Background: Insulin-like growth factor 1 (IGF1)/IGF1R signaling promotes cardiomyocyte survival and undergoes down-regulation after birth.
Results: Cardiac expression of miR-378 is induced after birth. IGF1R is a direct target of miR-378.
Conclusion: miR-378 promotes cardiac apoptosis.
Significance: Inhibition of miR-378 would be beneficial in promoting cardiac cell survival in an ailing heart.
Keywords: Aging, Apoptosis, Cardiac Muscle, Insulin-like Growth Factor (IGF), MicroRNA
Abstract
Postnatal cardiac remodeling is characterized by a marked decrease in the insulin-like growth factor 1 (IGF1) and IGF1 receptor (IGF1R) expression. The underlying mechanism remains unexplored. This study examined the role of microRNAs in postnatal cardiac remodeling. By expression profiling, we observed a 10-fold increase in miR-378 expression in 1-week-old neonatal mouse hearts compared with 16-day-old fetal hearts. There was also a 4–6-fold induction in expression of miR-378 in older (10 months) compared with younger (1 month) hearts. Interestingly, tissue distribution analysis identified miR-378 to be highly abundant in heart and skeletal muscles. In the heart, specific expression was observed in cardiac myocytes, which was inducible by a variety of stressors. Overexpression of miR-378 enhanced apoptosis of cardiomyocytes by direct targeting of IGF1R and reduced signaling in Akt cascade. The inhibition of miR-378 by its anti-miR protected cardiomyocytes against H2O2 and hypoxia reoxygenation-induced cell death by promoting IGF1R expression and downstream Akt signaling cascade. Additionally, our data show that miR-378 expression is inhibited by IGF1 in cardiomyocytes. In tissues such as fibroblasts and fetal hearts, where IGF1 levels are high, we found either absent or significantly low miR-378 levels, suggesting an inverse relationship between these two factors. Our study identifies miR-378 as a new cardioabundant microRNA that targets IGF1R. We also demonstrate the existence of a negative feedback loop between miR-378, IGF1R, and IGF1 that is associated with postnatal cardiac remodeling and with the regulation of cardiomyocyte survival during stress.
Introduction
MicroRNAs (miRNAs)2 are endogenous, small noncoding RNAs that have emerged as powerful negative regulators of gene expression. By targeting specific mRNAs mostly in the 3′ untranslated regions, miRNAs either destabilize target mRNAs and/or inhibit their translation. Bioinformatics predicts that almost 30% of all human mRNAs are regulated by miRNAs (1, 2). Recently these regulatory molecules have been suggested to be involved in several biological processes ranging from development and metabolism to apoptosis and signaling pathways. Among the almost 1000 human miRNAs described so far, only a handful are found to be abundant in the heart, such as miR-1, miR-133, miR-208a, miR-208b, and miR-499. These cardioabundant miRNAs have been shown to play fundamental roles in regulating development as well as disease processes (3).
Recent emerging data indicate that miRNAs also play a role in the cardiac remodeling that occurs during the postnatal period. Global inhibition of miRNA processing by cardiac-specific deletion of Dicer was shown to cause postnatal lethality from progressive heart failure (4). Cardiac-specific conditional deletion of Dicer in the myocardium after birth was also shown to produce spontaneous cardiac remodeling, and cardiac dysfunction resulting in premature death within 1 week (5). More recently, an elegant study from Olson's group (6) demonstrated that the loss of the proliferative potential of cardiomyocytes after birth is related to the up-regulation of miR-15 family members. Knockdown of a single member of this family, miR-195, was sufficient to increase the number of mitotic cardiomyocytes and de-repress Check1 (6).
Another important aspect of cardiac remodeling during the postnatal period is the regulatory role played by insulin-like growth factor 1 (IGF1), a key survival factor in the heart. It is released into the bloodstream by the liver, or synthesized locally by muscles and neural cells. Acting in an autocrine or paracrine fashion, IGF1 via the IGF1R triggers a signaling cascade that plays many essential regulatory roles in multiple aspects of cardiac biology. During the postnatal period, the switch in cardiac metabolism and withdrawal of the cardiomyocyte from the cell cycle is characterized by a marked decrease in the expression of IGF1 and IGF1R (7, 8). The down-regulation of IGF/IGF1R is also observed in other organ systems, such as the brain (9). The mechanism of IGF1R down-regulation has not yet been described.
In this study we demonstrate that postnatal repression of cardiac IGF1R is associated with the up-regulation of miR-378. Our data illustrate the high abundance of miR-378 in the heart and its stress-inducible and specific expression in cardiomyocytes. We found that miR-378 promotes cardiomyocyte apoptosis by directly targeting IGF1R and consequently inhibiting IGF1-mediated activation of Akt. In addition, ligand IGF1 was found to act as an inhibitor of miR-378 expression. Indeed IGF1 abundance correlated with the depression of miR-378 in cardiac fibroblasts in culture, and in vivo in the fetal heart and vice versa after birth. Together, our findings define a functional role of miR-378 in the heart and unravel a unique reciprocal molecular circuit between miR-378, IGF1R, and IGF-1, which appears to play a significant regulatory role during postnatal cardiac remodeling.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Reagents used in the study were purchased as follows: Dulbecco's modified Eagle's medium (DMEM), Lipofectamine 2000, TRIzol, T4 DNA ligase, restriction enzymes, SuperScript III reverse transcriptase kit, DH5α cells, RNA oligos (378-mimic, 378-anti-miR, scramble, and mimic control), Opti-MEM transfection media, and Lipofectamine were purchased from Invitrogen; fetal bovine serum (FBS) from Gemini Inc.; collagenase type 3, trypsin, and soybean inhibitor from Worthington Biochemicals; DNA probes for miR1, miR-378, miR-378*, miR-133a, U6, and miR-208a were synthesized from IDT; the gel extraction kit and DNA Maxi prep kit from Qiagen; Fast SYBR Green Master Mix was from Applied Biosystems; ECL Western blot detection kit, chemiluminescence films, and HyBond N+ nylon membrane were purchased from Amersham Biosciences; nitrocellulose transfer membrane, RC/DC assay, and Precision Plus Dual protein ladder from Bio-Rad; Restore Western blot stripping buffer was from Thermo Science; PQ401, horseradish peroxidase-conjugated anti-rabbit antibody, laminin, and other general chemicals were purchased from Sigma; hybridization buffer and Nuc Away columns from Ambion; the In Situ Cell Death (TUNEL) detection kit was from Roche Diagnostics; pmiRGLO vector, Caspase Glow assay, ApoTox Glo Triplex Assay kit, and Dual Luciferase assay kit were purchased from Promega; and [γ-32P]ATP from PerkinElmer Life Sciences. Specific antibodies were obtained from following sources: Akt, anti-pAkt (Thr308 and Ser478), pERK, FoxO3, pFoxO3a, and p-IGF1Rβ were purchased from Cell Signaling; FasL, TRAIL, Bim, GAPDH, ERK2, β-actin, and HRP-conjugated anti-mouse or anti-goat antibodies were from Santa Cruz; anti-human insulin-like growth factor 1 and anti-rabbit HRP antibody were from Sigma; IGF1R (β subunit) antibody from Millipore; α-actinin and phospho-ERK1/2 were from Abcam Inc.; fluorescent labeled Alexa Fluor 488 (green) and 594 (red) antibodies, ToPro nuclei labeling reagent, and ProLong Gold antifade reagent were purchased from Molecular Probes.
Animals
All animal experiments were performed in accordance with currently prescribed guidelines and under a protocol approved by the Institutional Animal Care and Use Committee at University of Illinois at Chicago.
Cell Culture
Primary cardiomyocyte cultures were isolated from 1-day-old neonatal rats (Harlan Sprague-Dawley) as per our published procedures (10). For experiments requiring non-muscle cells, cells were obtained during the pre-plating step and cultured for appropriate density. H9C2 cells were grown in DMEM supplemented with 10% FBS. Seventy-two h prior to transfection, cells were switched to differentiation medium (DMEM + 2% FBS) and maintained in this medium throughout the experimental period.
Northern Analysis
Total RNA was isolated using TRIzol reagent and 10–20 μg of total RNA resolved in 12% urea-PAGE, transferred onto HyBond N+ nylon membrane by electroblotting in 0.5 × TBE buffer (89 mm Tris base, 89 mm boric acid, 2 mm EDTA). Pre-hybridization for 2 h at 42 °C and hybridization in the presence of the [γ-32P]ATP-labeled specific oligo probe overnight was carried out in hybridization buffer. The membranes were washed in 2× SSC buffer (150 mm NaCl, 15 mm Na3 citrate). Images were captured by exposing to the phosphorimaging screen followed by scanning on a Storm 860 scanner. Signals were quantified using the Image J program. The U6-labeled radioactive probe was used as control for all samples after membranes were stripped in 1× SSC, 0.1% SDS buffer at 72 °C.
Western Analysis
Cell lysates were prepared in urea/thiourea sample buffer (8 m urea, 2 m thiourea, 0.05 m Tris, pH 6.8, 75 mm DTT, 3% SDS, 0.05% bromphenol blue) by brief sonication, protein concentrations were determined using the RC/DC protein assay kit and equalized proteins were resolved by SDS-PAGE gel (30 μg/lane unless otherwise indicated). Western blotting was performed using standard protocols. Each membrane was stripped and re-probed with either GAPDH or β-actin primary antibody for loading control. Signal intensities were quantified using ImageJ version 1.37 software (NIH).
Caspase 3/7 Activity Assay
Caspase activity was measured by luminescent assay for caspase 3/7 detection as per the manufacturer's instructions in a 96-well plate format. Signal was measured at 491 nm using a Promega luminometer microplate reader. Values are expressed as relative light units (blank subtracted).
DNA Fragmentation by TdT-mediated dUTP Nick End Labeling (TUNEL) Staining and Viability Assay
Cardiomyocytes were grown on glass coverslips. After 72 h of transfection, cells were permeabilized and the TUNEL reaction was performed as per the manufacturer's instructions. With each assay run, a negative control (by omitting dUTP or TdT) was included. TUNEL positive cells were identified by fluorescence microscopy using an excitation wavelength of 488 nm. Cells were defined as apoptotic when the TUNEL (green) labeled nuclei were detected with a sharply demarcated and condensed morphology. ToPro reagent was used for nuclei visualization, according to the manufacturer's protocol. For quantitative data expression, TUNEL positive nuclei were counted from 10 random fields with 30–50 nuclei per field and average values were calculated from 20 different images in each group. Cell viability was measured using a MultiTox-Glo Assay kit (Promega) as per the manufacturer's instructions.
Simulated Ischemia/Reperfusion Treatment
To simulate in vivo I/R conditions, 48 h after transfections, cardiomyocytes were exposed to hypoxia and oxygenation as described previously (11). Briefly, cells plated in DMEM complete medium were placed in a Plexiglas chamber, and exposed to a constant stream of water-saturated 2% O2, 93% N2, and 5% CO2 for the indicated times. Maintenance of the desired O2 concentration was constantly monitored during incubation using a microprocessor-based oxygen sensor. Following exposure to hypoxia, the cells were re-oxygenated in a humidified tissue culture incubator with 5% CO2, 95% air. For the inhibitor experiments, the IGF-1R inhibitor PQ401 was added 30 min prior to the exposure of hypoxia.
Immunofluorescent Staining and Confocal Microscopy
Cardiomyocytes were grown on glass coverslips and processed for confocal microscopy according to our published procedures (12) using FoxO3 primary antibody and additionally with α-actinin antibody for myocyte visualization. Cells were then washed and incubated with fluorescently labeled Alexa Fluor 488- and Alexa Fluor 568-conjugated antibodies. Nuclei were visualized by ToPro reagent as per the manufacturer's protocol. Cells were washed and mounted on glass slides using ProLong Gold antifade reagent. Imaging was performed on a Bio-Rad Laser Sharp 2000 system (Bio-Rad) using a ×40 objective (Zeiss). For each experimental group, there was a minimum of three experiments with at least three replicates of each sample.
Dual Luciferase Reporter Assay
Firefly and Renilla luciferase activities were measured sequentially using a Dual Luciferase Reporter Assay System as per the manufacturer's instructions. The firefly luciferase signal was measured first at 480 nm and followed by Renilla luciferase at 560 nm in the same sample using a EG&G Berthold LUMAT LB9507 reader. Firefly luciferase activity was normalized using the Renilla luciferase signal and values were expressed as arbitrary relative light units.
DNA Constructs and Cell Transfection
A three-repeat sequence of the miR-378 predicted target region was synthesized from the 3′ UTR of human IGF1R and cloned into an Xho-Xba site in a dual luciferase reporter vector pmiRGLO. The control construct with mutations incorporated in the miR-378 seed region was generated similarly. These constructs were sequence verified. Cells were transfected with various DNA constructs and RNA oligos using Opti-MEM and Lipofectamine 2000 as per the manufacturer's protocol. All transfections were performed in triplicate in 3 independent experiments.
Real-time Polymerase Chain Reaction
Total RNA was isolated and treated with DNase I to eliminate residual genomic DNA. MicroRNA expression profiling was performed according to the methods described in Ref. 13. Briefly, a multiplex cDNA reaction was carried out where 1 μg of total RNA was primed by a pool of 24 oligo nucleotides and reverse transcribed using SuperScript III reverse transcription reagent. Real-time amplification of individual microRNAs was then performed in a SYBR Green-based assay reaction using a Fast ABI cycler and microRNA-specific F and R primers. U6 was used as a normalizing control. Real-time PCR primers for mouse Igf1R and Pgc1β were designed using Real-time PCR Primer Design, the sequence is available upon request. 2.5 μg of DNase-digested total RNA was reverse transcribed using the SuperScript III kit and random hexamers. In a 20-μl PCR, 5 ng of cDNA template was mixed with primers to a final concentration of 200 nm and 10 μl of Fast SYBR Green master mix. Amplification was carried out in a 7500 Fast Real-time PCR system by first incubating the reaction mixture at 95 °C for 20 s, followed by 40 cycles of 95 °C for 20 s and 60 °C for 30 s. For quality control purposes, at the end of each run, dissociation curves were generated by incubating the reactions at 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s, and 60 °C for 15 s. Primer pairs used in the study were free of primer dimer artifacts. Expression ratios were calculated by the ΔΔCT method, where CT is the cycle threshold, using β-actin as a reference gene.
Statistical Analysis
Data are expressed as the mean ± S.D. of at least 3 independent experiments. Control and treatment groups were matched in sets containing cells isolated and cultured on the same day to eliminate variability due to a cell batch. The two-tailed Student's t test was used for performing analysis of variance in Excel software. A p value of 0.05 or less was considered statistically significant.
RESULTS
Several MicroRNAs Are Induced in Heart after Birth
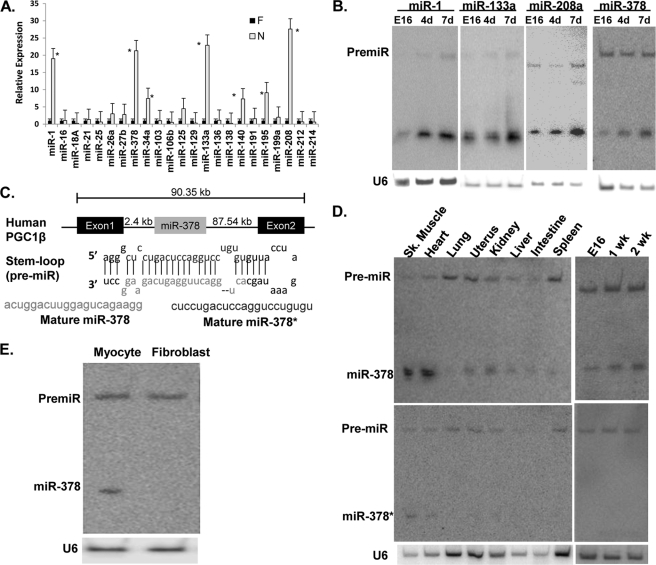
Real-time quantitative PCR of 23 randomly selected microRNAs showed increased expression of 8 microRNAs by 4-fold or more in the mouse neonatal heart (7 days after birth) when compared with fetal hearts at 16 days gestation. Among these, expression of miR-1, miR-133, miR-208a, and miR-378 was found to be increased by more than 10-fold (Fig. 1A). The findings were verified by Northern analysis (Fig. 1B), which also showed different kinetics of induction after birth (supplemental Fig. S1A), suggesting their differential roles in postnatal cardiac remodeling.
FIGURE 1.
Differential expression of microRNAs in postnatal heart. A, expression of randomly selected microRNAs in fetal (16-day gestation) and neonatal (7-day postnatal) mouse hearts by quantitative RT-PCR, asterisks mark p < 0.05 when compared with the fetal heart. B, validation of real-time PCR data by Northern analysis. C, schematic presentation and location of miR-378 in the first intron of the Pgc1β gene, stem-loop structure of pre-miR-378 and its processing into miR-378 and miR-378*. D, tissue distribution and development analysis of miR-378 and miR-378* by Northern blot analysis. E, expression of miR-378 in primary cultures of cardiomyocyte and fibroblasts obtained from the same culture (20 μg of RNA/lane). Each experiment was repeated a minimum of 3 times. U6 was used as the loading control.
High Abundance and Cardiomyocyte-specific Expression of miR-378 in Heart
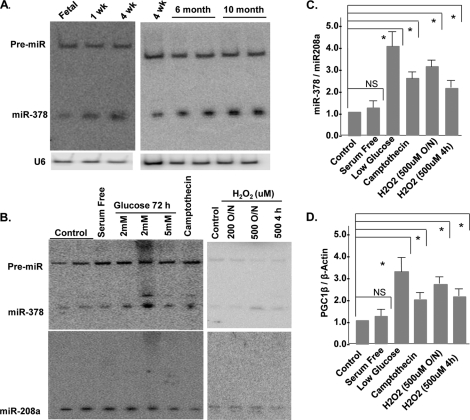
Several previous studies have suggested that miR-1, miR-133a, and miR-208a are abundantly expressed in the heart and play regulatory roles in various aspects of cardiac pathophysiology, but the role of miR-378 in the heart has never been investigated. We became interested in miR-378 because it is not only induced during the postnatal period, but is also derived from the Pgc1β gene, which has been shown to play critical roles in regulating cardiac cell metabolism (14). The first intron of the Pgc1β gene contains a stem-loop structure that gets processed into two microRNAs: miR-378 and miR-378* (Fig. 1C). We first performed a tissue distribution analysis and found that miR-378 is highly expressed in heart and skeletal muscles, low levels were found in kidney, liver, and small intestine, minimal in spleen and uterus, and none in the lung (Fig. 1D and supplemental Fig. S1B). We next examined the relative expression of miR-378* by stripping and re-probing the same membrane and found very weak cardiac expression of miR-378*. We also examined miR-378* in fetal and neonatal hearts and found that it is not inducible after birth (Fig. 1D). Therefore this study focused on miR-378 for further characterization in the heart. Intriguingly, miR-378 was found expressed only in the cardiac myocytes and not in non-muscle cells obtained during the pre-plating step of the same culture (Fig. 1E). We even used 3 times more RNA than used for cardiac myocytes and still did not find miR-378 in non-muscle cells, treatment of these cells with angiotensin II, a well known stimulator of fibroblast cell proliferation, also did not induce miR-378 (data not shown). Additionally, we observed that cardiac expression of miR-378 increased continuously as animals aged from 4 weeks to 6 months and further onto 10 months (Fig. 2A). Next, we examined if the age-related increase in miR-378 originates from its induction in aging fibroblasts, by culturing fibroblasts from the aged hearts and again found no expression (not shown), suggesting that the age-related increase of miR-378 occurs in the cardiomyocytes.
FIGURE 2.
miR378 is a stress inducible microRNA in rat neonatal cardiomyocytes. A, developmental and age-related expression of miR-378 by Northern blotting. B, expression of miR-378 following treatment of neonatal cardiomyocytes with various stressors for the indicated times and durations. Lower panel shows the expression of a non-related microRNA, miR-208, in the same membrane after stripping and hybridization with radiolabeled miR-208. C, quantification of miR-378 normalized to miR-208a, each bar is the mean ± S.D. of a minimum of 3 independent experiments. D, real-time PCR analysis of Pgc1β mRNA in the same samples as in B. Relative expression was calculated by the ΔΔCT method using β-actin as a reference gene. Asterisks mark the statistical significance (p < 0.05). NS, non-significant.
Up-regulation of miR-378 by Stress
One of the characteristic features of the aged myocardium is its greater susceptibility to stress-related injuries. Because miR-378 showed an age-related increase in expression, we asked whether it responds to stress-inducing agents. Cardiomyocytes were treated with camptothecin, which induces cell stress by inhibiting DNA topoisomerase (15), or with H2O2, a well known inducer of oxidative stress. Camptothecin and higher doses of H2O2 significantly enhanced miR-378 expression (Fig. 2, B and C). In contrast, growth promoting stimuli such as serum and the α-adrenergic receptor agonist phenylephrine (not shown) had no effect on miR-378 expression. We also examined the influence of nutrient stress on miR-378 and found that cardiomyocytes grown in low glucose (2 or 5 mm) medium had an almost 3-fold higher expression as compared with control media (25 mm glucose). The expression of the miR-378 precursor or the unrelated microRNA, miR-208a, was not influenced by any of the tested stressors. To probe into the mechanism of this stress-related induction of miR-378 expression, we examined the mRNA levels of its parent gene Pgc1β in the same RNA preparations and found a similar pattern of induction (Fig. 2D). Taken together, our results indicate that miR-378 is a stress-responsive miRNA in cardiac myocytes whose expression is co-induced with its parent gene by stressors.
IGF1R Is an Endogenous Target of miR-378-mediated Repression
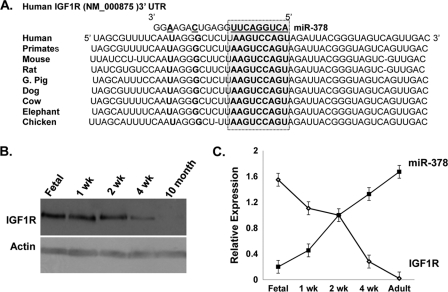
Using a bioinformatics approach, we searched for candidate miR-378 targets among genes that are known to be down-regulated during postnatal cardiac remodeling. We used prediction tools, TargetScan and Microcosm Targets, and both identified a match of a miR-378 “seed” sequence along with the flanking nucleotides within the 3′ UTR of IGF1R. This region was found highly conserved across several vertebrate species (Fig. 3A).
FIGURE 3.
IGF1R as a predicted target of miR-378. A, sequence alignment of IGF1R 3′ UTR with the miR-378 seed sequence (highlighted in the box) showing the position of the predicted binding site and species conservation. B, Western blot analysis of IGF1R in the fetal heart and after birth (50 μg of protein/lane). C, inverse expression pattern of miR-378 and IGF1R protein levels in cardiac tissues at the indicated development time. Blots are representative of a minimum of 3.
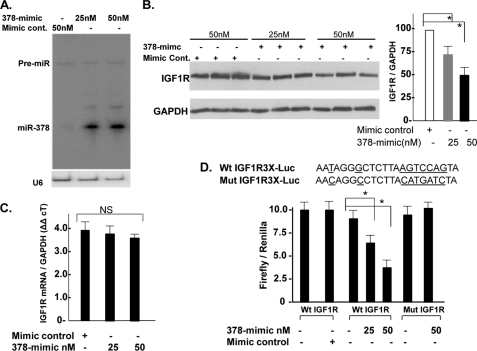
The IGF1R is comprised of two α-subunits and two β-subunits. Both α- and β-subunits are synthesized from a single mRNA precursor. The α-chains are located extracellularly, whereas the β-subunits span the membrane and are responsible for intracellular signal transduction upon ligand stimulation. Using an antibody against the β-subunit of IGF1R, we first examined the relationship between the expression levels of IGF1R and miR-378 (Figs. 2A and 3B) in 16-day-old fetal hearts versus 1 week, 2 weeks, 4 weeks, and 10 months after birth and observed an inverse relationship between miR-378 expression and IGF1R protein levels at all time points (Fig. 3C). This was also evident in fibroblasts where IGF1R is highly expressed, consistent with previous reports (16), whereas expression of miR-378 is not detected in cardiac fibroblasts (Fig. 1E). The inverse expression pattern suggested that IGF1R could serve as a target gene for miR-378. For experimental validation, gain-in-function studies of miR-378 for endogenous IGF1R were performed in cardiomyocytes. For overexpression, a synthetic, commercially available double-stranded miR-378 mimic (378-mimic) was used. For controls, a double-stranded mimic control designed with no homology to any of the miRNA sequences available in the mouse, rat, or human miRNA data base was used. Transfection of primary cultures of cardiomyocytes with 25 and 50 nm 378-mimic resulted in 10- and 15-fold higher expression of mature miR-378 with no effect on its precursor level (Fig. 4A). Western analysis showed a dose-dependent decline in IGF1R expression by 30 and 52% with 25 and 50 nm 378-mimics (Fig. 4B). A comparable decline in IGF1R expression in 378-mimic-transfected cells was also observed when we compared it against an additional control group where cells were transfected with a custom RNA oligo with the mutated seed region of miR-378 (not shown). We also evaluated IGF1R mRNA by real-time PCR and found no change with 378-mimic (Fig. 4C). Taken together our data suggest that miR-378 produces translation inhibition and not the mRNA stability of IGF1R.
FIGURE 4.
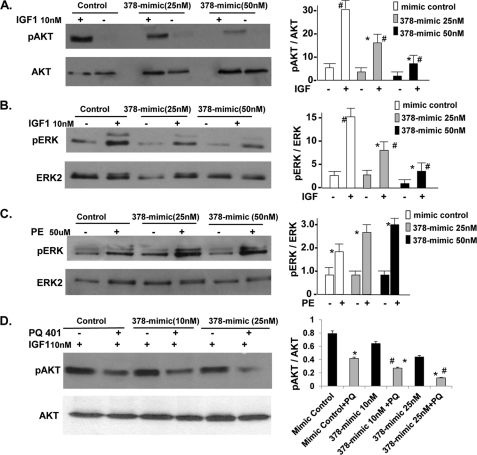
miR-378 reduces endogenous IGF1R expression by direct targeting of 3′ UTR. A, Northern analysis showing increased expression of mature miR-378 48 h following transfection of the 378-mimic in cardiac myocytes, U6 was used as a loading control. B, IGF1R expression by Western blot in triplicate (50 μg of protein/lane) in the presence of mimic control or increasing amounts of 378-mimic, GAPDH used as a loading control. Right graph is derived from 3 additional experiments. C, real-time PCR analysis of IGF1R mRNA levels in mimic control and 378-mimic transfected cells (n = 2). D, functional assay of the IGF1R 3′ UTR in H9C2 cells using a dual luciferase reporter system following transfection of various DNA constructs in the presence of 378-mimic or mimic control. The sequence shown is the predicted target sequence of IGF1R 3′ UTR, three repeats of this sequence (WtIGF1R3X-luc) or mutated sequence (underlined nucleotide mutIGF1R3X-luc) were cloned downstream of the luciferase reporter, Renilla luciferase activity was used for normalizing data. Data are derived from triplicate transfectants of 3 independent experiments. *, p < 0.05.
We next evaluated whether the predicted 3′ UTR sequence of IGF1R could be targeted by miR-378 in a luciferase-3′ UTR functional assay. To eliminate the influence of endogenous miR-378 targeting luciferase reporter, we first utilized cardiac fibroblasts, which do not express miR-378, but these cells could not survive with 378-mimic transfection. Subsequently, we analyzed miR-378 expression in H9C2 cells, a cell line derived from the embryonic rat heart. Consistent with low expression levels of miR-378 in the fetal heart, the H9C2 cells were found to express very low levels of miR-378, we had to use 3 times more RNA than from fetal heart to detect a minimal miR-378 signal (not shown). Co-transfection of wtIGF1R-luc with 378-mimic resulted in a dose-dependent reduction in luciferase activity, whereas mimic control had no effect. There was no effect on luciferase reporter, when 378-mimic was co-transfected with empty vector or with mutIGF1R3X-luc (with mutated IGF1R 3′ UTR sequence) (Fig. 4D). The incorporated mutation significantly disrupted the miR-378 seed region to form a hybrid as shown (supplemental Fig. S2) with increased minimum free energy from −27.5 kcal/mol (WT IGF1R/miR378 hybrid) to −10.7 kcal/mol (mutant IGF1R/miR-378 hybrid). Taken together, these results suggest IGF1R 3′ UTR as a bona fide miR-378 target.
It has been demonstrated that IGF1/IGF1R signaling involves activation of PI 3-kinase, the serine-threonine kinase Akt, and ERK1/2 (17). To examine the influence of miR-378 on IGF1-induced cardiac response, cardiomyocytes were either mock transfected or transfected with mimic control or 378-mimic (25 and 50 nm) in duplicates. After 48 h, cells were serum starved overnight, one set of plates was treated with IGF1 (10 nm) the other received vehicle for 15 min. IGF1 caused Akt activation (Thr308), which was comparable in mimic control and mock-transfected cells (not shown). Transfection of 378-mimic on the other hand produced a dose-dependent decline in Akt and ERK activation by IGF1 (Fig. 5, A and B). In contrast, when cells were treated with phenylephrine there was no inhibition in pERK but rather an increase was observed with 378-mimic compared with the mimic control group (Fig. 5C). It is therefore apparent from these findings that miR-378 specifically targets IGF1R-mediated signaling. To further establish a link between miR-378 and IGF1R-mediated signaling we used a specific inhibitor of IGF1R, N-(5-chloro-2-methoxyphenyl)-N′-(2-methylquinolin-4-yl) urea, also known as PQ-401, which is shown to suppress IGF1-stimulated autophosphorylation of IGF1R with an IC50 value of 12 μm (18). Cardiomyocytes were transfected and serum starved as above in duplicates, after 48 h, one set of plates was treated with PQ-401 and the other with vehicle for 90 min followed by IGF for 15 min. As shown in Fig. 5D, the inhibition of pAkt by PQ401 was augmented by miR-378. PQ401 produced about 48% reduction in pAkt in the mimic control group, whereas it produced 62 and 75% reduction when combined with 10 and 25 nm 378-mimic as compared with non-treated 378-mimic-transfected cells. The compounded effect of PQ-401 with miR-378 overexpression further suggests that this microRNA acts as a repressor of IGF1R.
FIGURE 5.
Inhibition of IGF1 but not PE-induced signaling and augmentation of PQ-401 effect on pAkt by miR-378. A, Western analysis showing pAkt (Thr308) and total Akt in neonatal rat cardiomyocytes following transfection with mimic control or 378-mimic for 48 h, cells were then serum starved overnight and treated with IGF1 (10 nm) for 15 min. B, same cell lysates as in A analyzed for pERK and total ERK2. C, cells were prepared as in A, but treated with phenylephrine (50 μm) for 5 min and analyzed for pERK and total ERK. D, cells were prepared as in A, but treated with PQ-401 (10 μm) 90 min prior to stimulation with IGF1. All results are representative of at least three independent experiments. p < 0.05, Significant when compared with the non-treated group (*) or when compared with the corresponding treated mimic control group (#).
Expression of miR-378 Sensitizes Cardiomyocytes to Cell Death Stimuli
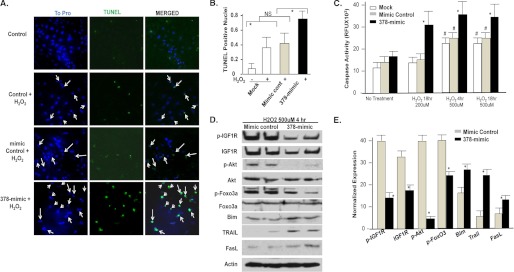
Activated Akt is a well known survival signal in cardiomyocytes particularly during stress. Data presented in Fig. 2 show that miR-378 is a stress responsive microRNA and those in Fig. 5 demonstrate that overexpression of miR-378 reduces pAkt. With this in mind, we examined the influence of 378-mimic on cardiomyocyte apoptosis induced by H2O2. Primary cultures of cardiomyocytes were either mock transfected with mimic control or 378-mimic. Evaluation of DNA fragmentation showed that a 4-h treatment with H2O2 produced a 2–2.5-fold more increase in TUNEL-positive nuclei with 378-mimic as compared with mimic control or mock-transfected cells (Fig. 6, A and B). Caspase 3/7 activity following different durations and doses of H2O2 treatment in cardiomyocytes was also found to be significantly higher with 378-mimic at all data points (Fig. 6C). More importantly, at suboptimal doses of H2O2 (200 μm, 18 h) when there was no caspase activation in control groups, overexpression of miR-378 resulted in twice as much caspase activation as observed in mock or mimic control-transfected cells. We next evaluated apoptosis inducing signaling cascade by examining the expression of FoxO3 and its transcriptional targets such as Bim, Trail, and FasL in the presence of 378-mimic or mimic control. As shown in Fig. 6, D and E, along with the reduced expression of p-IGF1R, IGF1R, and pAkt, 378-mimic also produced significantly lower levels of pFoxO3 accompanied by increased expression of Bim, Trail, and FasL (Fig. 6, D and E). These findings collectively suggest that enhanced expression of miR-378 makes cardiomyocytes more susceptible to caspase activation and DNA fragmentation in response to stress inducing agents, consistent with increased cell death. This may have an in vivo implication in imparting greater susceptibility of the aged myocardium (where miR-378 is found increased) to stress-related injuries.
FIGURE 6.
Effect of miR378 on cell survival. A, immunofluorescence images of cardiomyocytes 72 h after either mock transfection (control), mimic control or 378-mimic, and following a 4-h treatment with H2O2 (500 μm) showing ToPro-stained nuclei (blue), after TUNEL staining (green), and the two images merged together. Arrows mark the nuclei considered as TUNEL positive. B, quantification of TUNEL-positive nuclei as described for the different treatment groups. C, caspase 3/7 activity in cardiomyocytes transfected as in A and treated with H2O2 as indicated. #, significantly different from non-treated group; *, significant when compared with mock or mimic control transfected groups treated with H2O2 for the same period. D, Western analysis of IGF1R and signaling cascade in cardiomyocytes transfected as in A and after a 4-h H2O2 treatment. E, quantification of the signal intensity of D. Expression levels of IGF1R, pIGF1R, Bim, Trail, and FasL were normalized with β-actin, whereas pAkt and pFoxO3 were normalized with their non-phospho counterparts. *, p < 0.05. Each bar is a mean ± S.D. of a minimum of 3 independent experiments.
Inhibition of Endogenous miR-378 Induces IGF1R and pAkt Levels
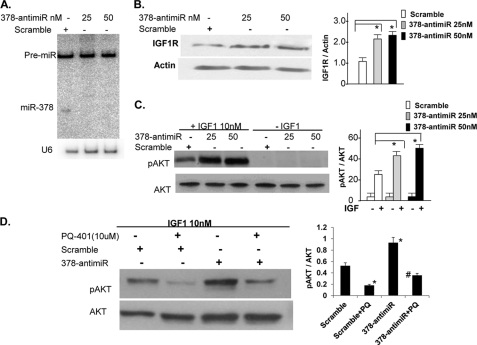
We also took a complementary approach where we knocked down miR-378 in cultured cardiomyocytes and evaluated Akt activation by IGF1. The knockdown of miR-378 was achieved by a commercially available 378-anti-miR and verified by Northern analysis (Fig. 7A). For controls, we used a single-stranded scramble oligo, which was designed not to target any known miR sequences. Evaluation of IGF1R expression by Western blots showed about 2–2.5-fold induction with 378-anti-miR in relationship to the scramble control group (Fig. 7B). The pAkt levels by IGF1 activation were also found significantly higher (9–10-fold) in the 378-anti-miR group than the scramble control group (only 5-fold-induction) in relationship to the corresponding non-treated cells (Fig. 7C). To analyze the role of IGF1R in 378-anti-miR-mediated enhancement of Akt activation, we performed the same experiment but in the presence of a IGF1R inhibitor, PQ-401. As shown in Fig. 7D, 378-anti-miR antagonized the inhibition of pAkt caused by PQ-401. The role of IGF1R signaling in 378-anti-miR-mediated Akt activation was also evident when we used a combined inhibition of IGF1R and downstream PI3K, the 378-anti-miR again minimized the effect of these inhibitors on Akt activation (supplemental Fig. S3). Taken together our results support that knockdown of miR-378 enhances IGF1R and its mediated signaling.
FIGURE 7.
miR-378 knockdown enhances IGF1R expression and IGF1-induced AKT activation. A, Northern blot showing knockdown of miR-378 in cardiac myocytes 48 h after transfection with either scramble control or 378-anti-miR, U6 represents the loading control on the same membrane. B, Western analysis of IGF1R 72 h after transfection of cardiomyocytes with scramble or increasing amounts of 378-anti-miR. C, expression of pAkt and total Akt in cardiomyocytes transfected with scramble control or 378-anti-miR. After a 48-h transfection, cells were incubated in serum-free media overnight and then treated with IGF1 for 15 min. D, cells were prepared as in C and treated with the IGF1R inhibitor PQ-401 (10 μm) 90 min prior to IGF1 treatment. p < 0.05, *, compared with non-PQ treated scramble control; #, compared with the PQ-401-treated scramble control (n = 2).
Inhibition of miR-378 Prevents H2O2-induced Activation of Apoptosis Signaling Cascade and Hypoxia/Reoxygenation-induced DNA Damage in IGF1R-dependent Manner
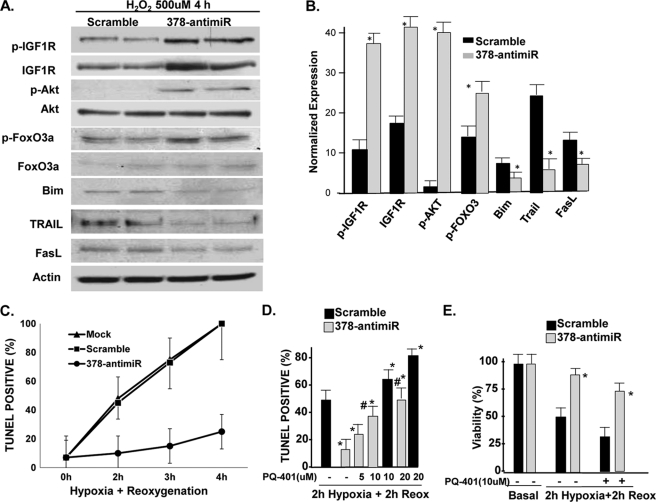
Several groups have shown that one of the mechanisms by which pAkt promotes cell survival under conditions of stress is by phosphorylating the FoxOs, promoting their nuclear exportation, and thereby inhibiting their ability to induce the apoptosis gene program (19). We examined 378-anti-miR in the context of pFoxO and its downstream apoptosis-inducing signaling cascade, such as Bim, Trail, and FasL in cardiomyocytes following H2O2 treatment (500 μm, 4 h). As shown in Fig. 8, A and B, 378-anti-miR not only induced the expression levels of pIGF1R, IGF1R, and pFoxO3, but significantly reduced the expression of Bim, Trail, and FasL. We also tested 378-anti-miR in another oxidative injury model of hypoxia/reoxygenation. As shown in Fig. 8C and supplemental Fig. S4, sequential exposure of cardiomyocytes to hypoxia and reoxygenation for increasing durations (2, 3, and 4 h) increased the number of TUNEL-positive nuclei in mock-transfected cells, which was similar to the scramble control group. Transfection of 378-anti-miR drastically reduced cell death at each time point such that at 4 h of hypoxia and 4 h of reoxygenation, there was almost 75% reduction in the number of TUNEL-positive nuclei in the 378-anti-miR group compared with scramble control. To examine the involvement of IGF1R in 378-anti-miR-mediated cardioprotection, cells were transfected as above with scramble or 378-anti-miR for 48 h and then exposed to varying amounts of PQ-401 for 90 min followed by 2 h hypoxia and 2 h reoxygenation. As shown in Fig. 8, D and E and supplemental Fig. S5, the presence of PQ-401 antagonized the 378-anti-miR-mediated cardioprotection on both viability as well as the number of TUNEL-positive cells. These results suggest that IGF1R plays a critical role in 378-anti-miR-mediated inhibition of cell death.
FIGURE 8.
miR-378 knockdown prevents the H2O2-induced apoptosis program and hypoxia/reoxygenation-induced cell death in an IGF1R-dependent manner. A, Western analysis (duplicates) of pIGF1R, IGF1R, and downstream signaling cascade in response to oxidative stress following transfection with either scramble control or 378-anti-miR. The same membrane was used again and again for probing with different antibodies after stripping. B, quantification of the signal intensity of A normalized essentially as described in the legend to Fig. 6E. C, time course response of TUNEL-positive nuclei in response to varying periods of hypoxia/reoxygenation in the presence of either scramble control or 378-anti-miR (50 nm). Each bar is a mean ± S.D. of a minimum of 3 independent experiments. D, quantification of TUNEL positive nuclei as described for the indicated treatment groups after 2 h of hypoxia followed by 2 h of reoxygenation injury to cardiac myocytes. The IGF1R inhibitor PQ-401(PQ) produced a dose-dependent inhibition of protective effects of 378-anti-miR. E, 378-anti-miR enhanced cardiomyocyte viability. Significant p < 0.05 (*) when compared with scramble control group and (#) when compared with the scramble PQ-treated group.
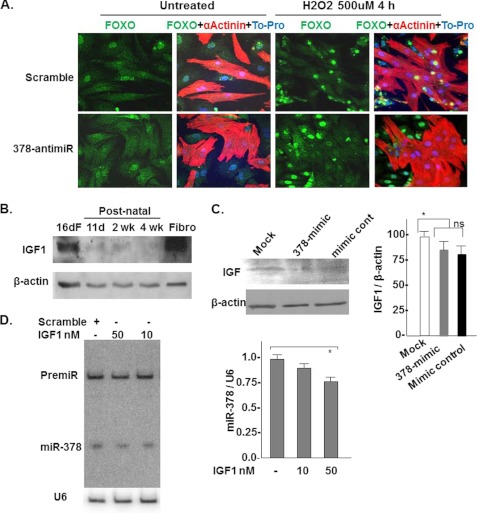
We also examined whether 378-anti-miR would influence the subcellular distribution of FoxO3 under oxidative stress conditions induced by H2O2 (500 μm, 4 h). Confocal microscopy was used to visualize FoxO3 distribution in co-cultures of cardiac myocytes and cardiac fibroblasts. Our reason for using co-culture for this particular experiment was related to the myocyte-specific expression of miR-378 (Fig. 1E). Therefore, following oxidative stress one would expect to see interference of the nuclear presence of FoxO3 in myocytes and not in non-muscle cells. Myocytes were identified by counterstaining with α-actinin. As shown in Fig. 9A, under basal non-treated conditions, transfection of cells with scramble or 378-anti-mR did not change the distribution in muscle or non-muscle cells in FoxO3. But when these cells were treated with H2O2, in the scramble control group nuclear translocation of FoxO3 was observed in both muscle as well as non-muscle cells, in contrast, with the 378-anti-miR transfection, FoxO3 immunofluorescence was not detected in nuclei of α-actinin positive (muscle) cells, whereas non-muscle cells showed nuclear FoxO3. Together these results indicate that in the presence of oxidative stress, 378-anti-miR interferes with FoxO3-mediated induction of the apoptosis signaling cascade preferentially in cardiomyocytes.
FIGURE 9.
miR-378 inhibition interferes with FoxO3 translocation in the cardiomyocyte nuclei. IGF1 is a negative regulator of miR378. A, confocal imaging showing FoxO3 subcellular distribution in co-cultures of neonatal cardiomyocyte and non-muscle cells 72 h after transfection with scramble control or 378-anti-miR with and without H2O2 treatment for 4 h. FoxO3 can be visualized as green immunofluorescence, nuclei as blue, myocytes as red immunofluorescence α-actinin positive cells, and non-muscle cells as absence of red fluorescence. Note, with 378-anti-miR transfection and H2O2 treatment FoxO3 was excluded only from the nuclei (seen as purple-stained nuclei in the merged image) of red-stained muscle cells, whereas its presence was detected in the nuclei of the scramble control transfected muscle cells (seen as intense yellow staining in the merged image). B, IGF1 expression and time course by Western blot analysis in fetal and postnatal heart tissues and in cultured neonatal cardiac fibroblasts. C, miR-378 overexpression does not significantly affect IGF1 expression in cardiomyocytes. D, IGF1 acts as a negative regulator of miR-378. Primary cultures of cardiomyocytes were treated with increasing doses of IGF1 for 72 h. miR-378 expression was analyzed by Northern. A modest but significant reduction was noted with a higher dose of IGF1 (n = 2).
IGF1 Acts as a Negative Regulator of miR-378
We also examined if miR-378 plays a role in the down-regulation of IGF1 seen during the postnatal period (Fig. 9B). For this cardiomyocytes were either mock transfected or with mimic control or 378-mimics, and the cell lysates were analyzed for IGF1 expression by Western. As shown in Fig. 9C and supplemental Fig. S6, although lower expression of IGF1 was noted when the 378-mimic group was compared with the mimic control group the difference did not reach statistical significance. The IGF1 expression was significantly different between two controls (mock and mimic control cells). We next asked whether IGF1, which is a ligand for IGF1R would regulate the miR-378 expression. For this, cardiac myocytes were treated with increasing doses of IGF1 (10 and 50 nm) for 48 h and miR-378 expression was evaluated by Northern blot analysis. IGF1 caused a dose-dependent reduction in miR-378 expression (Fig. 9D). These results together indicate that miR-378 does not influence the expression of IGF1 significantly but IGF1 acts as an inhibitor of miR-378 suggesting that the high levels of IGF1 before birth could be contributing to the reduced level of miR-378 observed in the fetal heart.
DISCUSSION
It has been long appreciated that the IGF1/IGF1R system is down-regulated during maturation of the heart after birth, which appears to be coupled to the attenuation of cardiomyocyte proliferation and the reduced uptake and metabolism of glucose by cardiomyocytes. The underlying mechanism of IGF1/IGF1R down-regulation remains unknown. In this study we provide the first evidence of down-regulation of IGF1R in cardiac myocytes by miR-378. The data supporting this conclusion are as follows. First, cardiac expression of miR-378 is significantly lower in the fetal heart and induction of miR-378 after birth was parallel with the diminution of IGF1R expression starting 1 week after birth and continuing thereafter. Second, a highly conserved target sequence of miR-378 is present in the 3′ UTR of IGF1R. When cloned downstream of the luciferase reporter, this sequence led to reduced luciferase activity in the presence of miR-378. Third, when overexpressed, miR-378 reduced the expression level of endogenous IGF1R without influencing IGF1R mRNA stability, whereas inhibition of miR-378 by 378-anti-miR resulted in the increased endogenous level of IGF1R beyond normal. From these data it is apparent that IGF1R is a direct target of miR-378, and that up-regulation of miR-378 after birth could contribute to postnatal repression of IGF1R.
It is known that miR-378 is the product of the guide strand of a miRNA hairpin located in the first intron of Pgc1β, the complementary strand also has the potential of processing into miR-378*. We found that in the heart, miR-378 is the predominant species, whereas the expression level of miR-378* is minimal. This is in contrast to a breast cancer cell line, BT-474, where both miR-378 and miR-378* mature forms were equally detected following transfection with 378-pre-miR (20) suggesting cell-type specificity in the processing of 378-pre-miR. Cell type-specific processing of miR-378 is also suggested by our data where the mature form of miR-378 is only in cardiomyocytes and not detected in non-muscle cells, whereas pre-miR expression was equally detected in both types of cells. In addition, our tissue distribution analysis of miR-378 also showed comparable or in some tissues even higher levels of 378-pre-miR expression, yet, the mature form of miR-378 was predominantly observed only in striated muscles, further supporting the role of tissue-specific factors in 378-pre-miR processing. A recent study also suggested cell type-specific factors in eliciting transactivation of miR-378 by c-Myc, and its oncogenic potential in c-Myc-driven cell transformation (21). Our findings of induced miR-378 expression during the postnatal period suggests that processing of 378-pre-miR in the heart is also subject to regulation by developmental factors. Previously, in carcinoma cell lines U87 and MT-1, as well as in pooled cells, transfection of the 378-pre-miR hairpin was reported to promote tumor growth and angiogenesis. This study showed the miR-378* seed sequence (addressed in this report as miR-378) targets transcription factors SuFu and Fus-1 (22) and promotes cell survival, whereas in our study we observed an apoptosis-inducing role of miR-378 in cardiomyocytes. Additionally, miR-378* has been shown to promote a metabolic switch from OXPHOS to glycolysis in the BT-474 breast cancer cell line (20). The role of miR-378 was not investigated in this study. The findings of our study demonstrate that miR-378 directly targets IGF1R (which is known to promote glucose uptake and metabolism) and suggests that miR-378 may also have a role in the metabolic switch that occurs in the postnatal heart. If so, miR-378 must work in collaboration with its parent gene, which is known to stimulate the expression of genes involved in OXPHOS metabolism rather than in a contradictory manner as seen for miR-378* in the BT-474 breast cancer cell line. This speculation needs to be examined in future experiments.
Previous studies have shown that cardiac myocytes possess IGF1R and are also capable of synthesizing and secreting IGF1. The local IGF1/IGF1R system plays a regulatory role in cardiomyocyte growth and survival. In transgenic mice, cardiac-specific overexpression of IGR1R triggered myocyte hypertrophy and enhanced systolic function (23). IGF1R signaling is also found to be essential in inducing physiological growth associated with exercise-induced cardiac hypertrophy (24). Exogenous administration of IGF1 in vivo reduces cardiomyocyte apoptosis in response to stress injury (25–28). Intriguingly, genetic studies conducted in murine models indicate that an excess of IGF1 signaling may also trigger cardiac dysfunction (29). This emphasizes the importance of gaining a better understanding of the mechanisms controlling IGF1/IGF1R regulation and gene transcription.
We show that miR-378 targets IGF1R, and the ligand IGF1 acts as a negative regulator of miR-378. This is evident in vitro as treatment of cardiomyocytes with IGF1 reduced the expression of endogenous miR-378. In vivo, we observed a negative correlation between high levels of IGF1 with reduced levels of miR-378 in fetal heart and no expression in fibroblasts. Thus, our study provides evidence of the existence of a feedback loop where miR-378 is reducing the expression of IGF1R and the ligand IGF1 is inhibiting miR-378. Our findings show that miR-378 does not significantly affect IGF1 expression in cardiomyocytes. Previous studies have shown miR-1 as a repressor of IGF1 and IGF1R in skeletal muscle during differentiation as well as in the heart (30) and down-regulation of miR-1 is linked to IGF1 induction in myocardial infarction (31). We found that miR-1 expression is induced in the heart after birth; therefore it is likely that the repression of IGF1/IGF1R signaling during the postnatal period must be the result of concerted activity of more than one microRNA in the heart.
The regulation of programmed cell death is crucial to normal physiology of almost all multicellular organisms. Increasing evidence demonstrates that apoptosis contributes to tissue injury in several cardiac disorders including myocardial infarction, atherosclerosis, myocarditis, cardiac failure, and during cardiac transplantation. In rodent and humans it is shown that about 5 to 30% of cardiac myocytes undergo apoptosis within hours of reperfusion injury (32–36), and higher apoptosis can be observed even after several months of ischemic insult (37, 38). Genetic approaches using rodents have determined that inhibition of apoptosis in cardiac myocytes reduces infarct size by more than 50% (39, 40). In line with these reports, short-term treatment with IGF-1, and cardiomyocyte-specific transgene expression of IGF1 has been reported to improve cardiac function and counteract the occurrence of apoptosis in experimental myocardial infarction and in heart failure of a murine model (41, 42). Because IGF1 is a well accepted survival factor, our data suggest that one of the mechanisms by which IGF1 protects cardiomyocytes may involve its ability to inhibit miR-378, thereby enhancing the expression of its receptor to trigger cell survival signals. This is evident from our data, which showed that during oxidative stress induced by H2O2 and by simulated ischemia/reperfusion injury, elimination of miR-378 by 378-anti-miR led to enhanced cell survival by promoting IGF1R and pAkt expression and reducing levels of apoptosis inducing factors, Bim, Trail, and FasL. Interestingly, in line with our observation of cardiomyocyte-specific expression of miR-378, we observed that following H2O2 challenge, inhibition of miR-378 resulted in cytoplasmic retention of FoxO3 only in cardiomyocytes, whereas the nuclear presence of FoxO3 was detected in non-muscle cells in the same culture. Therefore it would be possible to promote cardiomyocyte survival with 378-anti-miR implicating its therapeutic potential in conditions of ischemic injury where cardiomyocyte apoptosis contributes significantly to the disease process.
During the embryonic period, IGF1/IGF1R signaling is associated with the proliferation of cardiac myocytes. In cultures, stimulation of the IGF1/IGF1R system is known to activate DNA synthesis in neonatal cardiomyocytes (43) and promote cell cycle progression and cell proliferation in fibroblasts (16). The antisense inhibition of IGF1R markedly attenuates cell proliferation (8). Although we have not examined the influence of miR-378 on cell proliferation or cell cycle progression, our finding of induction of miR-378 during the postnatal period suggests that similar to miR-195, which is also induced in the postnatal heart and suppresses cardiac cell proliferation (6), miR-378 may exert similar effects by inhibiting IGF1R signaling. Another interesting aspect of our finding is the increased miR-378 expression in aged hearts. This would be important because cardiomyocytes isolated from IGF1 transgenic mice show attenuated response of age-related increases in markers of growth arrest and senescence, such as p27kip1, p16INK4a, p53, and p19ARF (44), suggesting that 378-anti-miR may also block such senescence markers by enhancing IGF1R signaling specifically in cardiac myocytes.
Our data presented in this study provide the first evidence of a role of miR-378 in the heart and indicates that 378-anti-miR could provide resistance to cardiac myocytes against stress-mediated cell death. Future studies directed toward examining the beneficial effect of miR-378 inhibition in conditions of pathological stresses at the whole organ level will provide further support to the therapeutic potential of 378-anti-miR. In addition, such interventions in young versus old animals are likely to shed more light on its role in age-related susceptibility of cardiomyocyte to stress-related injury.
Supplementary Material
Acknowledgment
We thank Dr. Shahab Akhtar, University of Chicago, for the use of the hypoxia-oxygenation chamber.
This work was supported, in whole or in part, by National Institutes of Health Grant Multi P.I. Grants 2RO1 HL 22231 (to M. G. and R. J. S.), PO1 HL 062626-Project 1 (to R. J. S.), RO1 HL 83423 (to M. P. G.), and T32 HL 07692 (to I. K.).

This article contains supplemental Figs. S1–S6.
- miRNA
- microRNA
- IGF
- insulin-like growth factor.
REFERENCES
- 1. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 2. Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., Cuppen E. (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120, 21–24 [DOI] [PubMed] [Google Scholar]
- 3. Small E. M., Olson E. N. (2011) Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J. F., Murchison E. P., Tang R., Callis T. E., Tatsuguchi M., Deng Z., Rojas M., Hammond S. M., Schneider M. D., Selzman C. H., Meissner G., Patterson C., Hannon G. J., Wang D. Z. (2008) Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. U.S.A. 105, 2111–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. da Costa Martins P. A., Bourajjaj M., Gladka M., Kortland M., van Oort R. J., Pinto Y. M., Molkentin J. D., De Windt L. J. (2008) Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 118, 1567–1576 [DOI] [PubMed] [Google Scholar]
- 6. Porrello E. R., Johnson B. A., Aurora A. B., Simpson E., Nam Y. J., Matkovich S. J., Dorn G. W., 2nd, van Rooij E., Olson E. N. (2011) MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 109, 670–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng W., Reiss K., Kajstura J., Kowal K., Quaini F., Anversa P. (1995) Down-regulation of the IGF-1 system parallels the attenuation in the proliferative capacity of rat ventricular myocytes during postnatal development. Lab. Invest. 72, 646–655 [PubMed] [Google Scholar]
- 8. Anversa P., Kajstura J., Cheng W., Reiss K., Cigola E., Olivetti G. (1996) Insulin-like growth factor-1 and myocyte growth, the danger of a dogma. Part I. Postnatal myocardial development, normal growth. Cardiovasc. Res. 32, 219–225 [DOI] [PubMed] [Google Scholar]
- 9. Torres-Aleman I., Pons S., Arévalo M. A. (1994) The insulin-like growth factor I system in the rat cerebellum. Developmental regulation and role in neuronal survival and differentiation. J. Neurosci. Res. 39, 117–126 [DOI] [PubMed] [Google Scholar]
- 10. Sulaiman M., Matta M. J., Sunderesan N. R., Gupta M. P., Periasamy M., Gupta M. (2010) Resveratrol, an activator of SIRT1, up-regulates sarcoplasmic calcium ATPase and improves cardiac function in diabetic cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 298, H833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malhotra R., Valuckaite V., Staron M. L., Theccanat T., D'Souza K. M., Alverdy J. C., Akhter S. A. (2011) High-molecular weight polyethylene glycol protects cardiac myocytes from hypoxia- and reoxygenation-induced cell death and preserves ventricular function. Am. J. Physiol. Heart Circ. Physiol. 300, H1733–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sundaresan N. R., Gupta M., Kim G., Rajamohan S. B., Isbatan A., Gupta M. P. (2009) Sirt3 blocks the cardiac hypertrophic response by augmenting FoxO3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 119, 2758–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X. (2009) A PCR-based platform for microRNA expression profiling studies. RNA 15, 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sonoda J., Mehl I. R., Chong L. W., Nofsinger R. R., Evans R. M. (2007) PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. U.S.A. 104, 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260, 14873–14878 [PubMed] [Google Scholar]
- 16. Reiss K., Cheng W., Kajstura J., Sonnenblick E. H., Meggs L. G., Anversa P. (1995) Fibroblast proliferation during myocardial development in rats is regulated by IGF-1 receptors. Am. J. Physiol. 269, H943–951 [DOI] [PubMed] [Google Scholar]
- 17. Bueno O. F., De Windt L. J., Tymitz K. M., Witt S. A., Kimball T. R., Klevitsky R., Hewett T. E., Jones S. P., Lefer D. J., Peng C. F., Kitsis R. N., Molkentin J. D. (2000) The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19, 6341–6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gable K. L., Maddux B. A., Penaranda C., Zavodovskaya M., Campbell M. J., Lobo M., Robinson L., Schow S., Kerner J. A., Goldfine I. D., Youngren J. F. (2006) Diarylureas are small-molecule inhibitors of insulin-like growth factor I receptor signaling and breast cancer cell growth. Mol. Cancer Ther. 5, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 19. Ronnebaum S. M., Patterson C. (2010) The FoxO family in cardiac function and dysfunction. Annu. Rev. Physiol. 72, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eichner L. J., Perry M. C., Dufour C. R., Bertos N., Park M., St-Pierre J., Giguère V. (2010) miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-β/ERRγ transcriptional pathway. Cell Metab. 12, 352–361 [DOI] [PubMed] [Google Scholar]
- 21. Feng M., Li Z., Aau M., Wong C. H., Yang X., Yu Q. (2011) Myc/miR-378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene 30, 2242–2251 [DOI] [PubMed] [Google Scholar]
- 22. Lee D. Y., Deng Z., Wang C. H., Yang B. B. (2007) MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. U.S.A. 104, 20350–20355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McMullen J. R., Shioi T., Huang W. Y., Zhang L., Tarnavski O., Bisping E., Schinke M., Kong S., Sherwood M. C., Brown J., Riggi L., Kang P. M., Izumo S. (2004) The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110α) pathway. J. Biol. Chem. 279, 4782–4793 [DOI] [PubMed] [Google Scholar]
- 24. Kim J., Wende A. R., Sena S., Theobald H. A., Soto J., Sloan C., Wayment B. E., Litwin S. E., Holzenberger M., LeRoith D., Abel E. D. (2008) Insulin-like growth factor I receptor signaling is required for exercise-induced cardiac hypertrophy. Mol. Endocrinol. 22, 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Samarel A. M. (2002) IGF-1 Overexpression rescues the failing heart. Circ. Res. 90, 631–633 [DOI] [PubMed] [Google Scholar]
- 26. Morales M. P., Gálvez A., Eltit J. M., Ocaranza P., Díaz-Araya G., Lavandero S. (2000) IGF-1 regulates apoptosis of cardiac myocyte induced by osmotic stress. Biochem. Biophys. Res. Commun. 270, 1029–1035 [DOI] [PubMed] [Google Scholar]
- 27. Ren J., Samson W. K., Sowers J. R. (1999) Insulin-like growth factor I as a cardiac hormone. Physiological and pathophysiological implications in heart disease. J. Mol. Cell Cardiol. 31, 2049–2061 [DOI] [PubMed] [Google Scholar]
- 28. Wang L., Ma W., Markovich R., Chen J. W., Wang P. H. (1998) Regulation of cardiomyocyte apoptotic signaling by insulin-like growth factor I. Circ. Res. 83, 516–522 [DOI] [PubMed] [Google Scholar]
- 29. Delaughter M. C., Taffet G. E., Fiorotto M. L., Entman M. L., Schwartz R. J. (1999) Local insulin-like growth factor I expression induces physiologic, then pathologic, cardiac hypertrophy in transgenic mice. FASEB J. 13, 1923–1929 [DOI] [PubMed] [Google Scholar]
- 30. Elia L., Contu R., Quintavalle M., Varrone F., Chimenti C., Russo M. A., Cimino V., De Marinis L., Frustaci A., Catalucci D., Condorelli G. (2009) Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 120, 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shan Z. X., Lin Q. X., Fu Y. H., Deng C. Y., Zhou Z. L., Zhu J. N., Liu X. Y., Zhang Y. Y., Li Y., Lin S. G., Yu X. Y. (2009) Up-regulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem. Biophys. Res. Commun. 381, 597–601 [DOI] [PubMed] [Google Scholar]
- 32. Fliss H., Gattinger D. (1996) Apoptosis in ischemic and reperfused rat myocardium. Circ. Res. 79, 949–956 [DOI] [PubMed] [Google Scholar]
- 33. Bialik S., Geenen D. L., Sasson I. E., Cheng R., Horner J. W., Evans S. M., Lord E. M., Koch C. J., Kitsis R. N. (1997) Myocyte apoptosis during acute myocardial infarction in the mouse localizes to hypoxic regions but occurs independently of p53. J. Clin. Invest. 100, 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olivetti G., Quaini F., Sala R., Lagrasta C., Corradi D., Bonacina E., Gambert S. R., Cigola E., Anversa P. (1996) Acute myocardial infarction in humans is associated with activation of programmed myocyte cell death in the surviving portion of the heart. J. Mol. Cell Cardiol. 28, 2005–2016 [DOI] [PubMed] [Google Scholar]
- 35. Palojoki E., Saraste A., Eriksson A., Pulkki K., Kallajoki M., Voipio-Pulkki L. M., Tikkanen I. (2001) Cardiomyocyte apoptosis and ventricular remodeling after myocardial infarction in rats. Am. J. Physiol. Heart Circ. Physiol. 280, H2726–2731 [DOI] [PubMed] [Google Scholar]
- 36. Wencker D., Chandra M., Nguyen K., Miao W., Garantziotis S., Factor S. M., Shirani J., Armstrong R. C., Kitsis R. N. (2003) A mechanistic role for cardiac myocyte apoptosis in heart failure. J. Clin. Invest. 111, 1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerra S., Leri A., Wang X., Finato N., Di Loreto C., Beltrami C. A., Kajstura J., Anversa P. (1999) Myocyte death in the failing human heart is gender dependent. Circ. Res. 85, 856–866 [DOI] [PubMed] [Google Scholar]
- 38. Olivetti G., Abbi R., Quaini F., Kajstura J., Cheng W., Nitahara J. A., Quaini E., Di Loreto C., Beltrami C. A., Krajewski S., Reed J. C., Anversa P. (1997) Apoptosis in the failing human heart. N. Engl. J. Med. 336, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 39. Chen Z., Chua C. C., Ho Y. S., Hamdy R. C., Chua B. H. (2001) Overexpression of Bcl-2 attenuates apoptosis and protects against myocardial I/R injury in transgenic mice. Am. J. Physiol. Heart Circ. Physiol. 280, H2313–2320 [DOI] [PubMed] [Google Scholar]
- 40. Miao W., Luo Z., Kitsis R. N., Walsh K. (2000) Intracoronary, adenovirus-mediated Akt gene transfer in heart limits infarct size following ischemia-reperfusion injury in vivo. J. Mol. Cell Cardiol. 32, 2397–2402 [DOI] [PubMed] [Google Scholar]
- 41. Davis M. E., Hsieh P. C., Takahashi T., Song Q., Zhang S., Kamm R. D., Grodzinsky A. J., Anversa P., Lee R. T. (2006) Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc. Natl. Acad. Sci. U.S.A. 103, 8155–8160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Welch S., Plank D., Witt S., Glascock B., Schaefer E., Chimenti S., Andreoli A. M., Limana F., Leri A., Kajstura J., Anversa P., Sussman M. A. (2002) Cardiac-specific IGF-1 expression attenuates dilated cardiomyopathy in tropomodulin-overexpressing transgenic mice. Circ. Res. 90, 641–648 [DOI] [PubMed] [Google Scholar]
- 43. Kardami E. (1990) Stimulation and inhibition of cardiac myocyte proliferation in vitro. Mol. Cell. Biochem. 92, 129–135 [DOI] [PubMed] [Google Scholar]
- 44. Torella D., Rota M., Nurzynska D., Musso E., Monsen A., Shiraishi I., Zias E., Walsh K., Rosenzweig A., Sussman M. A., Urbanek K., Nadal-Ginard B., Kajstura J., Anversa P., Leri A. (2004) Cardiac stem cell and myocyte aging, heart failure, and insulin-like growth factor-1 overexpression. Circ. Res. 94, 514–524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.