Background: The activity of the proton pumping vacuolar ATPase is regulated by reversible enzyme dissociation.
Results: The ATPase and proton channel domains are held together by several intermediate affinity interactions.
Conclusion: Intermediate affinity subunit-subunit interactions may play an important role in reversible enzyme dissociation.
Significance: Targeting subunit-subunit interactions may be a means of modulating aberrant V-ATPase activity involved in human disease.
Keywords: ATPases, Biophysics, Protein Structure, Proton Pumps, Vacuolar ATPase, Enzyme Mechanism, Isothermal Titration Calorimetry, Protein-protein interaction, Reversible disassembly, Small angle X-ray scattering
Abstract
Eukaryotic vacuolar ATPase (V-ATPase) is regulated by a reversible dissociation mechanism that involves breaking and reforming of protein-protein interactions at the interface of the V1-ATPase and Vo-proton channel domains. We found previously that the head domain of the single copy C subunit (Chead) binds one subunit EG heterodimer with high affinity (Oot, R.A. and Wilkens, S. (2010) J. Biol. Chem. 285, 24654–24664). Here we generated a water-soluble construct of the N-terminal domain of the Vo “a” subunit composed of amino acid residues 104–372 (aNT(104–372)). Analytical gel filtration chromatography and sedimentation velocity analysis revealed that aNT(104–372) undergoes reversible dimerization in a concentration-dependent manner. A low-resolution molecular envelope was calculated for the aNT(104–372) dimer using small angle x-ray scattering data. Isothermal titration calorimetry experiments revealed that aNT(104–372) binds the Cfoot and EG heterodimer with dissociation constants of 22 and 33 μm, respectively. We speculate that the spatial closeness of the aNT, Cfoot, and EG binding sites in the intact V-ATPase results in a high-avidity interaction that is able to resist the torque of rotational catalysis, and that reversible enzyme dissociation is initiated by breaking either the aNT(104–372)-Cfoot or aNT(104–372)-EG interaction by an as-yet unknown signaling mechanism.
Introduction
The vacuolar ATPase, or V-ATPase,2 is a rotary molecular motor found on the endomembrane system of all eukaryotic cells. The enzyme actively transports protons across membranes using the energy of ATP hydrolysis. V-ATPase plays a fundamental role in pH homeostasis, vesicular trafficking, endocytosis/exocytosis, cellular housekeeping, membrane energization, and secondary transport processes (1–4). The V-ATPase is composed of two structurally and functionally coupled sectors, a membrane integral proton pore, Vo, and a soluble catalytic sector, V1. The Vo sector is composed of subunits a, d, e, c, c′, and c″, and the V1 is made up of subunits A3B3(C)DE3FG3H (5, 6). ATP hydrolysis occurs in three catalytic sites located on the A3B3 hexamer, and the free energy released during that process drives rotation of a central rotor domain composed of subunits D, F, d, and the proteolipid c subunits. Proton translocation occurs at the interface between the proteolipid ring and subunit a. Like in the related F-ATP synthase, subunit a is thought to form a pair of water-accessible hemichannels, with one channel open to the lumen of the intracellular compartment and one exposed to the cytosol (7). In contrast to the F-type ATPase, however, the V-ATPase subunit a is the largest V-ATPase subunit (∼96 kDa), with a soluble N-terminal cytoplasmic domain (aNT) linked to the C-terminal membrane-embedded proton translocation domain (aCT) (8). During catalysis, subunits E, G, C, H, and aNT, known collectively as the peripheral stator, serve to absorb the torque of rotation by holding the A3B3 hexamer static so that rotation can be productive. In addition, the stator subunits act as a structural bridge between V1 and Vo, physically linking the soluble and membrane subunits. This feature of the stator subunits may indicate a role in the unique mode of enzyme regulation known as reversible dissociation. It has been shown that under conditions of metabolic stress, the enzyme undergoes a (fully reversible) structural rearrangement wherein the soluble V1 domain disengages from the membrane integral domain (9–11) and both functional units are silenced (12, 13). During this time, a single copy subunit, C, is released from the enzyme and reincorporated upon enzyme reassembly (9). As subunit C has been shown to both bind to and form cross-links with other stator subunits, namely E, G, and aNT, a number of potential interactions must be broken during regulation, thus enabling the release of subunit C from the enzyme (14–17). As these interactions must be broken and reformed in a controlled fashion, characterization of the interactions between subunit C and the other subunits at the V1-Vo interface is critical for an understanding of the mechanism of regulated enzyme disassembly and reassembly. Subunit C is an elongated protein with the N- and C termini forming a globular “foot” (Cfoot) domain that is separated from the globular “head” (Chead) domain by a coiled coil (18). From electron microscopic images and consistent with chemical cross-linking, subunit C is in proximity to two of the subunit EG peripheral stators and the soluble N terminus of the Vo subunit a (14, 19). We have shown recently that the interaction between Chead and EG occurs with high affinity, whereas no significant interaction could be detected between EG and Cfoot in vitro (17).
In this study, we have expanded the characterization of interactions between subunits that form the V1-Vo interface focusing on those involving the cytoplasmic N-terminal domain of the Vo a subunit (aNT). To examine the binding interactions between subunit a and subunits C and EG, we generated a water-soluble construct of aNT comprising residues 104–372 (aNT(104–372)). The construct was designed to include the predicted coiled-coil domain of aNT as well as the residues observed previously to form cross-links to the EG heterodimer (20). Biochemical and biophysical experiments showed that aNT(104–372) is folded and that the construct undergoes reversible dimerization at higher concentrations. Using isothermal titration calorimetry, we find that aNT(104–372) interacts with both the Cfoot and EG heterodimer with intermediate affinities of 22 and 33 μm, respectively. Although the individual affinities between aNT(104–372) and Cfoot and EG appear to be moderate, we speculate that the avidity of these combined interactions in the assembled V-ATPase is high, resulting in a stable interaction between V1 and Vo that can withstand the torque generated during rotational catalysis. We further reason that destabilizing or stabilizing one or both of the individual interactions involving aNT, EG, and C in vivo (by a yet unknown mechanism) is a key feature of reversible disassembly in eukaryotic V-ATPase.
EXPERIMENTAL PROCEDURES
Materials
All reagents were of analytical grade. Restriction endonucleases were from New England Biolabs. PreScission protease and the QuikChange kit were from GE Healthcare and Stratagene, respectively.
Plasmid Construction
The C subunit foot domain (Cfoot) was constructed as described previously (17). Briefly, the gene encoding subunit C from Saccharomyces cerevisiae (Vma5) in a pMal-c2e plasmid (New England Biolabs) with a Prescission protease cleavage site in place of the native enterokinase site (Vma5pMalPPase) was used as a template for construction of Cfoot using fusion PCR. The regions of the gene corresponding to the N and C termini of subunit C (amino acids 1–151 and 278–392) were linked by a four-amino acid flexible linker (GAAA).
The gene encoding the soluble N-terminal domain of subunit a (VPH1), residues 1–406, in the pMalPPase vector was a kind gift of Dr. Patricia Kane (State University of New York Upstate Medical University). This plasmid was used as a template for site-directed mutagenesis using the Stratagene QuikChange kit and the following primers: vph1d1–103F, CCC AAG GTA CCG GAA TTC GGA TCC GGT TCA GTG ATA GAT GAT TAT GTC CGG; vph1d1–103R, GCG TTC CGG ACA TAA TCA TCT ATC ACT GA ACC GGA TCC GAA TTC CGG TAC; T373stopF, CAT CCA TTA TCC AAG TCC TGG AT TAG AAC CAC ACT CCA CCT ACC TTC CAC AG; and T373stopR, AGT TCT GTG GAA GGT AGG TGG AGT GTG GTT CTA ATC CAG GAC TTG GAT AAT G. The resulting protein product encompasses amino acids 104–372, and the sites of truncation were chosen on the basis of secondary structure alignments with Stv1p as well as available cross-linking data indicating possible protein interaction sites (20). Secondary structure prediction was carried out using PSIPRED (21), and alignments were done using Clustal W (22). Construction of the plasmid for expression of subunits E and G will be described elsewhere.3 Briefly, the open reading frames for Vma10p (G) and Vma4p (E) separated by a Shine-Dalgarno sequence in a pBAD vector (kindly provided by Dr. Patricia Kane, SUNY Upstate Medical University) was used as a template for introduction of 5′ KpnI and 3′ PstI restriction sites for subsequent ligation into the pMalPPase vector. Expression from this construct results in an N-terminally tagged G subunit and untagged E subunit. Sequences of all constructs were verified by DNA sequencing at the SUNY Upstate Medical University Core DNA sequencing facility using the MalE primer (New England Biolabs).
Protein Expression and Purification
Subunits Cfoot and EG were expressed and purified as described previously (17). Briefly, all proteins used in this work were expressed in Escherichia coli strain Rosetta2 (Novagen) grown to mid-log phase in Rich Broth (Luria-Bertani-Lennox, Zogie, 20g/l, glucose) medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). Expression of Cfoot and EG was induced for 6 h at 30 °C with 0.5 mm or 1 mm isopropyl 1-thio-β-d-galactopyranoside, respectively. Expression of Vph1p 104–372 was at 20 °C overnight (∼16 h) and 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. All proteins were purified using amylose affinity chromatography followed by cleavage with Prescission protease. Cells were harvested by centrifugation, resuspended in 200 mm NaCl, 20 mm Tris, and 1 mm EDTA (pH 7.4) and lysed by sonication. Lysate was cleared by centrifugation at 12,000 × g and passed over an amylose affinity column. Bound protein was eluted in 25 ml of 200 mm NaCl, 20 mm Tris, 1 mm EDTA, and 10 mm maltose, and the maltose binding protein tag was cleaved with Prescission protease for 2 h in the presence of 5 mm DTT according to the manufacturer's instructions. For Cfoot and EG, the pH was adjusted to the pI by dialysis and the protease cleavage product passed over an anion exchange (diethyl amino ethyl (Cfoot)) or cation exchange (carboxymethyl (EG)) column for removal of maltose binding protein. The isolated protein was then dialyzed to readjust the pH away from the pI, concentrated, and subjected to size exclusion chromatography for further purification. In the case of EG, the cation exchange step was followed by passage over a DEAE column for removal of any excess G subunit before refinement by gel filtration. The Vph1p 104–372 cleavage product was brought to 35% w/v ammonium sulfate, pelleted by centrifugation at 13,000 × g and resuspended in and then dialyzed overnight into buffer A (25 mm Tris-HCl, 0.5 mm EDTA (pH 7.0)). The dialyzed sample was then passed over a MonoQ (anion exchanger) column attached to an AKTA FPLC in buffer A and subjected to a 30-ml linear gradient elution in buffer B (0–100%, buffer A plus 500 mm NaCl), concentrated, and further refined by size exclusion chromatography. Protein concentrations were determined by measuring absorbance at 280 nm ± guanidinium-HCl on a Varian Cary spectrophotometer. Wavelength scans were recorded from 400–200 nm with baseline correction to test for sample purity. Prediction of the pI and estimation of protein charge versus pH was done using the Scripps Protein Calculator.
Circular Dichroism Spectroscopy
Far UV CD spectra of 2 μm Vph1p 104–372 were recorded on an Aviv 420 spectrometer in 25 mm sodium phosphate buffered to pH 7.0 in a 2-mm path length cuvette at 20 °C. In the spectrum shown, mean molar ellipticity (θ) was plotted as a function of wavelength.
Analytical Gel Filtration Chromatography
Gel filtration chromatography was run using an H/R S75 1.6 × 50 cm column attached to an AKTA FPLC (GE Healthcare). The column was calibrated using blue dextran, albumin, GFP, RNase A, and insulin.
Analytical Ultracentrifugation
Because Vph1p 104–372 eluted from the gel filtration column as two peaks with molecular masses corresponding to monomeric and dimeric species, sedimentation velocity analytical ultracentrifugation was carried out to examine the concentration dependence of Vph1p 104–372 dimerization. Three different concentrations of Vph1p 104–372 (32 μm, 16 μm, and 3.2 μm) in 25 mm sodium phosphate, 0.5 mm EDTA, 1 mm TCEP (pH 7.0) were examined. Analytical ultracentrifugation was carried out in a Beckman Coulter ProteomeLabTM XL-A analytical ultracentrifuge in an eight-hole An-50 Ti rotor at 10 °C and 50,000 rpm (200,000 × g). Sedimentation was monitored at a wavelength of 235 nm with 300 scans taken per sample. A path length of 1.2 cm was used for the 3.2 μm sample, whereas a 0.3-cm path length was used for the 16 and 32 μm samples. The absorbance was corrected for this difference during data analysis. Data were analyzed using the SEDNTERP (23) and SEDFIT software (24).
Small Angle X-ray Scattering (SAXS)
Small angle x-ray scattering data were collected at Cornell High Energy Synchrotron Source F2 beam line using an Area Detector Systems Corporation Quantum 1 detector. Measurements were made at a wavelength of 1.2524 Å at 4 °C. SAXS measurements of Vph1p 104–372 were carried out in 25 mm sodium phosphate, 0.5 mm EDTA, and 1 mm TCEP (pH 7.0) at three sample concentrations (1.5, 3, and 6 mg/ml). Data acquisition was performed by taking three successive 300-s frames of each 30-μl sample. Lysozyme and glucose isomerase were used as standards. Initial data processing, including buffer subtraction and Guinier analysis, was done using bioxtas RAW (25). GNOM (26), DAMMIF (27), and DAMAVER (28) from the ATSAS software package were used for model calculation and averaging. The final filtered model represents the average of 20 individual models. The model represented as a mesh was generated using SITUS (29), and the fit of the M. ruber INT structure (PDB code 3RRK) (30) into the model was done using CHIMERA (31).
Isothermal Titration Calorimetry (ITC)
The interactions between Vph1p 104–372 and Cfoot or EG were quantified using a Microcal VP-ITC isothermal titration calorimeter in 25 mm sodium phosphate, 0.5 mm EDTA, and 1 mm TCEP (pH 7.0) at 10 °C. Briefly, a known concentration of ligand (Cfoot or Vph1p 104–372) was titrated into a fixed concentration of EG or Vph1p 104–372. Blank experiments (Cfoot and Vph1p 104–372 into buffer) were subtracted from titration experiments to negate any heats of dilution. Data were analyzed using the VP-ITC programs in Originlab.
RESULTS
The mechanism of V-ATPase regulation by reversible dissociation was first described in yeast (9) and insects (11) but has since been found to be a general mechanism for enzyme regulation in higher eukaryotes (32–34). Although this unique mode of regulation is well documented, the molecular mechanism remains poorly understood. The subunit-subunit interactions likely to be broken during regulated disassembly must also be sufficiently strong to maintain the structural integrity of the enzyme and resist the torque generated during catalysis in the assembled enzyme. Fig. 1A presents a schematic of the subunit-subunit interactions in the V1-Vo interface. In the following sections we have quantitatively examined a subset of subunit interactions that would be central to the linking of the soluble V1 sector with the membrane integral Vo. The many interactions known to exist between aNT and soluble subunits may indicate that this domain is a major point of interaction, linking V1 to the membrane (14, 20, 35, 36) (Fig. 1A). In addition, the release of subunit C from the enzyme upon regulated disassembly requires the breaking of at least one high-affinity interaction between the Chead domain and EG3 (Fig. 1C) (in the following, peripheral EG stators are numbered 1–3 as given in Ref. 6; see Fig. 1A), whereas the affinity and nature of the Cfoot domain interaction remains unclear (17). Here, we have constructed and characterized a soluble, folded subdomain of the cytoplasmic N-terminal domain of yeast V-ATPase subunit a (aNT) and quantified the affinity of its interactions with both Cfoot and EG heterodimer.
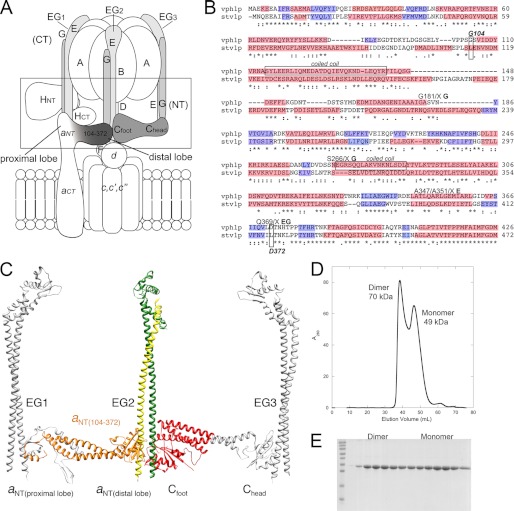
FIGURE 1.
Design of vph1 N-terminal domain construct. A, schematic representation of the V-ATPase with the subunits at the V1-Vo interface outlined by the box. The region of subunit a involved in the construct used in this study is shaded. Peripheral EG stators are numbered as in Ref. 6. B, sequence alignment shaded according to the secondary structure prediction for the N-terminal domains of the two yeast subunit a isoforms, Vph1p and Stv1p. The primary sequence is shaded in pink and blue, representing α helices and β sheet, respectively. The sites of cross-linking to the EG heterodimer are marked on the alignment. The N- and C-terminal boundaries for aNT(104–372) were chosen to preserve conserved structural elements as well as potential sites of interaction with other subunits. C, structural model of the peripheral stalk domain of yeast V-ATPase. Shown are structures of the EG heterodimer (from Thermus thermophilus, 3k5b), subunit C from yeast V-ATPase (1u7l), and a structural model of yeast aNT (the primary sequence of the Vph1p N-terminal domain threaded into the crystal structure of the archaeal ATPase INT domain, 3rrk. See supplemental Fig. S1 for details). As can be seen, construct aNT(104–372) constitutes the distal lobe and flanking coiled-coil segments (highlighted in orange in the aNT structural model), thus preserving the binding sites to both EG2 and Cfoot (colored in green, yellow, and red, respectively). D, size exclusion chromatography of construct aNT(104–372). The protein elutes as two species corresponding to monomer and dimer. The predicted molecular mass of the monomer is 31.5 kDa. The observed molecular masses are marked on the chromatograph. The larger observed molecular masses for the construct likely indicate that the molecule is elongated in solution. E, SDS-PAGE gel of purified aNT(104–372) from the size exclusion chromatography run in D. The two peaks can be seen on the gel, and the purity of both species indicates that no contaminants are contributing to the two species observed.
Subunit aNT Construct Design
Secondary structure prediction and sequence alignment was used to identify conserved structural elements in the N-terminal domains of yeast V-ATPase a subunit isoforms Vph1p and Stv1p (Fig. 1B). Earlier studies have shown that constructs including more or all the N-terminal residues proved difficult to work with because of the tendency of the proteins to aggregate and susceptibility to degradation (37, 38). We therefore decided to generate N-and C-terminal truncations to identify minimal soluble and folded domains that could serve in subunit-subunit interaction studies. Two of such constructs comprising the distal lobe and small regions of the predicted coiled coil (residues 142–274 and 136–280), although water-soluble, appeared to be unstructured based on 15N-Heteronuclear Single Quantum Coherence NMR spectroscopy (data not shown). A longer construct encompassing amino acids 104–406, on the other hand, was unstable (rapidly degraded). Construct Vph1p (104–372), which was designed to preserve the entire predicted coiled-coil domain and the reported sites of cross-linking to EG (20), appeared both highly soluble and much less susceptible to degradation, and this construct was therefore chosen for subsequent structural and subunit interaction studies. Vph1p (residues 104–372, highlighted in orange in Fig. 1C), is hereafter called aNT(104–372).
Subunit aNT(104–372) Purification
aNT(104–372) was expressed in E. coli as an N-terminal fusion with MBP connected by a cleavable linker. Upon amylose purification, soluble aNT(104–372) was cleaved from MBP and purified to homogeneity by ion exchange and size exclusion chromatography. Purification of aNT(104–372) is summarized in Fig. 1, D and E. Purified aNT(104–372) was applied to an analytical gel filtration column for estimation of sample homogeneity as well as molecular mass. Although the predicted molecular mass of aNT(104–372) is 31.5 kDa, it elutes as two species, one with an apparent molecular size of 49 kDa and one at 70 kDa. We interpret the peak at 49 kDa to represent aNT(104–372) eluting as an elongated monomer, a shape that is in accord with secondary structure predictions, the recent crystal structure of the equivalent domain of the archaeal homolog of V-ATPase subunit a and EM reconstruction data (30, 36). The 70 kDa species likely corresponds to a dimeric species that, as it is not twice the apparent mass of the monomer, is eluting as a more compact entity, suggesting a lateral association of the two monomers. Interestingly, this dimerization appears to be both concentration-dependent and reversible. The fractions containing the dimeric species were collected, concentrated, and reapplied to the column, and again, both monomeric and dimeric species were observed (data not shown).
Far UV Circular Dichroism Spectroscopy
An analysis of the primary sequence of aNT(104–372) predicts the presence of 63.5% α-helical and 16% β-strand secondary structure. To determine the secondary structure content of E. coli-expressed aNT(104–372), we employed CD spectroscopy. Fig. 2A shows a representative CD spectrum of 2 μm aNT(104–372) in 25 mm sodium phosphate buffer (pH 7.0). The characteristic minima at 208 and 222 nm in the spectrum indicate the presence of an α helical secondary structure. The CD data show that aNT(104–372) is folded in aqueous solution, with the spectrum dominated by helical character as predicted.
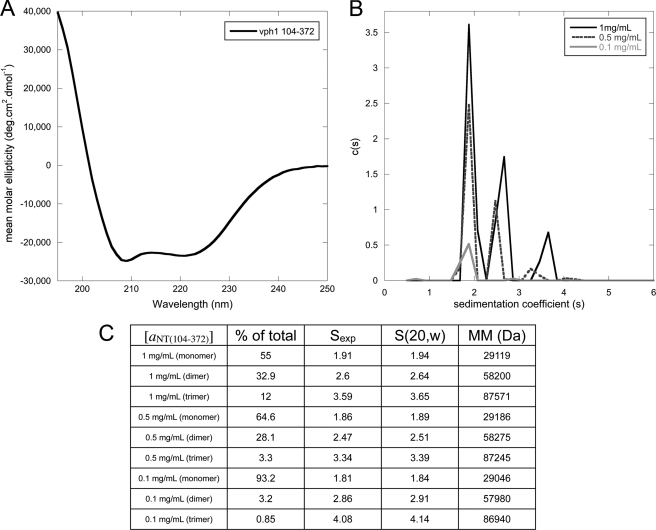
FIGURE 2.
Circular dichroism spectroscopy and analytical ultracentrifugation of aNT(104–372). A, far UV circular dichroism spectrum of aNT(104–372). The spectrum indicates that the protein is folded, with the characteristic minima at 208 and 222 nm indicating α helix. B, sedimentation velocity analytical ultracentrifugation experiments were used to examine the concentration dependence of aNT(104–372) dimerization. The c(s) distribution is shown for the construct at three protein concentrations (3.2 μm, 16 μm, and 32 μm). A clear concentration dependence on the oligomeric state can be seen, with an ∼93% monomer at 3.2 μm and an ∼55% dimer at 32 μm. As the samples were all concentrated and then diluted for analytical ultracentrifugation experiments, the concentration-dependent oligomerization is reversible. C, table of data for each peak in the overlaid c(s) distribution in B. The molecular mass and s (20, w) values of each species were calculated using the SEDNTERP and SEDFIT software.
Analytical Ultracentrifugation
As stated above, aNT(104–372) exhibits a reversible, concentration-dependent dimerization. We employed sedimentation velocity analytical ultracentrifugation to examine this concentration dependence more closely and to determine whether any higher oligomers were present in our preparations (Fig. 2, B and C). A clear concentration dependence on the oligomeric state was observed, with an ∼93% (1.843 s) monomer observed at 3.2 μm (∼0.1 mg/ml) concentration and a steady increase in dimer with increasing concentration. At 32 μm (∼1 mg/ml), 55% (1.94 s) of the sample is monomeric, 33% (2.64 s) dimeric, and 12% (3.648 s) trimeric. The increase in s value for the individual species with increasing concentration indicates that the monomer-oligomer equilibrium is in fast exchange, likely contributing to the reversibility of self-association. Importantly, no higher oligomers or aggregates were observed at any concentration. In addition it should be noted that the protein sample was concentrated and then diluted to the concentrations tested, illustrating the reversibility of dimerization as already observed from gel filtration experiments.
SAXS
Small angle x-ray scattering was used to examine the structural features of aNT(104–372) in solution. SAXS data were collected at the F2 beam line at Cornell High Energy Synchrotron Source, Cornell University, Ithaca, NY. SAXS data were collected at three protein concentrations (1.5, 3, 6 mg/ml; no concentration-dependent changes in data quality were observed). Data were analyzed using the RAW, GNOM, DAMMIF, and DAMAVER software packages (25–28). SAXS data analysis for aNT(104–372) is summarized in Fig. 3. The Guinier fit (Fig. 3B) yielded a radius of gyration (Rg) of 52.65 Å, in agreement with the value (53.08 ± 0.306 Å) derived from the GNOM software. The data range used for envelope calculation was qmin 0.017- qmax 0.2807 Å−1. The distance distribution function, P(r), indicates an elongated species with a maximum intraparticle distance (Dmax) of 185 Å (Fig. 3C). Using this value for Dmax and the data range specified above resulted in a total estimate (TE) score of 0.843, a score classified as a “good” solution by the GNOM program. Twenty individual models were calculated using DAMMIF and averaged using DAMAVER, with an average normalized spatial discrepancy (NSD) value of 1.011. The observed molecular mass, calculated using lysozyme and glucose isomerase as standards, was found to be 67 kDa (63 kDa predicated for dimer). Together, the P(r) plot, Dmax of 185 Å, and calculated molecular mass indicate an elongated dimeric species in solution.
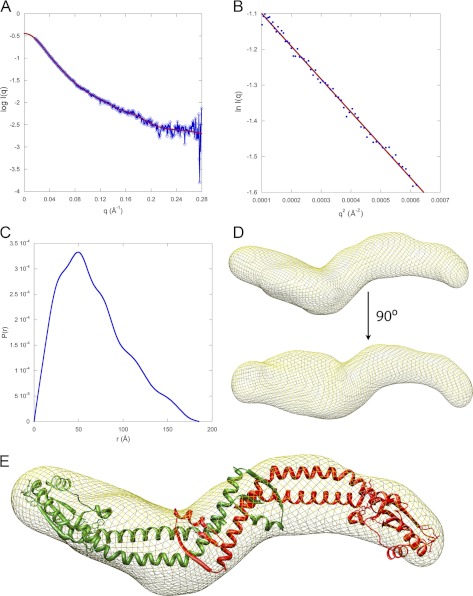
FIGURE 3.
SAXS analysis of aNT(104–372). A, small angle x-ray scattering curve of aNT(104–372). The data is plotted as the log of the scattering intensity as a function of momentum transfer, q (q = 4πsin(θ)/λ, where 2θ is the scattering angle and λ is the x-ray wavelength, 1.2524 Å). The fit to the scattering data, calculated using GNOM software, can be seen in the plot. B, Guinier plot of the data showing a linear fit (qRg = 1.3, where Rg is the radius of gyration of the particle). C, plot of the distance distribution function, P(r). The shape of the P(r) plot indicates an elongated protein with a maximum intraparticle distance of 185 Å. D, SAXS model of aNT(104–372). Twenty individual models were calculated using the DAMMIF program and averaged using DAMAVER. The final filtered model represented by a mesh was generated using SITUS. Fitting of the INT x-ray structure (minus the equivalent residues not present in aNT(104–372)) was done in CHIMERA. The SAXS model shows that the protein has a curved, elongated shape. The molecular mass of the protein was calculated from the SAXS data using glucose isomerase and lysozyme as standards and was found to be 67 kDa, indicating a dimer (predicted mass of the dimer is 63 kDa). The nature of the protrusion seen in the lower part of D is unknown but may be due to presence of a trimeric species in fast exchange with the likely dominant dimeric population at the concentration used for SAXS analysis.
The SAXS data, coupled with our biochemical characterization of aNT(104–372), demonstrates that this construct is folded, soluble, and, although reversible concentration dependent dimerization occurs, no aggregation or higher oligomerization is observed. The “S”-shaped SAXS envelope (Fig. 3D) shows that the protein is elongated, in agreement with secondary structure prediction, gel filtration, and EM reconstructions (36). Fig. 3E shows fitting of the recent x-ray crystal structure of the archaeal subunit I N-terminal domain (INT) into the molecular envelope of yeast V-ATPase aNT(104–372) obtained from SAXS measurements. As can be seen, two INT molecules fill the density well. For the fit, the N-terminal 80 residues of the INT crystal structure were removed, as the equivalent residues are not present in the aNT(104–372) construct analyzed here. The best fit was obtained with the distal lobe of INT exposed at the tips of the dimer, where they would be accessible to bind EG or Cfoot.
Overall, the SAXS data confirm that aNT(104–372) is folded and shows no tendency to aggregate beyond a stable dimer (and possibly some trimer) at the concentrations tested. Furthermore, the SAXS-derived molecular envelope suggests that the dimer interface is formed by an interaction of the proximal lobes of the two monomers.
ITC
As stated previously, although subunit C appears to interact with two EG heterodimers in vivo, a 1:1:1 stoichiometry is observed in vitro (14–17). We have previously quantified the thermodynamics of the interactions between EG and C and Cfoot and Chead using ITC (17). The in vitro interaction between EG and C was found to occur via Chead, with experimentally determined values for Kd, ΔH, and ΔG similar to those with the full-length subunit. In this study, we have examined the interactions between aNT(104–372) and Cfoot and the EG heterodimer. All experiments were carried out in 25 mm sodium phosphate, 0.5 mm EDTA, 1 mm TCEP (pH 7.0) at 10 °C (283 K). The interaction between Cfoot and aNT(104–372) occurs in a 1:1 stoichiometry as expected and is driven by enthalpy. The Kd was found to be 22 μm, ΔH = −15370 cal/mol, and ΔS = −32.9 cal/mol·K (Fig. 4A). These values yield a ΔG of association of −25.3 kJ/mol. The interaction between aNT(104–372) and EG was found to be entropically driven, with a stoichiometry of 1:1:1 and a Kd of 33 μm. Values of ΔH = +14000 cal/mol, ΔS = +70 cal/mol·K were used to calculate a ΔG = −24.3 kJ/mol (Fig. 4B). Fig. 4B also shows that titrating concentrated aNT(104–372) into buffer only produces very little heat of dilution, again consistent with weakly interacting monomers in the aNT(104–372) dimer. To summarize, the ITC data show that aNT(104–372) interacts with EG and Cfoot in a specific manner and with about equally moderate affinity.
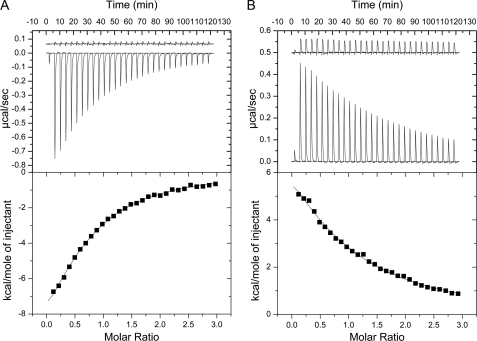
FIGURE 4.
Isothermal titration calorimetry of aNT(104–372) Cfoot and EG interactions. A, ITC was used to quantify the interactions between the foot domain of subunit C (Cfoot) and construct aNT(104–372). The top panel shows the heat evolved upon titration of Cfoot into aNT(104–372) and indicates that the interaction is exothermic in nature. The trace above the titration data is Cfoot titrated into buffer and shows no significant heat of dilution of the protein. The area under the peaks in the top panel were integrated and plotted as a function of molar ratio in the bottom panel. The curve was fit to a one-sites model and yielded a Kd of 22 μm. B, binding isotherm of subunit aNT(104–372) titration into the EG heterodimer. The plots and layout are as in A. As can be seen in the top panel, the interaction is endothermic in nature. The titration of aNT(104–372) into buffer showed only little heat of dilution, suggesting that the aNT(104–372) dimer is only weakly associated and that its dissociation does not interfere with ITC data analysis. The curve in the bottom panel was fit to a one-sites model yielding a Kd of 33 μm. Originlab software was used for ITC data analysis and plotting.
DISCUSSION
The proton pumping activity of the yeast V-ATPase is regulated by reversible enzyme disassembly, a mechanism that involves the breaking (and reforming) of protein-protein interactions in the V1-ATPase-Vo-proton channel interface (9, 39). The V1-Vo interface is comprised of a rotor connection, formed by V1 subunits DF binding to the Vo subunit d, and a stator connection, composed of the single copy C subunit, the N-terminal domains of the three subunit EG heterodimers (peripheral stators EG1–3), the soluble cytoplasmic domain of Vo subunit a (aNT), and subunit H (see Fig. 1A) (14, 35, 36, 40, 41). A critical role in the reversible disassembly process is played by subunit C, the only subunit that does not bind to either V1 or Vo after enzyme dissociation, and subunit a, whose soluble cytoplasmic domain provides an anchor to the membrane for the other stator subunits in the assembled complex.
Crystal structures of yeast subunit C and the bacterial homolog of aNT show the polypeptides folded as two globular domains, one formed by the middle of the protein sequence and the other by their N- and C termini (18, 30). The two domains (called Chead and Cfoot for subunit C and distal and proximal lobes for aNT, respectively) are each connected by an α helical coiled-coil, giving the proteins an elongated, curved appearance. EM reconstructions and photochemical cross-linking experiments showed that subunit C provides a critical connection between V1 and Vo by linking peripheral stator EG3 (via Chead) to a ternary subunit junction formed by Cfoot, the distal lobe of aNT and peripheral stator EG2 (14, 16, 19, 20, 36) (Fig. 1A). A third connection is formed by EG1 linking V1 and subunit H to the proximal lobe of aNT. As the cross-linking experiments and low resolution EM envelopes do not provide information about the affinities of these interactions, we have begun to produce purified recombinant subunits and subunit domains for in vitro binding studies aimed at delineating the individual contributions of the subunit-subunit interactions to the functional V1-Vo interface.
From experiments with EG heterodimer and subunit C, we have recently shown that Chead binds one equivalent of EG heterodimer with high affinity (17). No significant interaction was observed between EG and Cfoot, suggesting that the ternary subunit junction at the V1-Vo interface is formed by binding of the distal lobe of aNT to both Cfoot and EG2. To characterize the subunit-subunit interactions in this ternary interface, we have generated a subdomain of yeast V-ATPase subunit a (aNT(104–372)) containing the distal lobe and adjacent coiled coil domain including the sites shown to form cross-links to subunit EG heterodimers (20). We found that including the entire coiled-coil domain appears to help stabilize aNT(104–372), as constructs with shorter portions of the coiled coil appeared unstructured (data not shown). E. coli expressed aNT(104–372) is water-soluble and folded and can be purified to homogeneity in milligram quantities. Sedimentation velocity analytical ultracentrifugation and gel filtration experiments indicated that the protein is monomeric at low concentrations but undergoes reversible dimerization at higher concentrations (>∼3 μm). SAXS analysis of aNT(104–372) at concentrations between 47.5 and 190 μm confirmed the dimeric nature of the construct and allowed calculation of a molecular envelope that can accommodate a dimer of the crystal structure of archaeal A-ATPase INT (the homolog of yeast aNT) minus the N-terminal region not present in aNT(104–372). In addition, the elongated, curved shape of the SAXS envelope agrees well with the overall shape of the crystal structure (30). Indeed, secondary structure predictions of V-ATPase aNT indicate a high level of conservation with the archaeal INT, suggesting that the related subunits possess a similar overall topology (see supplemental Fig. S1).
At this point we cannot say whether the observed dimerization of aNT(104–372) has any physiological relevance. However, the proposed role for the Vo sector of the enzyme in vesicular fusion may indicate some function of a dimeric N-terminal domain of subunit a, as it has been shown previously that both presence of subunit a and dimerization of Vo sectors is required for fusion to occur (42, 43).
After the biochemical and biophysical characterization of construct aNT(104–372), we used the subdomain to determine the affinities of the interactions in the ternary subunit interface involving subunit aNT, Cfoot, and EG heterodimer. Although mixtures of the purified proteins did not comigrate in gel permeation chromatography or native gel electrophoresis experiments (suggesting lack of a high affinity interaction, data not shown), isothermal titration calorimetry revealed that both Cfoot and EG bind to aNT(104–372) in a 1:1 stoichiometry with dissociation constants of 22 and 33 μm, respectively. According to EM reconstructions of the eukaryotic and bacterial enzymes (16, 36, 42), aNT binds two EG heterodimers in the intact complex. In the bacterial enzyme, one of these interactions is with high affinity (Kd ∼ 150 nm) (44), consistent with the observation that bacterial EG copurifies with the membrane domain (45). As can be seen from Fig. 1C (see also supplemental Fig. S1), our construct aNT(104–372) is missing most of the proximal lobe, which means that the affinities we have measured are between the distal lobe of subunit a and EG or Cfoot. As the second EG binding site on subunit ais provided by the aNT proximal lobe, it is possible that this second interaction occurs with higher affinity than the affinity between EG and the aNT distal lobe of the yeast proteins reported here. However, there is currently no evidence of such high affinity interaction between EG and aNT for the eukaryotic V-ATPase. First, upon dissociation of the complex, all three EG heterodimers copurify with V1 (5) and second, no stable interaction between bacterially expressed Vph1p(1–406) and V1 could be detected using affinity chromatography assays (35). As there is currently no indication that the bacterial enzyme is regulated by reversible dissociation, it is possible that the interaction(s) between yeast aNT and EG1,2 and Cfoot have evolved to be of moderate affinity to enable rapid and regulated dissociation of the eukaryotic V-ATPase.
The relatively moderate affinities between aNT, EG, and Cfoot raise the question of how these interactions can support stable assembly of the V-ATPase in the cell. Along with the Chead-EG3 (17) and aNT EG2 and Cfoot interactions, V-ATPase is likely stabilized by interactions between rotor subunits DF and d (41) and by H and EG1 binding to the proximal lobe of aNT (35) (see Fig. 1A). Although the dissociation constants of these additional interactions have not been measured, we expect them also to be of moderate affinity to facilitate rapid and efficient dissociation of the complex when needed. In addition and perhaps counterintuitively, the sum of these interactions would be required to be strong enough to withstand the torque of rotational catalysis and to maintain the structural integrity of the complex. To reconcile these two conditions, we speculate that the spatial proximity of the distal lobe of aNT to its binding sites on Cfoot and EG2 in the assembled complex results in a ternary binding site with high “avidity” rather than affinity, whereby the avidity constant can be estimated from the product of the individual affinities for the aNT-EG2 and aNT-Cfoot interactions. In the assembled enzyme, the interactions between aNT, C, and EG2 will result in a ternary binding interface with an avidity in the low nanomolar range, an interaction surely strong enough to maintain the assembled state of the enzyme during turnover. At the same time, it is known that for enzyme dissociation to occur, V-ATPase must be engaged in steady-state turnover, suggesting that once the signal for dissociation has been received, the torque generated during ATP hydrolysis is sufficient for breaking the V1-Vo interface (46). The advantage of a V1-Vo interface formed by multiple weak interactions is that it can be easily disrupted by perturbing one (or some) of the individual interactions. Thus, by destabilizing the aNT-Cfoot or aNT-EG2 interaction, the V1-Vo interface may become susceptible to dissociation induced by the torque of rotational catalysis.
A broad range of cellular cues could potentially lead to destabilization of these interactions and, thus, initiation of reversible disassembly. Indeed, a number of environmental signals have been linked to enzyme dissociation, mainly related to metabolism and specifically glycolysis (10). Changes in enzyme association have been linked to phosphorylation (47), extracellular conditions (48), pH changes (34, 48), and subcellular localization (33, 49). Further complicating the understanding of this mechanism are the number of cellular binding partners for subunits likely to be involved in enzyme regulation. Subunit C, for example, has been shown to interact with kinases (47, 50), actin (51), ATP (52), and the Regulator of the H+-ATPase of vacuoles and endosomal membranes complex (53). Interestingly, the C subunit x-ray crystal structure was solved in two different conformations, but it remains unclear whether these changes may be linked to release of subunit C from the enzyme during regulated disassembly (18). In addition, subunit a has been proposed at act as a pH sensor (54) by sensing lumenal pH with its membrane integral C-terminal domain and transmitting this information to cellular binding partners via the soluble aNT. These findings are on the basis of the intraendosomal pH dependence of the interaction of subunit aNT with the ADP-ribosylation factor guanine nucleotide exchange factor (Arf-GEF), ADP ribosylation factor nucleotide site opener (55), and the glycolytic enzyme aldolase (56).
In this study, we have generated a water-soluble subdomain of subunit a that is suitable for biophysical studies. Furthermore, we have characterized the interaction of the subdomain with subunit C and EG heterodimer, and, on the basis of the results, we speculate that modulating the affinity of these interactions in vivo likely plays a key role in the mechanism of reversible enzyme dissociation. Although the nature of the signal(s) leading to reversible dissociation may differ between cell types, subcellular localization and/or function of the enzyme, the dissociation of V1 from Vo appears to be a conserved mechanism for V-ATPase regulation. As the structure and subunit composition of the enzyme are highly conserved among eukaryotes, the interactions that must be broken leading to regulated dissociation are likely the same. A detailed understanding of the nature and affinities of these interactions may not only lend insight into the structural basis for regulation by reversible dissociation but also offer new avenues for discovering molecules that may be used to modulate aberrant V-ATPase activity involved in human disease.
Supplementary Material
Acknowledgments
We thank Drs. Patricia Kane and Stewart Loh for helpful discussions, Drs. Michael Cosgrove and Anamika Patel for use of the analytical ultracentrifuge and for assistance with data processing and interpretation, and Dr. Richard Gillilan (MacCHESS) for help with SAXS data collection and processing.
This work was supported, in whole or in part, by National Institutes of Health Grant GM058600 (to S. W.). CHESS is supported by the National Science Foundation (NSF) and National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) via NSF award DMR-093684, and the MacCHESS resource is supported by NIGMS award GM-103485.

This article contains supplemental Fig. S1.
L. S. Parsons and S. Wilkens, unpublished data.
- V-ATPase
- vacuolar ATPase
- EM
- electron microscopy
- SAXS
- small angle x-ray scattering
- ITC
- isothermal titration calorimetry
- V1
- soluble domain of the vacuolar ATPase
- Vo
- membrane bound domain of the vacuolar ATPase
- Cfoot
- foot domain of yeast V-ATPase subunit C
- Chead
- head domain of yeast V-ATPase subunit C
- aNT
- N-terminal domain of V-ATPase subunit a
- aCT
- C-terminal domain of V-ATPase subunit a.
REFERENCES
- 1. Li S. C., Kane P. M. (2009) The yeast lysosome-like vacuole. Endpoint and crossroads. Biochim. Biophys. Acta 1793, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forgac M. (2007) Vacuolar ATPases. Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 3. Marshansky V., Futai M. (2008) The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function, Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohsumi Y., Anraku Y. (1983) Calcium transport driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 258, 5614–5617 [PubMed] [Google Scholar]
- 5. Kitagawa N., Mazon H., Heck A. J., Wilkens S. (2008) Stoichiometry of the peripheral stalk subunits E and G of yeast V1-ATPase determined by mass spectrometry. J. Biol. Chem. 283, 3329–3337 [DOI] [PubMed] [Google Scholar]
- 6. Muench S. P., Trinick J., Harrison M. A. (2011) Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 [DOI] [PubMed] [Google Scholar]
- 7. Toei M., Toei S., Forgac M. (2011) Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manolson M. F., Proteau D., Preston R. A., Stenbit A., Roberts B. T., Hoyt M. A., Preuss D., Mulholland J., Botstein D., Jones E. W. (1992) The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267, 14294–14303 [PubMed] [Google Scholar]
- 9. Kane P. M. (1995) Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270, 17025–17032 [PubMed] [Google Scholar]
- 10. Parra K. J., Kane P. M. (1998) Reversible association between the V1 and Vo domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 18, 7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sumner J. P., Dow J. A., Earley F. G., Klein U., Jäger D., Wieczorek H. (1995) Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 [DOI] [PubMed] [Google Scholar]
- 12. Parra K. J., Keenan K. L., Kane P. M. (2000) The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275, 21761–21767 [DOI] [PubMed] [Google Scholar]
- 13. Zhang J., Myers M., Forgac M. (1992) Characterization of the Vo domain of the coated vesicle (H+)-ATPase. J. Biol. Chem. 267, 9773–9778 [PubMed] [Google Scholar]
- 14. Inoue T., Forgac M. (2005) Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J. Biol. Chem. 280, 27896–27903 [DOI] [PubMed] [Google Scholar]
- 15. Féthière J., Venzke D., Madden D. R., Böttcher B. (2005) Peripheral stator of the yeast V-ATPase. Stoichiometry and specificity of interaction between the EG complex and subunits C and H. Biochemistry 44, 15906–15914 [DOI] [PubMed] [Google Scholar]
- 16. Diepholz M., Venzke D., Prinz S., Batisse C., Flörchinger B., Rössle M., Svergun D. I., Böttcher B., Féthière J. (2008) A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: possible implications for regulatory disassembly. Structure 16, 1789–1798 [DOI] [PubMed] [Google Scholar]
- 17. Oot R. A., Wilkens S. (2010) Domain characterization and interaction of the yeast vacuolar ATPase subunit C with the peripheral stator stalk subunits E and G. J. Biol. Chem. 285, 24654–24664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drory O., Frolow F., Nelson N. (2004) Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5, 1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Z., Inoue T., Forgac M., Wilkens S. (2006) Localization of subunit C (Vma5p) in the yeast vacuolar ATPase by immuno electron microscopy, FEBS Lett. 580, 2006–2010 [DOI] [PubMed] [Google Scholar]
- 20. Qi J., Forgac M. (2008) Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J. Biol. Chem. 283, 19274–19282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 22. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science, pp. 19–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 24. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nielsen S. S., Toft K. N., Snakenborg D., Jeppesen M. G., Jacobsen J. K., Vestergaard B., Kutter J. P., Arleth L. (2009) BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Cryst. 42, 959–964 [Google Scholar]
- 26. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria., J. Appl. Cryst. 25, 495–503 [Google Scholar]
- 27. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wriggers W. (2010) Using SITUS for the Integration of Multi-Resolution Structures. Biophys. Rev. 2, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srinivasan S., Vyas N. K., Baker M. L., Quiocho F. A. (2011) Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase. J. Mol. Biol. 412, 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 32. Trombetta E. S., Ebersold M., Garrett W., Pypaert M., Mellman I. (2003) Activation of lysosomal function during dendritic cell maturation. Science 299, 1400–1403 [DOI] [PubMed] [Google Scholar]
- 33. Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., van der Goot F. G. (2008) Regulation of the V-ATPase along the endocytic pathway occurs through reversible subunit association and membrane localization, PLoS ONE 3, e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dechant R., Binda M., Lee S. S., Pelet S., Winderickx J., Peter M. (2010) Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diab H., Ohira M., Liu M., Cobb E., Kane P. M. (2009) Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J. Biol. Chem. 284, 13316–13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Zheng Y., Mazon H., Milgrom E., Kitagawa N., Kish-Trier E., Heck A. J., Kane P. M., Wilkens S. (2008) Structure of the yeast vacuolar ATPase. J. Biol. Chem. 283, 35983–35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dechant R., Peter M. (2011) The N-terminal domain of the V-ATPase subunit “a” is regulated by pH in vitro and in vivo. Channels 5, 4–8 [DOI] [PubMed] [Google Scholar]
- 38. Merkulova M., McKee M., Dip P. V., Grüber G., Marshansky V. (2010) N-terminal domain of the V-ATPase a2-subunit displays integral membrane protein properties. Protein Sci. 19, 1850–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kane P. M., Parra K. J. (2000) Assembly and regulation of the yeast vacuolar H(+)-ATPase. J. Exp. Biol. 203, 81–87 [DOI] [PubMed] [Google Scholar]
- 40. Jones R. P., Durose L. J., Findlay J. B., Harrison M. A. (2005) Defined sites of interaction between subunits E (Vma4p), C (Vma5p), and G (Vma10p) within the stator structure of the vacuolar H+-ATPase. Biochemistry 44, 3933–3941 [DOI] [PubMed] [Google Scholar]
- 41. Saijo S., Arai S., Hossain K. M., Yamato I., Suzuki K., Kakinuma Y., Ishizuka-Katsura Y., Ohsawa N., Terada T., Shirouzu M., Yokoyama S., Iwata S., Murata T. (2011) Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proc. Natl. Acad. Sci. U.S.A. 108, 19955–19960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peters C., Bayer M. J., Bühler S., Andersen J. S., Mann M., Mayer A. (2001) Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409, 581–588 [DOI] [PubMed] [Google Scholar]
- 43. Bayer M. J., Reese C., Buhler S., Peters C., Mayer A. (2003) Vacuole membrane fusion. Vo functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel, J. Cell Biol. 162, 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamamoto M., Unzai S., Saijo S., Ito K., Mizutani K., Suno-Ikeda C., Yabuki-Miyata Y., Terada T., Toyama M., Shirouzu M., Kobayashi T., Kakinuma Y., Yamato I., Yokoyama S., Iwata S., Murata T. (2008) Interaction and stoichiometry of the peripheral stalk subunits NtpE and NtpF and the N-terminal hydrophilic domain of NtpI of Enterococcus hirae V-ATPase. J. Biol. Chem. 283, 19422–19431 [DOI] [PubMed] [Google Scholar]
- 45. Yokoyama K., Nagata K., Imamura H., Ohkuma S., Yoshida M., Tamakoshi M. (2003) Subunit arrangement in V-ATPase from Thermus thermophilus. J. Biol. Chem. 278, 42686–42691 [DOI] [PubMed] [Google Scholar]
- 46. Huss M., Wieczorek H. (2007) Influence of ATP and ADP on dissociation of the V-ATPase into its V(1) and V(O) complexes. FEBS Lett. 581, 5566–5572 [DOI] [PubMed] [Google Scholar]
- 47. Voss M., Vitavska O., Walz B., Wieczorek H., Baumann O. (2007) Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A. J. Biol. Chem. 282, 33735–33742 [DOI] [PubMed] [Google Scholar]
- 48. Diakov T. T., Kane P. M. (2010) Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J. Biol. Chem. 285, 23771–23778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kawasaki-Nishi S., Nishi T., Forgac M. (2001) Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276, 17941–17948 [DOI] [PubMed] [Google Scholar]
- 50. Hong-Hermesdorf A., Brüx A., Grüber A., Grüber G., Schumacher K. (2006) A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett. 580, 932–939 [DOI] [PubMed] [Google Scholar]
- 51. Vitavska O., Wieczorek H., Merzendorfer H. (2003) A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J. Biol. Chem. 278, 18499–18505 [DOI] [PubMed] [Google Scholar]
- 52. Armbrüster A., Hohn C., Hermesdorf A., Schumacher K., Börsch M., Grüber G. (2005) Evidence for major structural changes in subunit C of the vacuolar ATPase due to nucleotide binding. FEBS Lett. 579, 1961–1967 [DOI] [PubMed] [Google Scholar]
- 53. Smardon A. M., Kane P. M. (2007) RAVE is essential for the efficient assembly of the C subunit with the vacuolar H(+)-ATPase. J. Biol. Chem. 282, 26185–26194 [DOI] [PubMed] [Google Scholar]
- 54. Marshansky V. (2007) The V-ATPase a2-subunit as a putative endosomal pH-sensor. Biochem. Soc. Trans. 35, 1092–1099 [DOI] [PubMed] [Google Scholar]
- 55. Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 56. Merkulova M., Hurtado-Lorenzo A., Hosokawa H., Zhuang Z., Brown D., Ausiello D. A., Marshansky V. (2011) Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am. J. Physiol. Cell Physiol. 300, C1442–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.