG-induced increment of the IRF1 protein as well as the acetylation level (Fig. 4, D–F). Mutation assays indicated that Lys-78 was the key residue acetylated by CBP responsible for IRF1 accumulation (Fig. 4G). Thus, these results demonstrated that IRF1 can be acetylated in vivo. CBP facilitated IRF1 acetylation that increased protein stabilization and transcriptional activity after MNNG treatment. It has been shown that IRF2, IRF-3, and IRF-7 can be acetylated in vivo and in vitro (44–46). IRF2, which has been thought to function by competing with IRF1, is acetylated by p300 in a cell growth-regulated manner; however, acetylation of IRF-2 does not alter DNA binding activity in vitro. Although mutation of Lys-75 diminishes the IRF-2-dependent activation of histone H4 promoter activity, mutation of Lys-78 of IRF2 leads to the abrogation of DNA binding activity (44), and acetylation of Lys-92 (the equivalent residue of IRF1-Lys-78) also negatively modulates IRF7 DNA binding (46). These differences among IRF family members may be related to the components and topology of the transcription complexes when they perform different cellular functions.
Polη has a loose active center and lacks proofreading ability when it performs TLS passing over various types of DNA lesion. However, this is often at the cost of error-prone replication, and especially, DNA synthesis by Polη on undamaged DNA is highly mutagenic (11, 47). Too much error-prone synthesis could lead to a cellular catastrophe (35). TLS DNA polymerases are regulated at different levels based on the type of DNA damage. It is well known that UV irradiation-induced DNA damage immediately recruits Polη to the stalled replication forks with the polymerase sliding clamp proliferating cell nuclear antigen and other proteins to start TLS (49, 50). p53 and p21 promote the recruitment of Polη by ubiquitinating proliferating cell nuclear antigen (51, 52). Phosphorylation is also involved in the translocation of Polη to stalled replication forks, and both ATR and protein kinase C function in the process (53). After TLS in Caenorhabditis elegans, Polη undergoes DNA damage-induced proteolysis for removal of Polη from replication forks (30). However, little is known so far about the transcriptional regulation of Polη in response to DNA damage. It has been reported that DNA breaks induced by ionizing radiation or camptothecin increase POLH expression in a p53-dependent manner, and in turn Polη modulates the DNA damage checkpoint and p53 activation (33). Here, we demonstrated that MNNG-induced IRF1 to transactivate the expression of POLH. MNNG exposure or overexpression of IRF1 increased TLS synthesis and the frequency of nontargeted mutation. Furthermore, siRNA-Polη or the IRF1 4-mutant abolished these effects (Tables 1 and 2). Therefore, our findings provide a new transcriptional regulatory mechanism of the POLH gene, resulting in mutagenesis and carcinogenesis in MNNG-exposed cells. Although these consequences were induced by a one-time transient treatment with MNNG (half-life about 40 min in solution) in our experiments, it would be expected that repeated and persistent stimuli have more complicated and long-term effects in exposed cells (54).
It is known that, to perform the TLS function, Y-family DNA polymerase members Polη, Polι, and Rev1 locate to replication factories with proliferating cell nuclear antigen and other proteins associated with replication in the nucleus, whereas different Y family polymerases may pass diverse types of DNA damage with different competency (1, 2). We previously found that Polι is transcriptionally up-regulated by MNNG exposure in an Sp1-dependent manner (55). In the current study, we demonstrated that MNNG exposure can stimulate up-regulation of Polη through IRF1 transactivation. These data indicate that both Polη and Polι are transactivated in respond to MNNG but by different transcriptional factors. Furthermore, the Polη knockdown experiments demonstrate that Polη was the key TLS DNA polymerase responsible for the increased mutations, although other Y-family DNA polymerases and related proteins may work together to perform the TLS function.
We previously showed that MNNG induces nontargeted mutations in mammalian cells, which are associated with aberrant regulation of TLS DNA polymerases (13–16). Our genome-wide high-throughput screening further demonstrated that MNNG exposure induces a wide expression change of various genes, indicating that transcriptional regulation is an important modulating event in the exposed cells (20). In this study, we revealed how MNNG induced nontargeted mutations by transactivating the low-fidelity TLS DNA polymerase Polη via the transcription factor IRF1. This finding further confirms that in addition to directly causing DNA damage, MNNG can induce harmful effects indirectly via transcriptional regulation of important genes such as the TLS DNA polymerase η in exposed cells.
Although it is known that IRF1 is involved in DNA damage responses, the mechanism by which genotoxic stress induces IRF1, the context of the signaling components during the responses to DNA damage, and the biological consequences remain largely unclear. It is proposed that IRF1 is controlled by two distinct signaling pathways: a JAK/STAT pathway in virus-infected cells, and an ATM pathway in DNA-damaged cells (37). IRF1 is activated by the STAT1 pathway in response to IFN-β or IFN-γ, and by the NF-κB pathway in response to retinoic acid or IFN-α (23, 56, 57). IRF1 can associate with NF-κB and CBP to form part of a multiprotein enhanceosome that assembles on the IFN-β promoter induced by viral infection (48). In contrast, eukaryotic cells respond to DNA damage by activating damaged checkpoint pathways, which arrest cell cycle progression and alter induced gene expression to allow for repair and/or apoptosis. In response to DNA damage induced by ionizing radiation or the topoisomerase II inhibitor etoposide, IRF1 and p53 are coordinately up-regulated to induce p21 expression in an ATM-dependent manner in the melanoma cell line A375. ATM indirectly increases both the synthesis and half-life of the IRF1 protein, and at the same time, ATM-dependent phosphorylation up-regulates the p53 protein by inhibiting its degradation (37). It has been shown that ionizing radiation and poly(I:C) (which mimics viral infection by inducing IFNγ expression) induce the assembly of an IRF1-p300-p53 complex at the P21 promoter, where IRF1 enhances p300 to acetylate and activate p53. Probably, DNA damage-activated ATM-CHK2 signaling cascades phosphorylate p53, converting p53 from an MDM2-binding protein to a p300-binding protein and enhancing its acetylation by p300 (38). A potential p53-binding site is located in the promoter of the POLH gene (−1992 to −1967). A recent study reported that DNA breaks induced by camptothecin or ionizing radiation lead to p53 stabilization and activation through the ATM-Chk2 pathway; p53 in turn up-regulates Polη expression; and finally, elevated Polη enhances p53 activity via ATM/ChK2 and promotes p53-dependent apoptosis when DNA damage is irreparable (33). In this study, we found that IRF1 overexpression up-regulated both Polη and p53 (Fig. 3C), and MNNG treatment increased binding of the POLH promoter with both IRF1 and p53 (Fig. 3E), suggesting that IRF1 and p53 co-regulate Polη expression in response to MNNG. Moreover, in A549 and RKO cells, we also found that MNNG and camptothecin can up-regulate Polη, IRF1, and p53 (supplemental Fig. S2). Taken together, the above studies reveal a network among IRF1, Polη, and p53, as well as protein modifications such as phosphorylation and acetylation, during the cellular response to DNA damage. In addition, it seems that two pathways exist in the regulation of Polη in response to DNA damage in cells: 1) DNA damage activates the ATM/ATR-Chk1/2-p53 pathway, which leads to cell cycle arrest for DNA repair, including recruitment of basal Polη to block forks for TLS. This is a relatively fast response, usually within 30 min to 6 h as induced by UV irradiation; and 2) DNA damage stimulates new Polη expression between 6 and 24 h post-treatment by MNNG-induced IRF1 as shown in this study or by camptothecin-activated p53 (33), leading to mutagenesis or apoptosis, respectively. Furthermore, the finding that IRF1 plays an important role in activating the POLH gene hints at a relationship among cytokines, inflammation, and chemical carcinogenesis.
In summary, we propose a new regulatory mechanism for the TLS DNA polymerase Polη, as well as a new mutagenic and carcinogenic pathway by the environmental chemical carcinogen MNNG: MNNG-induced DNA damage facilitates CBP to acetylate IRF1, leading to its stabilization and activation; IRF1 in turn transactivates POLH expression by directly binding to the promoter; and finally, the aberrantly up-regulated Polη probably performs an error-prone TLS and loses template selectivity, leading to an overall increase in mutation load at both the DNA-damaged sites (targeted mutation) and at undamaged sequences (nontargeted mutation), resulting in genome instability and ultimately, carcinogenesis. In the future, more elaborate and meticulous investigations are needed to further clarify the upstream regulatory pathway that activates CBP to modify IRF1, the mechanism that stabilizes IRF1, and the mechanisms by which IRF1, p53, and Polη cooperate in DNA damage responses, which may provide new insights into the mechanism by which cells decide their fate after DNA damage.
Acknowledgments
We thank Dr. Iain C. Bruce and Dr. Jun Huang for review of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 30770831, 81000893, 30800413, 30873094, and 81090421, Ministry of Education of China Grant J20100041, and Zhejiang Provincial Science Foundation Grants R207153, 2008C23070, and J20110353.

This article contains supplemental Figs. S1 and S2 and Tables S1–S3.
- TLS
- translesion synthesis
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- IRF
- interferon regulatory factor
- TSA
- trichostatin A
- qRT
- quantitative real time
- TRITC
- tetramethylrhodamine isothiocyanate
- DMSO
- dimethyl sulfoxide
- DBD
- DNA-binding domain
- CBP
- CREB (cAMP response element binding protein) binding protein
- EdU
- 5-ethynyl-2′-deoxyuridine.
REFERENCES
- 1. Friedberg E. C., Wagner R., Radman M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 31, 1627–1630 [DOI] [PubMed] [Google Scholar]
- 2. Lehmann A. R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J. F., Kannouche P. L., Green C. M. (2007) Translesion synthesis. Y-family polymerases and the polymerase switch. DNA Repair 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 3. Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., Yang W. (2010) Structure and mechanism of human DNA polymerase η. Nature 24, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi J. H., Besaratinia A., Lee D. H., Lee C. S., Pfeifer G. P. (2005) The role of DNA polymerase η in UV mutational spectra. DNA Repair 4, 211–220 [DOI] [PubMed] [Google Scholar]
- 5. Johnson R. E., Prakash S., Prakash L. (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 6. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 7. Inui H., Oh K. S., Nadem C., Ueda T., Khan S. G., Metin A., Gozukara E., Emmert S., Slor H., Busch D. B., Baker C. C., DiGiovanna J. J., Tamura D., Seitz C. S., Gratchev A., Wu W. H., Chung K. Y., Chung H. J., Azizi E., Woodgate R., Schneider T. D., Kraemer K. H. (2008) Xeroderma pigmentosum variant patients from America, Europe, and Asia. J. Invest. Dermatol 128, 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaney S. G., Campbell S. L., Bassett E., Wu Y. (2005) Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol. Hematol. 53, 3–11 [DOI] [PubMed] [Google Scholar]
- 9. Kawamoto T., Araki K., Sonoda E., Yamashita Y. M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Takeda S. (2005) Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol. Cell 20, 793–799 [DOI] [PubMed] [Google Scholar]
- 10. Mcilwraith M. J., Vaisman A., Liu Y., Fanning E., Woodgate R., West S. C. (2005) Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20, 783–792 [DOI] [PubMed] [Google Scholar]
- 11. Matsuda T., Bebenek K., Masutani C., Hanaoka F., Kunkel T. A. (2000) Low fidelity DNA synthesis by human DNA polymerase-η. Nature 404, 1011–1013 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Yuan F., Wu X., Rechkoblit O., Taylor J. S., Geacintov N. E., Wang Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 28, 4717–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Z., Yu Y., Chen X. (1999) Effect of low concentration N-methyl-N-nitro-N-nitrosoguanidine on mRNA expression of DNA polymerases and topoisomerase II in HeLa cell. Chin. J. Pharmacol. Toxicol. 13, 217–220 [Google Scholar]
- 14. Jiang Y., Qian Y., Yu Y. (2006) Expression response to methyl-nitro-nitrosoguanidine and the bioinformative analysis of binding sites of transcription factors of POLK, POLH, and POLI genes in mammalian cells. Chin. J. Pathophysiol. 22, 1249–1255 [Google Scholar]
- 15. Luo Y. Q., Yang J., Yu Y. N. (2003) Establishment of a cell line with antisense-blocked POLH and the role of POLH in alkylating agent MNNG induced nontargeted mutagenesis. Zhejiang Da Xue Xue Bao Yi Xue Ban 32, 398–402 [DOI] [PubMed] [Google Scholar]
- 16. Shi B. S., Cai Z. N., Yang J., Yu Y. N. (2004) N-Methyl-N-nitro-N-nitrosoguanidine sensitivity, mutator phenotype and sequence specificity of spontaneous mutagenesis in FEN-1-deficient cells. Mutat. Res. 556, 1–9 [DOI] [PubMed] [Google Scholar]
- 17. Gao Z., Yang J., Huang Y., Yu Y. (2005) N-Methyl-N-nitro-N-nitrosoguanidine interferes with the epidermal growth factor receptor-mediated signaling pathway. Mutat Res. 570, 175–184 [DOI] [PubMed] [Google Scholar]
- 18. Wang Z., Wang G., Yang J., Guo L., Yu Y. (2003) Activation of protein kinase A and clustering of cell surface receptors by N-methyl-N-nitro-N-nitrosoguanidine are independent of genomic DNA damage. Mutat. Res. 528, 29–36 [DOI] [PubMed] [Google Scholar]
- 19. Liu G., Shang Y., Yu Y. (2006) Induced endoplasmic reticulum (ER) stress and binding of overexpressed ER-specific chaperone GRP78/BiP with dimerized epidermal growth factor receptor in mammalian cells exposed to low concentration of N-methyl-N-nitro-N-nitrosoguanidine. Mutat. Res. 596, 12–21 [DOI] [PubMed] [Google Scholar]
- 20. Li H., Shao J., Lu X., Gao Z., Yu Y. (2008) Identification of early responsive genes in human amnion epithelial FL cells induced by N-methyl-N-nitro-N-nitrosoguanidine using oligonucleotide microarray and quantitative real-time RT-PCR approaches. Mutat. Res. 644, 1–10 [DOI] [PubMed] [Google Scholar]
- 21. Shen J., Wu M., Yu Y. (2006) Proteomic profiling for cellular responses to different concentrations of N-methyl-N-nitro-N-nitrosoguanidine. J. Proteome Res. 5, 385–395 [DOI] [PubMed] [Google Scholar]
- 22. Shen J., Chen W., Yin X., Yu Y. (2008) Proteomic analysis of different temporal expression patterns induced by N-methyl-N′-nitro-N-nitrosoguanidine treatment. J. Proteome Res. 7, 2999–3009 [DOI] [PubMed] [Google Scholar]
- 23. Percario Z. A., Giandomenico V., Fiorucci G., Chiantore M. V., Vannucchi S., Hiscott J., Affabris E., Romeo G. (1999) Retinoic acid is able to induce interferon regulatory factor 1 in squamous carcinoma cells via a STAT-1 independent signaling pathway. Cell Growth Differ. 10, 263–270 [PubMed] [Google Scholar]
- 24. Lin Y., Zhu X., McLntee F. L., Xiao H., Zhang J., Fu M., Chen Y. E. (2004) Interferon regulatory factor-1 mediates PPAR γ-induced apoptosis in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 24, 257–263 [DOI] [PubMed] [Google Scholar]
- 25. Tanaka N., Taniguchi T. (2000) The interferon regulatory factors and oncogenesis. Semin. Cancer Biol. 10, 73–81 [DOI] [PubMed] [Google Scholar]
- 26. Tanaka N., Ishihara M., Lamphier M. S., Nozawa H., Matsuyama T., Mak T. W., Aizawa S., Tokino T., Oren M., Taniguchi T. (1996) Cooperation of the tumor suppressors IRF-1 and p53 in response to DNA damage. Nature 382, 816–818 [DOI] [PubMed] [Google Scholar]
- 27. Prost S., Bellamy C. O., Cunningham D. S., Harrison D. J. (1998) Altered DNA repair and dysregulation of p53 in IRF-1 null hepatocytes. FASEB J. 12, 181–188 [DOI] [PubMed] [Google Scholar]
- 28. Frontini M., Vijayakumar M., Garvin A., Clarke N. (2009) A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 37, 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirsch J. D., Eslamizar L., Filanoski B. J., Malekzadeh N., Haugland R. P., Beechem J. M. (2002) Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins. Uses for protein labeling, detection, and isolation. Anal. Biochem. 308, 343–357 [DOI] [PubMed] [Google Scholar]
- 30. Kim S. H., Michael W. M. (2008) Regulated proteolysis of DNA polymerase η during the DNA-damage response in C. elegans. Mol. Cell 26, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26, 365–369 [DOI] [PubMed] [Google Scholar]
- 32. Smith H. O., Nathans D. (1973) Letter, a suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J. Mol. Biol. 81, 419–423 [DOI] [PubMed] [Google Scholar]
- 33. Liu G., Chen X. (2006) DNA polymerase η, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 26, 1398–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Escalante C. R., Yie J., Thanos D., Aggarwal A. K. (1998) Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391, 103–106 [DOI] [PubMed] [Google Scholar]
- 35. McCulloch S. D., Kunkel T. A. (2008) The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Book M. M., Yu-Lee L. (2001) Prolactin activation of IRF-1 transcription involves changes in histone acetylation. FEBS Lett. 12, 91–94 [DOI] [PubMed] [Google Scholar]
- 37. Pamment J., Ramsay E., Kelleher M., Dornan D., Ball K. L. (2002) Regulation of the IRF-1 tumor modifier during the response to genotoxic stress involves an ATM-dependent signaling pathway. Oncogene 21, 7776–7785 [DOI] [PubMed] [Google Scholar]
- 38. Dornan D., Eckert M., Wallace M., Shimizu H., Ramsay E., Hupp T. R., Ball K. L. (2004) Interferon regulatory factor 1 binding to p300 stimulates DNA-dependent acetylation of p53. Mol. Cell. Biol. 24, 10083–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caron C., Boyault C., Khochbin S. (2005) Regulatory cross-talk between lysine acetylation and ubiquitination. Role in the control of protein stability. BioEssays 27, 408–415 [DOI] [PubMed] [Google Scholar]
- 41. Li K., Wang R., Lozada E., Fan W., Orren D. K., Luo J. (2010) Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. Plos One 5, e10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim S. H., Kang H. J., Na H., Lee M. O. (2010) Tsictleatin A enhances acetylation as well as protein stability of ERα through induction of p300 Protein. Breast Cancer Res. 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marsili G., Remoli A. L., Sgarbanti M., Battistini A. (2006) Role of acetylases and deacetylase inhibitors in IRF-1-mediated HIV-1 long terminal repeat transcription. Ann. N.Y. Acad. Sci. 13, 636–643 [DOI] [PubMed] [Google Scholar]
- 44. Masumi A., Yamakawa Y., Fukazawa H., Ozato K., Komuro K. (2003) Interferon regulatory factor-2 regulates cell growth through its acetylation. J. Biol. Chem. 278, 25401–25407 [DOI] [PubMed] [Google Scholar]
- 45. Suhara W., Yoneyama M., Kitabayashi I., Fujita T. (2002) Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 277, 22304–22313 [DOI] [PubMed] [Google Scholar]
- 46. Caillaud A., Prakash A., Smith E., Masumi A., Hovanessian A. G., Levy D. E., Marié I. (2002) Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J. Biol. Chem. 277, 49417–49421 [DOI] [PubMed] [Google Scholar]
- 47. Masutani C., Kusumoto R., Iwai S., Hanaoka F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 19, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merika M., Williams A. J., Chen G., Collins T., Thanos D. (1998) Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287 [DOI] [PubMed] [Google Scholar]
- 49. Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. (2001) Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21, 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edmunds C. E., Simpson L. J., Sale J. E. (2008) PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell 30, 519–529 [DOI] [PubMed] [Google Scholar]
- 51. Livneh Z. (2006) Keeping mammalian mutation load in check. Regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle 5, 1918–1922 [DOI] [PubMed] [Google Scholar]
- 52. Acharya N., Yoon J. H., Gali H., Unk I., Haracska L., Johnson R. E., Hurwitz J., Prakash L., Prakash S. (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Y. W., Cleaver J. E., Hatahet Z., Honkanen R. E., Chang J. Y., Yen Y., Chou K. M. (2008) Human DNA polymerase η activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 16578–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamashita S., Nomoto T., Abe M., Tatematsu M., Sugimura T., Ushijima T. (2004) Persistence of gene expression changes in stomach mucosae induced by short-term N-methyl-N′-nitro-N-nitrosoguanidine treatment and their presence in stomach cancers. Mutat. Res. 549, 185–193 [DOI] [PubMed] [Google Scholar]
- 55. Zhu H., Fan Y., Jiang H., Shen J., Qi H., Mei R., Shao J. (2010) Response of human DNA polymerase ι promoter to N-methyl-N′-nitro-N-nitrosoguanidine. Environ. Toxicol. Pharmacol. 29, 79–86 [DOI] [PubMed] [Google Scholar]
- 56. Pine R. (1997) Convergence of TNF-α and IFNγ signaling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/κB promoter element. Nucleic Acids Res. 254, 346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ohmori Y., Schreiber R. D., Hamilton T. A. (1997) Synergy between interferon-γ and tumor necrosis factor-α in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor κB. J. Biol. Chem. 272, 14899–14907 [DOI] [PubMed] [Google Scholar]