Background: Myo1a is an abundant membrane binding motor that targets to microvilli in intestinal epithelial cells.
Results: Myo1a interacts with acidic phospholipids using distinct motifs at the N and C terminus of the TH1 domain.
Conclusion: Membrane binding potential of Myo1a is distributed throughout TH1 rather than localized to a single motif.
Significance: Different class I myosins rely on different strategies for targeting to cellular membranes.
Keywords: Actin, Epithelial Cell, Membrane Lipids, Membrane Proteins, Myosin, Microvilli, Brush Border, Phosphatidylserine, Liposome, TIRF
Abstract
One of the most abundant components of the enterocyte brush border is the actin-based monomeric motor, myosin-1a (Myo1a). Within brush border microvilli, Myo1a carries out a number of critical functions at the interface between membrane and actin cytoskeleton. Proper physiological function of Myo1a depends on its ability to bind to microvillar membrane, an interaction mediated by a C-terminal tail homology 1 (TH1) domain. However, little is known about the mechanistic details of the Myo1a-TH1/membrane interaction. Structure-function analysis of Myo1a-TH1 targeting in epithelial cells revealed that an N-terminal motif conserved among class I myosins and a C-terminal motif unique to Myo1a-TH1 are both required for steady state microvillar enrichment. Purified Myo1a bound to liposomes composed of phosphatidylserine and phosphoinositol 4,5-bisphosphate, with moderate affinity in a charge-dependent manner. Additionally, peptides of the N- and C-terminal regions required for targeting were able to compete with Myo1a for binding to highly charged liposomes in vitro. Single molecule total internal reflection fluorescence microscopy showed that these motifs are also necessary for slowing the membrane detachment rate in cells. Finally, Myo1a-TH1 co-localized with both lactadherin-C2 (a phosphatidylserine-binding protein) and PLCδ1-PH (a phosphoinositol 4,5-bisphosphate-binding protein) in microvilli, but only lactaderin-C2 expression reduced brush border targeting of Myo1a-TH1. Together, our results suggest that Myo1a targeting to microvilli is driven by membrane binding potential that is distributed throughout TH1 rather than localized to a single motif. These data highlight the diversity of mechanisms that enable different class I myosins to target membranes in distinct biological contexts.
Introduction
Class I myosins are monomeric, membrane-binding, actin-based motor proteins that comprise a major fraction of the myosin superfamily (1). All myosin-1 isoforms contain at least three core domains: an N-terminal motor domain that coordinates ATP hydrolysis with actin binding and force generation; a central neck region made up of varying numbers of IQ motifs, which bind calmodulin or calmodulin-like proteins; and a tail region, which includes a highly basic C-terminal tail homology 1 (TH1)2 domain that is responsible for membrane binding (Fig. 1A) (2–5). Vertebrates express eight class I myosins, which are found in diverse cell types and tissues; because these motors simultaneously bind to actin and membrane, they provide a dynamic linkage between the cytoskeleton and the overlying plasma membrane in a variety of biological contexts (2–5). Although numerous studies have contributed to our understanding of the biochemical and mechanical properties of class I myosins, only recently have the detailed mechanisms of membrane binding become the focus of intense investigation (6–15).
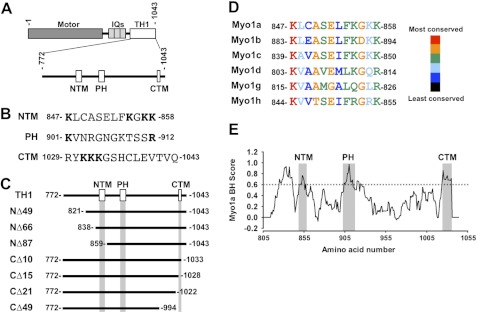
FIGURE 1.
Domain organization of Myo1a and constructs used in this study. A, bar diagram representing the structural domains of Myo1a and regions of interest in Myo1a-TH1. B, sequence from the NTM, PH domain, or CTM; residues in boldface type were mutated to alanine in structure-function studies of EGFP-Myo1a-TH1. C, bar diagrams representing EGFP-tagged Myo1a-TH1 N-terminal and C-terminal truncation constructs; boxes representing the NTM, PH, and CTM are drawn to scale. D, MegAlign alignment of the region representing the NTM from human short tailed class I myosins. E, a BH plot of the Myo1a-TH1 domain (amino acids 805–1043); regions corresponding to the NTM, PH domain, and CTM are indicated.
Myo1a is expressed in the enterocytes that line the small intestine, where it localizes to the apical brush border (16–18). This domain is defined by an extensive array of actin-supported membrane protrusions, referred to as microvilli, which extend into the lumen of the gut (19). In addition to maintaining brush border composition (20, 21) and structure (21), and regulating microvillar membrane tension (22), Myo1a also plays a role in powering the release of vesicles from the tips of microvilli, an activity that may hold important implications for gut host defense (23, 24). Although membrane binding is expected to be critical for all of these functions, little is known about the mechanisms underlying Myo1a membrane interactions. Previous studies with chicken Myo1a showed that TH1 is a bona fide membrane-binding domain that interacts with liposomes composed of negatively charged phospholipids with moderate affinity (25). Myo1a-TH1 is also sufficient for targeting to microvilli in cultured epithelial cells (26). However, the structural motifs that govern Myo1a-TH1 membrane binding and the lipid species relevant to these interactions in vivo have not been identified.
Reports published in the past several years have identified a myosin-1 pleckstrin homology (Myo1-PH) domain as a conserved region within the TH1 domain that may contribute to membrane binding (9–13). The Myo1-PH domain was originally identified in Myo1c, which binds stereospecifically to PI(4,5)P2 via interactions with two conserved signature basic residues that flank this motif (9, 10). When these signature residues are mutated to alanine in Myo1c, Myo1b, Myo1g, and Myo1f, targeting to the membrane is abolished (10–13). These studies highlight the importance of the Myo1-PH domain and, in some cases, specific interactions with phosphoinositides, such as PI(4,5)P2, in the membrane binding mechanism of these class I myosins. However, other characterized isoforms, such as Acanthamoeba Myo1c and vertebrate Myo1e, appear to target membranes using a mechanism that does not require stereospecific phosphoinositide recognition (8, 14); instead, these myosins appear to bind membranes through less specific electrostatic interactions with a variety of acidic phospholipids.
In this study, we used a cell-based targeting screen to identify putative membrane binding motifs in Myo1a-TH1. Microvillar targeting assays in cultured cells revealed that Myo1a-TH1 does not depend solely on the Myo1-PH domain for enrichment to microvilli. However, we identified two other regions that are essential for targeting to microvilli at steady state: an N-terminal targeting motif (NTM) that is conserved across short-tailed class I myosins and a C-terminal targeting motif (CTM) that is less well conserved. Cosedimentation studies revealed that purified Myo1a bound to liposomes composed of the acidic phospholipids PS and PI(4,5)P2 with moderate affinity in a charge-dependent manner. In vitro, NTM and CTM peptides were able to compete with Myo1a for binding to highly charged liposomes, confirming that the motifs identified in our cell-based targeting screen represent bona fide lipid binding regions. Single molecule TIRF microscopy assays in live cells indicate that mutations to either the NTM or the CTM reduce the Myo1a-TH1 membrane-bound lifetime. Finally, although Myo1a-TH1 co-localizes with both the PS-binding protein, lactadherin C2 (27, 28), and the PI(4,5)P2-binding protein, PLCδ1-PH (29, 30), only lactadherin C2 is able to compete with Myo1a for targeting to microvilli. Together, our results suggest that Myo1a targeting to microvilli is driven by membrane binding potential that is distributed throughout TH1 rather than localized to a single motif. Importantly, these findings suggest that different class I myosins may rely on distinct membrane-targeting mechanisms, which could provide a basis for the diversity of cellular functions carried out by these motors.
EXPERIMENTAL PROCEDURES
Molecular Biology
Truncation constructs and point mutants were cloned using the previously described pEGFP-tagged human Myo1a-TH1 (residues 772–1043) as a template (GenBankTM AF009961) (26); each new construct was confirmed by sequencing before expression in cells. Truncations included the following amino acids: NΔ49, residues 821–1043; NΔ66, residues 838–1043; NΔ87, residues 859–1043; CΔ49, residues 772–994; CΔ21, residues 772–1022; CΔ15, residues 772–1028; CΔ10, residues 772–1033. Point mutagenesis was performed using a QuikChange Lightning Mutagenesis Kit (Qiagen), and mutations were verified by sequencing. EGFP-lactadherin-C2, developed by the Grinstein laboratory (27), was obtained from Addgene. PLCδ1-PH-GFP was a generous gift from Tamas Balla (29, 30).
Cell Culture
LLC-PK1-CL4 (CL4) cells were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium with high glucose (Invitrogen), 2 mm l-glutamine (Invitrogen), and 10% defined fetal bovine serum (Atlantic Biosciences) as described previously (26). SF9 insect cells were cultured at 27 °C in Grace's insect medium (Invitrogen) with 1% pluronic F-68 solution (Sigma) and 1% antibiotic-antimycotic (Invitrogen).
Confocal Microscopy
CL4 cells were transfected using Lipofectamine 2000 according to the manufacturer's protocols (Invitrogen). After 4–5 h of incubation in transfection reagent with DNA, CL4 cells were processed for microscopy. For steady state targeting assays, cells were washed two times in warm PBS, fixed for 15 min in 4% paraformaldehyde, permeabilized for 6 min in 0.1% Triton X-100 (Sigma) in PBS, and stained with AlexaFluor 568-labeled phalloidin (Invitrogen) at 1:100 in PBS. For phosphatidylserine antibody labeling, cells were fixed as above, permeabilized for 10 min in 0.2% saponin in PBS, blocked in 5% BSA in PBS for 30 min, incubated in anti-PS (1:50; Millipore) overnight at 4 °C, washed three times for 5 min each in PBS, incubated at room temperature for 30 min in donkey anti-mouse Alexa 568 secondary antibody (1:200) (Invitrogen), and then washed again. Coverslips with fixed cells were mounted on slides using Prolong Gold with DAPI (Invitrogen) and sealed using clear nail polish. Cells were imaged using a Leica TCS SP5 laser-scanning confocal microscope equipped with ×63 and ×100 objectives. Images were smoothed, contrast-enhanced, and pseudocolored using ImageJ version 1.43u (National Institutes of Health).
Fold Enrichment in Microvilli Quantification
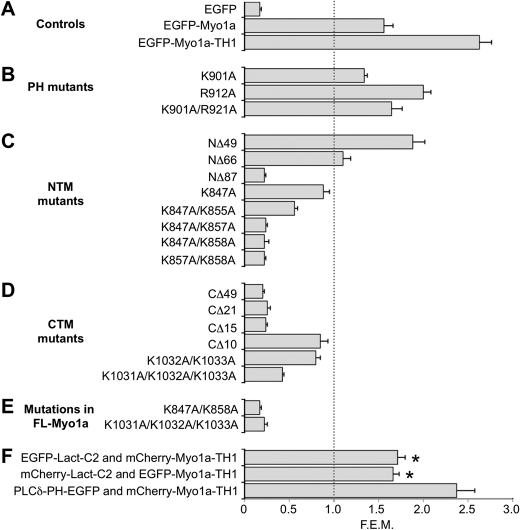
x-z (i.e. vertical) section images were used to quantify EGFP-Myo1a-TH1 construct targeting to microvilli. For each construct, 6–13 cells from 2–4 experiments were analyzed to obtain “-fold enrichment in microvilli” (FEM) values. FEM values were generated as follows: for a given cell, microvilli were first localized using the F-actin (phalloidin) channel. In the corresponding EGFP channel image, the intensity of Myo1a-TH1 was measured at 5–7 points that mapped to the position of microvillar F-actin labeling. Another 5–7 intensity points were acquired from a region that corresponded to non-nuclear cytosol. Intensity values from microvillar and cytosolic regions were averaged separately, and the resulting means were used to obtain microvilli/cytosol intensity ratios (i.e. FEM). Errors for FEM values were calculated as S.D./√n, where n corresponds to the number of averaged FEM values for a given construct. A FEM value of 1 (see Fig. 3, dashed line) is the enrichment threshold, the value for a protein that partitions equally between the microvilli and cytosol (11). Constructs with FEM values above this threshold are enriched in microvilli, whereas constructs with FEM values below 1 are enriched in the cytosol. Similar quantification methods were used previously to assay the structural basis of membrane enrichment for Myo1g (11), mDia2 (31), and lactadherin-C2 (32).
FIGURE 3.
FEM for various Myo1a-TH1 mutant constructs. Control constructs (A), EGFP-Myo1a-TH1 constructs with point mutations to the PH region (B), EGFP-Myo1a-TH1 constructs with truncations or mutations to the NTM (C) or CTM (D), or EGFP-Myo1a full-length molecules with mutations to the key lysine residues identified in TH1 (E) were expressed individually in CL4 cells and analyzed for microvillar targeting. F, Myo1a-TH1 was coexpressed with Lact-C2 or PLCδ1-PH, and TH1 targeting was quantified in the presence of each construct. Lact-C2, but not PLCδ1-PH, reduced TH1 targeting to microvilli. *, p ≪ 0.0001 versus EGFP-Myo1a-TH1 alone. Plotted FEM values represent the average microvillar intensity/cytosolic intensity ratio. The gray dashed line represents the 1:1 enrichment threshold; constructs with FEM values below this level are not enriched in microvilli. Error bars, S.E.
Live Cell Single Molecule TIRF Microscopy
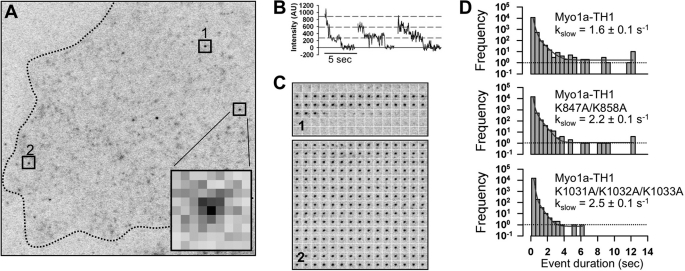
Myo1a-TH1 and selected Myo1a-TH1 mutants were tagged with three tandem copies of mCitrine (3x-mCitrine) using a vector previously described (a generous gift from K. Verhey) (33) and verified by sequencing. CL4 cells expressing 3x-mCitrine, 3x-mCitrine-Myo1a-TH1, 3x-mCitrine-Myo1a-TH1-K847A/K858A, or 3x-mCitrine-Myo1a-TH1-K1031A/K1032A/K1033A were plated sparsely in glass bottom imaging dishes. Cells were gently washed two times in Ringer's buffer (10 mm HEPES/KOH, 155 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 2 mm NaH2PO4, 10 mm glucose, pH 7.2) (33, 34) and kept in the same buffer for imaging experiments. TIRF microscopy was performed on a Nikon TiE inverted light microscope equipped with a Nikon TIRF illuminator, a ×100/1.49 numerical aperture TIRF objective (used in combination with a ×1.5 optivar), a Hamamatsu ImagEM-CCD camera, and MetaMorph software to control image acquisition. Calibrated pixel size was 75 nm/pixel. Fluorescence was excited using a 50-milliwatt 491-nm diode laser operating between 20 and 30% of maximum power. Single molecule membrane binding events appeared as bright diffraction limited spots on the ventral surface of the cell. The presence of tandem fluorescent proteins was confirmed by observations of multistep photobleaching at high laser power (see Fig. 6B). Cells expressing exceedingly low levels of each construct were chosen for analysis to improve the signal/noise ratio of single molecule detection; image stacks consisting of 250 frames (12.5 s) were captured at 20 frames/s and then subject to contrast enhancement and two-frame rolling average using ImageJ. Image stacks were then processed using DiaTrack (35) to extract the lifetimes of spots with intensities above background, which typically exhibited a grayscale value of 70–80. Using a TIRF system such as this, spot lifetimes will be limited by the detachment rate of 3x-mCitrine fusion proteins or the photobleaching of mCitrine fluorescent proteins. Because photobleaching is a stochastic event, the likelihood of observing a simultaneous triple photobleaching event is small (33, 34). Moreover, imaging was carried out at reduced laser power to minimize the impact of mCitrine photobleaching on observed spot lifetimes (i.e. kinetics were dominated by detachment from the membrane rather than photobleaching). In support of this point, we frequently observed events that persisted for the full 12.5-s recording (see Fig. 6C, 2). Importantly, long lived events were difficult to resolve in cells expressing 3x-mCitrine empty vector (supplemental Movie S1).
FIGURE 6.
Single molecule TIRF microscopy reveals that the NTM and CTM contribute to long lived membrane binding events. A, a single time point from a representative 250-frame (12.5-s) image stack of a single CL4 cell expressing low levels of 3x-mCitrine-Myo1a-TH1. The dashed line traces the periphery of the cell. Inset, enlarged diffraction-limited spot representing a single molecule binding event (75 nm/pixel). B, multistep photobleaching is observed for individual 3x-mCitrine molecules. C, montage of individual frames demonstrate the lifetimes of single molecules highlighted in A (see boxes 1 and 2). D, unitary membrane binding event lifetime distribution for 3x-mCitrine-Myo1a-TH1, 3x-mCitrine-Myo1a-TH1-K847A/K858A, and 3x-mCitrine-Myo1a-TH1-K1031A/K1032A/K1033A (KKK>AAA). The solid lines show fits to a biexponential decay; slow rate constants ± S.E. of the fit parameter are given in each case.
Protein Purification
Full-length Myo1a tagged with a Myc-FLAG epitope (36) and calmodulin (CaMΔall, a generous gift from K. Trybus) (37), were coexpressed in SF9 insect cells, and Myo1a-Myc-FLAG was purified using affinity chromatography as described previously (36). Briefly, infected cells were collected by centrifugation at 300 × g and lysed using a Dounce homogenizer in lysis buffer (10 mm Tris-Cl, pH 7.5, 200 mm KCl, 1 mm EGTA, 1 mm EDTA, 1 mm MgCl2, 2 mm ATP, 0.1% Nonidet P-40, 5% sucrose, 1 mm dithiothreitol, pefabloc, leupeptin, phenylmethanesulfonyl fluoride). Lysate was then centrifuged at 186,000 × g in a Type 45 Ti rotor using a Beckman ultracentrifuge. Myo1a-Myc-FLAG was collected from the resulting supernatant using immobilized M2 FLAG resin (Sigma), washed with 25 column volumes of wash buffer (10 mm Tris-Cl, pH 7.5, 200 mm KCl, 1 mm EGTA, 1 mm EDTA, 1 mm MgCl2, 2 mm ATP, 1 mm dithiothreitol, pefabloc, leupeptin, phenylmethanesulfonyl fluoride) containing 5 μg/ml purified bovine calmodulin (Calbiochem), eluted using FLAG peptide (Sigma) in calmodulin-free wash buffer, and dialyzed overnight into binding buffer (10 mm HEPES, 100 mm KCl, 1 mm EGTA). Purified protein was either flash frozen and stored at −80 °C or stored on ice for up to 1 week.
Liposome Preparation
Liposomes were prepared by extrusion according to slightly modified published protocols (8, 9, 38). Lipids (1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoenthanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), and PI(4,5)P2; Avanti Polar Lipids) in ChCl3 were mixed in glass vials to desired concentrations and dried under a stream of nitrogen with gentle vortexing. Lipid mixtures were resuspended in binding buffer and subjected to five freeze-thaw cycles. We noted that sucrose-loaded liposomes, which are frequently used for cosedimentation studies, appeared multilamellar by negative stain electron microscopy. Additionally, sucrose-loaded liposomes require dialysis to remove sucrose from the resuspension buffer, which resulted in an extensive loss of liposome product. Therefore, liposomes for cosedimentation experiments were prepared in the absence of sucrose. Mixtures were then pushed through an extruder fit with a 100-nm filter 16–20 times. Total loss of lipid during the extrusion process was calculated in control experiments using liposomes containing 2% β-BODIPY-500/510 hexadecanoyl-sn-glycero-3-phosphate (Invitrogen), and final concentrations were adjusted accordingly. Liposomes were stored on ice and used for up to 3 days.
Liposome Binding Assays
Cosedimentation assays for measuring Myo1a binding to liposomes were performed as described previously (13). To clear any protein aggregates, purified Myo1a (2–5 μm) was centrifuged at 4 °C for 30 min at 352,000 × g using a TL100 Beckman centrifuge and a TLA100 rotor. Centrifuge tubes were precoated with 100 μm DOPC and 100 μg/ml BSA. Myo1a-Myc-FLAG (50 nm) was then incubated at room temperature with increasing concentrations (0–100 μm) of liposomes composed of varying mol % of either DOPC/DOPS, DOPC/DOPE, or DOPC/DOPE with PI(4,5)P2 in a final volume of 200 μl of buffer. After incubation, liposome-protein complexes were centrifuged at 352,000 × g for 30 min at 25 °C. Then 170 μl were removed, and the remaining 30-μl “pellet” was resuspended in 10 μl of boiling sample buffer. Although liposomes were not sucrose-loaded, using fluorescent liposomes, we determined >90% of the liposomes sedimented under these conditions. Protein was analyzed by SDS-PAGE and Coomassie Blue (Bio-Rad) staining and then visualized with an Odyssey infrared imaging system (LI-COR Biosciences). All individual data points were normalized to the percentage of Myo1a bound to 100 μm 80% DOPS and are expressed as a function of lipid concentration. Data points are averages of 2–6 experiments. Competition experiments were performed using liposomes composed of 20% DOPC, 80% DOPS (50 μm). Peptides (GenScript) corresponding to the NTM (KLCASELFKGKKASYPQSVP, residues 847–866), the CTM (GDNSKLRYKKKGSHCLEVTV, residues 1023–1042), or a region from the motor domain (negative control, LLEGSVGVEDLVLLEPLVEE, residues 3–22) at varying concentrations (25–400 μm) were incubated with liposomes and Myo1a and then processed as described above. The total percentage of bound Myo1a in the presence of control peptide was equal to the percentage bound in the absence of peptide; therefore, for each sample containing NTM or CTM peptide, the percentage of Myo1a bound was calculated using the control peptide condition as 100% bound. Binding data were fit to hyperbolae using SigmaPlot (Systat Software Inc.).
RESULTS
Dissecting Mechanism of Myo1a-TH1 Targeting in Cultured Epithelial Cells
Our initial goal was to identify regions within Myo1a-TH1 (defined here as amino acids 772–1043 of human Myo1a) that are needed for targeting to brush border microvilli. Targeting is expected to be at least partially driven by direct interactions with the apical membrane (25). We reasoned that once critical targeting motifs were identified, we could then use other more direct in vitro biochemical methods to assess their contribution to membrane binding. To this end, we developed a microvillar targeting assay using the CL4 epithelial cell model system (26). CL4 cells polarize quickly and build a brush border with densely packed apical microvilli; they are also highly transfectable, which makes them ideal for large scale structure-function studies and for screening numerous Myo1a-TH1 mutants and truncation constructs. Previous studies employed CL4 cells successfully to investigate the targeting determinants of full-length Myo1a and other microvillar components (26, 39, 40).
Using confocal microscopy, we acquired x-y and x-z images of cells expressing EGFP-tagged Myo1a-TH1 constructs. In the x-y orientation, constructs that target to microvilli appear punctate in the apical domain. Images in the x-z plane provided simultaneous access to microvillar and cytosolic intensity levels and were used to quantify FEM, the ratio of microvillar/cytosolic signal. The lower limit for enrichment, or FEM value for a protein that partitions equally between microvilli and cytosol, is 1 (see Fig. 3, dashed line) (11). As controls, we expressed EGFP-Myo1a, EGFP-Myo1a-TH1, and EGFP in CL4 cells and analyzed localization (see Fig. 2, A–C). EGFP-Myo1a and EGFP-Myo1a-TH1 were both enriched in microvilli as reported previously (26) (FEM = 1.56 ± 0.10, 2.63 ± 0.14, respectively; see Fig. 3A), whereas EGFP was soluble and showed no microvillar targeting (FEM = 0.17 ± 0.02; see Fig. 3A). To further validate this approach, we examined the FEM scores of other proteins that are expected to be enriched in the brush border. For example, F-actin, which forms the core of the microvillus, exhibits extremely high enrichment in this assay (FEM = 4.14 ± 0.36). Moreover, the PLCδ1-PH domain was also highly enriched in microvilli (FEM = 2.61 ± 0.21), whereas a mutant deficient in membrane-binding, PLCδ1-PH-R40L, was not (FEM = 0.47 ± 0.02) (supplemental Fig. S1) (41, 42). Thus, the FEM assay provides a simple and robust readout on the microvillar enrichment of constructs of interest.
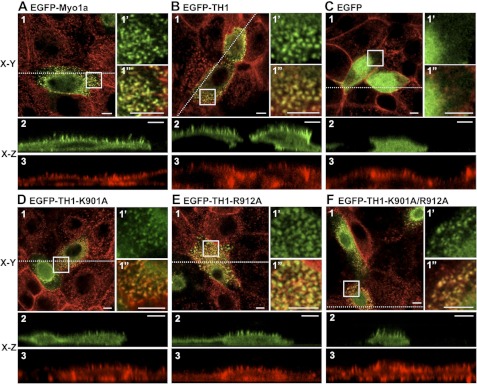
FIGURE 2.
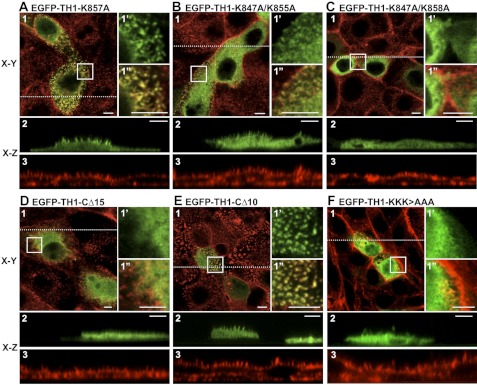
Analysis of EGFP-Myo1a, EGFP-Myo1a-TH1, and EGFP-Myo1a-TH1-PH domain mutant targeting to microvilli. A–C, representative confocal images show that EGFP-Myo1a (A) and EGFP-Myo1a-TH1 (B) are enriched in microvilli, but EGFP (C) is soluble. D–F, representative confocal images show that EGFP-Myo1a-TH1-K901A (D), EGFP-Myo1a-TH1-R912A (E), and EGFP-Myo1a-TH1-K901A/R912A (F) are enriched in microvilli. Here, 1 represents an apical en face view of EGFP-tagged construct (green) merged with Alexa568-phalloidin-labeled F-actin (red); the boxed area in 1 is shown in 1′ (EGFP or EGFP-tagged construct) and 1″ (merge with phalloidin). x-z (vertical) sections show EGFP-tagged constructs (green) (2) or F-actin (phalloidin; red) (3). The dashed line in 1 represents the location in the cell where the x-z section was imaged. Bars, 5 μm.
Myo1-PH Domain Is Not Essential for Enrichment of Myo1a-TH1 in Microvilli
Previous studies on multiple vertebrate class I myosins show that when the signature basic residues in the Myo1-PH are mutated to alanine, targeting to membrane is abolished (10–13). To determine if the Myo1-PH was necessary for Myo1a-TH1 targeting, we mutated the equivalent conserved residues, Lys-901 and Arg-912, to alanine (Fig. 1, A and B) and expressed the mutant constructs in CL4 cells. Although Myo1-PH point mutants K901A and R912A and double point mutant K901A/R912A reduced microvillar targeting of EGFP-Myo1a-TH1, all three constructs were still above the enrichment threshold (FEM = 1.34 ± 0.03, 2.00 ± 0.08, and 1.64 ± 0.12, respectively; Figs. 2, D–F, and 3B). This result is reminiscent of experiments with vertebrate Myo1e and Acanthamoeba Myo1c, which revealed that mutations to signature PH residues do not significantly impact subcellular targeting (14) or binding to liposomes in vitro (8, 14). From these experiments, we conclude that a functional Myo1-PH is not necessary for Myo1a-TH1 enrichment in microvilli.
Discovery of Novel Microvillar Targeting Motifs in Myo1a-TH1
Because mutations to the signature PH domain residues in Myo1a-TH1 did not abolish the microvillar enrichment, we sought to identify other motifs that might contribute to the targeting of Myo1a to the brush border. To this end, we truncated Myo1a-TH1 from the N terminus (Fig. 1C), expressed EGFP-tagged truncation constructs in CL4 cells, and assessed microvillar targeting. EGFP-Myo1a-TH1-NΔ49 and EGFP-Myo1a-TH1-NΔ66 both targeted at levels above the enrichment threshold (FEM = 1.88 ± 0.13, 1.10 ± 0.08, respectively; Fig. 3C). However, when we further truncated Myo1a-TH1 by an additional 20 amino acids (EGFP-Myo1a-TH1-NΔ87), targeting to microvilli was completely lost (FEM = 0.22 ± 0.05; Fig. 3C). We compared the Myo1a-TH1 primary sequence with other short tailed myosin-1 TH1 domains to identify conserved residues between NΔ66 and NΔ87 that might contribute to targeting. We found several residues that appear conserved from humans to Dictyostelium (Fig. 1D and supplemental Fig. S2A). We also used the basic-hydrophobic (BH) scale to examine the predicted membrane binding potential of this region (7). Recent development of the BH algorithm followed the identification of a potentially unstructured non-PH domain membrane-binding site, rich in basic and hydrophobic residues, in the Acanthamoeba Myo1c TH1 domain (8). BH analysis of Myo1a-TH1 revealed a moderate peak corresponding to this region (Fig. 1E). In light of these analyses, we chose to focus additional mutagenesis on the basic residues in this region for two reasons: (i) recent studies show that in the context of Myo1g, the conserved basic residues that flank this region are required for membrane targeting in Jurkat cells (11), and (ii) early studies with chicken Myo1a determined that binding to charged liposomes is attenuated as ionic strength is increased, implying an electrostatic mechanism (25). We made single or double alanine substitutions to conserved lysine residues Lys-847, Lys-855, Lys-857, and Lys-858 in order to determine their significance in Myo1a-TH1 targeting (Fig. 1B). The single point mutant K847A had a minor impact on enrichment (FEM = 0.88 ± 0.07), whereas K847A/K855A and K847A/K858A abolished targeting to microvilli (FEM = 0.56 ± 0.03 and 0.22 ± 0.02, respectively; Figs. 3C and 4, A–C). We refer to this region here as the N-terminal targeting motif (NTM; Myo1a-TH1 residues 847–858).
FIGURE 4.
NTM and CTM are required for Myo1a-TH1 targeting to microvilli. A–C, representative confocal images of a subset of expressed NTM mutant constructs. EGFP-Myo1a-TH1-K857A is enriched in microvilli (A), whereas EGFP-Myo1a-TH1-K847A/K855A (B) and EGFP-Myo1a-TH1-K847A/K858A (C) do not target to microvilli. D–F, representative confocal images of a subset of expressed CTM mutant constructs. EGFP-Myo1a-TH1-CΔ15 does not target to microvilli (D), whereas EGFP-Myo1a-TH1-CΔ10 exhibits reduced enrichment (E). F, a construct with alanine mutations to the three lysine residues in the CTM, EGFP-Myo1a-TH1-K1031A/K1032A/K1033A (KKK>AAA), does not localize to microvilli. Here, 1 represents an apical, en face view of EGFP-tagged construct (green) merged with Alexa568-phalloidin labeled F-actin (red); the boxed area in 1 is shown in 1′ (EGFP or EGFP-tagged construct) and 1″ (merge with phalloidin). x-z sections show EGFP-tagged constructs (green) (2) or F-actin (phalloidin; red) (3). The dashed line in 1 represents the location in the cell where the x-z section was imaged. Bars, 5 μm.
To explore targeting contributions from the C terminus of Myo1a-TH1, we carried out a similar truncation analysis on this end of the molecule (Fig. 1C). Truncations CΔ49 and CΔ21 both abolished microvillar targeting (FEM = 0.20 ± 0.02 and 0.25 ± 0.03, respectively; Fig. 3D). Interestingly, the region encompassing the last 15 residues of Myo1a-TH1 scored highly on the BH scale (Fig. 1E). Deletion of the C-terminal hydrophobic residues in this region (CΔ10; Fig. 1C) reduced targeting to just below the enrichment threshold (FEM = 0.85 ± 0.08; Figs. 3D and 4, D–F). Further truncation, which removed a cluster of four basic residues (CΔ15; Fig. 1C), abolished targeting (FEM = 0.24 ± 0.02; Figs. 3D and 4, D–F). For this reason, we focused site-directed mutagenesis on these residues, making alanine substitutions to one, two, or three of the lysines in this region. Only the triple point mutation, K1031A/K1032A/K1033A (Fig. 1B), abolished enrichment of EGFP-Myo1a-TH1 in microvilli (FEM = 0.42 ± 0.02; Figs. 3D and 4, D–F). Sequence alignments revealed that the C-terminal region is well conserved among vertebrate Myo1a isoforms but not class I myosins in general (supplemental Fig. S2B). Thus, these experiments identify a second region required for Myo1a-TH1 microvillar targeting, the C-terminal targeting motif (CTM; Myo1a-TH1 residues 1029–1043), which appears unique to Myo1a (supplemental Fig. S2B).
Whereas the structure-function studies described above were carried out using the Myo1a-TH1, we next sought to determine if the targeting motifs identified with this approach also played a role in the microvillar enrichment of full-length Myo1a (FL-Myo1a). We created EGFP-tagged variants of FL-Myo1a-expressing mutations in the NTM or CTM, which eliminated Myo1a-TH1 enrichment in microvilli (K847A/K858A or K1031A/K1032A/K1033A, respectively; Fig. 1B). FEM analysis of both mutants revealed major reductions in steady state microvillar targeting (FEM = 0.17 ± 0.02 and 0.22 ± 0.03, respectively; Fig. 3E). This suggests that similar to Myo1a-TH1, FL-Myo1a also targets to the microvillus using a mechanism that involves the NTM and CTM.
Myo1a Binds to Acidic Phospholipids in Vitro
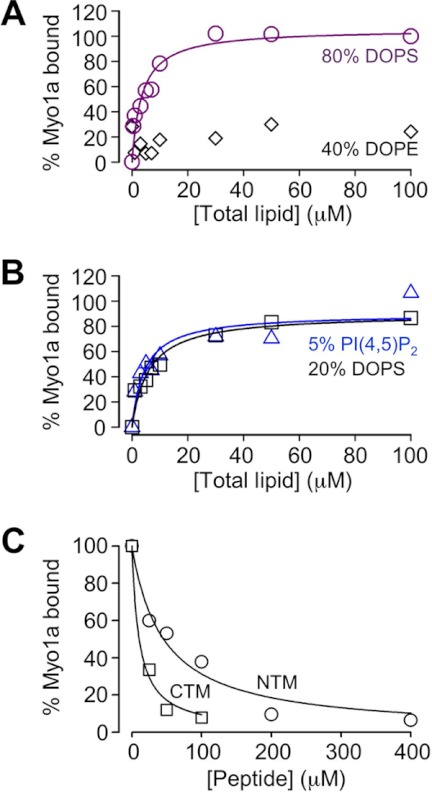
Previous studies have shown that several class I myosins from lower and higher eukaryotes, including chicken Myo1a (25), are bona fide membrane-binding proteins (5). We sought to determine if this is also the case for human Myo1a. To this end, we examined the lipid binding ability of purified full-length human Myo1a using co-sedimentation assays (Fig. 5, A and B). Myo1a bound with moderate affinity to liposomes composed of DOPS at a physiologically relevant concentration (20% DOPS; KD = 6.0 ± 1.4 μm; Fig. 5B). Increasing the concentration of DOPS 4-fold increased the affinity slightly (80% DOPS; KD = 3.6 ± 0.9 μm; Fig. 5A). Moreover, liposomes containing 5% PI(4,5)P2 produced binding similar to that observed with 20% DOPS vesicles, which are equivalent in total charge (Fig. 5B). No significant binding was observed with liposomes containing 2% PI(4,5)P2 (supplemental Fig. S3). These results suggest that, in contrast to vertebrate Myo1b and Myo1c (9, 13), Myo1a does not demonstrate a binding preference for PI(4,5)P2. Instead, Myo1a probably binds to membranes via non-stereospecific electrostatic interactions driven by the density of negative charge, similar to vertebrate Myo1e and Acanthamoeba Myo1c (8, 14).
FIGURE 5.
NTM and CTM compete with Myo1a for binding to negatively charged liposomes in vitro. A, cosedimentation assays using Myo1a-Myc-FLAG (50 nm) and liposomes composed of varying lipid species (0–100 μm). Myo1a binds to liposomes composed of 20% DOPC, 80% DOPS with moderate affinity (purple symbols and line; KD = 3.6 ± 0.9 μm). Additionally, Myo1a did not show significant binding to liposomes composed of 60% DOPC, 40% DOPE (black diamonds). B, Myo1a bound similarly to liposomes composed of either 40% DOPE, 55% DOPC, and 5% PI(4,5)P2 (blue symbols and line; KD = 4.5 ± 1.7 μm) or 80% DOPC, 20% DOPS (black symbols and line; KD = 6.0 ± 1.4 μm), suggesting that the interaction between Myo1a and membrane is electrostatic rather than stereospecific. Data were normalized to the percentage bound value for 100 μm 80% DOPS and fit to hyperbolae using SigmaPlot. C, peptides corresponding to a control sequence from the Myo1a motor domain, the NTM, or the CTM were incubated with 50 nm Myo1a-Myc-FLAG and 50 μm liposomes composed of 80% DOPS, 20% DOPC. In the presence of NTM or CTM peptides, but not the control peptide, Myo1a binding to liposomes was significantly reduced. The percentage of bound Myo1a was calculated by dividing total bound Myo1a for each condition by total bound in the presence of control peptide. Data are averages from two (NTM peptide, 200 and 400 μm) or three independent experiments. Fits to hyperbolic decays yield IC50 values of 47 ± 9 μm for the NTM and 10 ± 2 μm for the CTM. Errors represent S.E. values of the fit parameter.
NTM and CTM Peptides Compete with Myo1a for Binding to Acidic Phospholipids in Vitro
Because mutations to NTM and CTM basic residues abolished Myo1a-TH1 and FL-Myo1a targeting to microvilli and our co-sedimentation data indicate that human Myo1a binds liposomes containing acidic phospholipids in vitro, we sought to determine if either of these newly discovered motifs possessed membrane binding activity. Purified Myo1a was mixed with 50 μm liposomes composed of 20% DOPC, 80% DOPS in the presence of NTM peptide, CTM peptide, or a peptide of comparable length obtained from a random region in the motor domain (negative control); co-sedimentation was used to assess the extent of Myo1a liposome binding in each reaction. Although Myo1a binding was unaffected by the control peptide, binding decreased in a dose-dependent manner when NTM or CTM peptides were included in reactions (Fig. 5B). Interestingly, the CTM peptide was a more potent inhibitor of Myo1a liposome binding (IC50 = 10 μm) compared with the NTM (IC50 = 47 μm), but this was not due to a higher charge density because both NTM and CTM peptides have a pI of ∼9.5. These results indicate that both NTM and CTM peptides interact directly with membranes composed of acidic phospholipids and further suggest that these motifs are capable of mediating Myo1a membrane binding in cells.
NTM and CTM Regulate Myo1a-TH1 Membrane Binding Dynamics in Live Cells
If the NTM and CTM are capable of binding directly to membrane lipids, mutations to these motifs probably impact steady state microvillar enrichment of Myo1a (Figs. 3 and 4) by reducing the amount of time this molecule spends in a membrane-bound state. To explore this possibility, we used single molecule TIRF microscopy to measure the lifetimes of Myo1a-TH1 molecules bound to the plasma membrane of live cells. For these studies, we tagged Myo1a-TH1 and two non-targeting Myo1a-TH1 mutants (K847A/K858A and K1031A/K1032A/K1033A) with three tandem copies of mCitrine (3x-mCitrine) and expressed these constructs in CL4 cells. Previous studies exploited this approach to examine the dynamics and directed movement of single kinesin molecules in live cells (33, 34). TIRF imaging was performed on the ventral surface of sparsely plated, unpolarized CL4 cells. Observing cells with extremely low expression levels of 3x-mCitrine-tagged constructs allowed us to image single molecule binding events, which appeared as bright diffraction-limited spots on the ventral cell surface (Fig. 6A, inset, and supplemental Movies S2–S4). Fluorescence spots in image stacks were tracked using a FIONA-based algorithm, which enabled us to measure membrane-bound lifetimes and assemble lifetime distributions (Fig. 6D) (43). Each record yielded thousands of individual binding events, the vast majority of which were rapid collisions between fluorescent molecules and the ventral cell surface. Single molecule lifetime measurements for 3x-mCitrine-Myo1a-TH1 revealed event durations that ranged from 100 ms (the shortest detectable event) to 12.5 s (the entire 250-frame record) (Fig. 6D). The longest events (>10 s) are consistent with lifetime estimates from previous studies (6, 26). We assumed that lifetime distributions consisted of two types of events: (i) short lived nonspecific interactions (fast component), and (ii) longer lived “productive” membrane binding events (slow component). Thus, fitting lifetime distributions to a biexponential decay revealed that Myo1a-TH1-K847A/K858A and Myo1a-TH1-K1031A/K1032A/K1033A both showed an increase in the rate of the slow component (kslow = 2.2 ± 0.1 s−1 and 2.5 ± 0.1 s−1, respectively; Fig. 6D) relative to 3x-mCitrine-Myo1a-TH1 (kslow = 1.6 ± 0.1 s−1; Fig. 6D). Other poorly targeting mutants with low FEM values (CΔ10 and K847A/K855A) produced similar results (data not shown). These experiments revealed that the NTM and the CTM probably contribute to membrane enrichment of Myo1a by slowing the rate at which Myo1a-TH1 detaches from the membrane.
Myo1a-TH1 Interacts with PS in Brush Border Microvilli
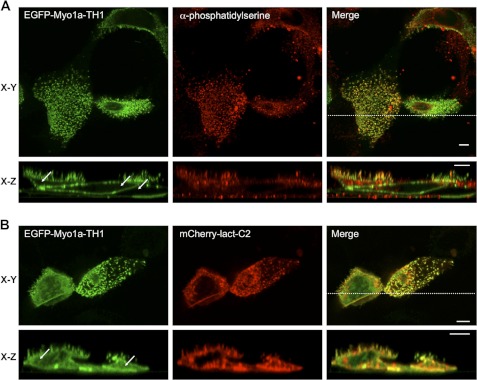
Our in vitro binding studies suggests that an electrostatic interaction with the plasma membrane may be the primary mechanism for Myo1a targeting to microvilli. Myo1a bound DOPS-containing liposomes in vitro, and PS is one of the most abundant negatively charged lipid species found in the inner leaflet of the plasma membrane (44, 45). Thus, we sought to determine if PS colocalizes with Myo1a-TH1 in microvilli. As probes for observing the PS distribution, we used an antibody raised against PS and the PS sensor, lactadherin-C2 (Lact-C2). Lact-C2 is a PS-specific binding module and has been used previously to analyze PS localization in cells (27, 28, 46). Both PS probes revealed that Myo1a-TH1 and PS colocalized in microvilli (Fig. 7). Whereas the anti-PS probe exhibited strong microvillar labeling with some minor staining of subapical material (probably biosynthetic vesicles; Fig. 7A), Lact-C2 targeted to plasma membrane and intracellular vesicles throughout the cell as reported previously (46). The wider distribution of the Lact-C2 probe is most likely due to the fact that the construct is overexpressed under these conditions. Surprisingly, these studies also revealed that in the presence of Lact-C2, Myo1a-TH1 appeared much more soluble (Fig. 7, compare arrows in A and B, EGFP x-z sections). FEM analysis of Myo1a-TH1 in cells coexpressing Lact-C2 revealed a significant decrease in microvillar enrichment, which was independent of the fluorescent protein tag used to visualize these molecules (EGFP-Myo1a-TH1, FEM = 1.66 ± 0.07; mCherry-Myo1a-TH1, FEM = 1.71 ± 0.08; Fig. 3F). Moreover, this effect was specific to the PS-binding protein; although Myo1a-TH1 co-localized with the PI(4,5)P2-binding protein, PLCδ1-PH, microvillar enrichment of Myo1a-TH1 was not impacted by coexpression of this probe (FEM = 2.37 ± 0.20; Fig. 3F) despite the fact that PLCδ1-PH appears to bind liposomes composed of 2% PI(4,5)P2 with higher affinity than Myo1a (supplemental Fig. S3). These data suggest that PI(4,5)P2 in the apical membrane makes only a minor contribution to Myo1a enrichment in microvilli. Instead, Myo1a-TH1 probably binds to PS in the brush border, and targeting of Myo1a to this subcellular domain is at least partially driven by this interaction.
FIGURE 7.
Myo1a-TH1 interacts with PS in microvilli. EGFP-Myo1a-TH1 colocalizes with antibody-stained PS (A) and coexpressed PS-binding protein, mCherry-Lact-C2 (B), in microvilli. The cytosolic pool of EGFP-Myo1a-TH1 increases in the presence of Lact-C2 (compare white arrows in x-z sections in A and B). Bars, 5 μm.
DISCUSSION
Dissecting the Membrane Binding Mechanism of Myo1a-TH1
In this study, we used a cell-based screen to discover two regions in Myo1a-TH1 that are essential for normal microvillar targeting: the NTM (Myo1a-TH1 residues 847–858) and the CTM (Myo1a-TH1 residues 1029–1043; Figs. 1, 3, and 4). Loss of charge mutations to basic residues in either of these regions resulted in a significant loss of Myo1a-TH1 targeting to microvilli at steady state. Moreover, in vitro biochemical experiments showed that Myo1a binds directly to acidic phospholipids with moderate affinity, and peptides encompassing either the NTM or CTM compete with Myo1a for binding to liposomes composed of 80% DOPS, 20% DOPC (Fig. 5), suggesting that these regions interact directly with membranes. Analysis of membrane binding dynamics in live cells using single molecule TIRF microscopy revealed that mutations to either of these motifs reduced the amount of time that Myo1a-TH1 spends in a membrane bound state (Fig. 6). This latter result provides a mechanism for the loss of steady state targeting observed in the initial structure-function experiments (Fig. 3). Finally, Myo1a-TH1 colocalizes with PS in the brush border; a PS-binding protein (Lact-C2), but not PI(4,5)P2-binding protein (PLCδ1-PH), reduces Myo1a-TH1 enrichment in microvilli. Taken together, the data presented here indicate that the NTM and CTM are bona fide membrane-binding motifs that contribute to electrostatic interactions between Myo1a-TH1 and acidic phospholipids, such as PS, in the inner leaflet of the microvillar membrane.
The NTM identified in Myo1a-TH1 is predicted to be α-helical (supplemental Fig. S4) (47, 48) and is well conserved across vertebrate short tailed class I myosins (Fig. 1D and supplemental Fig. S2A), suggesting that this motif might represent a common membrane-binding feature. In support of this proposal, a region equivalent to the NTM in Myo1g (referred to as the “pre-PH” domain) was shown to be critical for targeting of this motor in Jurkat cells; mutating either of the two conserved basic residues that flank this region (Lys-815 and Arg-826 in Myo1g, equivalent to Lys-847 and Lys-858 in Myo1a) significantly reduced the enrichment of Myo1g at the plasma membrane (11). Helical wheel analysis of the NTM in Myo1g (11) and Myo1a (not shown) reveals that the flanking basic residues are on the same face of a short α-helix and, thus, well positioned to form a lipid binding interface. A folded structure may help explain why a single point mutation (K847A) to the NTM impacts Myo1a-TH1 enrichment (Fig. 3C), despite the modest BH score for this region (Fig. 1E). Additional experiments will be needed to determine if the NTM functions as a general membrane binding motif in other class I myosins.
The CTM identified in Myo1a-TH1 exhibits poor sequence conservation with other class I myosins. Analysis of CTM secondary structure revealed a β-sheet prediction, although the confidence scores were weak, suggesting the region may be partially unstructured (supplemental Fig. S4). This would be consistent with the broad BH peak observed for this motif (Fig. 1E). Studies with BH motifs from Acanthamoeba and Dictyostelium myosins-1 revealed that mutations to either the basic or hydrophobic residues reduced binding to lipids (7, 8). Consistent with these findings, removal of the hydrophobic residues at the C terminus the CTM (Myo1a-TH1-CΔ10) or mutagenesis of the CTM basic cluster (Myo1a-TH1-K1031A/K1032A/K1033A), both had an impact on targeting to microvilli (Figs. 3D and 4, E and F). These results further support the proposal that the CTM may function as a Myo1a-specific, unstructured lipid binding motif.
We also employed single molecule TIRF microscopy to investigate how the NTM and CTM contribute to the kinetics of unitary Myo1a-TH1 membrane binding events. Measurements of membrane-bound lifetimes obtained from TIRF time lapse data provide direct access to detachment rates (koff = 1/bound lifetime). Imaging single molecule dynamics at the apical surface of polarized cells using TIRF is technically challenging due to the orientation of polarized cells grown in culture and the confinement of the TIRF excitation to the coverslip surface. Thus, we focused on the ventral membrane of unpolarized CL4 cells, which is amenable to TIRF illumination and expected to contain many of the major acidic phospholipid species, including PS. These imaging data revealed that mutations to the NTM or CTM increased the membrane detachment rate for Myo1a-TH1. Together with our steady state targeting data, these findings suggest that the NTM and CTM work together to produce long lived membrane binding events.
Myo1-PH Domain Is Not Essential for Myo1a-TH1 Targeting in Cells
Previous studies with short tailed class I myosins, Myo1b, Myo1c, and Myo1g, have implicated the conserved Myo1-PH domain in the targeting of these motors to cellular membranes (9–13). Biochemical studies with Myo1b and Myo1c have also revealed high affinity interactions with PI(4,5)P2 (9, 10, 13). Unexpectedly, we find that the Myo1-PH domain and interactions with PI(4,5)P2 are minor players in the mechanism that enables Myo1a-TH1 to become enriched on the microvillar membrane. This is supported by three principle findings: (i) mutations to signature Myo1-PH residues do not abolish the enrichment of Myo1a-TH1 in microvilli (Figs. 2 and 3B); (ii) Myo1a binds similarly to liposomes containing DOPS or PI(4,5)P2 at levels that provide equivalent negative charge (Fig. 5B), and (iii) PLCδ1-PH does not compete with Myo1a-TH1 for binding sites in microvilli (Fig. 3F). We note here that although Myo1a-TH1 variants with PH domain mutations are still enriched in microvilli, FEM values are reduced relative to control constructs (Fig. 3B). Thus, in addition to the NTM and CTM, basic residues in the PH domain probably contribute to general electrostatic interactions with the plasma membrane, similar to what has been proposed for Acanthamoeba Myo1c (8).
Our findings are consistent with other recent results, which indicate that TH1 structural elements outside the Myo1-PH domain probably contribute to membrane binding. For example, the Myo1c PH domain interacts tightly and specifically with PI(4,5)P2, yet transient kinetic and single molecule assays (15, 49) show that this interaction alone cannot account for the long membrane bound lifetimes observed with more physiological mixtures of lipids (15). Vertebrate long tailed Myo1e binds to liposomes composed of physiological concentrations of PI(4,5)P2 with high affinity, yet mutations to PH domain signature residues do not disrupt in vivo targeting or liposome binding (14). Acanthamoeba Myo1c also binds to acidic phospholipids using motifs outside the PH domain (8). Finally, although Myo1g membrane targeting depends on the signature residues of the PH domain, the conserved residues of the NTM also contribute to localization of this isoform (11, 12). Differences in the role of the PH domain noted for these different myosin-1 isoforms are probably related to the lack of uniform sequence conservation in this region, as previously pointed out by others (14). Together with our work, however, these studies support a model where membrane binding motifs outside the Myo1-PH contribute to proper targeting of class I myosins.
PS as Lipid Target for Myo1a-TH1 in Microvilli
Our studies suggest that Myo1a-TH1 binds directly to PS in microvilli (Figs. 3F and 7). PS is one of the most abundant phospholipid species found in the inner leaflet of the plasma membrane (44, 45) and is therefore a good candidate for supporting electrostatic interactions between Myo1a-TH1 and the microvillar membrane. Although the labeling methods used here show that PS and PI(4,5)P2 are both found in microvilli, it is still unclear if these lipids compartmentalize into subdomains along the microvillar axis. In bullfrog saccular hair cells, PI(4,5)P2 is found strictly at the tips of stereocilia, whereas PS is more widely distributed along the length of these protrusions (50). If such compartmentalization of lipid species is found in microvilli, it could provide one possible mechanism for polarizing the distribution of membrane-associated proteins throughout this structure. Intriguingly, Myo1d, a second class I myosin found in the enterocyte brush border, targets to microvillar tips, whereas Myo1a is distributed along the microvillar axis (51). Further experimentation is needed to determine if differential distribution observed for these two class I myosins is based on the sorting of highly charged phospholipids along the microvillar axis. Moreover, elucidation of the brush border membrane “lipidome” will allow us put the current results into context and help focus future studies on poorly characterized phospholipids that might also contribute to the targeting of class I myosins in this domain.
Functional Implications of Multiple Membrane Binding Motifs
Although the NTM and CTM both contribute to Myo1a-TH1 membrane binding, our studies suggest that the binding affinities generated by each individual motif are below the threshold needed to create enrichment on the membrane. Indeed, mutations to either the NTM or CTM abolish enrichment on microvillar membrane and increase the membrane detachment rate (Figs. 3 and 6), and peptides representing these motifs bind to acidic phospholipids with a lower affinity than full-length Myo1a (Fig. 5). These findings suggest that residues from both motifs are needed for full binding functionality. Multisite binding mechanisms have been described for a number of peripheral membrane binding proteins and are hypothesized to provide several functional advantages (52, 53). For example, multisite binding is able to create high binding affinities from relatively weak electrostatic interactions. This is because binding affinities are combined in a multiplicative (rather than additive) manner (54). Thus, two distinct sites that bind membrane weakly on their own demonstrate a much higher binding affinity (and, thus, longer membrane-bound lifetimes) when tethered together in the same protein or protein complex (52, 53, 55). Multisite binding might also enable a protein to sequester lipids, thus reducing its diffusional mobility while bound to the inner leaflet (55). This would be advantageous for a motor such as Myo1a, which functions in processes that require force generation between the actin cytoskeleton and fluid plasma membrane.
How the NTM and CTM are arranged in the context of a folded TH1 domain and how the spatial arrangement of these motifs impacts membrane binding remain unclear. One possibility is that these two motifs fold into a single membrane binding site, although this was not supported by the results of de novo modeling studies using I-TASSER (47), which reproducibly positioned these motifs on separate faces of the TH1 domain (not shown). Thus, a more likely possibility is that these motifs represent unique membrane-interacting regions. If the NTM and CTM are spatially separated, it is tempting to speculate that these different sites could facilitate membrane binding during different phases of the Myo1a mechano-chemical cycle. For example, one binding site might be optimally engaged with the lever arm in a prepower orientation, whereas the other binding site might be better oriented to bind membrane after the power stroke is complete. This would allow the TH1 domain to remain bound for the duration of the power stroke, despite the large conformational change that takes place during lever arm rotation (56). High resolution structural studies of the TH1 domain will be needed to develop our understanding of the spatial and functional relationships between the multiple membrane binding motifs identified in this and previous studies.
Supplementary Material
Acknowledgments
We thank all members of the Tyska laboratory for advice and support; Dr. Kristen Verhey, Dr. Chris Yengo, Dr. Kathy Trybus, Dr. Tamas Balla, Dr. Jim Goldenring, and Dr. Todd Graham for generously sharing reagents; and Dr. Ryoma Ohi, Chauca English Moore, and Dr. Elena Krementsova for assistance with SF9 culture and reagents. We also thank the Vanderbilt University Epithelial Biology Center Imaging Resource for generous support.
Note Added in Proof
Recently identified mutations in the Myo 1a CTM disrupt targeting to microvilli and are linked to colorectal tumor formation, underscoring the significance of this region of TH1 (57).
This work was supported, in whole or in part, by National Institutes of Health Grant R01-DK075555 (to M. J. T.). This work was also supported by American Heart Association Grants 09GRNT2310188 (to M. J. T.) and 11PRE7390066 (to J. N. M.), Vanderbilt University Medical Center Molecular Biophysics Training Program Training Grant T32-GM008320 (to Dr. Walter Chazin), and a Vanderbilt University IDEAs award (to M. J. T.).

This article contains supplemental Figs. S1–S4 and Movies S1–S4.
- TH1
- tail homology 1
- PH
- pleckstrin homology
- PI(4,5)P2
- phosphatidylinositol-4,5-bisphosphate
- NTM
- N-terminal targeting motif
- CTM
- C-terminal targeting motif
- PS
- phosphatidylserine
- CL4
- LLC-PK1-CL4
- EGFP
- enhanced green fluorescent protein
- FEM
- -fold enrichment in microvilli
- TIRF
- total internal reflection fluorescence
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoenthanolamine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- BH
- basic-hydrophobic
- Lact-C2
- lactadherin-C2.
REFERENCES
- 1. Odronitz F., Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McConnell R. E., Tyska M. J. (2010) Leveraging the membrane-cytoskeleton interface with myosin-1. Trends Cell Biol. 20, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nambiar R., McConnell R. E., Tyska M. J. (2010) Myosin motor function. The ins and outs of actin-based membrane protrusions. Cell Mol. Life Sci. 67, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krendel M., Mooseker M. S. (2005) Myosins. Tails (and heads) of functional diversity. Physiology 20, 239–251 [DOI] [PubMed] [Google Scholar]
- 5. Coluccio L. M. (2008) Myosin I, pp. 95–124, Springer-Verlag New York Inc., New York [Google Scholar]
- 6. Tang N., Lin T., Ostap E. M. (2002) Dynamics of Myo1c (myosin-Iβ) lipid binding and dissociation. J. Biol. Chem. 277, 42763–42768 [DOI] [PubMed] [Google Scholar]
- 7. Brzeska H., Guag J., Remmert K., Chacko S., Korn E. D. (2010) An experimentally based computer search identifies unstructured membrane-binding sites in proteins. Application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285, 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brzeska H., Hwang K. J., Korn E. D. (2008) Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J. Biol. Chem. 283, 32014–32023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hokanson D. E., Ostap E. M. (2006) Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc. Natl. Acad. Sci. U.S.A. 103, 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hokanson D. E., Laakso J. M., Lin T., Sept D., Ostap E. M. (2006) Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell 17, 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patino-Lopez G., Aravind L., Dong X., Kruhlak M. J., Ostap E. M., Shaw S. (2010) Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH). J. Biol. Chem. 285, 8675–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olety B., Wälte M., Honnert U., Schillers H., Bähler M. (2010) Myosin 1G (Myo1G) is a hematopoietic specific myosin that localizes to the plasma membrane and regulates cell elasticity. FEBS Lett. 584, 493–499 [DOI] [PubMed] [Google Scholar]
- 13. Komaba S., Coluccio L. M. (2010) Localization of myosin 1b to actin protrusions requires phosphoinositide binding. J. Biol. Chem. 285, 27686–27693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feeser E. A., Ignacio C. M., Krendel M., Ostap E. M. (2010) Myo1e binds anionic phospholipids with high affinity. Biochemistry 49, 9353–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenna J. M., Ostap E. M. (2009) Kinetics of the interaction of myo1c with phosphoinositides. J. Biol. Chem. 284, 28650–28659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Collins J. H., Borysenko C. W. (1984) The 110,000-dalton actin- and calmodulin-binding protein from intestinal brush border is a myosin-like ATPase. J. Biol. Chem. 259, 14128–14135 [PubMed] [Google Scholar]
- 17. Matsudaira P. T., Burgess D. R. (1979) Identification and organization of the components in the isolated microvillus cytoskeleton. J. Cell Biol. 83, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skowron J. F., Mooseker M. S. (1999) Cloning and characterization of mouse brush border myosin-I in adult and embryonic intestine. J. Exp. Zool. 283, 242–257 [PubMed] [Google Scholar]
- 19. Mooseker M. S., Tilney L. G. (1975) Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J. Cell Biol. 67, 725–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tyska M. J., Mooseker M. S. (2004) A role for myosin-1A in the localization of a brush border disaccharidase. J. Cell Biol. 165, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tyska M. J., Mackey A. T., Huang J. D., Copeland N. G., Jenkins N. A., Mooseker M. S. (2005) Myosin-1a is critical for normal brush border structure and composition. Mol. Biol. Cell 16, 2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nambiar R., McConnell R. E., Tyska M. J. (2009) Control of cell membrane tension by myosin-I. Proc. Natl. Acad. Sci. U.S.A. 106, 11972–11977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McConnell R. E., Tyska M. J. (2007) Myosin-1a powers the sliding of apical membrane along microvillar actin bundles. J. Cell Biol. 177, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McConnell R. E., Higginbotham J. N., Shifrin D. A., Jr., Tabb D. L., Coffey R. J., Tyska M. J. (2009) The enterocyte microvillus is a vesicle-generating organelle. J. Cell Biol. 185, 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayden S. M., Wolenski J. S., Mooseker M. S. (1990) Binding of brush border myosin I to phospholipid vesicles. J. Cell Biol. 111, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyska M. J., Mooseker M. S. (2002) MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 82, 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 28. Yeung T., Heit B., Dubuisson J. F., Fairn G. D., Chiu B., Inman R., Kapus A., Swanson M., Grinstein S. (2009) Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol. 185, 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szentpetery Z., Balla A., Kim Y. J., Lemmon M. A., Balla T. (2009) Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate. A comparative study. BMC Cell Biol. 10, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balla T., Varnai P. (2009) Visualization of cellular phosphoinositide pools with GFP-fused protein domains. Curr. Protoc. Cell Biol., Chapter 24, Unit 24.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorelik R., Yang C., Kameswaran V., Dominguez R., Svitkina T. (2011) Mechanisms of plasma membrane targeting of formin mDia2 through its amino-terminal domains. Mol. Biol. Cell 22, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fairn G. D., Hermansson M., Somerharju P., Grinstein S. (2011) Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 13, 1424–1430 [DOI] [PubMed] [Google Scholar]
- 33. Cai D., Verhey K. J., Meyhöfer E. (2007) Tracking single kinesin molecules in the cytoplasm of mammalian cells. Biophys. J. 92, 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai D., McEwen D. P., Martens J. R., Meyhofer E., Verhey K. J. (2009) Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 7, e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vallotton P., Ponti A., Waterman-Storer C. M., Salmon E. D., Danuser G. (2003) Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells. A fluorescent speckle microscopy study. Biophys. J. 85, 1289–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yengo C. M., Ananthanarayanan S. K., Brosey C. A., Mao S., Tyska M. J. (2008) Human deafness mutation E385D disrupts the mechanochemical coupling and subcellular targeting of myosin-1a. Biophys. J. 94, L5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krementsov D. N., Krementsova E. B., Trybus K. M. (2004) Myosin V. Regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 164, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P., Unger V. M. (2008) Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loomis P. A., Zheng L., Sekerková G., Changyaleket B., Mugnaini E., Bartles J. R. (2003) Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 163, 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagata K., Zheng L., Madathany T., Castiglioni A. J., Bartles J. R., García-Añoveros J. (2008) The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc. Natl. Acad. Sci. U.S.A. 105, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 42. Várnai P., Balla T. (1998) Visualization of phosphoinositides that bind pleckstrin homology domains. Calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toprak E., Kural C., Selvin P. R. (2010) Super-accuracy and super-resolution getting around the diffraction limit. Methods Enzymol. 475, 1–26 [DOI] [PubMed] [Google Scholar]
- 44. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Meer G., de Kroon A. I. (2011) Lipid map of the mammalian cell. J. Cell Sci. 124, 5–8 [DOI] [PubMed] [Google Scholar]
- 46. Fairn G. D., Schieber N. L., Ariotti N., Murphy S., Kuerschner L., Webb R. I., Grinstein S., Parton R. G. (2011) High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pyrpassopoulos S., Shuman H., Ostap E. M. (2010) Single-molecule adhesion forces and attachment lifetimes of myosin-I phosphoinositide interactions. Biophys. J. 99, 3916–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hirono M., Denis C. S., Richardson G. P., Gillespie P. G. (2004) Hair cells require phosphatidylinositol 4,5-bisphosphate for mechanical transduction and adaptation. Neuron 44, 309–320 [DOI] [PubMed] [Google Scholar]
- 51. Benesh A. E., Nambiar R., McConnell R. E., Mao S., Tabb D. L., Tyska M. J. (2010) Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol. Biol. Cell 21, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McLaughlin S., Aderem A. (1995) The myristoyl-electrostatic switch. A modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 20, 272–276 [DOI] [PubMed] [Google Scholar]
- 53. McLaughlin S., Wang J., Gambhir A., Murray D. (2002) PIP2 and proteins. Interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 [DOI] [PubMed] [Google Scholar]
- 54. Crothers D. M., Metzger H. (1972) The influence of polyvalency on the binding properties of antibodies. Immunochemistry 9, 341–357 [DOI] [PubMed] [Google Scholar]
- 55. Knight J. D., Lerner M. G., Marcano-Velázquez J. G., Pastor R. W., Falke J. J. (2010) Single molecule diffusion of membrane-bound proteins. Window into lipid contacts and bilayer dynamics. Biophys. J. 99, 2879–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jontes J. D., Wilson-Kubalek E. M., Milligan R. A. (1995) A 32º tail swing in brush border myosin I on ADP release. Nature 378, 751–753 [DOI] [PubMed] [Google Scholar]
- 57. Mazzolini R., Dopeso H., Mateo-Lozano S., Chang W., Rodrigues P., Bazzocco S., Alazzouzi H., Landolfi S., Hernández-Losa J., Andretta E., Alhopuro P., Espín E., Armengol M., Tabernero J., Ramón y Cajal S., Kloor M., Gebert J., Mariadason J. M., Schwartz S., Jr., Aaltonen L. A., Mooseker M. S., Arango D. (2012) Brush border Myosin Ia has tumor suppressor activity in the intestine. Proc. Natl. Acad. Sci. U.S.A. 109, 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.