Background: Aminoglycoside 2″-phosphotransferase IVa is an antibiotic resistance enzyme capable of using both ATP and GTP.
Results: An ordered solvent network and an interdomain linker loop coordinate nucleoside binding.
Conclusion: These structural elements are paralleled among related enzymes and are crucial in determining nucleotide specificity.
Significance: Knowledge of the functionally crucial features of the nucleoside-binding site advances rational inhibitor design against clinically relevant aminoglycoside phosphotransferases.
Keywords: Antibiotic Resistance, Dual Specificity Kinase, Enzyme Structure, Structural Biology, X-ray Crystallography, Aminoglycoside Phosphotransferase, Nucleotide Specificity
Abstract
Enzymatic phosphorylation through a family of enzymes called aminoglycoside O-phosphotransferases (APHs) is a major mechanism by which bacteria confer resistance to aminoglycoside antibiotics. Members of the APH(2″) subfamily are of particular clinical interest because of their prevalence in pathogenic strains and their broad substrate spectra. APH(2″) enzymes display differential preferences between ATP or GTP as the phosphate donor, with aminoglycoside 2″-phosphotransferase IVa (APH(2″)-IVa) being a member that utilizes both nucleotides at comparable efficiencies. We report here four crystal structures of APH(2″)-IVa, two of the wild type enzyme and two of single amino acid mutants, each in complex with either adenosine or guanosine. Together, these structures afford a detailed look at the nucleoside-binding site architecture for this enzyme and reveal key elements that confer dual nucleotide specificity, including a solvent network in the interior of the nucleoside-binding pocket and the conformation of an interdomain linker loop. Steady state kinetic studies, as well as sequence and structural comparisons with members of the APH(2″) subfamily and other aminoglycoside kinases, rationalize the different substrate preferences for these enzymes. Finally, despite poor overall sequence similarity and structural homology, analysis of the nucleoside-binding pocket of APH(2″)-IVa shows a striking resemblance to that of eukaryotic casein kinase 2 (CK2), which also exhibits dual nucleotide specificity. These results, in complement with the multitude of existing inhibitors against CK2, can serve as a structural basis for the design of nucleotide-competitive inhibitors against clinically relevant APH enzymes.
Introduction
Aminoglycosides form an important class of bactericidal antibiotics in therapeutic use today. However, bacterial resistance against nearly all known aminoglycosides persistently emerges, which poses a serious clinical threat in cases of pathogenic species (1). A major mechanism of resistance to aminoglycoside antibiotics is the covalent addition of functional groups by a large repertoire of proteins collectively referred to as the aminoglycoside-modifying enzymes. Among them, aminoglycoside O-phosphotransferases specialize in the phosphorylation of specific hydroxyl groups and thereby prevent this class of antibiotics from effectively binding to their intended ribosomal target (2).
Several subfamilies of APH3 enzymes have been discovered, and together they are capable of detoxifying aminoglycosides of diverse chemical structures. Members within the same subfamily, although generally sharing significant sequence similarity and structural homology, often exhibit distinctly different aminoglycoside preferences (3). Despite diverging antibiotic substrate profiles, all known APHs share a limited set of phosphate donors, namely ATP or GTP. Structurally, this implies that although the aminoglycoside-binding site shows wide variations among the APHs, the nucleotide-binding site is comparatively more conserved. Therefore, the latter has been considered an attractive target for the development of small molecule inhibitors that would ideally be active against a broad range of APHs and could thus serve as adjuvants in combination therapy with existing aminoglycosides (4).
Although some nucleotide-competitive kinase inhibitors have also been shown to inhibit a number of APHs, the prognostic for developing a pan-APH inhibitor is poor due to significant structural divergences among different APH subfamilies (5). This does not imply, however, that a common inhibitor against a smaller subset of APHs, such as those belonging to either the APH(3′) or the APH(2″) subfamily, is impossible to find. Such an inhibitor would harbor significant clinical potential because these two subfamilies, beside comprising over half of all known APH enzymes, are both characterized by a broad antibiotic substrate spectrum and contain some of the most prevalent resistance enzymes found in clinical isolates worldwide (6–8).
A divide between APH(3′) and APH(2″) enzymes lies in their nucleotide specificity. Members of the APH(3′) subfamily are ATP-specific, whereas members of the APH(2″) subfamily are also able to use GTP. Currently, the crystal structures of several APHs in complex with ATP-analogues have been determined (9–11), but APH structures with a bound GTP analog remain elusive. Among aminoglycoside phosphotransferases, APH(2″)-IVa stands out with its nearly identical catalytic efficiencies with either ATP or GTP, which presents the opportunity of contrasting ATP versus GTP binding at the same active site and elucidating key structural features that influence nucleotide specificity.
We have previously reported the apo and aminoglycoside-bound structures of APH(2″)-IVa (12), and kinetic parameters had also been established for this enzyme (13). Here, we present four nucleoside-bound crystal structures of wild type and mutant APH(2″)-IVa. These structures shed light on the detailed binding patterns of the different nucleoside substrates and explain the nucleotide specificity of this enzyme. These results, especially in complement with existing data on the nucleotide-bound structures of other APH enzymes, inform avenues for the rational design of small molecule inhibitors that can potentially target multiple subfamilies of aminoglycoside phosphotransferases.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
F95M and F95Y mutants of APH(2″)-IVa were constructed using the PCR method with the oligonucleotides 5′-GAAACGTACCAAATGTCTTTCGCAGGTATGACAAAAATTAAAGGAGTACCATTG-3′ and 5′-GAAACGTACCAAATGTCTTTCGCAGGTTATACAAAAATTAAAGGAGTACCATTG-3′, and their appropriate reverse complements, respectively. Each 50-μl PCR reaction contained 5 μl of 10 × Pfu X7 buffer, 60 ng of template DNA (wild type aph(2″)-IVa in a pET 22b(+) plasmid), 0.125 μg of each mutagenic primer, 10 mm dNTPs, and 2.5 units of Pfu X7 DNA polymerase. The PCR product was digested by DpnI restriction endonuclease for 1 h at 37 °C, and mutant plasmids were recovered by transformation of Escherichia coli DH5α cells. Successful introduction of the desired mutations was verified by sequencing of plasmid DNA.
Crystallization and Data Collection
Wild type as well as mutant APH(2″)-IVa, containing a C-terminal His6 tag, were expressed and purified as previously described (12). Crystals of both binary complexes were grown at 4 °C via the sitting-drop vapor diffusion method. The reservoir solution used was composed of 100 mm HEPES at pH 7.5, 150 mm potassium nitrate, 17% (w/v) polyethylene glycol 3350, 6% 2-propanol and 10% glycerol. Initially, irregular, jagged-shaped crystals were obtained by equilibrating a 3-μl drop against 40 μl of reservoir solution, where the drop consisted of 50% reservoir solution and 50% protein solution, which was composed of 6 mg/ml APH(2″)-IVa and 3.2 mm nucleoside substrate in 50 mm Tris-HCl at pH 8.5 and 300 mm sodium chloride. Such crystals were transferred to 50 μl of reservoir solution, broken up, and used as seeds. In subsequent iterations of crystallization, each 3-μl drop was supplemented with 0.5 μl of the seeding solution at 120-fold dilution. After four cycles of crystallization, prism-shaped crystals with approximate dimensions of 0.1 × 0.1 × 0.2 mm were obtained. Diffraction data were collected under cryogenic conditions (−180 °C) on a Rigaku rotating copper anode x-ray generator with a Saturn 300-mm charge-coupled device detector. For the wild type guanosine-bound structure, a data set of 180 images with an oscillation angle of 1° was collected, and for all other structures, data sets of 360 images with an oscillation angle of 1° were collected. All data sets were processed with the HKL2000 program suite (14), with the results summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
See supplemental Table S1 for a more comprehensive summary of data collection and refinement statistics.
| Wild type + adenosine | Wild type + guanosine | F95M + adenosine | F95Y + guanosine | |
|---|---|---|---|---|
| Resolution range (Å)a | 41.9–2.15 (2.23–2.15) | 34.0–2.10 (2.18–2.10) | 30.0–2.35 (2.41–2.35) | 30.0–2.10 (2.18–2.10) |
| Completeness (%) | 99.9 (98.3) | 99.0 (89.0) | 99.9 (99.6) | 99.3 (91.2) |
| Redundancy | 7.3 (6.8) | 3.3 (3.1) | 6.9 (6.1) | 7.3 (5.8) |
| Rsymb | 0.075 (0.30) | 0.074 (0.32) | 0.089 (0.35) | 0.086 (0.31) |
| Rcrystc/Rfreed | 0.190/0.245 | 0.185/0.242 | 0.198/0.257 | 0.195/0.246 |
| Number of non-hydrogen atoms | ||||
| Protein | 4843 | 4819 | 4865 | 4958 |
| Substrate | 38 | 40 | 38 | 40 |
| Solvent | 199 | 200 | 89 | 185 |
a Values in parentheses refer to reflections in the highest resolution shell and apply to the entire table.
b Rsym = Σhkl Σi |Ii(hkl)- <I(hkl)>|/Σhkl Σi Ii(hkl), where <I(hkl)> is the average intensity of equivalent reflections and the sum is extended over all measured observations for all unique reflections.
c Rcryst = Σhkl (|Fi| − |Fc|)/Σhkl |Fo|, where |Fi| is the observed and |Fc| is the calculated structure factor amplitude of a reflection.
d Rfree was calculated by randomly omitting 5% of the observed reflections from the refinement.
Structure Determination and Refinement
The crystal structure of adenosine-bound APH(2″)-IVa was solved by molecular replacement with Phaser from the CCP4 program suite (15), using protein chain A of the APH(2″)-IVa-tobramycin complex (Protein Data Bank (PDB) entry 3SG8) as the search model, and difference Fourier methods were used to obtain the phases for the remaining structures. Refinement of all models consisted of successive iterations of reciprocal space refinement with REFMAC (16), incorporating isotropic temperature factor and torsion-libration-screw refinement, alternated with manual model building with the program Coot (17). Torsion-libration-screw refinement was computed with seven subsegments per protein molecule, with each subsegment chosen based on secondary structural features. Adenosine and guanosine molecules were added to the respective model based on unambiguous density shown in the difference electron density maps (Fo − Fc and 2Fo − Fc) and refined on the basis of stereochemical constraints obtained from the PRODRG2 server (18). Solvent molecules were subsequently inserted until no significant improvements could be achieved as judged by decreases in the Rfree value. Final refinement statistics pertaining to the four models are given in Table 1.
Kinetic Assay
A continuous spectrophotometric assay that couples aminoglycoside phosphorylation with lactate production was used to measure the consumption of ATP or GTP as a function of NADH oxidation (19). Data were collected at 37 °C with a SpectraMax 190 absorbance microplate reader, where each assay was performed with a total volume of 200 μl, containing 50 mm Tris-HCl at pH 7.5, 40 mm potassium chloride, 10 mm magnesium chloride, 3.0 mm phosphoenolpyruvate, 3.0 mm tobramycin, 360 mm β-nicotinamide adenine dinucleotide (reduced), 20 units/ml pyruvate kinase, and 25 units/ml lactate dehydrogenase. Enzyme at 0.10 mg/ml (final concentration) was added to the reaction mixture 5 min before the reaction was initiated by the addition of NTP at variable concentrations. A total of 16 concentrations between 0.03 and 2.0 mm were assayed in triplicates for each nucleotide substrate. Steady state kinetic parameters were analyzed using Prism 5.0 (GraphPad Software) and are summarized in Table 2. Values for kcat, Km, and kcat/Km were obtained by fitting the kinetic data nonlinearly with the Michaelis-Menten equation,
 |
where kcat = Vmax/[E], Vmax is the maximum velocity, [E] is the enzyme concentration, and [S] and Km are the concentration and the Michaelis-Menten constant of the nucleotide substrate.
TABLE 2.
Steady state kinetic parameters for APH(2″)-IVa
| Protein | NTP | kcat | Km | kcat/Km |
|---|---|---|---|---|
| s−1 | μm | m−1s−1 | ||
| Wild type | ATP | 4.37 ± 0.04 | 69 ± 3 | (6.3 ± 0.3) × 104 |
| GTP | 5.03 ± 0.04 | 168 ± 5 | (3.0 ± 0.2) × 104 | |
| F95M | ATP | 3.06 ± 0.04 | 55 ± 3 | (5.5 ± 0.4) × 104 |
| GTP | 4.35 ± 0.04 | 156 ± 5 | (2.8 ± 0.2) × 104 | |
| F95Y | ATP | 1.06 ± 0.01 | 72 ± 3 | (1.5 ± 0.1) × 104 |
| GTP | 1.15 ± 0.01 | 49 ± 2 | (2.3 ± 0.2) × 104 |
RESULTS
Overall Structural Characteristics
The wild type APH(2″)-IVa-adenosine complex has been refined to 2.15 Å with an Rcryst of 0.192 and an Rfree of 0.244, whereas the guanosine-bound structure of wild type APH(2″)-IVa has been refined to 2.10 Å with an Rcryst of 0.187 and an Rfree of 0.247. These represent the first nucleoside-bound structures of APH(2″)-IVa and the first crystal structure of an aminoglycoside phosphotransferase in complex with a bound GTP-like substrate. Both binary structures were solved in a monoclinic space group with two protein molecules per asymmetric unit and with cell dimensions nearly identical to the previously reported values of aminoglycoside-bound structures of APH(2″)-IVa (12). In fact, superpositions of each of the nucleoside-bound structures of APH(2″)-IVa with the tobramycin-bound structure show that the two forms of binary structures are virtually identical, with r.m.s. deviations of 0.52 and 0.25 Å for the adenosine- and guanosine-bound structures, respectively. Superposition of the two nucleoside-bound structures yielded an r.m.s. deviation of 0.43 Å, demonstrating that the protein adopts an invariable binary structure irrespective of the identity of its substrate.
Nucleoside Binding
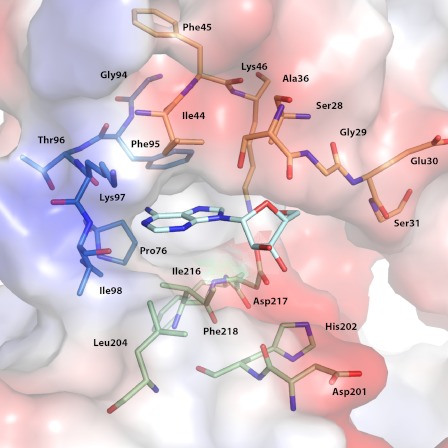
Traditionally, structures of aminoglycoside phosphotransferases have been divided into two structural components, termed the N-terminal and C-terminal lobe, in analogy to the structurally related eukaryotic protein kinases (20). The C-terminal lobe is itself further divided into the core region and the thumb region. The two lobes are joined by a linker loop of 10 amino acids, and the interface between the two lobes forms the nucleotide-binding pocket, with the linker loop serving as the base of the binding site. The nucleoside-binding pocket of APH(2″)-IVa is mainly described by 20 residues from the N-terminal lobe and the core subdomain. These 20 residues can be deconstructed into three main structural components (Fig. 1). A portion of the β-sheet from the N-terminal lobe forms region 1, the top face of the binding pocket. Specifically, residues Ser-28–Gly-31 fold over the ribose moiety and are poised to interact with the triphosphate group. These residues lead into a flexible loop that has been implicated in catalysis for both APH enzymes and eukaryotic protein kinases (21). Also, residues Ile-44–Lys-46 are positioned above the purine base and contribute to the hydrophobic character of this pocket. Region 2 is part of the linker loop that connects strand β5 of the N-terminal lobe with helix α3 of the core subdomain. In addition to affording the only hydrogen bonds with the purine base, residues Gly-94–Ile-98 also form the interior side of the binding cavity. Region 3 describes the bottom face of the cleft and consists of a group of five residues from two loops of the core-subdomain.
FIGURE 1.
Key components of nucleoside-binding site. Residues forming the nucleoside-binding site of APH(2″)-IVa are shown. A bound adenosine molecule is shown in cyan stick representation. Key residues are divided into three regions based on secondary structure elements and color-coded as follows: the N-terminal β-strands forming the top face of the cleft are shown in orange, the linker loop is shown in blue, and the loops from the core subdomain forming the bottom face of the cleft are shown in green.
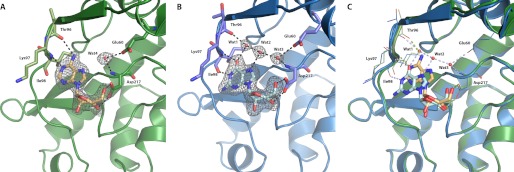
In the APH(2″)-IVa-adenosine complex, the nucleoside is buried deeply in a cleft between the two lobes (Fig. 2A). The adenine plane is sandwiched between hydrophobic side chains of residues from both lobes, notably Ile-44 and Ile-216. The base is stabilized by a hydrogen-bonding pattern reminiscent of Watson-Crick base pairing, consisting of two hydrogen bonds between N6 and the backbone oxygen of Thr-96 and between N1 and the amide nitrogen of Ile-98. In addition, the ribose ring is further stabilized by a pair of interactions between the side chain carboxyl group of Asp-217 and O3′ and O5′ of the sugar moiety. These last interactions are also observed in the guanosine-bound structure (Fig. 2B), but the ribose ring there adopts a slightly different puckered conformation, thus leading to a shift in the position of the base. Atoms of the guanine base are offset by up to 2.4 Å as compared with their counterparts in adenine, such that the O6 atom of guanosine replaces not the N6 but the N1 atom of adenosine and thus interacts with the amide nitrogen of Ile-98 (Fig. 2C). Such a dislocation, which has been previously termed a “hydrogen-bonding frameshift” (22), is necessary because the N6 atom of adenine acts as a hydrogen donor, whereas the O6 atom of guanine is a hydrogen acceptor. Thus, the guanine base must shift along the linker loop until it can favorably interact with the appropriate hydrogen-bonding partners, resulting in a lack of interaction with Thr-96, which is energetically compensated by a new hydrogen bond observed between the N1 atom of guanosine and the carbonyl oxygen of Ile-98. This shift along the linker region has previously been predicted by in silico modeling (13). The Watson-Crick base pairing-like bonding pattern observed for adenine is incomplete for guanine because no interaction partner is in position to accept a hydrogen from N2. The linker loop itself shows minimal differences between the two nucleoside-bound structures, with a total displacement of about 0.6–0.8 Å for the key residues that form the base of the binding site, likely an adaptation to more favorably interact with the altered position of the purine base.
FIGURE 2.
Adenosine versus guanosine binding for APH(2″)-IVa. A, APH(2″)-IVa-adenosine complex, showing the enzyme in green graphic representation and the 2Fo − Fc electron density (gray, 1.0 σ) for the adenosine molecule in light orange stick representation. Hydrogen-bonding interactions are represented as black dashed lines. Residues that directly interact with the substrate are show in stick representation and colored light green. B, APH(2″)-IVa-guanosine complex, showing the enzyme in blue graphic representation and the 2Fo − Fc electron density (gray, 1.0 σ) for the adenosine molecule in cyan stick representation. Ordered water molecules (Wat1–Wat4) forming a solvent network are highlighted as red spheres. Residues involved in interacting with the substrate are show in stick representation and colored purple. C, structural superposition of the APH(2″)-IVa-guanosine structure (blue) onto the APH(2″)-IVa-adenosine structure (green), showing the displacement of the purine base. Hydrogen-bonding interactions between the protein and each nucleoside are shown in the color of the protein molecule. The conformation of the backbone is slightly shifted to optimize binding with the respective nucleoside. The solvent network only applies to the guanosine-bound structure because it would clash with the adenosine molecule.
Globally, the displacement observed for guanosine positions the base less deeply in the nucleotide-binding cleft as compared with adenosine and thus vacates a pocket at the inside of the cleft. Fascinatingly, this pocket is occupied by a network of three clearly defined water molecules (Wat1, Wat2, and Wat3 in Fig. 2B) that serve as bridges between the guanine base and the residues lining the interior of the binding cleft. One of these water molecules (Wat1) is less than 1 Å away from the position occupied by the N6 atom in the adenosine structure, thereby acting as an intermediary that connects the O6 atom of guanosine with the carbonyl group of Thr-96. Similarly, the other two water molecules indirectly connect the N7 atom of guanosine with the backbone amide of Asp-217 and the side chain carboxyl group of Glu-60. In contrast, because of the deeper insertion of the adenine plane into the nucleotide-binding cleft, there is insufficient space for the formation of an effective solvent network. Although a water molecule linking the amide nitrogen of Asp-17 and the terminal carboxyl oxygen of Glu-60 is clearly visible (Wat4 in Fig. 2A), it is too far removed from the N7 atom of adenosine to form a hydrogen bond. Also, no water molecules occupying the equivalent positions of guanine atoms such as N1 were evident in the APH(2″)-IVa-adenosine structure at the given resolution.
Structure-Function Studies
As detailed below, the mutants F95M and F95Y were created based on our structural analysis of the nucleoside-binding pocket and a sequence comparison with related APH enzymes to explore the influence of this key residue on nucleotide specificity, and kinetic data were collected on the wild type as well as the mutant variants. The steady state kinetic parameters of wild type APH(2″)-IVa confirm that its activity is comparable in the presence of either ATP or GTP (Table 2). The kcat/Km values determined here deviate somewhat from previously reported parameters that range between 3 × 103 and 8 × 103 m−1 s−1 (3, 13). This is likely due to small differences in the experimental conditions. In general, F95M and F95Y mutations both result in a small decrease in catalytic efficiency. The F95M mutant does not show significantly different binding affinities as compared with the wild type, whereas the F95Y mutation shifts the nucleotide selectivity from a 2.5-fold preference for ATP to a 1.5-fold preference for GTP.
To complement kinetic studies, crystal structures of both mutants were determined. The F95M APH(2″)-IVa-adenosine complex has been refined to 2.4 Å with an Rcryst of 0.198 and an Rfree of 0.257, and the guanosine-bound structure of F95Y APH(2″)-IVa has been refined to 2.10 Å with an Rcryst of 0.195 and an Rfree of 0.246. No global changes in conformation are brought about by either mutation, and the overall r.m.s. deviations for both mutants are less than 0.4 Å as compared with the wild type enzyme. For the F95M mutant, electron density of the side chain of Met-95 is not clearly defined, suggesting that it is flexible and may partially project toward the nucleotide-binding pocket, thereby impacting the formation of an ordered solvent network (supplemental Fig. S1A). For F95Y, the two protein molecules in the asymmetric unit show varying conformations for the mutated residue. In one instance, the mutated residue adopts an identical conformation as compared with the wild type phenylalanine, with the additional hydroxyl group forming a new hydrogen bond with the side chain carboxyl group of Glu-60. Although this conformation does not result in a direct interaction with the substrate, the increased hydrophilicity change to the local environment could favor the formation of an ordered solvent network (supplemental S1B). For the second protein molecule, the electron density for Tyr-95 is poorly defined, and an alternative conformation of the side chain can be modeled, where the terminal hydroxyl group replaces a water molecule and forms a hydrogen bond with the N7 atom of the guanine moiety. The solvent network is absent, and only one water molecule (Wat3) could be modeled (supplemental Fig. S1C).
DISCUSSION
Structural Determinants for Nucleotide Specificity
The crystal structure of APH(2″)-IVa bound to guanosine represents the first instance where an aminoglycoside resistance enzyme was successfully co-crystallized with a GTP analog. Together with the APH(2″)-IVa-adenosine structure, it offers a detailed perspective of the nucleoside-binding site architecture of this enzyme. The number and nature of hydrogen bonds between each nucleoside and the protein are quite similar. Adenosine, being more deeply inserted in the binding cleft, likely benefits from more significant van der Waals interactions with hydrophobic residues found in the interior of the binding cavity, but guanosine compensates by having a stabilizing hydrogen-bonding network despite incurring the entropy cost of at least three localized water molecules. These results rationalize the kinetic data showing that ATP and GTP can bind with comparable efficiencies.
Previous studies have shown that nucleotide preference deviates among the four main APH(2″) enzymes, with APH(2″)-Ia and APH(2″)-IIIa being GTP-specific, APH(2″)-IIa capable of binding GTP but preferring ATP, and APH(2″)-IVa willing to accept both nucleotides to a near equal extent (3). It is instructive to consider the structural elements that give rise to this incongruity, especially because they may also help explain the nucleotide specificity of more distantly related APH enzymes. Based on a multiple amino acid sequence alignment, it is clear that the overall sequence similarity among this subfamily of enzymes is relatively low (all below 30%). However, the three major regions that form the nucleotide-binding site show conspicuous conservation, with 16 out of 20 residues either perfectly conserved or strongly similar (Fig. 3). The only region that shows considerable divergence is region two despite its essential role in stabilizing both the adenine and the guanine ring. The strong conservation of the other regions and the lack of conservation in this loop are suggestive of the presence of key residues that differ among the four enzymes and thus act as determinants of nucleotide specificity. This area of the binding pocket is outlined mainly by the curvature of the backbone of residues Gly-94–Ile-98, with the majority of side chains pointing away from the nucleotide. A notable exception is Phe-95, the phenyl group of which is a major constituent of the hydrophobic interior wall of the cleft. This amino acid is replaced by a methionine residue in APH(2″)-IIa (Met-85) and by tyrosine residues in both APH(2″)-Ia and APH(2″)-IIIa (Tyr-100 and Tyr-92, respectively). To assess the impact of this amino acid, we created the F95M and F95Y mutants, determined their kinetic parameters, and solved their crystal structures in complex with adenosine or guanosine in the same conditions as with the wild type enzyme. Kinetic parameters of the F95M mutant do not deviate significantly from wild type APH(2″)-IVa. The F95Y mutant, on the other hand, shows an increase in affinity for GTP and a decrease in affinity for ATP, such that the ratio between Km(ATP) and Km(GTP) is 1.4 as opposed to 0.4 for the wild type enzyme. The additional hydroxyl group of F95Y increases the polar character of a largely hydrophobic pocket and stabilizes Glu-60, a residue important for interacting with the water network. More importantly, the crystal structure shows that the conformation of Tyr-95 is flexible and that its side chain hydroxyl group is able to replace a water molecule from the solvent network. In this conformation, guanosine binding is favored because of the additional hydrogen bond, and adenosine binding would be discouraged due to steric hindrance. Guanosine selectivity would be enhanced if the tyrosine residue could be trapped in this conformation, and the partial occupancy observed in the model is evidence that the surrounding residues in the binding pocket could impact nucleotide selectivity through their effect on residue 95.
FIGURE 3.
Partial multiple sequence alignment of selected APH enzymes and CK2. The aligned enzymes are ordered based on increasing preference for GTP as the second substrate. Secondary structural elements are indicated below the alignment. Residues forming the nucleoside-binding site in APH(2″)-IVa are separated into three regions. Phe-95 of APH(2″)-IVa and its corresponding amino acids in the other enzymes are highlighted by the red box. The alignment for APH(2″) enzymes was created with Clustal Omega (32). APH(3′)-IIIa and CK2 were aligned based on a manual structural alignment between representative structures (PDB accession numbers 1L8T and 1LP4) and the APH(2″)-IVa-adenosine complex. The kinetic parameters for the six enzymes were taken from literature (3, 33, 34). The Km(ATP)/Km(GTP) ratio for APH(2″)-IVa varies among three independent studies due to small differences in specific experimental conditions (3, 13).
Therefore, despite the effects of Phe-95 upon nucleotide specificity among APH(2″) enzymes as indicated by these mutagenesis studies, the complete picture is clearly more complex, and nucleotide specificity is not dictated by a single amino acid. It is probable that the composite of residues around the nucleotide-binding site collectively forms a space that is adapted for the specific substrate preference of the given APH and that although individual residues such as Phe-95 play a critical role, other differences around the binding pocket also have a crucial complementary function. This is highlighted when comparing the nucleoside-binding site of APH(2″)-IVa with more distantly related enzymes, such as the ATP-specific APH(3′)-IIIa. When the ADP-bound structure of APH(3′)-IIIa (PDB code 1L8T) is superimposed onto the adenosine-bound and guanosine-bound structures of APH(2″)-IVa, most residues around the binding site show relatively strong positional conservation given the limited overall structural homology. However, two discrepancies are noteworthy. A substantial difference is the conformation of the interdomain linker. Although the beginning of this loop, including Ser-91 (equivalent to Thr-96 in APH(2″)-IVa) that interacts with the adenine base, is well conserved, the loop makes a sharp turn in APH(3′)-IIIa thereafter, such that the backbone oxygen of residue Ala-93 (equivalent to Ile-98 in APH(2″)-IVa) is over 4 Å away from the N1 atom of the guanine moiety and therefore unavailable for hydrogen bonding (Fig. 4A). Secondly, the aforementioned Phe-95 of APH(2″)-IVa is replaced by a methionine residue (Met-90) in APH(3′)-IIIa, which adopts a conformation such that the terminal thiomethyl group is directed toward the nucleotide and would sterically hinder the formation of a solvent network in this pocket (Fig. 4B). The finding that a similar orientation is not adopted by the F95M mutant of APH(2″)-IVa indicates that although residue 95 has the potential to control nucleoside binding, its effect is dependent on its conformation, which is in turn dictated by the surrounding residues forming the local microenvironment. Taken together, the inability to form an ordered solvent network and the lack of sufficient hydrogen-bonding interactions due to an altered conformation of the linker loop act synergistically to prohibit APH(3′)-IIIa from binding GTP, and neither contributing factor is controlled by individual amino acids. That other ATP-selective APHs cannot bind GTP for similar reasons is substantiated by comparisons with available structural data, such as the nucleotide-bound structure of APH(9)-Ia (11).
FIGURE 4.
Structural basis for ATP-selectivity of APH(3′)-IIIa. A, nucleotide-binding clefts of APH(2″)-IVa-guanosine (blue with cyan ligand) and APH(3′)-IIIa-ADP (brown with dark red ligand), showing that the key residue Ala-93 for coordinating the guanine ring is positioned too far away from the base for effective interactions in APH(3′)-IIIa. B, alternative view of the nucleotide-binding clefts, showing that the conformation of residue Met-90 of APH(3′)-IIIa clashes with a solvent network that would be required for guanosine binding. The ADP molecule aligns with the adenosine in the APH(2″)-IVa-adenosine complex, which is shown in semitransparent orange stick representation.
Comparison with CK2
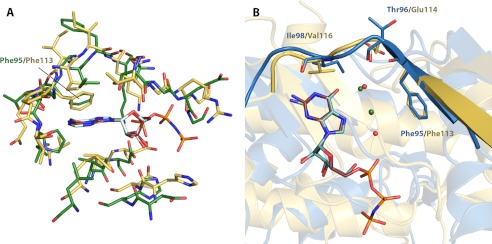
Structural parallels between aminoglycoside phosphotransferases and eukaryotic protein kinases have been described since the solution of the first APH crystal structure (20). The ability to accept different types of nucleotides as a phosphate source is uncommon among protein kinases, and only very few examples have been reported to date (23, 24). The best studied case is CK2 (formerly casein kinase 2), which distinguishes itself from other eukaryotic protein kinases by its dual nucleotide specificity as well as its constitutive activity, in both counts similar to APH(2″)-IVa (22, 25). A structural comparison between adenosine-bound APH(2″)-IVa and the AMPPNP-bound catalytic subunit of CK2 from Zea mays (PDB code 1LP4), which is regarded as a reference structure among the over 40 deposited structures of CK2 to date (25, 26), shows clear structural divergence. However, the nucleoside-binding site is remarkably well conserved (r.m.s. deviation 0.96 Å), with almost every one of the 20 relevant residues in APH(2″)-IVa having a counterpart in CK2 (Fig. 5A). Notably, the conformations of the linker loop in general and Phe-95 (equivalent to Phe-113 in CK2) in particular closely resemble each other in the two proteins. In addition, the basis for dual nucleotide specificity is virtually identical between the two enzymes, as evidenced by the close resemblance of the GMPPNP-bound structure of CK2 (PDB code 1DAY) and the APH(2″)-IVa-guanosine complex. The same hydrogen-bonding pattern between the purine moiety and the interdomain loop is present in both structures, and GMPPNP binding in CK2 is also supported by a solvent network in the interior of the binding pocket (Fig. 5B). One difference is that for CK2, AMPPNP and GMPPNP are each stabilized by two water molecules (22), whereas for APH(2″)-IVa, three waters are associated with guanosine binding and none are associated with adenosine binding. Such variations are not unexpected considering the structural and sequence disparity. In fact, it is intriguing to see that nature has convergently evolved the same molecular architecture supporting dual nucleotide specificity in two enzymes that are phylogenetically only distantly related.
FIGURE 5.
Structural comparison between nucleoside-bound complexes of APH(2″)-IVa and CK2. A, structural superposition of residues forming the nucleoside-binding pocket of AMPPNP-bound CK2α (yellow with red ligand) onto those of adenosine-bound APH(2″)-IVa (green with light cyan ligand). Despite significant discrepancies in the overall protein structure, this region shows strong structural conservation. B, structural superposition of GMPPNP-bound CK2α structure (yellow with red ligand) onto guanosine-bound APH(2″)-IVa (blue with dark cyan ligand). The conformation of the interdomain linker, highlighted in graphic representation, and the position of key residues, shown in stick representation, are conserved. Also conserved between the two structures is a solvent network, consisting of two water molecules for CK2 (green spheres) and three water molecules for APH(2″)-IVa (red spheres, with one overlapping and occluded by a green sphere).
Implications for Inhibitor Design
Some ATP-competitive inhibitors originally developed for eukaryotic protein kinases have been shown to inhibit several aminoglycoside phosphotransferases (27). Notably, CKI-7, an inhibitor of the isoquinolinesulfonamide family, is able to bind APH(3′)-IIIa, APH(9)-Ia, and APH(2″)-Ia (5), which sets a precedent for an inhibitor active against both ATP-specific and GTP-specific APHs. If the discrepancies of the nucleotide-binding sites between APH enzymes should be too extensive to permit the optimization of a common inhibitor, then the guanine-bound structure of APH(2″)-IVa can still serve as a point of departure for the development of inhibitors against GTP-binding APHs. A number of inhibitors against CK2, belonging to diverse chemical families such as anthraquinones, coumarins, and pyrazolotriazines (28–30), promise improved steric and chemical complementarity for a nucleotide-binding site adapted to accommodate GTP. Such molecules provide structural frameworks for the development of adjuvants to complement broad spectrum aminoglycosides currently rendered ineffective by APH(2″) enzymes. This is corroborated by a recent inhibitor profile study of various aminoglycoside kinases, which generated similar chemical scaffolds (31).
Conclusions
In summary, our structural and kinetic analyses of the adenosine- and guanosine-bound complexes of wild type and mutant APH(2″)-IVa reveal the basis for nucleotide promiscuity and highlight the importance of the integrity of a solvent network in the interior of the binding cleft and the conformation of the interdomain linker in determining nucleotide specificity. These contributions to our understanding of nucleotide binding of APH enzymes serve as the first step for the structure-guided design of competitive inhibitors derived from chemical classes that have not previously been employed in the study of aminoglycoside phosphotransferases, with the ultimate aim of developing both potent and specific inhibitors against these resistance factors.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Chow for providing us with the aph(2″)-IVa gene. We also thank past and present members of the Berghuis laboratory, in particular John Ozcelik, for assistance and helpful discussions.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) (Grant MOP-13107) (to A. M. B.).
This article was selected as a Paper of the Week.

This article contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (codes 4DT8, 4DT9, 4DTA, and 4DTB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- APH
- aminoglycoside O-phosphotransferase
- APH(2″)-IVa
- aminoglycoside 2″-phosphotransferase IVa
- APH(3′)-IIIa
- 3′,5″-aminoglycoside phosphotransferase type IIIa
- CK2
- casein kinase 2
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate
- GMPPNP
- guanosine 5′-(β,γ-imino)triphosphate
- r.m.s.
- root mean square.
REFERENCES
- 1. Smith C. A., Baker E. N. (2002) Aminoglycoside antibiotic resistance by enzymatic deactivation. Curr. Drug Targets Infect. Disord. 2, 143–160 [DOI] [PubMed] [Google Scholar]
- 2. Chow J. W. (2000) Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31, 586–589 [DOI] [PubMed] [Google Scholar]
- 3. Toth M., Chow J. W., Mobashery S., Vakulenko S. B. (2009) Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2"-phosphotransferases. J. Biol. Chem. 284, 6690–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burk D. L., Berghuis A. M. (2002) Protein kinase inhibitors and antibiotic resistance. Pharmacol. Ther. 93, 283–292 [DOI] [PubMed] [Google Scholar]
- 5. Fong D. H., Xiong B., Hwang J., Berghuis A. M. (2011) Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor. PLoS ONE 6, e19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yadegar A., Sattari M., Mozafari N. A., Goudarzi G. R. (2009) Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb. Drug Resist. 15, 109–113 [DOI] [PubMed] [Google Scholar]
- 7. Chandrakanth R. K., Raju S., Patil S. A. (2008) Aminoglycoside resistance mechanisms in multidrug-resistant Staphylococcus aureus clinical isolates. Curr. Microbiol. 56, 558–562 [DOI] [PubMed] [Google Scholar]
- 8. Zarrilli R., Tripodi M. F., Di Popolo A., Fortunato R., Bagattini M., Crispino M., Florio A., Triassi M., Utili R. (2005) Molecular epidemiology of high-level aminoglycoside-resistant enterococci isolated from patients in a university hospital in southern Italy. J. Antimicrob. Chemother. 56, 827–835 [DOI] [PubMed] [Google Scholar]
- 9. Young P. G., Walanj R., Lakshmi V., Byrnes L. J., Metcalf P., Baker E. N., Vakulenko S. B., Smith C. A. (2009) The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2"-phosphotransferase-IIa (APH(2")-IIa) provide insights into substrate selectivity in the APH(2") subfamily. J. Bacteriol. 191, 4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nurizzo D., Shewry S. C., Perlin M. H., Brown S. A., Dholakia J. N., Fuchs R. L., Deva T., Baker E. N., Smith C. A. (2003) The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327, 491–506 [DOI] [PubMed] [Google Scholar]
- 11. Fong D. H., Lemke C. T., Hwang J., Xiong B., Berghuis A. M. (2010) Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila. J. Biol. Chem. 285, 9545–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi K., Houston D. R., Berghuis A. M. (2011) Crystal structures of antibiotic-bound complexes of aminoglycoside 2"-phosphotransferase IVa highlight the diversity in substrate binding modes among aminoglycoside kinases. Biochemistry 50, 6237–6244 [DOI] [PubMed] [Google Scholar]
- 13. Toth M., Frase H., Antunes N. T., Smith C. A., Vakulenko S. B. (2010) Crystal structure and kinetic mechanism of aminoglycoside phosphotransferase-2"-IVa. Protein Sci. 19, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 19. McKay G. A., Thompson P. R., Wright G. D. (1994) Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification, and substrate specificity. Biochemistry 33, 6936–6944 [DOI] [PubMed] [Google Scholar]
- 20. Hon W. C., McKay G. A., Thompson P. R., Sweet R. M., Yang D. S., Wright G. D., Berghuis A. M. (1997) Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89, 887–895 [DOI] [PubMed] [Google Scholar]
- 21. Burk D. L., Hon W. C., Leung A. K., Berghuis A. M. (2001) Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry 40, 8756–8764 [DOI] [PubMed] [Google Scholar]
- 22. Niefind K., Pütter M., Guerra B., Issinger O. G., Schomburg D. (1999) GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6, 1100–1103 [DOI] [PubMed] [Google Scholar]
- 23. Gschwendt M., Kittstein W., Kielbassa K., Marks F. (1995) Protein kinase C δ accepts GTP for autophosphorylation. Biochem. Biophys. Res. Comm. 206, 614–620 [DOI] [PubMed] [Google Scholar]
- 24. Schinkmann K., Blenis J. (1997) Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J. Biol. Chem. 272, 28695–28703 [DOI] [PubMed] [Google Scholar]
- 25. Niefind K., Raaf J., Issinger O. G. (2009) Protein kinase CK2 in health and disease. Protein kinase CK2: from structures to insights. Cell Mol. Life Sci. 66, 1800–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yde C. W., Ermakova I., Issinger O. G., Niefind K. (2005) Inclining the purine base-binding plane in protein kinase CK2 by exchanging the flanking side chains generates a preference for ATP as a co-substrate. J. Mol. Biol. 347, 399–414 [DOI] [PubMed] [Google Scholar]
- 27. Daigle D. M., McKay G. A., Wright G. D. (1997) Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J. Biol. Chem. 272, 24755–24758 [DOI] [PubMed] [Google Scholar]
- 28. Battistutta R., Sarno S., De Moliner E., Papinutto E., Zanotti G., Pinna L. (2000) The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2. J. Biol. Chem. 275, 29618–29622 [DOI] [PubMed] [Google Scholar]
- 29. Chilin A., Battistutta R., Bortolato A., Cozza G., Zanatta S., Poletto G., Mazzorana M., Zagotto G., Uriarte E., Guiotto A., Pinna L. A., Meggio F., Moro S. (2008) Coumarin as attractive casein kinase 2 (CK2) inhibitor scaffold: an integrate approach to elucidate the putative binding motif and explain structure-activity relationships. J. Med. Chem. 51, 752–759 [DOI] [PubMed] [Google Scholar]
- 30. Nie Z., Perretta C., Erickson P., Margosiak S., Almassy R., Lu J., Averill A., Yager K. M., Chu S. (2007) Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2. Bioorg. Med. Chem. Lett. 17, 4191–4195 [DOI] [PubMed] [Google Scholar]
- 31. Shakya T., Stogios P. J., Waglechner N., Evdokimova E., Ejim L., Blanchard J. E., McArthur A. G., Savchenko A., Wright G. D. (2011) A small molecule discrimination map of the antibiotic resistance kinome. Chem. Biol. 18, 1591–1601 [DOI] [PubMed] [Google Scholar]
- 32. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shakya T., Wright G. D. (2010) Nucleotide selectivity of antibiotic kinases. Antimicrob. Agents Chemother. 54, 1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gatica M., Hinrichs M. V., Jedlicki A., Allende C. C., Allende J. E. (1993) Effect of metal ions on the activity of casein kinase II from Xenopus laevis. FEBS Lett. 315, 173–177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.