Background: The T cell receptor (TCR) triggers signaling in T cells via an unknown mechanism.
Results: The structure of the signaling subunit, CD3ϵ, is unchanged by signal-inducing antibodies, and mutations that would block intersubunit rearrangements do not affect signaling.
Conclusion: Antibodies trigger TCR signaling without inducing large structural rearrangements.
Significance: TCR triggering might generally occur in the absence of large structural rearrangements.
Keywords: Crystallography, Protein Structure, Signal Transduction, Site-directed Mutagenesis, T cell Receptor, Receptor Triggering
Abstract
Native and non-native ligands of the T cell receptor (TCR), including antibodies, have been proposed to induce signaling in T cells via intra- or intersubunit conformational rearrangements within the extracellular regions of TCR complexes. We have investigated whether any signatures can be found for such postulated structural changes during TCR triggering induced by antibodies, using crystallographic and mutagenesis-based approaches. The crystal structure of murine CD3ϵ complexed with the mitogenic anti-CD3ϵ antibody 2C11 enabled the first direct structural comparisons of antibody-liganded and unliganded forms of CD3ϵ from a single species, which revealed that antibody binding does not induce any substantial rearrangements within CD3ϵ. Saturation mutagenesis of surface-exposed CD3ϵ residues, coupled with assays of antibody-induced signaling by the mutated complexes, suggests a new configuration for the complex within which CD3ϵ is highly exposed and reveals that no large new CD3ϵ interfaces are required to form during antibody-induced signaling. The TCR complex therefore appears to be a structure that is capable of initiating intracellular signaling in T cells without substantial structural rearrangements within or between the component subunits. Our findings raise the possibility that signaling by native ligands might also be initiated in the absence of large structural rearrangements in the receptor.
Introduction
Understanding the assembly, overall structure, and triggering mechanism of the TCR5 complex remains one of the major challenges in molecular immunology. Comprised of six different type I membrane proteins, the TCR complex is an unusually complicated assembly of membrane surface proteins and glycoproteins. In contrast to receptors for soluble ligands triggered by tyrosine autophosphorylation, such as growth factor receptors, the TCR lacks directly associated kinase activity, so signal transduction is dependent on the phosphorylation of immunoreceptor tyrosine activation motifs (1, 2) in the cytoplasmic domains of the TCR by extrinsic Src kinases (3, 4).
At the level of individual receptors, each TCR complex is comprised of single antigen-binding αβ heterodimers and invariant CD3-ϵδ, -ϵγ, and -ζζ dimers responsible for signal transduction (5–8). It has been suggested that, on a larger scale, the TCR is organized into “protein islands” containing 8–20 freely diffusing complexes, which concatenate into microclusters upon activation, prior to formation of the immunological synapse (9–11), but whether this truly reflects the “resting” organization of the complex has been questioned (12). Assembly of the TCR complex is tightly controlled and depends on the formation of disulfide bonds between the α and β chains and between the CD3ζ subunits and on highly conserved trimolecular, charge-charge interactions involving the transmembrane helices of (i) TCRα, CD3δ, and CD3ϵ; (ii) TCRβ, CD3γ, and CD3ϵ; and (iii) TCRα and the two copies of CD3ζ (13–15). Superimposed upon these interactions are likely noncovalent contacts between the extracellular regions of the TCR subunits, involving both the “connecting peptides” (16) and the immunoglobulin superfamily domains. Initial mutational data suggested that the DE loop of the constant (Cα) region of TCRα contacts CD3ϵδ and that CD3ϵγ contacts the TCR via the CC′ loop of Cβ (17, 18), ruling out the “bunch of balloons” arrangement of nonassociated extracellular domains implied by a lack of detectable interactions in solution (19–22). Otherwise, we have limited understanding of the three-dimensional organization of the assembled complex.
Another important question concerns whether the TCR changes structure during triggering or is essentially rigid. Janeway (23) first mooted the idea of conformational change after observing a poor correlation between the signaling effects of anti-TCR antibodies and their affinities. A ligand-dependent structural rearrangement of one TCR has been proposed to occur in solution and in crystal lattices but has yet to be directly linked to receptor triggering in vivo (24). Much of the recent impetus for the conformational change hypothesis, however, comes from studies of CD3 cytoplasmic domains. A conformational alteration in CD3ϵ was claimed to initiate TCR signaling by inducing recruitment of the Nck adaptor protein to a proline-rich region of the cytoplasmic domain of CD3ϵ (25, 26), although this is controversial (27–29). More recently, Wucherpfennig and co-workers (30) have proposed that the cytoplasmic domain of CD3ϵ associates with acidic phospholipids, suggesting that triggering requires its release from the membrane, but this is also controversial (31).
If intracellular changes were to occur, the forces driving them would derive from the extracellular domains of the TCR subunits, where ligand binding takes place. In deconstructing the form and function of the TCR, Kuhns et al. (19) suggested that this could involve large intra- or intersubunit conformational changes. However, if a set of specific structural rearrangements were necessary for “transmission” of the signal across the membrane to the cytoplasmic domains, then such changes would have to be invoked by all triggering ligands, that is, by peptide-MHC and by mitogenic anti-TCR/CD3 antibodies. We have looked for the signatures of large structural rearrangements induced in the extracellular region of CD3ϵ by mitogenic antibody ligation, which has for many years been a widely used surrogate of ligand-induced signaling. Mitogenic mAbs directed against the CD3ϵ chain (32) were shown to induce Ca2+ release (33), IL-2 secretion (34), and immune synapse formation (35) through the activation of the same pathways, with similar kinetics as those induced by agonist peptide-MHC binding. Anti-CD3ϵ mAbs have also been used as immunomodulating agents in the treatment of autoimmune diseases (36, 37).
The crystal structure of a murine CD3ϵ-mitogenic antibody complex described herein allows the first direct analysis of the structural effects of mitogenic anti-CD3ϵ antibodies. Previously, comparisons could only be made between the structures of apo and antibody-liganded forms of CD3ϵ from different species, which were substantially different (20, 21, 38, 39). The new structure now shows that significant intrasubunit changes in CD3ϵ structure do not accompany antibody binding, at least in solution. Furthermore, mutational analysis of the cell surface-expressed complex suggests that large new intersubunit contacts involving CD3ϵ do not form during antibody-induced receptor triggering either. Our findings therefore indicate that the TCR complex is configured in such a way that substantial structural rearrangements of its component subunits are not a prerequisite of signaling by antibodies and raise the possibility that native ligands could initiate signaling via a mechanism involving relatively minor or no structural rearrangements in the complex.
MATERIALS AND METHODS
Expression of Stable CD3ϵ Homodimer
Chimeric genes comprising the globular ectodomain fragments of mouse CD3ϵ, δ, and γ subunits fused to residues 213–450 of the mouse IgG1 heavy chain (Fc) were designed for the expression of soluble mouse CD3 subunits as noncovalent dimers (mCD3Fc) in mammalian cells, as described previously for the production of soluble ectodomain dimers (40, 41). Purified soluble mouse CD3ϵϵ retained native topology for several anti-CD3ϵ mAb epitopes, as determined by binding to a panel of anti-CD3 mAbs. Specifically, mitogenic mAb 2C11 and the conformation-dependent mAb 17A2 bound soluble CD3ϵϵ material in a highly specific, dose-dependent manner (supplemental Fig. S1A). Detailed procedures for the purification of CD3 homodimer are described in the supplemental “Experimental Procedures.”
Preparation of 2C11 Fab and Purification of mCD3ϵϵ-2C11 Fab Complex
Procedures for Fab and Fab complex preparation are described in the supplemental “Experimental Procedures.”
Crystallization and Data Collection for 2C11 Fab and mCD3ϵϵ-2C11 Complex
The proteins were crystallized by sitting drop vapor diffusion at 8 and 10 mg/ml for Fab 2C11 and the mCD3ϵϵ-Fab 2C11 complex, respectively. Each drop contained 0.45 μl of protein mixed with an equal volume of precipitate solution. Crystals of Fab 2C11 were obtained from a solution containing 0.4 m potassium nitrate, 0.1 m HEPES, pH 7.5, 23% PEG 4000 and flash cooled to 100 K following immersion in a cryoprotectant comprising precipitant supplemented with 20% PEG 200. Crystals of mCD3ϵϵ-Fab 2C11 were obtained from a solution containing 1.7 m ammonium sulfate, 0.1 m MES, pH 5.2, 10% (w/v) dioxane at 16 °C and flash cooled to 100 K following immersion in a cryoprotectant comprising the precipitant supplemented with 20% glycerol. The data sets were collected at 2.5 Å for the Fab alone and at 4.1 Å for the complex from single crystals at the Advanced Photon Source Beamline 23-ID and integrated and scaled using Denzo and Scalepack (42), as implemented in the HKL2000 suite. Unit cell dimensions, data collection and processing statistics are detailed in supplemental Table S1. Structural determination, refinement and validation of 2C11 Fab and the mCD3ϵϵ-2C11 complex are described in detail in the supplemental “Experimental Procedures.” Atomic coordinates and structure factors for 2C11 Fab and the mCD3ϵ-2C11 Fab complex have been deposited in the Protein Data Bank with accession numbers 3R06 and 3R08, respectively.
T Cell Activation and NFAT/IL-2 Promoter Reporter Assay
Jurkat T cells stably expressing wild-type or mutated HA-tagged proteins were transduced with a 3× IL-2 Renilla luciferase reporter construct using lentivirus. Forty-eight hours after infection, the cells were plated at 1 × 105/well in 100 μl of RPMI in 96-well flat bottomed tissue culture plates previously treated with a 25 μg/ml solution of donkey anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) overnight at 4 °C followed by a second overnight incubation at 4 °C with anti-CD2 (Miltenyi Biotech, Bergisch Gladbach, Germany) and anti-CD28 (7.3B6) plus either anti-CD3 (OKT3), anti-HA (HA-7; Sigma), or anti-Thy1 (OX7) at 10 μg/ml each. After 6 h of antibody stimulation, luciferase activity was measured by adding coelenterazine-h (Lux Biotechnology, Edinburgh, UK) at a final concentration of 10 μm to cells before reading total emission on a microplate analyzer. The results were plotted as a ratio between appropriate antibodies after background subtraction. During the 6-h stimulation, cell surface expression of HA-tagged proteins and GFP levels expressed under an IRES regulator were quantified by FACS, as described in the supplemental “Experimental Procedures.” Procedures for identification of residues for mutation, choice of drastic mutations, constructs, flow cytometry, and lentiviral transduction of cell lines are described in detail in the supplemental “Experimental Procedures.”
RESULTS
Expression and Crystallization of mCD3ϵ as a Soluble Homodimer-Fab Complex
CD3-ϵ, -δ, and -γ subunits are present on the surface of T cells as noncovalent heterodimers, and dimerization appears to be required for their expression as soluble proteins (20, 21, 38, 39). We tested for new pair-wise interactions of murine (m) CD3 ectodomains by co-expressing the noncovalently associated ectodomains in all six combinations (i.e. as ϵϵ, ϵδ, ϵγ, δδ, γγ, and γδ pairs) as chimeras with IgG heavy chain Fc regions in mammalian cells. No combination of intact extracellular regions yielded nonaggregated chimeric protein, and of the forms expressed with cysteine-to-serine mutations of the membrane-proximal Cys-Xaa-Xaa-Cys-Xaa-Glu motif, only mCD3ϵFc yielded soluble protein. Following removal of the Fc region, the mCD3ϵ ectodomain formed a soluble homodimer that bound strongly to anti-mCD3ϵ mAbs (supplemental Fig. S1A) and was resistant to dissociation at micromolar concentrations (supplemental Fig. S1B). Whereas the mCD3ϵ homodimer failed to form reproducible crystals, complexes formed with the 2C11 Fab readily crystallized in cubic space group I4132 at pH 5.2, with unit cell dimensions a = b = c = 263.2 Å, α = β = γ = 90°. Preliminary phases were determined by molecular replacement (see supplemental “Experimental Procedures”). The 2C11 Fab-mCD3ϵ complex structure was refined to 4.1 Å resolution to Rcryst (22.2) and Rfree (27.7) values within the acceptable range for this resolution. The 2Fo − Fc electron density was well defined for Cα atoms throughout the complex (supplemental Fig. S2). The apo 2C11 Fab structure at 2.5 Å resolution facilitated refinement and validation. Despite its modest resolution, the availability of high resolution crystal structures of human CD3ϵ (hCD3ϵ; Protein Data Bank code 1SY6; 2.1 Å) (39) and the 2C11 Fab (2.5Å), as well as the apo mCD3ϵ NMR structures (Protein Data Bank codes 1JBJ and 1XMW) (20, 21), allowed confident interpretation of the data. Data collection and refinement statistics are given in supplemental Table S1.
Overall Structure of mCD3ϵ-2C11 Fab Complex
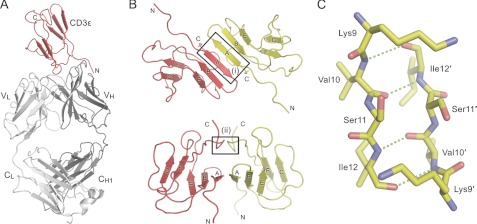
The mCD3ϵ-2C11 Fab structure comprises a new, putatively “homodimeric” CD3ϵ interaction and allows the first direct comparisons of mitogenic antibody-bound and unbound forms of CD3ϵ. The crystallographic asymmetric unit consists of one mCD3ϵ ectodomain bound to a single 2C11 Fab (Fig. 1A), with the “homodimerization” interface straddling a 2-fold symmetry axis of the crystal. The mCD3ϵ ectodomain has an immunoglobulin superfamily fold comprised of a sandwich of two anti-parallel β-sheets with GFCC′/EBA topology, rather than the GFCC′/DEBA β-strand arrangement of human CD3ϵ (20, 21, 38, 39). The homodimer is formed by a head-to-tail arrangement of the monomers mediated by anti-parallel pairing of the A strands of each subunit (Tyr8-Ser13; box (i) in Fig. 1B), thereby forming a continuous β-sheet that spans the interface, composed of the A, B, and E strands of each monomer (Fig. 1B). This interaction is stabilized by the reciprocal H-bonding of Val10 and Ile12 (Fig. 1C). A β-sheet also traverses the interfaces of CD3-ϵδ and -ϵγ heterodimers, although it is formed by parallel interactions of the G-strands rather than the A-strands (20, 21). A second, smaller region of association involves Ala77 and Arg78 of the C-terminal loops of each monomer (box (ii) in Fig. 1B).
FIGURE 1.
Structure of the mCD3ϵ-Fab 2C11 complex. A, cartoon representation of the asymmetric unit containing one copy each of mCD3ϵ (red) and the Fab fragment from the CD3ϵ-specific monoclonal antibody 2C11 (gray). B, crystallographic homodimer of mCD3ϵ. The two CD3ϵ monomers are colored red and yellow, respectively. The top panel shows the contiguous β-sheet formed by anti-parallel pairing of the A-strands of the two monomers. The bottom panel shows a view after a 90° rotation about the horizontal axis. Boxes (i) and (ii) highlight the two main regions of interaction. C, ball-and-stick representation of the interaction between the A strands in box (i) from B. Potential hydrogen bonds are shown as dashed green lines. See also supplemental Figs. S1 and S2 and supplemental Table S1.
Is mCD3ϵ homodimerization compatible with its heterodimerization with CD3γ and with CD3δ? Initially, it appeared possible to dock CD3γ and CD3δ with either of the CD3ϵ domains in the CD3ϵ homodimer, because the heterodimeric interfaces are distinct from the homodimeric interface (Fig. 2A). However, formation of the small interface involving Ala77 and Arg78 of the C-terminal loops of CD3ϵ would be precluded by prior heterodimerization of CD3ϵ with either CD3γ or CD3δ (circled in Fig. 2A). If this was avoided by the C terminus adopting another conformation in the presence of CD3γ and CD3δ, formation of a CD3γ-CD3δ-(CD3ϵ)2 heterotetramer via CD3ϵ homodimerization would nevertheless be precluded by steric clashes between the AB loops of CD3γ and CD3δ and between the EF loop of each CD3ϵ monomer and the EF loop of the CD3γ and CD3δ subunit bound to the other CD3ϵ monomer (Fig. 2B). In addition, the C termini of the four subunits would be buried in the center of the CD3γ-CD3δ-(CD3ϵ)2 heterotetramer, with the only route of exit being a tiny hole (∼2 Å in diameter) too small to accommodate all four stalks connecting the extracellular regions to the membrane. Finally, as discussed below, mutations of the region of hCD3ϵ equivalent to that mediating mCD3ϵ homodimerization do not prevent TCR complex assembly. Overall, the mCD3ϵ homodimer seems to be nonphysiological, despite its stability in solution.
FIGURE 2.
The CD3ϵ crystallographic homodimer is unlikely to mediate CD3 heterotetramerization. A, the crystallographic CD3ϵ homodimer is shown in red and yellow, with CD3δ (blue; Protein Data Bank code 1XMW) and CD3γ (green; Protein Data Bank code 1JBJ) shown docked with the red CD3ϵ monomer in the positions that CD3δ and CD3γ occupy in the NMR structures of the CD3ϵδ and CD3ϵγ heterodimers (20, 21). Prior heterodimerization of CD3ϵ with either CD3γ or CD3δ creates a clash (circled) that prevents the formation of the small homodimerization interface involving the C-terminal loops of CD3ϵ (box (ii) in Fig. 1B). B, upper panel, a putative heterotetrameric CD3 complex, with CD3δ (blue) and CD3γ (green) each docked with one of the CD3ϵ monomers in the CD3ϵ homodimer (red and yellow), in the manner observed in the NMR structures of the CD3ϵδ and CD3ϵγ heterodimers. Most of the yellow CD3ϵ monomer is hidden behind CD3γ. Lower panel, a section through the middle of the heterotetramer revealing the positions of the subunit surfaces; the section does not incorporate any part of the second (yellow) CD3ϵ monomer. Subunit regions that clash are labeled. These clashes would likely prevent formation of the heterotetramer in vivo. See also supplemental Fig. S1.
2C11 Fab Epitope and Comparison with Other CD3-Fab Complex Structures
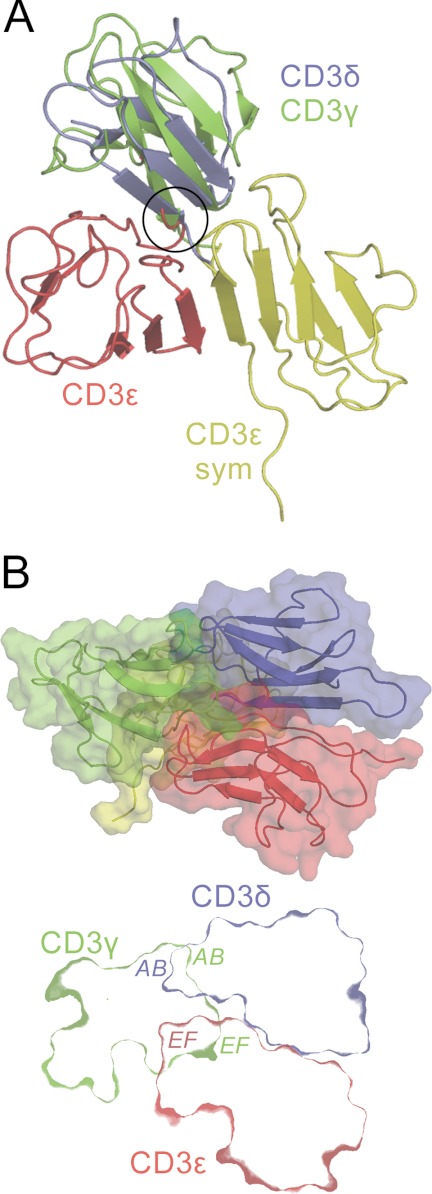
The 2Fo − Fc electron density is of high quality in the region of the Fab 2C11 combining site, allowing reliable identification of the interacting elements despite the modest resolution of the data. 2C11 binds across the BC and FG loops at the “top” of the mCD3ϵ ectodomain, perpendicular to the dimerization interface of mCD3ϵδ and mCD3ϵγ (Figs. 1A and 3A). The 2C11 epitope is centered on Leu23–Asn28 and Lys30 of the BC loop, Tyr63–Asn70 of the FG loop, and Asp1–Asn5 at the N terminus of mCD3ϵ. The major part (i.e. 79%) of the Fab surface buried in the complex (total 720 Å2) is within the VH region of 2C11, a bias commonly observed in antibody/antigen interactions. The Fab-buried surface area is typical for antibody-protein antigen complexes (43) and comparable with that of the Fab UCHT1-hCD3ϵ complex (660 Å2; Protein Data Bank code 1XIW) (38) but much larger than that of the Fab OKT3-hCD3ϵ complex (445Å2; Protein Data Bank code 1SY6; Ref. 39; Fig. 3A). Comparison of the 2C11, OKT3, and UCHT1 complexes (Fig. 3A) reveals considerable overlap in the regions of CD3ϵ contacted by the mAbs, despite (i) the significant differences between the mouse and human structures and (ii) that the UCHT1 and OKT3 epitopes are centered on the FG loop rather than the BC loop. Together these epitopes define a contiguous area of ∼1200 Å2 likely to be exposed in CD3ϵ prior to and during antibody triggering. For a CD3 heterodimer positioned with its pseudo-symmetry axis orthogonal to the membrane, the positions of the Fabs differ by ∼60° rotation around this axis and by a ∼50° rotation away from it toward the membrane, with 2C11 the most “upright” of the Fabs (Fig. 3B). Although the regions of CD3ϵ available for antibody binding within the complex appear restricted, there is nonetheless considerable variation in the binding orientations and dispositions of these three mitogenic anti-CD3 antibodies.
FIGURE 3.
Mitogenic antibody binding to CD3ϵ. A, surfaces of mouse and human CD3ϵ that bind mitogenic mAbs. The left panel shows the epitope buried by 2C11 (blue) on mCD3ϵ, with a subset of the interface residues labeled. In the right panel, the epitopes of hCD3ϵ (from Protein Data Bank code 1XIW) that are buried by OKT3 (green, Protein Data Bank code 1SY6), UCHT1 (orange, Protein Data Bank code 1XIW), or both antibodies (purple) are shown. The human and mouse structures were superimposed so that each is seen from a similar viewpoint roughly orthogonal to the 2C11 epitope. B, the location of the 2C11 Fab (blue) on mCD3ϵ is shown compared with that of OKT3 (green) and UCHT1 (orange) Fab fragments following superposition of hCD3ϵ from these complexes onto the mCD3ϵ structure. For UCHT1, the OKT3 Fab is used but superpositioned on the UCHT1 Fv fragment that is present in the structure. The location of mCD3γ (yellow) is also shown following superposition of the CD3γϵ complex (Protein Data Bank code 1JBJ) onto the mouse CD3ϵ crystal structure. C, superposition of the Cα traces of the NMR structures of unbound mCD3ϵ (from CD3ϵδ, Protein Data Bank code 1XMW, blue; and from CD3ϵγ, Protein Data Bank code 1JBJ, green) onto 2C11-bound mCD3ϵ (red). Two orthogonal views are shown with the N and C termini, and the FG loop referred to in the text, all labeled.
Influence of Mitogenic Antibody Binding on CD3ϵ
At the present resolution (4.1 Å), it is only possible to be confident about the orientation, location, and trace of the Cα backbones in the complex. With this caveat, comparison of the crystal structure of the Fab 2C11-mCD3ϵ complex with the NMR structures of mCD3ϵγ (Protein Data Bank code 1JBJ) and mCD3ϵδ (Protein Data Bank code 1XMW) reveals that the effects of antibody binding on the architecture of the CD3ϵ ectodomain are remarkably limited (Fig. 3C). In particular, mCD3ϵ from the mCD3ϵδ and mCD3ϵγ heterodimers is largely superimposable with the Fab-complexed mCD3ϵ monomer. Minor variation in the conformations of loops in all three structures is indicative of limited inherent flexibility in these regions. The only clear difference in conformation within the domain on binding is limited to a very minor change to the FG loop and the top of the G strand (Fig. 3C), presumably because of local interactions of this loop with the Fab. Outside the folded part of the Ig superfamily domain, the N and C termini are substantially different. The extended N terminus of mCD3ϵ, in particular Asp1–Ile6, is fixed by interactions with Fab 2C11 in the 2C11-mCD3ϵ complex (Fig. 1A), but the equivalent residues are not constrained in the NMR structures given that their positions vary considerably between models (Fig. 3C). The largest definitive differences involve Lys76–Arg78, which produce a hairpin-like structure at the C terminus of 2C11-bound mCD3ϵ that is stabilized by contacts in the homodimerization interface (marked C in Fig. 3C). These differences reflect considerable variability of the N and C termini of mCD3ϵ, as in the case of hCD3ϵ (Protein Data Bank code 1SY6). No significant backbone conformational changes in the variable regions of the Fab accompany CD3ϵ binding (root mean square deviations for Fv-equivalent Cα atoms are between 0.6 and 1.0 Å). 2C11 thus engages mCD3ϵ using a rigid docking mechanism and induces only small changes in mCD3ϵ restricted to one of the loops (Fig. 3C).
Saturation Mutagenesis-based Subunit Interface Mapping
Because significant structural rearrangements within CD3ϵ did not accompany mitogenic antibody binding, we investigated whether the interactions of the CD3ϵ subunit with the rest of the TCR complex change substantially during antibody triggering. Assuming an intimately assembled complex that allows the transmission of structural changes between subunits (19), such rearrangements would be predicted to bury surfaces previously exposed in the assembled complex. Mutations that prevent these surfaces from becoming buried would be expected to block signaling.
To identify CD3ϵ residues that are exposed in the assembled TCR complex prior to triggering, we made use of the fact that the TCR complex is an obligate hetero-oligomer, that is, surface expression is dependent on the assembly of the entire complex (44, 45). Thus, “drastic” mutations of residues lining subunit interfaces, but not residues exposed in the fully assembled complex, should prevent assembly and expression of the complex at the cell surface, allowing the exposed and buried surfaces of the subunit to be mapped. By mutating residues exposed to solvent according to the crystal structures of CD3ϵ (38, 39), we sought to avoid buried residues whose mutation might prevent assembly via effects on folding. After identifying the exposed and buried surfaces of CD3ϵ, we went on to determine whether initially exposed residues are buried during receptor triggering, by testing whether drastic mutations of these residues prevent signaling.
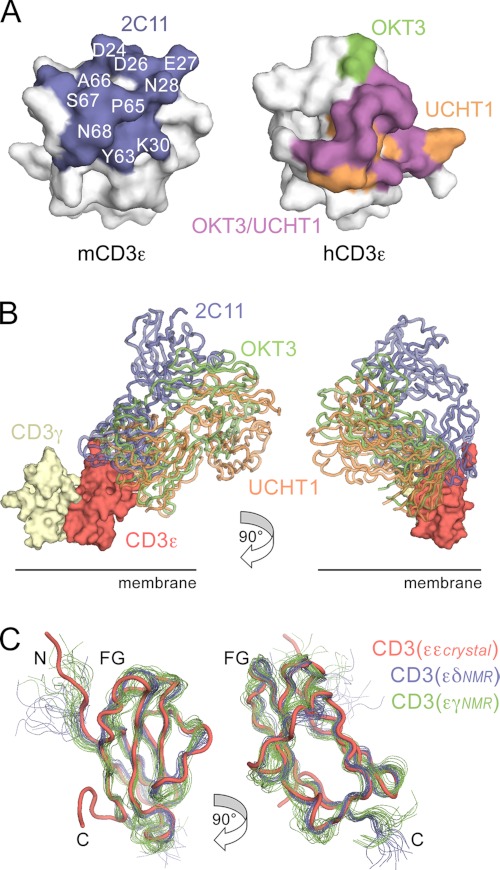
To show that the mutational approach was capable of identifying surface-exposed and buried residues in obligate complexes, we first attempted to classify the surface residues of the human CD8αα homodimer in this way (46, 47). All residues in the Ig superfamily V-set domain of CD8α whose side chains were >50% solvent-exposed (using NACCESS; (48)) were “drastically” mutated to Arg, except for basic residues, which were mutated to Glu. Expression of 80 HA-tagged mutant forms of CD8α in Jurkat T cells using a lentivirus-based expression system (as described in the supplemental “Experimental Procedures”) was tested by flow cytometry with anti-HA (HA-7; Fig. 4A) and anti-CD8 antibodies (OKT8, DK25, and SK1; supplemental Fig. S3). Anti-HA antibody reactivity, which directly measured CD8 expression, was reduced to <15% of wild-type levels for 13 mutant proteins and to 15–40% of wild-type levels for four other mutants. All 17 mutations also reduced anti-CD8 antibody reactivity, confirming that these were bona fide effects on expression (supplemental Fig. S3). Eight of the mutations that reduced expression to <15% of wild-type levels, mutations of Ser34, Leu36, Phe48, Try91, Phe93, Ser95, Met102, and Phe104, form a cluster comprising a contiguous surface (Fig. 4B) corresponding to the core of the known CD8αα homodimer interface (Fig. 4C). Ala42 is also located at the known subunit interface, whereas the other four mutations with large effects (Pro7, Glu16, Arg67, and Glu89) are distributed at single sites and are interpreted as having folding effects. Mutations disrupting antibody binding also mapped to clusters of four to five residues, presumably identifying their core epitopes (supplemental Fig. S3). We conclude from the analysis of the CD8 homodimer that subunit interface residues and exposed residues in obligate complexes are identifiable using saturation mutagenesis and drastic mutations, guided by known structures of the subunits.
FIGURE 4.
Identification of surface-exposed and buried residues in the human CD8αα homodimer. A, histogram showing relative surface expression levels of drastically mutated, lentiviral expressed, HA-tagged CD8 proteins. The surface expression ratio is calculated as the median fluorescence of anti-HA antibody binding divided by the median fluorescence of IRES-encoded GFP (as an indicator of the efficiency of viral transduction), each measured by FACS. The surface expression ratio is normalized against the value obtained for the wild-type protein in the same experiment, and the average values for at least two experiments, expressed as percentages, are shown along with standard errors of the mean. The bars are colored according to whether mutation of the indicated residue reduced CD8 expression by more than 85% (red), by 60–85% (orange), or by less than 60% (green) versus wild-type HA-tagged CD8 expression (blue). B–D, the surface of CD8α (from Protein Data Bank code 1AKJ) (46) is shown. In each panel, the upper view looks onto the GFCC′C″ face, and the lower view is rotated 180° about the vertical axis (hence looking onto the DEBA sheet). In B, mutated residues are colored as described in A. In C, residues shown by crystallographic analysis to mediate CD8αα homodimerization are colored blue. In D, residues whose drastic mutation affects cell surface expression are colored yellow, and the remaining residues are colored according to whether they drastically reduce (>85%) the binding of the anti-CD8 mAbs DK25 (blue), SK1 (purple), or OKT8 (red) or have no effect on expression or antibody binding (green). See also supplemental Fig. S3.
CD3ϵ Is Fully Exposed in the TCR Complex
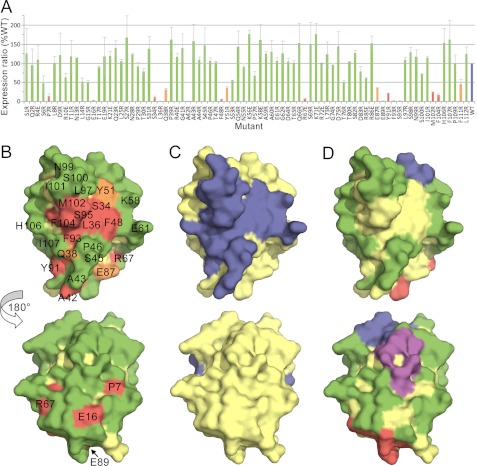
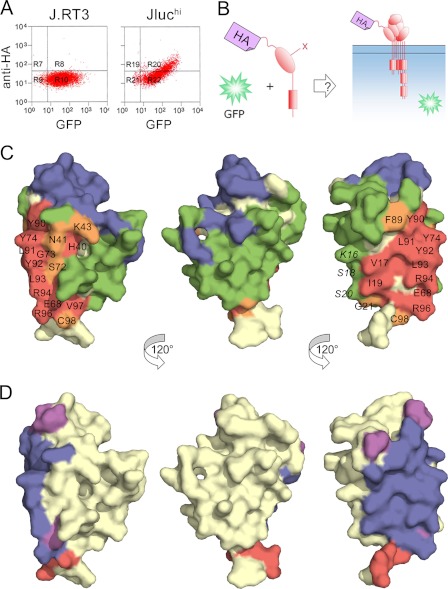
Applying this approach to the CD3ϵ subunit of the TCR complex, we first established an assay for CD3ϵ incorporation into the complex. J.RT3-T3.5 cells, which lack a functional TCRβ chain (45), fail to detectably express lentiviral transduced HA-tagged human CD3ϵ (Fig. 5A, left panel). J.RT3-T3.5 cells stably expressing a TCRβ/luciferase chimera (referred to as Jluchi cells), however, do express HA-tagged CD3ϵ at the cell surface (Fig. 5A, right panel). Expression of HA-tagged CD3ϵ at the surface of Jluchi cells was therefore used to assay CD3ϵ incorporation into the TCR complex (Fig. 5B). Quantitative flow cytometric analysis revealed that the ratio of HA-tagged to endogenous CD3ϵ at the cell surface was 9:1 (data not shown), implying that >80% of the assembled receptors carried two HA-tagged CD3ϵ subunits. Using the CD3ϵδ and CD3ϵγ crystal structures as a guide, we drastically mutated the surface-exposed residues of CD3ϵ. For completeness, we mutated any residue whose side chain contained an atom whose solvent-exposed surface area was >5Å2 according to NACCESS (54 HA-tagged CD3ϵ mutants in total). However, the known UCHT1 (38) and OKT3 (39) antibody epitopes exposed in the complex were not mutated. Trp38 and Ile45 buried in the core of the domain were mutated as misfolding controls.
FIGURE 5.
Identification of surface-exposed and buried residues in human CD3ϵ in the TCR-CD3 complex. A, FACS analysis of cells stably transduced with a lentiviral vector encoding both HA-tagged CD3ϵ and GFP, separated by an IRES sequence. The level of GFP fluorescence (x axis) indicates the efficiency of transduction. Anti-HA antibody staining (y axis) for transduced J.RT3-T3.5 cells lacking a functional TCRβ chain and for transduced J.RT3-T3.5 cells stably expressing a TCRβ/luciferase chimera (Jluchi cells) is shown in the left and right panels, respectively. B, a schematic showing the assay for CD3ϵ incorporation. C, three views of the surface of CD3ϵ (from Protein Data Bank code 1XIW), each related by a 120° rotation about the vertical axis. Residues colored blue are buried by the monoclonal antibodies UCHT1 (as in Protein Data Bank code 1XIW) or OKT3 (as in Protein Data Bank code 1SY6). Other residues are colored according to whether their mutation reduces CD3ϵ expression by more than 85% (red), by 60–85% (orange), or by less than 60% (green) versus wild-type CD3ϵ expression. D, the same views of CD3ϵ as in C colored according to whether the residues are buried in the heterodimeric interfaces with CD3γ (purple; as in Protein Data Bank code 1SY6), CD3δ (red; as in Protein Data Bank code 1XIW), or both (blue). See also supplemental Fig. S4.
Forty-one of the mutant CD3ϵ proteins were detectable at the cell surface with anti-HA antibody at levels similar to wild-type HA-tagged CD3ϵ (supplemental Fig. S4); the Trp38 and Ile45 misfolding controls were not expressed. Residues mutated in 12 of the 13 other nonexpressing mutants (Val17, Ile19,Glu68, Gly73, Tyr74, Tyr90, Leu91, Tyr92, Leu93, Arg94, Arg96, and Val97) form a contiguous surface (Fig. 5C) exhibiting remarkable overlap with the shared interface that CD3ϵ forms with CD3δ or CD3γ (Fig. 5D; Refs. 38 and 39). Mutation of His40, which lies immediately “behind” the interface, prevents expression, presumably by perturbing neighboring residues Ser72, Gly73, and Tyr74 at the interface with CD3δ and CD3γ. Smaller effects were observed for residues at the edges of the interface (Fig. 5C). These results imply that, apart from the surface buried with CD3δ and CD3γ, CD3ϵ is completely exposed in the TCR complex. Mutations of residues in the A strand that are not involved in γ/δ association (Lys16, Ser18, and Ser20) do not affect expression, suggesting that there is no obligate homodimerization of human CD3ϵ in the manner observed in the mCD3ϵ crystals. However, this result could also reflect the largely main chain/main chain character of the contacts at this interface.
Subunit Interface Analysis of TCRαβ, CD3δ, and CD3γ
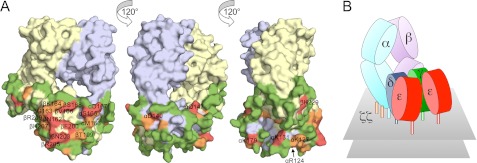
The analysis was extended to the rest of the complex. All of the residues whose side chains were >50% solvent-exposed in the TCR-α and -β constant (C) domains and in CD3δ and CD3γ according to NACCESS (apart from those buried by CD3ϵ) were mutated and tested for expression in Jluchi cells. Most mutations of TCR-α and -β were without effect, indicating that large regions of the αβ heterodimer are exposed in the TCR complex (supplemental Fig. S5). Unclustered mutations that reduced anti-HA-detectable expression were attributed to folding effects; these included one residue in Cβ (Lys229 in the FG loop) and five others scattered throughout Cα (Arg124, Lys131, Asp140, Gln147, and Lys179; Fig. 6A). Other mutations with large effects on surface expression clustered in a single, relatively small contiguous surface comprised of two Cα DE loop residues (Ser165 and Asp167) and six Cβ CD (Ser168, Val166, and Asn162) and EF (Phe200, Asn203, and Arg205) loop residues. Mutations of six other residues surrounding this region, i.e. αMet166, βGly163, βLys164, βThr199, βHis207, and βArg209, had weaker effects. This set of 14 residues, i.e. αSer165, αAsp167, βSer168, βVal166, βAsn162, βPhe200, βAsn203, βArg205, αMet166, βGly163, βLys164, βThr199, βHis207, and βArg209, likely comprises a surface forming the single point of contact of the αβ heterodimer with the CD3 signaling subunits.
FIGURE 6.
Surface mutation of TCRαβ and model for the TCR-CD3 complex. A, three views of the surface of the TCRαβ heterodimer (from Protein Data Bank code 1OGA (71)), each related by a 120° rotation about the vertical axis. The α chain is colored blue-gray, and the β chain is yellow. Mutated residues are colored according to whether their mutation reduces TCR expression by more than 85% (red), by 60–85% (orange), or by less than 60% (green) versus wild-type TCR expression. B, cartoon illustration of the proposed quaternary arrangement of extracellular domains within the TCR-CD3 complex based on all of the mutagenesis data presented here. It remains unclear whether both CD3γ and δ contact the TCRαβ heterodimer or whether one (most likely CD3δ (17)) forms the major contact and stabilizes the association of the other in the complex in the absence of direct contacts with TCRαβ. See also supplemental Figs. S5 and S6.
For CD3δ and CD3γ, the analysis was less clear-cut. Incorporation of HA-tagged CD3δ and CD3γ into the TCR complex was less efficient than for CD3ϵ, and only the most highly transfected cells could be analyzed. In contrast to CD3ϵ and TCRαβ, the majority of surface mutants of CD3δ and approximately half those of CD3γ failed to reach the surface of Jluchi cells (supplemental Fig. S6, A and B). The few expressed mutants nevertheless identify a contiguous surface at a membrane distal position apparently exposed at the top of the subunits (supplemental Fig. S6, C and D). To determine whether the sensitivity of CD3δ, and presumably CD3γ, to mutation was likely due to folding effects, we established a CD3δ folding assay based on the observation that in 293T cells, CD3ϵ surface expression requires only CD3δ co-expression (supplemental Fig. S6E; CD3γ does not rescue CD3ϵ expression). Only two of the CD3δ mutants that failed to reach the surface of Jluchi cells (Leu8 and Lys61) and one that reduced expression (Lys41) rescued CD3ϵ expression in 293T cells (supplemental Fig. S6, F and G), indicating that CD3δ folding, and presumably also CD3γ folding, is extremely sensitive to mutation. The mutational data nevertheless suggest a new interpretation for the configuration of the quaternary structure of the TCR complex (Fig. 6B), wherein CD3ϵ is fully exposed and the αβ heterodimer associates asymmetrically via contacts with CD3γ and CD3δ only.
Mutations of Exposed CD3ϵ Surface Residues Do Not Block Triggering
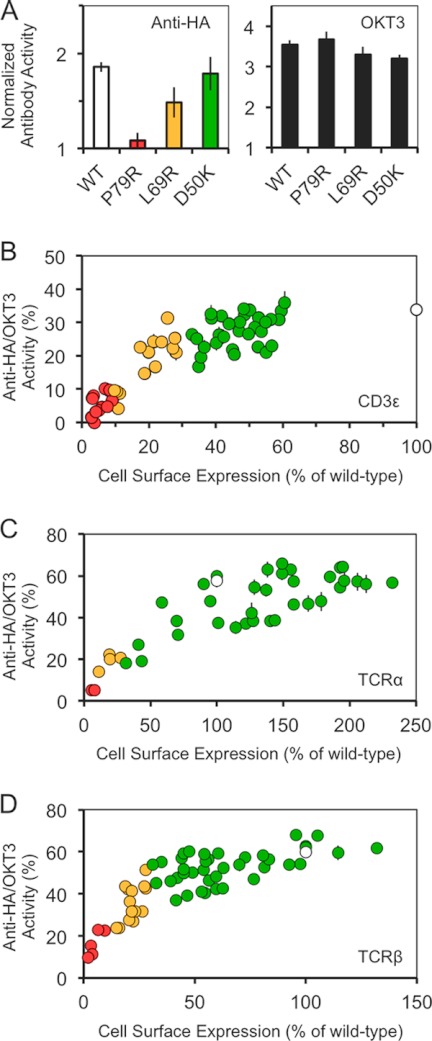
Having identified surfaces in CD3ϵ and the αβ heterodimer that are exposed in the fully assembled TCR complex on the basis that drastic mutation of these surfaces did not completely prevent receptor expression, we then determined whether structural rearrangements during antibody triggering bury these exposed surfaces by testing whether triggering was prevented by any of the mutations of the exposed surfaces of CD3ϵ and the αβ heterodimer. The HA-tagged mutant subunits were expressed in Jurkat cells, and the mutant TCR complexes were triggered with plate-bound anti-HA antibody. Activation was measured using a reporter assay for NFAT (nuclear factor of activated T cells) promoter activity in which Renilla luciferase is expressed under the control of three elements from the IL-2 promoter (49). After correcting for nonspecific activation, we determined the ratio of IL-2 promoter activity induced by the anti-HA antibodies versus that induced by OKT3 and plotted this ratio against the levels of surface expression of the mutants (Fig. 7). Importantly, all of the CD3ϵ mutant complexes initiated IL-2 promoter activity in direct proportion to their levels of expression (Fig. 7, A and B), which varied up to 4-fold versus the wild-type complex. This implies that receptor triggering is not dependent on the burial of any particular surface and is insensitive to the structural effects of mutations that reduce expression up to 4-fold. Similar data were obtained for TCRα (Fig. 7C) and β (Fig. 7D). Rossjohn and co-workers (24) have argued that the AB loop of Cα (Arg124-Lys131) is involved in conformational rearrangements important for receptor triggering, but we found that drastic mutations of this loop have essentially no effect on expression or signaling, apart from the Lys131 mutant, which failed to reach the cell surface.
FIGURE 7.
Effect of CD3ϵ mutations on TCR triggering. Jurkat T cells expressing a luciferase reporter construct for NFAT/IL-2 promoter activation, and either wild-type or mutant HA-tagged CD3ϵ was activated with plate-bound anti-CD2 and anti-CD28 antibodies, plus either an anti-HA antibody, an anti-CD3 antibody (OKT3), or an irrelevant antibody (OX7, anti-Thy-1). A, example of data obtained for cells expressing wild-type or Pro79, Leu69 or Asp50-mutated HA-tagged CD3ϵ. Anti-HA IgG (left panel) and OKT3 (right panel) responses were normalized against the responses obtained with OX7. Responses for wild-type CD3ϵ (open bar) or the CD3ϵ mutants (colored by expression level as in Fig. 5) are shown. B, normalized IL-2 promoter activity plotted against the normalized cell surface expression level of wild-type or mutant HA-tagged CD3ϵ. Normalized IL-2 promoter activity was calculated as the specific response (that is, following subtraction of the response to OX7) of each cell line to anti-HA antibody, divided by the specific response to OKT3, expressed as a percentage. Cell surface expression levels were normalized using the geometric mean of anti-HA antibody staining expressed as percentages of that obtained for wild-type HA-tagged CD3ϵ. Circles corresponding to the mutant CD3ϵ responses are colored by surface expression level (as in Fig. 5) with wild-type shown as an open circle. C and D, similar results were obtained for wild-type and mutant forms of TCRα (C) and TCRβ (D). The data shown are representative of two separate experiments; the means of triplicates and standard deviations are shown.
DISCUSSION
Implications for Receptor Triggering
An unusual feature of antigen receptors is that their ligand recognition and signal-initiating functions are performed by separate subunits. There is much interest, therefore, in the possibility that for the TCR, information is transferred between the αβ and CD3 subunits in the form of concerted structural rearrangements during receptor triggering (24, 26). Functionally linked structural transformations in proteins are well documented. At different ends of the spectrum are the relatively subtle intra- and intersubunit rearrangements of hemoglobin (50) and the radical secondary structural changes in influenza hemagglutinin trimers (51). The important point is that very specific rearrangements are required for these proteins to perform their functions via the structural rearrangements. If changes of this type comprise the TCR triggering mechanism, it could reasonably be expected that all triggering ligands would have to effect the same structural rearrangements in the receptor. With this premise, we investigated whether antibodies, which are convenient and widely used but poorly understood proxies of native TCR ligands, induce large structural rearrangements in the TCR complex. We find that antibodies are capable of initiating signaling without inducing such changes, which indicates that the TCR is not configured in such a way that it is always reliant on conformational rearrangements in the manner of hemoglobin or hemagglutinin. These findings raise the possibility that native ligands might also trigger signaling by the TCR via a mechanism involving relatively minor or no structural rearrangements in the complex.
To identify putative structural rearrangements induced within CD3ϵ by mitogenic antibodies, we crystallized Fab fragments of the mitogenic anti-mouse CD3ϵ antibody 2C11 bound to murine CD3ϵ for comparison with the previously determined apo murine CD3ϵ structures. CD3ϵ crystallized as a homodimer that is unlikely to be physiological because its formation would be blocked by δ and γ heterodimerization unless significant rearrangements occur in some of the surface loops in CD3-ϵ, -δ, and -γ on heterotetrameric assembly. Moreover, drastic mutations of the equivalent residues in the human structure, including a subset of residues that are not involved in main chain interactions in the homodimer, do not prevent complex assembly, which also argues against CD3ϵ homodimerization.
The details of CD3ϵ binding by mitogenic antibodies are substantially different. 2C11 binds mouse CD3ϵ in a region partially overlapping with the region on human CD3ϵ bound by OKT3 and UCHT1, but with the long axis of the anti-mouse Fab displaying a greater angle with respect to the plane of the membrane than OKT3 or UCHT1. Conformational rearrangements induced by antibody binding, which are generally restricted to flexible regions of protein antigens (see, for example, Ref. 52), are usually antibody specific, even for antibodies with overlapping epitopes (53). If a structural rearrangement in CD3ϵ was required for signaling, it therefore seems highly unlikely that all three antibodies would induce it. Because the antibodies are all mitogenic, it follows that a unique conformational rearrangement is very unlikely to drive signaling. But do the anti-CD3ϵ antibodies induce conformational changes at all? This new structure now allows the question to be addressed directly. In the past, comparisons could only be made between the structures of the apo and antibody-liganded forms of murine and human CD3ϵ, respectively, which are substantially different, most notably in that murine CD3ϵ lacks β-strand D (20, 21, 38, 39). With the caveat that, at 4.1 Å resolution, we can only be confident about the overall conformation of the protein backbone, the folded regions of the apo and 2C11-bound structures of murine CD3ϵ are largely identical, ruling out large scale, antibody-induced rearrangements. At this resolution, we cannot exclude more subtle conformational rearrangements, such as the “stiffening” effects observed in molecular dynamics simulations of antibody binding to CD3ϵ (26). It is worth noting, however, that configurational entropy (flexibility) losses are frequently observed for antibodies interacting with the globular regions of protein antigens (52) and that such changes may not necessarily have any functional significance.
It has been proposed (19) that there are at least five ways in which large scale intersubunit movements within the extracellular region of the TCR complex could initiate signaling. In large subunit rearrangements of these types, a subset of residues previously exposed in the folded complex would have to become buried to some extent during triggering. We identified what are likely to be all of the surface-exposed residues in cell surface-expressed CD3ϵ and in the constant regions of TCRαβ and found that receptor triggering by antibodies is not prevented by mutation of any of these residues. This suggests that large intersubunit structural rearrangements are not a prerequisite for triggering.
The TCR complex therefore appears to be configured in such a way that substantial structural rearrangements of its component subunits are not invariably required for the initiation of signaling. The idea that native ligands might induce triggering via a mechanism involving relatively minor or no structural rearrangements in the receptor is not a new one. Following detailed comparisons of a number of complexes, Garcia et al. (54) concluded that “no large-scale conformational changes are obvious in the complex structures that might have an impact on signal transduction.” Similarly, for pMHC antigens that induce qualitatively distinct signals and focusing particularly on the well ordered variable regions, Ding et al. (55) proposed that “the lack of correlation between structural changes and the type of T cell signals induced provides direct evidence that different signals are not generated by different ligand-induced conformational changes in the αβ TCR.” The early structural work thus implied that ligand recognition by TCRαβ involves rigid body interactions with pMHC. It would now seem that the TCR is not alone in being capable of signaling via rigid body interactions: the extracellular region of CTLA-4, an inhibitory receptor also phosphorylated by extrinsic kinases (56), is unchanged by ligand binding, at least in solution (57).
Implications for Receptor Organization
We previously employed systematic “drastic” mutations to identify the ligand binding surfaces of the adhesion proteins CD2 and CD48 (58–60). We now extend the approach to identifying surface and buried residues in obligate complexes, having verified the method by identifying the known site of subunit homodimerization in CD8. The importance of transmembrane interactions in TCR assembly is well established (61), but the extent to which extracellular interactions stabilize the complex is less well understood (19, 62). We hypothesized that drastic mutagenesis might yield a relatively simple pattern of buried subunit interfaces limiting the number of possible arrangements of subunits within the complex. A single buried surface on CD3ϵ corresponding almost perfectly with the region buried in CD3ϵδ and CD3ϵγ heterodimers revealed by crystallography and a single, relatively small contiguous surface on TCRαβ that influences assembly of the TCR complex were identified. Kuhns et al. (18) proposed that murine TCR complexes may dimerize via a surface formed by the AB loop and the C and F β strands, but we were unable to identify an equivalent interface in the human complex, because drastic mutations of residues in this region failed to block complex assembly or triggering. The existence of stable dimers is also incompatible with single-molecule analyses of both murine (7) and human (6) TCRs showing that the dominant form of the TCR complex comprises single TCRαβ heterodimers.
What can be concluded about the organization of the TCR complex? First, apart from the interface that it forms with CD3δ and CD3γ, CD3ϵ is apparently completely exposed in the complex. This observation explains the greater antigenicity of CD3ϵ versus CD3δ and CD3γ but is inconsistent with early (63, 64) and more recent (18) studies. The expectation that CD3ϵ interacts directly with TCRαβ came from early studies in which CD3ϵ was chemically cross-linked to both TCRα and TCRβ (63, 65) and from the apparent ability of CD3ϵ to rescue TCRβ expression in co-transfections (62, 64). However, other cross-linking studies suggest closer association of TCRβ with CD3γ than with the other CD3 chains (66), and the expression of chimeric proteins suggests preferential association of TCRα with CD3ϵδ (67, 68). Preferential associations must involve CD3δ and CD3γ chains directly, because CD3ϵ is present in both heterodimers. Because of their extreme sensitivity to mutation, we are unable to assign docking sites for TCRαβ on CD3δ and CD3γ. It seems clear, however, that similar regions at the top of both CD3δ and γ, where all four of the conserved glycosylation sites are found in primates (at the start of the C and G strands in CD3γ and in the BC and FG loops of CD3δ), are exposed in the complex.
A second conclusion is that TCRαβ contacts the CD3 chains at a single site formed by the Cβ CD and EF loops plus several Cα DE loop residues. An asymmetric arrangement of closely associated CD3 heterodimers was first suggested by the dependence of CD3ϵγ docking on CD3ϵδ binding (13, 19) and then confirmed by whole loop mutagenesis of the Cβ CD and Cα DE loops (17). Our data suggest that the “tips” of these loops probably do not make contact with the CD3 subunits and that CD3ϵγ and CD3ϵδ make contact at the base of TCRαβ constant regions. Like Kuhns and Davis (17), we were able to exclude several regions of Cα, including the C and F strands, considered previously to be potential docking sites because of their unusual flexibility (69), electrostatic properties (39), conservation (17), or overlap with the epitope of mAb H28 whose binding is blocked by CD3 association (70) or on the basis of in silico modeling (21). Although the patch on TCRαβ is relatively small, it protrudes from the structure in such a way that both CD3γ and CD3δ could form small contacts with TCRαβ. What remains to be determined is how such a complex initiates signaling without, in the case of antibodies at least, substantial changes in the overall structure and disposition of its component subunits.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI042266. This work was also supported by the Wellcome Trust, the United Kingdom Medical Research Council, the Fundação para a Ciência e a Tecnologia of Portugal, and the Skaggs Institute for Chemical Biology.

This article contains supplemental text, Table S1, and Figs. S1–S6.
- TCR
- T cell receptor
- m
- murine
- h
- human.
REFERENCES
- 1. Irving B. A., Chan A. C., Weiss A. (1993) Functional characterization of a signal transducing motif present in the T cell antigen receptor ζ chain. J. Exp. Med. 177, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reth M. (1989) Antigen receptor tail clue. Nature 338, 383–384 [PubMed] [Google Scholar]
- 3. Letourneur F., Klausner R. D. (1992) Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3ϵ. Science 255, 79–82 [DOI] [PubMed] [Google Scholar]
- 4. Straus D. B., Weiss A. (1992) Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell 70, 585–593 [DOI] [PubMed] [Google Scholar]
- 5. Call M. E., Pyrdol J., Wucherpfennig K. W. (2004) Stoichiometry of the T-cell receptor-CD3 complex and key intermediates assembled in the endoplasmic reticulum. EMBO J. 23, 2348–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunne P. D., Fernandes R. A., McColl J., Yoon J. W., James J. R., Davis S. J., Klenerman D. (2009) DySCo. Quantitating associations of membrane proteins using two-color single-molecule tracking. Biophys. J. 97, L5–L7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. James J. R., White S. S., Clarke R. W., Johansen A. M., Dunne P. D., Sleep D. L., Fitzgerald W. J., Davis S. J., Klenerman D. (2007) Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl. Acad. Sci. U.S.A. 104, 17662–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Punt J. A., Roberts J. L., Kearse K. P., Singer A. (1994) Stoichiometry of the T cell antigen receptor (TCR) complex. Each TCR/CD3 complex contains one TCRα, one TCRβ, and two CD3ϵ chains. J. Exp. Med. 180, 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lillemeier B. F., Mörtelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M. (2010) TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Varma R., Campi G., Yokosuka T., Saito T., Dustin M. L. (2006) T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A., Tokunaga M., Dustin M. L., Saito T. (2005) Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 12. James J. R., McColl J., Oliveira M. I., Dunne P. D., Huang E., Jansson A., Nilsson P., Sleep D. L., Gonçalves C. M., Morgan S. H., Felce J. H., Mahen R., Fernandes R. A., Carmo A. M., Klenerman D., Davis S. J. (2011) The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J. Biol. Chem. 286, 31993–32001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Call M. E., Pyrdol J., Wiedmann M., Wucherpfennig K. W. (2002) The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Call M. E., Schnell J. R., Xu C., Lutz R. A., Chou J. J., Wucherpfennig K. W. (2006) The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127, 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Call M. E., Wucherpfennig K. W. (2004) Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol. Immunol. 40, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu C., Call M. E., Wucherpfennig K. W. (2006) A membrane-proximal tetracysteine motif contributes to assembly of CD3δϵ and CD3γϵ dimers with the T cell receptor. J. Biol. Chem. 281, 36977–36984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhns M. S., Davis M. M. (2007) Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity 26, 357–369 [DOI] [PubMed] [Google Scholar]
- 18. Kuhns M. S., Girvin A. T., Klein L. O., Chen R., Jensen K. D., Newell E. W., Huppa J. B., Lillemeier B. F., Huse M., Chien Y. H., Garcia K. C., Davis M. M. (2010) Evidence for a functional sidedness to the αβTCR. Proc. Natl. Acad. Sci. U.S.A. 107, 5094–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhns M. S., Davis M. M., Garcia K. C. (2006) Deconstructing the form and function of the TCR/CD3 complex. Immunity 24, 133–139 [DOI] [PubMed] [Google Scholar]
- 20. Sun Z. J., Kim K. S., Wagner G., Reinherz E. L. (2001) Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3ϵγ heterodimer. Cell 105, 913–923 [DOI] [PubMed] [Google Scholar]
- 21. Sun Z. Y., Kim S. T., Kim I. C., Fahmy A., Reinherz E. L., Wagner G. (2004) Solution structure of the CD3ϵδ ectodomain and comparison with CD3ϵγ as a basis for modeling T cell receptor topology and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wegener A. M., Hou X., Dietrich J., Geisler C. (1995) Distinct domains of the CD3-γ chain are involved in surface expression and function of the T cell antigen receptor. J. Biol. Chem. 270, 4675–4680 [DOI] [PubMed] [Google Scholar]
- 23. Janeway C. A. (1995) Ligands for the T-cell receptor. Hard times for avidity models. Immunol. Today 16, 223–225 [DOI] [PubMed] [Google Scholar]
- 24. Beddoe T., Chen Z., Clements C. S., Ely L. K., Bushell S. R., Vivian J. P., Kjer-Nielsen L., Pang S. S., Dunstone M. A., Liu Y. C., Macdonald W. A., Perugini M. A., Wilce M. C., Burrows S. R., Purcell A. W., Tiganis T., Bottomley S. P., McCluskey J., Rossjohn J. (2009) Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity 30, 777–788 [DOI] [PubMed] [Google Scholar]
- 25. Gil D., Schamel W. W., Montoya M., Sánchez-Madrid F., Alarcón B. (2002) Recruitment of Nck by CD3ϵ reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912 [DOI] [PubMed] [Google Scholar]
- 26. Martínez-Martín N., Risueño R. M., Morreale A., Zaldívar I., Fernández-Arenas E., Herranz F., Ortiz A. R., Alarcón B. (2009) Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3ϵ. Sci. Signal. 2, ra43. [DOI] [PubMed] [Google Scholar]
- 27. Mingueneau M., Sansoni A., Grégoire C., Roncagalli R., Aguado E., Weiss A., Malissen M., Malissen B. (2008) The proline-rich sequence of CD3ϵ controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat. Immunol. 9, 522–532 [DOI] [PubMed] [Google Scholar]
- 28. Szymczak A. L., Workman C. J., Gil D., Dilioglou S., Vignali K. M., Palmer E., Vignali D. A. (2005) The CD3ϵ proline-rich sequence, and its interaction with Nck, is not required for T cell development and function. J. Immunol. 175, 270–275 [DOI] [PubMed] [Google Scholar]
- 29. Tailor P., Tsai S., Shameli A., Serra P., Wang J., Robbins S., Nagata M., Szymczak-Workman A. L., Vignali D. A., Santamaria P. (2008) The proline-rich sequence of CD3ϵ as an amplifier of low-avidity TCR signaling. J. Immunol. 181, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu C., Gagnon E., Call M. E., Schnell J. R., Schwieters C. D., Carman C. V., Chou J. J., Wucherpfennig K. W. (2008) Regulation of T cell receptor activation by dynamic membrane binding of the CD3ϵ cytoplasmic tyrosine-based motif. Cell 135, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandes R. A., Yu C., Carmo A. M., Evans E. J., van der Merwe P. A., Davis S. J. (2010) What controls T cell receptor phosphorylation? Cell 142, 668–669 [DOI] [PubMed] [Google Scholar]
- 32. Burns G. F., Boyd A. W., Beverley P. C. (1982) Two monoclonal anti-human T lymphocyte antibodies have similar biologic effects and recognize the same cell surface antigen. J. Immunol. 129, 1451–1457 [PubMed] [Google Scholar]
- 33. Weiss A., Imboden J., Shoback D., Stobo J. (1984) Role of T3 surface molecules in human T-cell activation. T3-dependent activation results in an increase in cytoplasmic free calcium. Proc. Natl. Acad. Sci. U.S.A. 81, 4169–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weiss A., Wiskocil R. L., Stobo J. D. (1984) The role of T3 surface molecules in the activation of human T cells. A two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J. Immunol. 133, 123–128 [PubMed] [Google Scholar]
- 35. Ilani T., Vasiliver-Shamis G., Vardhana S., Bretscher A., Dustin M. L. (2009) T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat. Immunol. 10, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Belghith M., Bluestone J. A., Barriot S., Mégret J., Bach J. F., Chatenoud L. (2003) TGF-β-dependent mechanisms mediate restoration of self-tolerance induced by antibodies to CD3 in overt autoimmune diabetes. Nat. Med. 9, 1202–1208 [DOI] [PubMed] [Google Scholar]
- 37. Nishio J., Feuerer M., Wong J., Mathis D., Benoist C. (2010) Anti-CD3 therapy permits regulatory T cells to surmount T cell receptor-specified peripheral niche constraints. J. Exp. Med. 207, 1879–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arnett K. L., Harrison S. C., Wiley D. C. (2004) Crystal structure of a human CD3-ϵ/δ dimer in complex with a UCHT1 single-chain antibody fragment. Proc. Natl. Acad. Sci. U.S.A. 101, 16268–16273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kjer-Nielsen L., Dunstone M. A., Kostenko L., Ely L. K., Beddoe T., Mifsud N. A., Purcell A. W., Brooks A. G., McCluskey J., Rossjohn J. (2004) Crystal structure of the human T cell receptor CD3ϵγ heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl. Acad. Sci. U.S.A. 101, 7675–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van der Merwe P. A., Bodian D. L., Daenke S., Linsley P., Davis S. J. (1997) CD80 (B7–1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J. Exp. Med. 185, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Evans E. J., Esnouf R. M., Manso-Sancho R., Gilbert R. J., James J. R., Yu C., Fennelly J. A., Vowles C., Hanke T., Walse B., Hünig T., Sørensen P., Stuart D. I., Davis S. J. (2005) Crystal structure of a soluble CD28-Fab complex. Nat. Immunol. 6, 271–279 [DOI] [PubMed] [Google Scholar]
- 42. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 43. Wilson I. A., Stanfield R. L. (1993) Antibody-antigen interactions. Curr. Opin. Struct. Biol. 3, 113–118 [DOI] [PubMed] [Google Scholar]
- 44. Frank S. J., Niklinska B. B., Orloff D. G., Merep M., Ashwell J. D., Klausner R. D. (1990) Structural mutations of the T cell receptor ζ chain and its role in T cell activation. Science 249, 174–177 [DOI] [PubMed] [Google Scholar]
- 45. Ohashi P. S., Mak T. W., Van den Elsen P., Yanagi Y., Yoshikai Y., Calman A. F., Terhorst C., Stobo J. D., Weiss A. (1985) Reconstitution of an active surface T3/T-cell antigen receptor by DNA transfer. Nature 316, 606–609 [DOI] [PubMed] [Google Scholar]
- 46. Gao G. F., Tormo J., Gerth U. C., Wyer J. R., McMichael A. J., Stuart D. I., Bell J. I., Jones E. Y., Jakobsen B. K. (1997) Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature 387, 630–634 [DOI] [PubMed] [Google Scholar]
- 47. Leahy D. J., Axel R., Hendrickson W. A. (1992) Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6 A resolution. Cell 68, 1145–1162 [DOI] [PubMed] [Google Scholar]
- 48. Hubbard S. J., Thornton J. M., Campbell S. F. (1992) Substrate recognition by proteinases. Faraday Discuss. 93, 13–23 [DOI] [PubMed] [Google Scholar]
- 49. Northrop J. P., Ullman K. S., Crabtree G. R. (1993) Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem. 268, 2917–2923 [PubMed] [Google Scholar]
- 50. Perutz M. F. (1990) Mechanisms regulating the reactions of human hemoglobin with oxygen and carbon monoxide. Annu. Rev. Physiol. 52, 1–25 [DOI] [PubMed] [Google Scholar]
- 51. Skehel J. J., Wiley D. C. (2000) Receptor binding and membrane fusion in virus entry. The influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 52. Mohan S., Kourentzi K., Schick K. A., Uehara C., Lipschultz C. A., Acchione M., Desantis M. E., Smith-Gill S. J., Willson R. C. (2009) Association energetics of cross-reactive and specific antibodies. Biochemistry 48, 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davies D. R., Cohen G. H. (1996) Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. U.S.A. 93, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garcia K. C., Teyton L., Wilson I. A. (1999) Structural basis of T cell recognition. Annu. Rev. Immunol. 17, 369–397 [DOI] [PubMed] [Google Scholar]
- 55. Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., Wiley D. C. (1999) Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11, 45–56 [DOI] [PubMed] [Google Scholar]
- 56. Shiratori T., Miyatake S., Ohno H., Nakaseko C., Isono K., Bonifacino J. S., Saito T. (1997) Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 6, 583–589 [DOI] [PubMed] [Google Scholar]
- 57. Yu C., Sonnen A. F., George R., Dessailly B. H., Stagg L. J., Evans E. J., Orengo C. A., Stuart D. I., Ladbury J. E., Ikemizu S., Gilbert R. J., Davis S. J. (2011) Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering. J. Biol. Chem. 286, 6685–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davis S. J., Davies E. A., Tucknott M. G., Jones E. Y., van der Merwe P. A. (1998) The role of charged residues mediating low affinity protein-protein recognition at the cell surface by CD2. Proc. Natl. Acad. Sci. U.S.A. 95, 5490–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evans E. J., Castro M. A., O'Brien R., Kearney A., Walsh H., Sparks L. M., Tucknott M. G., Davies E. A., Carmo A. M., van der Merwe P. A., Stuart D. I., Jones E. Y., Ladbury J. E., Ikemizu S., Davis S. J. (2006) Crystal structure and binding properties of the CD2 and CD244 (2B4)-binding protein, CD48. J. Biol. Chem. 281, 29309–29320 [DOI] [PubMed] [Google Scholar]
- 60. van der Merwe P. A., McNamee P. N., Davies E. A., Barclay A. N., Davis S. J. (1995) Topology of the CD2-CD48 cell-adhesion molecule complex. Implications for antigen recognition by T cells. Curr. Biol. 5, 74–84 [DOI] [PubMed] [Google Scholar]
- 61. Call M. E., Wucherpfennig K. W. (2005) The T cell receptor. Critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 23, 101–125 [DOI] [PubMed] [Google Scholar]
- 62. Manolios N., Kemp O., Li Z. G. (1994) The T cell antigen receptor α and β chains interact via distinct regions with CD3 chains. Eur. J. Immunol. 24, 84–92 [DOI] [PubMed] [Google Scholar]
- 63. Koning F., Maloy W. L., Coligan J. E. (1990) The implications of subunit interactions for the structure of the T cell receptor-CD3 complex. Eur. J. Immunol. 20, 299–305 [DOI] [PubMed] [Google Scholar]
- 64. Manolios N., Letourneur F., Bonifacino J. S., Klausner R. D. (1991) Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J. 10, 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koning F., Lew A. M., Maloy W. L., Valas R., Coligan J. E. (1988) The biosynthesis and assembly of T cell receptor α- and β-chains with the CD3 complex. J. Immunol. 140, 3126–3134 [PubMed] [Google Scholar]
- 66. Brenner M. B., Trowbridge I. S., Strominger J. L. (1985) Cross-linking of human T cell receptor proteins. Association between the T cell idiotype beta subunit and the T3 glycoprotein heavy subunit. Cell 40, 183–190 [DOI] [PubMed] [Google Scholar]
- 67. Bäckström B. T., Müller U., Hausmann B., Palmer E. (1998) Positive selection through a motif in the αβ T cell receptor. Science 281, 835–838 [DOI] [PubMed] [Google Scholar]
- 68. Gouaillard C., Huchenq-Champagne A., Arnaud J., Chen Cl C. L., Rubin B. (2001) Evolution of T cell receptor (TCR) αβ heterodimer assembly with the CD3 complex. Eur. J. Immunol. 31, 3798–3805 [DOI] [PubMed] [Google Scholar]
- 69. Garcia K. C., Degano M., Stanfield R. L., Brunmark A., Jackson M. R., Peterson P. A., Teyton L., Wilson I. A. (1996) An αβ T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 [DOI] [PubMed] [Google Scholar]
- 70. Karaivanova V., Suzuki C., Howe C., Kearse K. P. (1999) Characterization of the epitope on murine T-cell receptor (TCR) α proteins recognized by H28–710 monoclonal antibody. Hybridoma 18, 497–503 [DOI] [PubMed] [Google Scholar]
- 71. Stewart-Jones G. B., McMichael A. J., Bell J. I., Stuart D. I., Jones E. Y. (2003) A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4, 657–663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.