Background: NFATc1 is a necessary and sufficient transcription factor for osteoclastogenesis.
Results: JMJD5 negatively regulates NFATc1 protein level through its hydroxylase activity.
Conclusion: JMJD5 is a novel osteoclastogenic repressor that induces the degradation of NFATc1 protein.
Significance: This study revealed a novel mechanism that regulates NFATc1 activity during osteoclastogenesis.
Keywords: Cell Differentiation, Osteoclast, Post-translational Modification, Protein Degradation, Transcription Factors, Hydroxylation, JMJD5, NFATc1, Osteoclastogenesis
Abstract
Osteoclastogenesis is a highly regulated process governed by diverse classes of regulators. Among them, nuclear factor of activated T-cells calcineurin-dependent 1 (NFATc1) is the primary osteoclastogenic transcription factor, and its expression is transcriptionally induced during early osteoclastogenesis by receptor activation of nuclear factor κB ligand (RANKL), an osteoclastogenic cytokine. Here, we report the novel enzymatic function of JMJD5, which regulates NFATc1 protein stability. Among the tested Jumonji C (JmjC) domain-containing proteins, decreased mRNA expression levels during osteoclastogenesis were found for JMJD5 in RAW264 cells stimulated by RANKL. To examine the functional role of JMJD5 in osteoclast differentiation, we established stable JMJD5 knockdown cells, and osteoclast formation was assessed. Down-regulated expression of JMJD5 led to accelerated osteoclast formation together with induction of several osteoclast-specific genes such as Ctsk and DC-STAMP, suggesting that JMJD5 is a negative regulator in osteoclast differentiation. Although JMJD5 was recently reported as a histone demethylase for histone H3K36me2, no histone demethylase activity was detected in JMJD5 in vitro or in living cells, even for other methylated histone residues. Instead, JMJD5 co-repressed transcriptional activity by destabilizing NFATc1 protein. Protein hydroxylase activity mediated by the JmjC domain in JMJD5 was required for the observed functions of JMJD5. JMJD5 induced the association of hydroxylated NFATc1 with the E3 ubiquitin ligase Von Hippel-Lindau tumor suppressor (VHL), thereby presumably facilitating proteasomal degradation of NFATc1 via ubiquitination. Taken together, the present study demonstrated that JMJD5 is a post-translational co-repressor for NFATc1 that attenuates osteoclastogenesis.
Introduction
Mature osteoclasts are giant multinucleated cells that are responsible for bone resorption in bone remodeling and function in a regulated balance with bone-forming osteoblasts (1, 2). Commitment of hematopoietic stem cells to differentiate to osteoclasts is defined by two key factors, macrophage colony-stimulating factor and receptor activation of nuclear factor κB ligand (RANKL),3 factors that are produced by osteoblast lineage cells in bone tissues (3–6). During cell maturation, mononuclear precursor cells fuse to generate multinucleated osteoclasts. This multinucleation process is necessarily required for osteoclastic maturation to undergo efficient bone resorption (7, 8). In reorganizing genetic networks during osteoclastogenesis, diverse transcription factors play pivotal roles, and among them, nuclear factor of activated T-cells calcineurin-dependent 1 (NFATc1) is the necessary and sufficient transcription factor (9, 10). Osteoclastogenesis induced by RANKL is aborted in NFATc1−/− embryonic stem cells (9). Therefore, NFATc1 is considered to be an essential factor in osteoclastogenesis. Moreover, ectopic expression of NFATc1 in osteoclast precursor cells efficiently induces osteoclastogenesis even in the absence of RANKL stimulation (10). Supporting the importance of NFATc1 in osteoclastogenesis, NFATc1 is a transcriptional inducer of several osteoclast-specific genes (required for maturation and bone resorption activity of osteoclasts), including DC-STAMP (dendritic cell-specific transmembrane protein), OSCAR (osteoclast-associated receptor), Acp5 (acid phosphatase 5, tartrate-resistant), and Ctsk (cathepsin K) (9, 11, 12).
Gene expression can be regulated during transcriptional, post-transcriptional, translational, and post-translational processes (13). Most previous studies of NFATc1-related gene expression have been focused on uncovering the molecular mechanisms at the transcriptional level (9, 10, 14–18). Nfatc1 was cloned as a RANKL-induced gene, and its expression is transcriptionally induced by transcription factors NFκB and c-Fos, which are activated by RANKL/RANK signaling (9, 16). Once the Nfatc1 gene is induced, an autoregulatory loop is established in which it binds to its own promoter region, leading to its robust induction (16). On the other hand, it was recently reported that NFATc1 protein is degraded during late stages of osteoclast differentiation by Cbl protein-mediated ubiquitination (19). Therefore, it is conceivable that NFATc1 activity is controlled by its protein stability through post-translational regulation.
The importance of epigenetic modification of histone tails has emerged in studies of the regulation of cellular differentiation, and a histone methylation signature generally appears upstream in epigenetic regulation of transcription. Among various histone methylation modifiers, Jumonji C (JmjC) domain-containing proteins have recently been reported to bear histone demethylase activity. Indeed, several kinds of JmjC domain-containing proteins have been identified as histone demethylases, including JHDM1 for the lysine 36 residue in histone H3 (H3K36), JHDM2 for H3K27, and so on (20–23). On the other hand, two JmjC domain-containing proteins, factor inhibiting hypoxia-inducible factor 1 (HIF1) (FIH1) and Jumonji domain-6 (JMJD6), were characterized as hydroxylases for non-histone proteins (24–27). FIH1 post-translationally hydroxylates HIF1α on its asparagine residue, and hydroxylated HIF1α was shown to be a transcriptionally inactive form because of inhibition of cognate co-activator interaction (24). Likewise, JMJD6 serves as a lysine hydroxylase, modulating the function of RNA splicing factors, suggesting its specific roles in the regulation of alternative RNA splicing (27). Taken together, it is now believed that JmjC domain-containing proteins exert enzymatic activity as protein hydroxylases besides histone demethylases. However, the functions of JmjC domain-containing proteins during osteoclastogenesis remain to be studied.
In this study, we show that one of the epigenetic regulators, a JmjC domain-containing protein, JMJD5, negatively regulates osteoclastogenesis by reducing the stability of NFATc1 protein via its hydroxylase activity. Moreover, JMJD5 was found to induce the association of NFATc1 with E3 ubiquitin ligase Von Hippel-Lindau tumor suppressor (VHL) in HEK293T cells. Thus, the present study demonstrates that the enzymatic function of a JmjC domain-containing protein acts as a post-translational co-repressor of a primary transcription factor during osteoclastogenesis.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length cDNA of mouse JMJD5 was obtained by RT-PCR from total RNA of mouse heart and cloned into the pcDNA3-FLAG or the pcDNA3.1-Myc mammalian expression plasmids (Invitrogen). To generate an expression plasmid of ZsGreen-tagged JMJD5, cDNA for ZsGreen protein was amplified from pZsGreen1-DR plasmid (Clontech) by PCR. Amplified cDNA was subcloned into the C terminus of the previously generated pcDNA3-FLAG-JMJD5 plasmid. The point mutant FLAG-JMJD5-K334A plasmid was generated from the wild-type plasmid by site-directed mutagenesis (28). GST-fused JMJD5 and mutant JMJD5-K334A expression plasmids were generated by subcloning into the pGEX-4T-1 plasmid. Full-length cDNAs of mouse NFATc1α and NFATc1β were obtained by RT-PCR from mouse spleen and cloned into the pcDNA3-FLAG or the pcDNA3.1-Myc mammalian expression vector. To generate GST-fused truncated NFATc1α constructs, amino acids 1–300, 301–596, and 589–827 were partially amplified by PCR from pcDNA-FLAG-NFATc1α plasmid and then subcloned into the pGEX-4T-1 plasmid. A full-length cDNA of mouse FIH1 was obtained by RT-PCR from mouse testis and cloned into the pGEX-4T-1 plasmid. A full-length cDNA of mouse p105 was obtained by RT-PCR from total RNA of mouse spleen and cloned into the pGEX-4T-1 plasmid. For the GST-fused p105-ARD expression plasmid, amino acids 537–809 were partially amplified by PCR from a full-length plasmid and then subcloned into the pGEX-4T-1 plasmid. Sequences of all cDNAs were verified by automated sequencing. For the NFATc1-response element (RE) reporter plasmid, the mouse Acp5 promoter (−485 to +525) was amplified from genomic DNA of RAW264 cells and cloned into the upstream region of the luciferase gene derived from pGL4.10 (luc2) vector (Promega). Oligonucleotide short hairpin RNA (shRNA) targeting mouse JMJD5 (shRNA 1; 5′-GGAAGGAAGTACATCCGATTG-3′) was inserted into the pSUPER-Retro vector (OligoEngine) according to the manufacturer's instructions.
Cell Culture and Transfection
HEK293T cells were maintained in Dulbecco's modified Eagle medium (Wako) supplemented with 10% fetal bovine serum (JHR Biosciences) and 1% antibiotic-antimycotic solution (Invitrogen). MG132 (Calbiochem) was used at a concentration of 10 μm. Transient transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. An oligonucleotide siRNA targeting human JMJD5 (siGENOME SMARTpool; 5′-AGCCAAGGGACGUCGGGUA-3′, 5′-UCAACGAGUUCAUCAGCAA-3′, 5′-GAAGUUGGUUCGAGGUACA-3′, and 5′-CCACUGAGCUCUUCUACGA-3′) and a control oligonucleotide siRNA (siGENOME Non-Targeting siRNA pool 1) were purchased from Thermo Scientific. For high knockdown efficiency, siRNA was transfected twice for 4 days.
Osteoclast Differentiation
RAW264 cells were maintained in α-minimum Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (JHR Biosciences), 1% antibiotic-antimycotic solution (Invitrogen), and 1% minimum Eagle's medium non-essential amino acids (Invitrogen). For osteoclastogenesis, RAW264 cells were seeded in 10-cm dishes at a density of 1 × 105 cells/dish or in 6-well plates at a density of 3 × 104 cells/well and were treated with 235 ng/ml GST-RANKL (Oriental Yeast Co., Ltd.) for 6 days (29). The medium was changed every 2nd day. Differentiated cells were fixed with 3.7% formaldehyde for 10 min at room temperature and visualized with an Olympus IX70 light microscope equipped with Zeiss AxioVision 4.6.
Establishment of Stable Cell Lines
RAW264 cells were transfected with the pSUPER-mJMJD5-shRNA plasmid by Lipofectamine 2000 (Invitrogen). After 24 h, 3 μg/ml puromycin (Sigma) was added, and cultivation was continued for approximately 3 weeks to select stably transfected cells (29). Culture media were changed every 2nd day. After detecting colony formation, several colonies were individually picked up, and their knockdown efficiency was confirmed by RT-PCR.
RNA Analysis
Total RNA was extracted with TRIzol reagent (Invitrogen) according to a previous report (30). For real time RT-PCR, 500 ng of total RNA was reverse transcribed into first strand cDNA with oligo(dT) and random hexamers using a PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the manufacturer's instructions. Real time PCR was performed with SYBR Premix Ex Taq (Takara Bio, Inc.) and the Thermal Cycler Dice Real Time System (Applied Biosystems). For an internal control, the Gapdh gene was used. The primers were purchased from Takara Bio, Inc. For RT-PCR, 1 μg of total RNA was reverse transcribed into first strand cDNA with oligo(dT) using SSIII reverse transcriptase (Invitrogen), and cDNA was amplified with the indicated primer sets. All primer sets used are indicated in the supplemental Experimental Procedures.
Luciferase Assay
HEK293T cells in 12-well plates were transfected with the NFATc1-RE reporter plasmid (500 ng) together with each expression vector (700 ng). Transfection efficiency was normalized by co-transfection with pGL4.75 (hRluc/CMV) (5 ng). After 36 h, a luciferase assay was performed with the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer's instructions.
2-OG Decarboxylation Assay
Decarboxylation assays of 14C-radiolabeled 2-OG were performed as reported previously (26). Briefly, 80 μm enzyme, 80 μm substrate, 24 μm 14C-radiolabeled 2-OG, 576 μm 2-OG, 8 mm sodium ascorbate, 200 μm (NH4)2Fe(SO4)2, and 2 mm DTT were mixed in a total volume of 100 μl with 50 mm Tris-Cl (pH 7.5). A 500-μl Eppendorf tube containing 200 μl of hyamine hydroxide (Fisher Scientific) was added to each tube, and tubes were closed with a rubber septum. After incubation at 37 °C for 30 min, the reaction was stopped by setting tubes on ice for 30 min. Then Eppendorf tubes containing the hyamine hydroxide were transferred to scintillation vials and mixed with OptiPhase Hisafe 2 Liquid Scintillation Mixture (Fisher Scientific). Total released 14C counts were quantified using an LSC-5100 scintillation counter (Aloka). Data were analyzed by two-tailed Student's t test, and p < 0.05 was accepted as significant.
GST-fused Protein Preparation
GST-fused proteins were prepared as described previously (31–33). Briefly, established plasmids for GST-fused proteins were transformed into Escherichia coli strain Rosetta (DE3) (Novagen), which was grown at 37 °C in LB medium containing 100 μg/ml ampicillin. GST-fused protein was induced by 0.5 mm isopropyl β-d-thiogalactoside and incubation at 25 °C for 5 h. Cell pellets were resuspended with LEW-Tween 20 buffer (50 mm NaH2PO4, 300 mm NaCl, 0.05% Tween 20, pH7.4) containing 1 mg/ml lysozyme (Sigma) and sonicated with a UD-200 ultrasonic disruptor (Tomy). After centrifugation, the supernatants were mixed with glutathione-Sepharose 4B (GE Healthcare), and bound proteins were eluted with 50 mm glutathione (Wako). Ion exchange chromatography (ÄKTA Mono Q column, GE Healthcare) yielded highly pure GST-fused proteins that were visualized by Coomassie Brilliant Blue staining.
Immunocytochemistry
HEK293T cells were transfected with pcDNA3-FLAG-JMJD5-ZsGreen plasmid. After 24 h of transfection, cells were plated in Lab-TekII chamber slides (Nalge Nunc International). After 48 h of transfection, cells were fixed with 2% paraformaldehyde in PBS and permeabilized in 0.05% Triton X-100 in PBS (PBST). Cells were blocked with 5% skim milk in PBST, and the indicated primary antibodies were bound overnight at 4 °C. Primary antibody signals were generated with a rhodamine-conjugated secondary antibody (Molecular Probes). The slides were mounted with mounting medium containing DAPI (Vector Laboratories, Inc.) and visualized with a fluorescence confocal microscope (LSM510, Carl Zeiss, Inc.) (28). The following primary antibodies were used: H3K4me2 (Upstate, 07-030), H3K4me3 (Abcam, ab8580), H3K9me1 (Upstate, 07-450), H3K9me2 (Abcam, ab1220), H3K27me1 (Upstate, 07-448), H3K27me2 (Upstate, 07-452), H3K27me3 (Upstate, 07-449), H3K36me1 (Abcam, ab9048), and H3K36me2 (Upstate, 07-369).
Immunoblotting and Immunoprecipitation
Cells were washed with PBS and resuspended with TNE buffer (10 mm Tris-Cl (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, and protease inhibitors). Total cell lysates were resolved in SDS-polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk in PBST, and primary antibody was bound overnight at 4 °C. After washing with PBST, horseradish peroxide (HRP)-conjugated secondary antibody (Dako) was bound for 1 h at RT. Immunoreactive signals were detected with ECL Western blotting detection reagent (GE Healthcare) (32). Antibodies against FLAG (clone M2) and Myc (clone 4A6) epitopes were purchased from Sigma and Millipore, respectively. NFATc1 (sc-7294) and β-actin (sc-1616) antibodies were purchased from Santa Cruz Biotechnology. JMJD5 (ab36104), p65 (ab790), and p105/50 (ab7971) antibodies were purchased from Abcam. Antibodies against VHL (2738), ubiquitin (Z0458), and GAPDH (MAB374) were purchased from Cell Signaling Technology, Dako, and Chemicon, respectively. The following primary antibodies were used to detect histone modifications: H3K4me1 (Upstate, 07-436), H3K4me2 (Abcam, ab7766), H3K4me3 (Abcam, ab1012), H3K9me1 (Upstate, 07-395), H3K9me2 (Upstate, 07-441), H3K9me3 (Abcam, ab6001), H3K27me1 (Upstate, 07-448), H3K27me2 (Upstate, 07-452), H3K27me3 (Upstate, 07-449), H3K36me1 (Abcam, ab9048), H3K36me2 (Upstate, 07-274), H3K36me3 (Abcam, ab9050), H3K79me1 (Abcam, ab2886), H3K79me2 (Abcam, ab3594), and H3K79me3 (Abcam, ab2621). For immunoprecipitation assays, total cell lysates were precleared with Protein G-Sepharose 4 fast flow (GE Healthcare) and IgG antibody (Sigma). Then immunoprecipitation was performed by incubating precleared lysates with 2 μg of anti-NFATc1 antibody overnight at 4 °C. After incubation with Protein G-Sepharose 4 fast flow for 1 h at 4 °C, immunoprecipitated proteins were washed five times with TNE buffer and analyzed by immunoblotting. For immunoprecipitation assays with anti-FLAG antibody, total cell lysates were incubated with anti-FLAG M2-agarose affinity gel (Sigma). After 5 h of incubation at 4 °C, immunoprecipitated proteins were washed five times with TNE buffer and eluted with 400 mg/ml FLAG peptide (Sigma). Eluted proteins were analyzed by immunoblotting.
RESULTS
JMJD5 Represses Osteoclastogenesis by Reducing Expression of Several Osteoclast-specific Genes
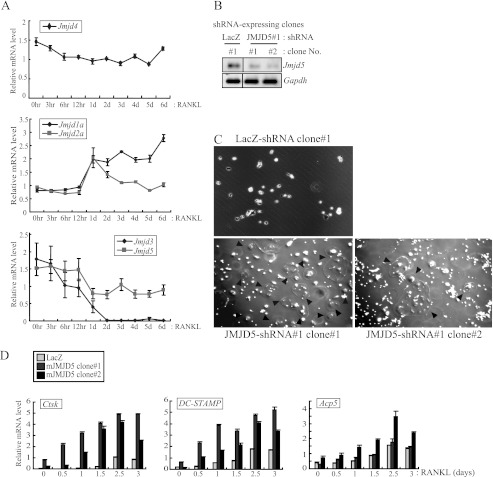
To clarify epigenetic regulation during osteoclastogenesis, we focused on JmjC domain-containing proteins, which are histone demethylases. Their physiological roles in bone metabolism have not been characterized. We first observed their expression profiles during osteoclastogenesis. RAW264 cells were differentiated to mature osteoclasts by RANKL treatment, and total RNA was prepared from each differentiation stage to assess the expression levels of JmjC domain-containing proteins. Through a real time RT-PCR analysis, we detected their various expression patterns. The expression levels of JMJD1a/2a transcripts increased, whereas those of JMJD5/3 decreased (Fig. 1A and supplemental Fig. S1). On the other hand, the expression level of JMJD4 transcripts was unaltered during osteoclastogenesis.
FIGURE 1.
JMJD5 repressed osteoclastogenesis with reduced expression of osteoclast-specific genes. A, mature osteoclasts were differentiated from RAW264 cells by RANKL treatment for 6 days. Total RNAs were extracted at the indicated time points and analyzed by real time RT-PCR. Relative mRNA levels are represented by values that were normalized to Gapdh transcript levels. The error bars represent S.D. values from at least triplicate experiments. B, stable cell lines that constitutively express JMJD5-specific shRNA were established in RAW264 cells by puromycin selection. Individual colonies were picked up, and the expression levels of the Jmjd5 gene were detected by RT-PCR. Two stable clones, which have shown high knockdown efficiency, were selected for further study. LacZ-specific shRNA was used for a negative control. C, stable clones were treated with RANKL for osteoclast formation. Differentiated cells were fixed at 3.5 days and assessed with an Olympus IX70 light microscope. Black arrowheads indicate multinucleated osteoclasts. D, during RANKL treatment, cells were harvested at the indicated time points, and total RNAs were prepared. Expression levels of osteoclast-specific genes (Ctsk, DC-STAMP, and Acp5) were analyzed by real time RT-PCR.
Although various factors, such as NFATc1, promote osteoclastogenesis and are induced during osteoclastogenesis (9), little is known about intracellular negative regulators of osteoclastogenesis. Because JMJD5/3 expression levels decreased during osteoclastogenesis, we reasoned that their functions were related to negative regulation. To test this idea, stable cell lines constitutively expressing JMJD5- or JMJD3-specific shRNAs with the puromycin-resistant gene were established from RAW264 cells by puromycin selection. Each cell line was then treated with RANKL for osteoclast formation. The shRNA knockdown was confirmed by attenuated expression levels of either JMJD3 or JMJD5 transcripts in the stable lines (Fig. 1B and supplemental Fig. S2). The stable RAW264 JMJD3 knockdown line differentiated normally into osteoclasts and displayed normal expression levels of osteoclast-specific genes Acp5, DC-STAMP, and Nfatc1 (supplemental Fig. S2). However, when JMJD5 was knocked down, osteoclastic maturation was accelerated (Fig. 1C and supplemental Fig. S3). The same results were observed in bona fide osteoclasts derived from murine bone marrow macrophages (supplemental Fig. S4). Consistent with the accelerated formation of osteoclasts, the induced expression of Ctsk, DC-STAMP, and Acp5 genes was also observed (Fig. 1D). These results implied that JMJD5 is a negative regulator for osteoclast differentiation.
JMJD5 Represses Transcriptional Activity of NFATc1 by Reducing Level of Its Protein
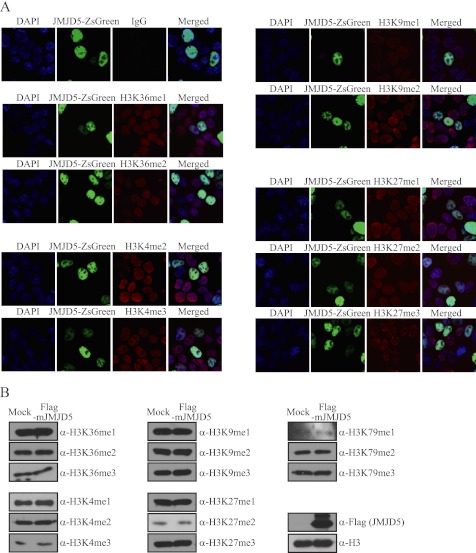
JMJD5 is reportedly an H3K36me2 demethylase (34). To verify its histone demethylase activity, we measured the methylation level of histone H3K36 in HEK293T cells overexpressing JMJD5 using immunofluorescence and immunoblotting analyses. However, we could not detect the expected alteration in the level of H3K36me2 (Fig. 2A). Although other methylated histone lysine residues were tested with several methylation-specific histone antibodies, we did not detect altered methylation levels on any tested lysine residues of histone H3 (Fig. 2, A and B). Thus, from these observations, we assumed that JMJD5 is not a histone demethylase.
FIGURE 2.
Overexpressed JMJD5 did not affect methylation levels on tested histone H3 lysine residues. A, HEK293T cells were transfected with JMJD5-ZsGreen. After 36 h of transfection, cells were fixed and analyzed by immunocytochemistry. JMJD5 was visualized by the tagged ZsGreen protein (green). Histone H3 lysine methylation levels were observed by staining with the indicated primary antibodies (red). IgG antibody was used for the negative control. DAPI was used to stain nuclei (blue). B, HEK293T cells were transfected with FLAG-mJMJD5. After 48 h of transfection, chromosomal extracts were prepared and analyzed by immunoblotting with the indicated primary antibodies. Histone H3 was used as loading control for immunoblotting.
To assess other enzymatic functions of JMJD5, we examined the cellular distribution of JMJD5 protein during osteoclastogenesis based on its solubility differences in lysis buffers. JMJD5 was present in the chromatin fraction as expected, and its distribution appeared to be unaffected by RANKL stimulation (supplemental Fig. S5). JMJD5 was also abundant in the soluble nuclear fraction, suggesting that JMJD5 serves as a soluble transcriptional co-regulator in the nucleus rather than a histone demethylase bound to chromatin.
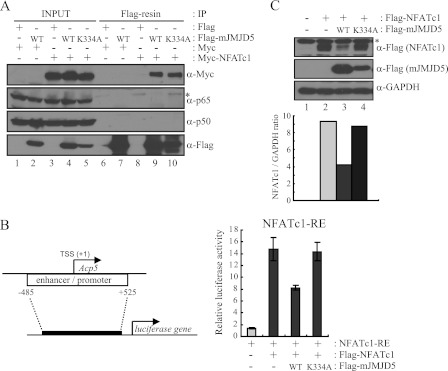
To test this idea, we examined the association of JMJD5 with the primary transcription factors for osteoclastogenesis, i.e. NFATc1, NFκB, and AP-1. We expressed and purified recombinant affinity-tagged full-length proteins from E. coli and then performed in vitro pulldown assays. His-tagged JMJD5 was an interactant with tested GST-fused transcription factors NFATc1α/β, c-Jun, and NFκB (p105/50) but not c-Fos (supplemental Fig. S6). To further address their association, we tested the in vivo interactions within cells. Total cellular extracts were prepared from HEK293T cells expressing FLAG-JMJD5 and Myc-NFATc1, and these proteins were immunoprecipitated by anti-FLAG antibody. The bound proteins were eluted with FLAG peptides and observed by immunoblotting with anti-Myc and anti-NFκB (p65/50) antibodies. Among the tested factors, only NFATc1 was detected (Fig. 3A). The interaction of JMJD5 with NFATc1 was still significant with a catalytically inactive mutant form of JMJD5 (JMJD5-K334A) in which the α-ketoglutarate (2-OG)-binding residue Lys-334 was substituted by alanine (Fig. 3A, last lane).
FIGURE 3.
JMJD5 repressed transcriptional activity of NFATc1 by reducing its protein level. A, HEK293T cells were transfected with Myc-NFATc1 together with FLAG-mJMJD5-WT or FLAG-mJMJD5-K334A. After 48 h of transfection, total cell lysates were prepared and subjected to immunoprecipitation (IP) with anti-FLAG M2-agarose. Bound proteins were eluted by FLAG peptides and detected by immunoblotting with the indicated antibodies. An asterisk indicates a nonspecific band. B and C, NFATc1-RE gene was established with the indicated region of the Acp5 gene promoter (B, left). HEK293T cells were transfected with NFATc1-RE together with FLAG-NFATc1 and FLAG-mJMJD5-WT or FLAG-mJMJD5-K334A. After 36 h of transfection, cells were analyzed with a luciferase assay (B, right) or by immunoblotting (C). The error bars represent S.D. values from at least triplicate experiments. GAPDH was used as loading control for immunoblotting. An asterisk indicates a nonspecific band. The signals obtained from the immunoblotting were quantified with the ImageJ program. TSS, transcription start site.
NFATc1 is a primary transcription factor for osteoclastogenesis, and it regulates the expression of osteoclast-specific genes (9, 16). From its interaction with JMJD5, we hypothesized that JMJD5 co-regulated the transcriptional activity of NFATc1. To examine this possibility, we constructed an NFATc1-RE reporter plasmid by inserting the −485 to +525 region of the Acp5 gene in front of the luciferase gene and measured the transcriptional activity of NFATc1 when JMJD5 was overexpressed. The luciferase activity was increased ∼15-fold when NFATc1 was expressed, whereas it was significantly decreased by co-expression of JMJD5 (Fig. 3B). To characterize the repressive function of JMJD5, we then analyzed the lysates used in the luciferase assay by immunoblotting, testing whether JMJD5 regulated an event after transcription. Unexpectedly, the NFATc1 protein level was significantly reduced by expression of JMJD5, indicating that decreased NFATc1 transcriptional activity is due to the reduced protein level of NFATc1 (Fig. 3C). Such JMJD5 action was not seen for NFκB (p50/65) (supplemental Fig. S7, A and B). Importantly, the suppressive actions of JMJD5 on the transcriptional activity and protein level of NFATc1 were not observed when the JMJD5-K334A mutant was expressed (Fig. 3C, last lane), suggesting that the repressive function of JMJD5 against NFATc1 depended on its 2-OG-dependent activity.
Post-translational Destabilization of NFATc1 Protein by JMJD5
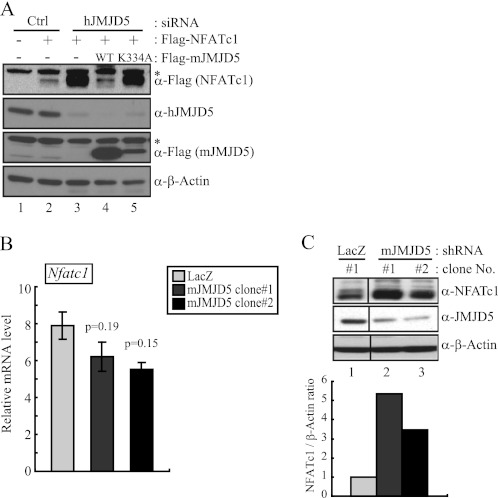
To determine whether JMJD5 was capable of modulating the stability of NFATc1 protein, the level of exogenous NFATc1 protein was monitored during the knockdown of JMJD5. Human JMJD5-specific siRNA was introduced into HEK293T cells, and the level of exogenously expressed NFATc1 was measured by immunoblotting. As anticipated, reduced expression of JMJD5 was seen, and the NFATc1 protein level was significantly elevated (Fig. 4A). Furthermore, when JMJD5 was overexpressed in these knockdown cells, the NFATc1 protein level was indistinguishable from that in the control cells. The JMJD5-K334A mutant was not effective in regulating the protein level of NFATc1 (Fig. 4A).
FIGURE 4.
JMJD5 reduced level of NFATc1 protein by post-translational regulation. A, HEK293T cells were transfected with human JMJD5-specific siRNA. After 48 h of siRNA transfection, cells were also transfected with human JMJD5-specific siRNA together with FLAG-NFATc1 and FLAG-JMJD5-WT or FLAG-JMJD5-K334A. After 48 h, total cell lysates were prepared and analyzed by immunoblotting with the indicated antibodies. β-Actin was used as a loading control for immunoblotting. An asterisk indicates a nonspecific band. B and C, total RNAs and cell lysates were prepared from two stable clones constitutively expressing JMJD5 shRNA in RAW264 cells. Expression levels of the Nfatc1 gene were detected by real time RT-PCR (left). Relative mRNA levels are represented by values that were normalized to Gapdh transcript levels. The error bars represent S.D. values from at least triplicate experiments. The p value for statistical difference was calculated using a two-tailed Student's t test. Also, the NFATc1 protein level was detected by immunoblotting with an anti-NFATc1 antibody (right). β-Actin was used as a loading control for immunoblotting. The signals obtained from the immunoblotting were quantified with the ImageJ program. Ctrl, control.
Next, we asked whether the level of endogenous NFATc1 protein was indeed regulated by JMJD5. Using the stable cell lines expressing JMJD5-specific shRNA, total RNA and cellular extracts were prepared for RT-PCR and immunoblotting. Although the level of NFATc1 transcripts was unaltered (Fig. 4B), the level of NFATc1 protein was greater than that in the control knockdown cells (Fig. 4C). Taken together, it appears that the mechanism of down-regulating the level of NFATc1 protein by JMJD5 is post-translational.
JMJD5 Hydroxylates NFATc1 Protein through Its Enzymatic Activity
Recently, some members of JmjC domain-containing proteins have been shown to serve as protein hydroxylases, for example JMJD6 and FIH1 (24, 27). Because JMJD5 exhibits sequence homology with JMJD6 and FIH1, it is conceivable that JMJD5 is a protein hydroxylase. This idea is further supported by the present findings that a catalytically inactive JMJD5 mutant was impaired in the regulation of the NFATc1 protein level.
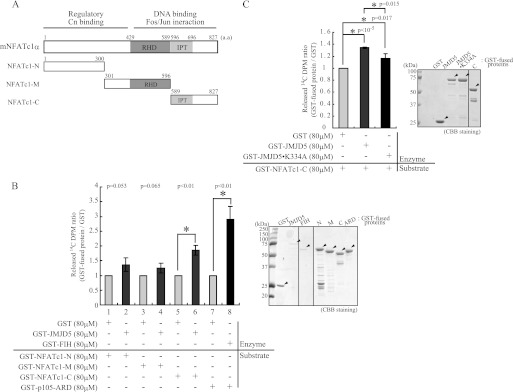
To address this idea, we performed an in vitro “2-OG decarboxylation assay” with GST-fused recombinant proteins (25, 26). Recombinant NFATc1 deletion mutant proteins were expressed and purified from E. coli (Fig. 5A), and each mutant was incubated with the purified GST-tagged JMJD5 to induce 14CO2 formation. After confirming FIH1 activity for the known hydroxylase substrate (p105) (Fig. 5B, lanes 7 and 8) (26), we tested whether NFATc1 is a hydroxylase substrate for JMJD5. The C-terminal domain of NFATc1 was found to serve as a substrate, although its 2-OG decarboxylation was less than that for p105 (Fig. 5B, lanes 5 and 6). To confirm that the observed decarboxylation activity of JMJD5 mediated by the JmjC domain, the JMJD5-K334A mutant was also used for this assay. As anticipated, a reduction in 14CO2 formation was seen (Fig. 5C). These findings suggest that JMJD5 has hydroxylase activity and that NFATc1 is its substrate.
FIGURE 5.
JMJD5 hydroxylated NFATc1 protein through its enzymatic activity. A, diagram of the full length and three generated fragments (NFATc1-N, -M, and -C) of the mNFATc1 protein. B and C, GST-fused proteins were expressed in E. coli and purified by a two-step purification, glutathione-Sepharose and then ion exchange chromatography (Mono Q column). Purified GST-JMJD5 and the indicated GST-fused substrate proteins were mixed and incubated together with 14C-radiolabeled 2-OG. 2-OG decarboxylation was measured by 14CO2 formation. Values are expressed as relative -fold induction compared with GST only. The error bars represent S.D. values from at least triplicate experiments. The p value for statistical difference was calculated using a two-tailed Student's t test and is indicated by asterisks. GST-fused proteins were visualized by Coomassie Brilliant Blue (CBB) staining. a.a, amino acids. Cn, calcineurin; RHD, Rel homology domain; IPT, immunoglobulin-like fold, Plexins, transcription factors.
Destabilization of NFATc1 by JMJD5 Mediates Ubiquitin-mediated Proteasomal Degradation
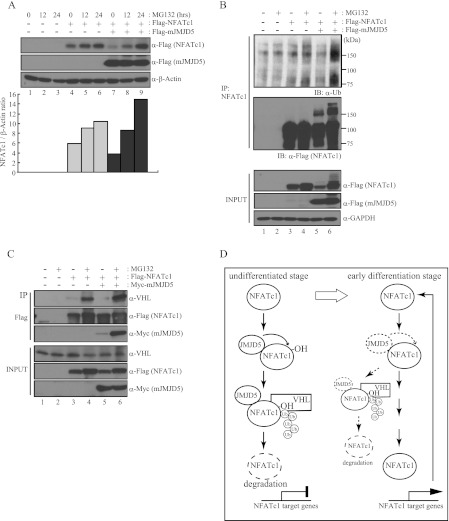
Here, we showed that JMJD5 destabilizes NFATc1 protein, presumably through its hydroxylase activity. To determine whether this process was related to protein degradation, we first examined the effect of a proteasome inhibitor (MG132). In HEK293T cells expressing exogenous NFATc1, NFATc1 protein did indeed accumulate in a time-dependent manner during MG132 treatment (Fig. 6A, lanes 4–6). Moreover, the reduced protein level of NFATc1 caused by co-expression of JMJD5 was completely reversed by MG132 treatment (Fig. 6A, lanes 7 and 8). We also confirmed in osteoclasts derived from bone marrow macrophages that endogenous NFATc1 protein accumulated during MG132 treatment to the same level as that seen when JMJD5 was knocked down (supplemental Fig. S8).
FIGURE 6.
JMJD5 induced ubiquitin-mediated proteasomal degradation of NFATc1. A, HEK293T cells were co-transfected with FLAG-NFATc1 and FLAG-mJMJD5 and then treated with MG132 (10 μm) for the indicated times. After 48 h of transfection, total cell lysates were prepared and analyzed by immunoblotting with anti-FLAG antibody. β-Actin was used as a loading control for immunoblotting. The signals obtained from the immunoblotting were quantified with the ImageJ program. B and C, HEK293T cells were co-transfected with FLAG-NFATc1 and Myc-mJMJD5 and then treated with MG132 (10 μm) for 16 h. After 48 h of transfection, total cell lysates were prepared and subjected to immunoprecipitation (IP) with an anti-NFATc1 antibody (B) or anti-FLAG M2-agarose (C). Bound proteins were detected by immunoblotting with the indicated antibodies. GAPDH was used as loading control for immunoblotting. D, suggested model.
Next, we examined whether the proteasomal degradation of NFATc1 was ubiquitin-dependent. Immunoprecipitation by anti-NFATc1 antibody was performed in HEK293T cells exogenously expressing NFATc1 and JMJD5 after which the ubiquitinated NFATc1 was detected with an anti-ubiquitin antibody. Ubiquitinated NFATc1 was barely detectable when MG132 was used (Fig. 6B, upper panel, lane 4), whereas it was significantly increased by co-expression of JMJD5 (Fig. 6B, upper panel, lane 6). In addition, when the immunoprecipitants were probed with anti-FLAG antibody, the band shifts representing ubiquitinated NFATc1 were made more evident by JMJD5 co-expression (Fig. 6B, middle panel, lanes 5 and 6). These findings suggest that JMJD5 facilitates the ubiquitin-dependent proteasomal proteolysis of NFATc1.
Because JMJD5 does not contain a specific domain to ubiquitinate, it appeared likely that ubiquitin ligases were required for NFATc1 degradation by JMJD5. Among them, we focused on VHL (a component of the multimeric E3 ubiquitin ligase) because this factor is known to directly bind to HIF1α protein through a hydroxylated proline residue on HIF1α for subsequent ubiquitination and proteasomal degradation (35–37). Therefore, we asked whether degradation of NFATc1 by JMJD5 was mediated by VHL. Total cellular extracts of HEK293T cells overexpressing NFATc1 and JMJD5 were immunoprecipitated with anti-FLAG antibody and probed with an anti-VHL antibody. VHL was visible when the cells were treated with MG132, indicating that it interacted with NFATc1 (Fig. 6C, lane 4). Furthermore, the interaction of NFATc1 with VHL was potentiated when JMJD5 was overexpressed (Fig. 6C, lane 6). Taken together, these results imply that JMJD5 facilitates the association of NFATc1 with VHL E3 ligase, and as a result, NFATc1 is prone to undergo ubiquitin-mediated proteasomal degradation (Fig. 6D).
DISCUSSION
JMJD5 Attenuates Osteoclastogenesis
In the present study, we showed that JMJD5 functions as a novel repressor for osteoclastogenesis. JMJD5 was found to serve as a protein hydroxylase for NFATc1, thereby facilitating NFATc1 protein degradation through association with VHL E3 ubiquitin ligase. This is a novel mechanism of regulating NFATc1 activity during osteoclastogenesis at a post-translational level. Interestingly, the expression level of JMJD5 was decreased the 1st day after RANKL treatment in contrast to NFATc1 expression during osteoclastic differentiation (Fig. 1A). It appears that JMJD5 functions to maintain the undifferentiated state of osteoclastic progenitor cells. Because JMJD5 appears to be a co-regulator of NFATc1 at the post-translational level, this factor represents a previously uncharacterized class of regulators in osteoclastogenesis. The present study also implies the possible presence of other co-regulators of NFATc1 at other levels. Additionally, it is quite conceivable from recent reports of epigenetic co-regulators of DNA-binding transcription factors that co-regulators of other important transcription factors directing osteoclastogenesis remain to be identified.
It is currently believed that epigenetic regulation through histone methylation/demethylation plays a central role in gene regulation during differentiation of many types of cells, presumably also bone cells. In particular, such enzymes (histone methyltransferases and demethylases) would transcriptionally co-regulate the function of transcription factors governing osteoclastic differentiation. For example, the JmjC domain-containing protein NO66 reportedly acts as a negative regulator of Osterix, an essential transcription factor in osteoblastogenesis (38). Consistent with biochemical characterization of NO66 as a direct interactant with Osterix, NO66 co-repressed the Osterix-mediated gene cascade through demethylation of active histone methylation marks (H3K4 and H3K36). Gene regulation by epigenetic marks is also evident during osteoclastic differentiation. Likewise, promoter activity of the Nfatc1 gene appears to be under negative control by methylation of histone H3K4me3 and H3K27me3 at an undifferentiated stage of osteoclasts (39). In contrast, the inactive methylation markers of H3K27me3 are removed for Nfatc1 gene induction along with RANKL-induced osteoclastogenesis (39). Therefore, it is quite conceivable that JmjC domain-containing proteins are critical for proper regulation of nuclear events during osteoclastic differentiation through histone demethylation.
JMJD5 Lacks Histone Demethylase Activity
It was recently reported that JMJD5 is a histone demethylase (34). However, in our experimental settings, no histone demethylase activity was detected for methylated H3K36 and the known methylated histone lysine residues (Fig. 2). Based on the abundant distribution of JMJD5 in the nucleoplasm, we next raised the possibility that its associating protein, NFATc1, is a substrate of JMJD5 demethylase. However, methylated lysine residues were not evident on NFATc1 protein when tested by immunoblotting with anti-Lys methylation antibody (data not shown). From the fact that JMJD5 is a member of the same JmjC domain-containing protein group as FIH1 and JMJD6 protein hydroxylases (24–27, 40), we next asked whether JMJD5 functioned as a hydroxylase of NFATc1. Indeed, hydroxylation of NFATc1 by JMJD5 was detected with an in vitro 2-OG decarboxylation assay (Fig. 5), suggesting that JMJD5 bears uncharacterized enzymatic function as an NFATc1 hydroxylase. As JMJD5 is found in the soluble nuclear fraction (supplemental Fig. 5), other non-histone proteins may be hydroxylation substrates for JMJD5.
These previous and present observations suggest that JMJD5 has dual functions, and the functions are very dependent on its cellular locations, functioning as a histone demethylase in chromatin and functioning as a protein hydroxylase in soluble nucleus. In our experimental system, although distribution of JMJD5 was detected in chromatin, it was more abundantly located in soluble nucleus (supplemental Fig. 5). Thus, we think that we could easily detect its function as a hydroxylase but not its histone demethylase activity, which may occur at a level below our detection.
Hydroxylation Is One Post-translational Modification Directing NFATc1 Functions
Post-translational modifications are important to regulate the function or expression of many kinds of proteins. It is well known that NFATc1 is modified by phosphorylation (41–43). In its phosphorylated form, NFATc1 is present in the cytoplasmic compartment and is imported into the nucleus when dephosphorylated by calcineurin. Recently, it was reported that NFATc1 is ubiquitinated in the cytoplasm for its degradation during late stages of osteoclastogenesis (19). However, it remained to be uncovered whether post-translational modifications regulated NFATc1 during osteoclastogenesis. In the present study, we provide evidence of a novel regulatory system for NFATc1 protein stability by post-translational modification. Specifically, NFATc1 is hydroxylated by JMJD5 and degraded via a ubiquitin-mediated proteasomal pathway. Hypoxia-specific transcription regulator HIF1α is constitutively synthesized at a high level even under normoxia. However, its level is repressed through hydroxylation on specific proline residues by prolyl hydroxylases and subsequent proteasomal degradation until hypoxia is induced. In this respect, it will be interesting to understand at the molecular level how JMJD5 enzymatic activity is controlled during osteoclastogenesis.
Supplementary Material
Acknowledgments
We thank Peter J. Ratcliffe and Ming Yang for expert technical advice concerning the 2-OG decarboxylation assay; E. Suzuki, Y. Okuno, K. Inoue, and I. Takada for helpful discussion; and M. Shibata for manuscript preparation.
This work was supported by a postdoctoral fellowship for foreign researchers (to M.-Y. Y.) and a grant for specially promoted research (to S. K.) from the Japan Society for the Promotion of Science and Grants-in-aid 23689066 and 23659712 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y. I.).

This article contains supplemental Experimental Procedures and Figs. S1–S8.
- RANKL
- receptor activation of nuclear factor κB ligand
- JmjC
- Jumonji C
- NFATc1
- nuclear factor of activated T-cells calcineurin-dependent 1
- RANK
- receptor activation of nuclear factor κB
- VHL
- Von Hippel-Lindau tumor suppressor
- DC-STAMP
- dendritic cell-specific transmembrane protein
- Acp5
- acid phosphatase 5, tartrate-resistant
- Ctsk
- cathepsin K
- FIH1
- factor inhibiting HIF1
- JMJD6
- Jumonji domain-6
- HIF1
- hypoxia-inducible factor 1
- 2-OG
- 2-oxoglutarate
- RE
- response element.
REFERENCES
- 1. Rodan G. A., Martin T. J. (2000) Therapeutic approaches to bone diseases. Science 289, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 2. Karsenty G., Wagner E. F. (2002) Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2, 389–406 [DOI] [PubMed] [Google Scholar]
- 3. Teitelbaum S. L., Ross F. P. (2003) Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 4. Baron R. (2004) Arming the osteoclast. Nat. Med. 10, 458–460 [DOI] [PubMed] [Google Scholar]
- 5. Novack D. V., Teitelbaum S. L. (2008) The osteoclast: friend or foe? Annu. Rev. Pathol. 3, 457–484 [DOI] [PubMed] [Google Scholar]
- 6. Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J. Q., Bonewald L. F., Kodama T., Wutz A., Wagner E. F., Penninger J. M., Takayanagi H. (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17, 1231–1234 [DOI] [PubMed] [Google Scholar]
- 7. Vignery A. (2000) Osteoclasts and giant cells: macrophage-macrophage fusion mechanism. Int. J. Exp. Pathol. 81, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bar-Shavit Z. (2007) The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J. Cell. Biochem. 102, 1130–1139 [DOI] [PubMed] [Google Scholar]
- 9. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 10. Hirotani H., Tuohy N. A., Woo J. T., Stern P. H., Clipstone N. A. (2004) The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J. Biol. Chem. 279, 13984–13992 [DOI] [PubMed] [Google Scholar]
- 11. Kim Y., Sato K., Asagiri M., Morita I., Soma K., Takayanagi H. (2005) Contribution of nuclear factor of activated T cells c1 to the transcriptional control of immunoreceptor osteoclast-associated receptor but not triggering receptor expressed by myeloid cells-2 during osteoclastogenesis. J. Biol. Chem. 280, 32905–32913 [DOI] [PubMed] [Google Scholar]
- 12. Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., Oike Y., Takeya M., Toyama Y., Suda T. (2005) DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrison P. R. (1990) Molecular mechanisms involved in the regulation of gene expression during cell differentiation and development. Immunol. Ser. 49, 411–464 [PubMed] [Google Scholar]
- 14. Ishida N., Hayashi K., Hoshijima M., Ogawa T., Koga S., Miyatake Y., Kumegawa M., Kimura T., Takeya T. (2002) Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 277, 41147–41156 [DOI] [PubMed] [Google Scholar]
- 15. Day C. J., Kim M. S., Stephens S. R., Simcock W. E., Aitken C. J., Nicholson G. C., Morrison N. A. (2004) Gene array identification of osteoclast genes: differential inhibition of osteoclastogenesis by cyclosporin A and granulocyte macrophage colony stimulating factor. J. Cell. Biochem. 91, 303–315 [DOI] [PubMed] [Google Scholar]
- 16. Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao B., Takami M., Yamada A., Wang X., Koga T., Hu X., Tamura T., Ozato K., Choi Y., Ivashkiv L. B., Takayanagi H., Kamijo R. (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nishikawa K., Nakashima T., Hayashi M., Fukunaga T., Kato S., Kodama T., Takahashi S., Calame K., Takayanagi H. (2010) Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J. H., Kim K., Jin H. M., Song I., Youn B. U., Lee S. H., Choi Y., Kim N. (2010) Negative feedback control of osteoclast formation through ubiquitin-mediated down-regulation of NFATc1. J. Biol. Chem. 285, 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 21. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 22. Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 23. Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 24. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., Ratcliffe P. J., Pugh C. W., Schofield C. J. (2002) Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 26. Cockman M. E., Lancaster D. E., Stolze I. P., Hewitson K. S., McDonough M. A., Coleman M. L., Coles C. H., Yu X., Hay R. T., Ley S. C., Pugh C. W., Oldham N. J., Masson N., Schofield C. J., Ratcliffe P. J. (2006) Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH). Proc. Natl. Acad. Sci. U.S.A. 103, 14767–14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Webby C. J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M. L., Schmitz C., Butler D. S., Yates J. R., 3rd, Delahunty C. M., Hahn P., Lengeling A., Mann M., Proudfoot N. J., Schofield C. J., Böttger A. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 [DOI] [PubMed] [Google Scholar]
- 28. Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C. A., Igarashi K., Kanno J., Brown M., Kato S. (2011) PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 13, 668–675 [DOI] [PubMed] [Google Scholar]
- 29. Youn M. Y., Takada I., Imai Y., Yasuda H., Kato S. (2010) Transcriptionally active nuclei are selective in mature multinucleated osteoclasts. Genes Cells 15, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 30. Miyamoto J., Matsumoto T., Shiina H., Inoue K., Takada I., Ito S., Itoh J., Minematsu T., Sato T., Yanase T., Nawata H., Osamura Y. R., Kato S. (2007) The pituitary function of androgen receptor constitutes a glucocorticoid production circuit. Mol. Cell. Biol. 27, 4807–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takada I., Mihara M., Suzawa M., Ohtake F., Kobayashi S., Igarashi M., Youn M. Y., Takeyama K., Nakamura T., Mezaki Y., Takezawa S., Yogiashi Y., Kitagawa H., Yamada G., Takada S., Minami Y., Shibuya H., Matsumoto K., Kato S. (2007) A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-γ transactivation. Nat. Cell Biol. 9, 1273–1285 [DOI] [PubMed] [Google Scholar]
- 32. Fujiki R., Chikanishi T., Hashiba W., Ito H., Takada I., Roeder R. G., Kitagawa H., Kato S. (2009) GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459, 455–459 [DOI] [PubMed] [Google Scholar]
- 33. Sawatsubashi S., Murata T., Lim J., Fujiki R., Ito S., Suzuki E., Tanabe M., Zhao Y., Kimura S., Fujiyama S., Ueda T., Umetsu D., Ito T., Takeyama K., Kato S. (2010) A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 24, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsia D. A., Tepper C. G., Pochampalli M. R., Hsia E. Y., Izumiya C., Huerta S. B., Wright M. E., Chen H. W., Kung H. J., Izumiya Y. (2010) KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 107, 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lisztwan J., Imbert G., Wirbelauer C., Gstaiger M., Krek W. (1999) The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13, 1822–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 37. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 38. Sinha K. M., Yasuda H., Coombes M. M., Dent S. Y., de Crombrugghe B. (2010) Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 29, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yasui T., Hirose J., Tsutsumi S., Nakamura K., Aburatani H., Tanaka S. (2011) Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J. Bone Miner. Res. 26, 2665–2671 [DOI] [PubMed] [Google Scholar]
- 40. Klose R. J., Kallin E. M., Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 41. Rao A., Luo C., Hogan P. G. (1997) Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 42. Porter C. M., Havens M. A., Clipstone N. A. (2000) Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 275, 3543–3551 [DOI] [PubMed] [Google Scholar]
- 43. Murphy L. L., Hughes C. C. (2002) Endothelial cells stimulate T cell NFAT nuclear translocation in the presence of cyclosporin A: involvement of the wnt/glycogen synthase kinase-3β pathway. J. Immunol. 169, 3717–3725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.