Background: The mammalian deubiquitinase that hydrolyzes the ubiquitin-PEX5 thioester conjugate was unknown.
Results: USP9X was found to be the most active deubiquitinase acting on ubiquitin-PEX5.
Conclusion: We propose that USP9X participates in the PEX5-mediated peroxisomal protein import pathway.
Significance: The unbiased biochemical strategy described here will be useful to identify deubiquitinases acting on other substrates.
Keywords: Deubiquitination, Enzyme Purification, Peroxisomes, Protein Translocation, Receptors, PEX5, USP9X, Monoubiquitination, Thioester
Abstract
Peroxin 5 (PEX5), the peroxisomal protein shuttling receptor, binds newly synthesized peroxisomal matrix proteins in the cytosol and promotes their translocation across the organelle membrane. During the translocation step, PEX5 itself becomes inserted into the peroxisomal docking/translocation machinery. PEX5 is then monoubiquitinated at a conserved cysteine residue and extracted back into the cytosol in an ATP-dependent manner. We have previously shown that the ubiquitin-PEX5 thioester conjugate (Ub-PEX5) released into the cytosol can be efficiently disrupted by physiological concentrations of glutathione, raising the possibility that a fraction of Ub-PEX5 is nonenzymatically deubiquitinated in vivo. However, data suggesting that Ub-PEX5 is also a target of a deubiquitinase were also obtained in that work. Here, we used an unbiased biochemical approach to identify this enzyme. Our results suggest that ubiquitin-specific protease 9X (USP9X) is by far the most active deubiquitinase acting on Ub-PEX5, both in female rat liver and HeLa cells. We also show that USP9X is an elongated monomeric protein with the capacity to hydrolyze thioester, isopeptide, and peptide bonds. The strategy described here will be useful in identifying deubiquitinases acting on other ubiquitin conjugates.
Introduction
Peroxisomal matrix proteins are synthesized on cytosolic ribosomes and post-translationally imported into the organelle (1). The vast majority of these proteins possess a peroxisomal targeting sequence type 1, a tripeptide with the sequence SKL, or similar, present at the C terminus (2). A minor fraction of matrix proteins contains instead a peroxisomal targeting sequence type 2, a degenerated nonapeptide located at their N termini (3). In mammals, plants, and many other organisms, both PTS1- and PTS2-containing proteins are targeted to the organelle by PEX5, the so-called peroxisomal shuttling receptor (4–7). The PEX5-PTS1 interaction is direct, whereas PTS2-containing proteins bind PEX5 via the adaptor protein PEX7 (8, 9).
PEX5 is a monomeric protein in solution (10, 11) displaying a dual subcellular localization, cytosolic and peroxisomal (12). Structurally, it can be divided into two domains: a C-terminal half containing seven tetratricopeptide repeats organized into a ring-like structure (13, 14) and a natively unfolded N-terminal half (11, 15). Although it has been shown that the C-terminal half of PEX5 has a crucial role in the interaction with the PTS1 (13, 14), it is becoming increasingly apparent that the N-terminal half of PEX5 also contributes to the interaction with cargo proteins (16–20). Interestingly, recent data suggest that PEX5 may also act as a chaperone/holdase at least for some peroxisomal matrix proteins (16).
According to current models (21–24), PEX5 recognizes newly synthesized proteins in the cytosol and targets them to the docking/translocation machinery (DTM),7 a multisubunit protein complex present in the peroxisomal membrane (25, 26). Here, the PEX5-cargo protein complex becomes inserted into the DTM with the concomitant translocation of the cargo protein across the organelle membrane (27, 28). In at least one case, that of the PTS2-containing protein thiolase, it is also at this step that PEX5 releases its cargo into the peroxisomal matrix (29). Notably, in vitro import experiments suggest that none of these steps requires hydrolysis of ATP, suggesting that the driving force for peroxisomal protein import relies on the strong protein-protein interactions that are established between PEX5 on one side and members of the DTM on the other side (29–32). PEX5 is then recycled back into the cytosol in a process that includes two steps as follows: first, PEX5 is monoubiquitinated at a conserved cysteine residue (cysteine 11 in the human protein) (33–35); second, this monoubiquitinated species (Ub-PEX5) is extracted in an ATP-dependent manner by the receptor export module, a protein complex comprising the ATPases PEX1 and PEX6 and the peroxisomal membrane protein PEX26 (30, 36, 37). Finally, Ub-PEX5 is deubiquitinated in the cytosol regenerating unmodified PEX5. A new cycle of protein transportation is then initiated.
We have recently shown that the Ub-PEX5 conjugate can be readily disrupted by physiological concentrations of glutathione (38). This finding led us to propose that a fraction of Ub-PEX5 may be deubiquitinated in vivo through a nonenzymatic mechanism. However, data suggesting that Ub-PEX5 is also deubiquitinated by a deubiquitinase (DUB)-mediated process were also obtained in that work. Here, we describe the identification and biochemical characterization of this enzyme.
EXPERIMENTAL PROCEDURES
Plasmids and Recombinant Proteins
cDNAs encoding the large isoform of PEX5 (4, 39), hereafter referred to as PEX5 for simplicity, and a mutated version of it possessing a lysine at position 11 (PEX5(C11K)) cloned into the pGEM-4 vector (Promega) have been described elsewhere (38, 40). A cDNA encoding a truncated version of PEX5(C11K) comprising its first 324 amino acid residues preceded by the T7 RNA polymerase promoter was obtained by PCR amplification using pGEM4-PEX5(C11K) plasmid as template and the primers described in Ref. 27. A plasmid encoding a ubiquitin-PEX5 fusion protein not cleavable by DUBs, Ub(G76V)-S-PEX5(12–639), was generated as follows. First, the ubiquitin cDNA was amplified from pDEST15-Ub (41) using primers (A) 5′-CTAGTCTAGAGCCACCATGCAGATCTTCGTGAAAACCCTTACC-3′ and (B) 5′-TGGCACCCCCGCTAACACCTCTCAGACGCAGGACCAG-3′, and the PEX5 cDNA sequence was amplified from pGEM4-PEX5 (40) using primers (C) 5′-CGTCTGAGAGGTGTTAGCGGGGGTGCCAACC-3′ and (D) 5′-CTAGTCTAGATCACTGGGGCAGGCCAAACATA-3′. In the second step, both PCR products were combined, and a second PCR was performed using primers A and D described above. The amplified cDNA was then cloned into the XbaI site of pGEM4 yielding pGEM4-Ub(G76V)-S-PEX5(12–639). This plasmid was then used as a template in a PCR with the primers 5′-GGCACCTCATATGCAGATCTTCGTGAAGAC-3′ and 5′-GCGCGGATCCTCATTAGTACCCCTTATCATAGGTAGCTG-3′. The DNA fragment obtained was digested with NdeI and BamHI and cloned into pET-28b (Novagen) yielding pET28b-His6-Ub(G76V)-S-PEX5(12–324). A plasmid encoding His6-Ub-PEX5(11–324), a DUB-cleavable recombinant protein, was obtained by site-directed mutagenesis (QuickChange® site-directed mutagenesis kit; Stratagene) using pET28b-His6-Ub(G76V)-S-PEX5(12–324) as a template and the primers 5′-GCGTCTGAGAGGTGGTTGCGGGGGTGCCAAC-3′ and 5′-GTTGGCACCCCCGCAACCACCTCTCAGACGC-3′. Bacterial expression constructs encoding GST-tagged versions of Ub-PEX5(11–324) and Ub(G76V)-S-PEX5(12–324) and pDEST15-Ub-PEX5(11–324) and pDEST15-Ub(G76V)-S-PEX5(12–324), respectively, were obtained by cloning the DNA fragment derived from BamHI/SalI digestion of the corresponding pET28b-based plasmids into pDEST15-Ub plasmid digested with the same restriction enzymes. A pDEST15 plasmid encoding GST-tagged Ub(G76V)S was obtained by site-directed mutagenesis of pDEST15-Ub(G76V)-S-PEX5(12–324) plasmid using the primers 5′-TCTGAGAGGTGTTAGCTAGGGTGCCAACCCGCTC-3′ and 5′-GAGCGGGTTGGCACCCTAGCTAACACCTCTCAGA-3′. To generate a plasmid encoding HA-tagged ubiquitin, pET28a-HA-Ub, the HA-Ub encoding sequence was amplified from pRK5-HA-Ub-WT (Addgene clone 17608) (42) and inserted into the NcoI/EcoRI sites of pET28a (Novagen). Plasmids encoding GST-LKS (used here as negative control GST protein; GST-Control) (27), a histidine-tagged protein comprising the first 324 amino acid residues of PEX5 (His6-PEX5(1–324)) (43), an HA-tagged ubiquitin fused to intein and a chitin-binding domain (pTYB-HAUb plasmid) (44), and V5-tagged USP9X (plasmid pDEST51-USP9X) (45), have been described before. Recombinant HA-Ub was expressed in the Escherichia coli BL21(DE3) strain by induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. HA-Ub was purified by anion exchange chromatography using a linear gradient of 0–500 mm NaCl in 10 mm Tris-HCl, pH 8.0, 1 mm EDTA-NaOH, pH 8.0, followed by size exclusion chromatography (SEC) using a Superose 12 HR 10/30 (GE Healthcare) column running with 10 mm Tris-HCl, pH 8.0, 1 mm EDTA-NaOH, pH 8.0, at 0.5 ml/min. All the other recombinant proteins were purified as described before (27, 46) and stored in buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, and 1 mm DTT) at −80 °C. We note that a small fraction of the ubiquitin-PEX5 fusion proteins used here is cleaved during the protein induction step at the ubiquitin-PEX5 junction. The E. coli protease responsible for this phenomenon is unknown, but ElaD is a good candidate (47). HA-tagged ubiquitin-aldehyde (HA-Ubal) and HA-tagged ubiquitin-vinyl methyl ester (HA-UbVME), two potent inhibitors of many DUBs (44, 48), were prepared as described before (44, 49), except that the ion exchange chromatography was omitted.
Preparation of Subcellular Fractions
Rat liver post-nuclear supernatants (PNS) were prepared in SE buffer (0.25 m sucrose, 20 mm MOPS-KOH, pH 7.2, 1 mm EDTA-NaOH, pH 8.0) supplemented with 2 μg/ml N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide, as described before (40), and were either frozen in liquid N2 and stored at −80 °C (for the in vitro import/export assays; see below) or immediately used for the preparation of organelle and cytosolic fractions. For this purpose, the PNS was centrifuged at 12,300 × g for 20 min at 4 °C, and the supernatant was further centrifuged at 100,000 × g for 1 h at 4 °C. The supernatant from this last centrifugation (cytosolic fraction) was removed, and the pellets from the two centrifugations were resuspended in SE buffer supplemented with 2 μg/ml N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide and combined (total organelle fraction). HeLa and HEK293T cells were resuspended in SE buffer containing 0.1% (w/v) Triton X-100 and sonicated on ice using a Bandelin electronic sonicator UW2200 equipped with a microtip. Insoluble material was removed by centrifugation at 100,000 × g for 30 min at 4 °C.
In Vitro Import and Export of PEX5 Proteins
35S-Labeled PEX5 proteins were synthesized in rabbit reticulocyte lysates using the TnT® quick-coupled transcription/translation system (Promega) in the presence of EasyTagTM l-[35S]methionine (specific activity, >1000 Ci/mmol; PerkinElmer Life Sciences) following the manufacturer's instructions. For the generation of soluble Ub-PEX5 proteins, rat liver PNS (600 μg of protein per 100-μl reaction) was primed for import by incubating in import buffer (0.25 m sucrose, 50 mm KCl, 20 mm MOPS-KOH, pH 7.2, 3 mm MgCl2, 20 μm methionine, and 2 μg/ml N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide) containing 5 μm bovine ubiquitin, and 0.3 mm ATP for 5 min at 37 °C, as described previously (30). 35S-Labeled PEX5 proteins (1–2 μl of reticulocyte lysate per 100-μl reaction) and 3 mm (final concentration) of AMP-PNP (Sigma) were added, and the reactions were incubated for 20 min at 37 °C. At the end of this incubation, reactions were diluted with an equal volume of ice-cold SEMK buffer (0.25 m sucrose, 80 mm KCl, 20 mm MOPS-KOH, pH 7.2, 1 mm EDTA-NaOH, pH 8.0), and the organelles were isolated by centrifugation (16,000 × g, 20 min, 4 °C). The organelle pellet was then carefully resuspended in import buffer containing 0.01% (w/v) bovine serum albumin and 5 mm ATP, and the suspension was incubated at 37 °C for 20 min. The organelles were then removed by centrifugation (16,000 × g, 20 min, 4 °C), and the supernatants containing monoubiquitinated 35S-labeled PEX5 proteins were used for the deubiquitination assays (see below). Where specified, 2 μm HA-Ubal was included in the export step.
SEC and Sucrose Gradient Centrifugation
Rat liver cytosolic proteins (4–6 mg) in 200 μl of 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, 1 mm DTT, and 10% (w/v) glycerol were injected into a Superose 6 10/300 GL column (GE Healthcare) running with the same buffer lacking DTT at a flow rate of 0.5 ml/min. The column was calibrated with the following protein standards (numbers in parentheses indicate the respective Stokes radius): thyroglobulin (8.5 nm), ferritin (6.1 nm), bovine serum albumin (3.6 nm), and soybean trypsin inhibitor (2.3 nm). Fractions of 1 ml were collected, aliquoted, frozen in liquid N2, and stored at −80 °C. For the sucrose gradient centrifugation analyses, 3 mg of rat liver cytosolic proteins in 300 μl of 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, and 1 mm DTT were loaded onto the top of a continuous 5–30% (w/v) sucrose gradient in the same buffer. After centrifugation at 38,000 rpm for 16 h at 4 °C in an SW41 rotor (Beckman), 14 aliquots of 0.8 ml were collected from the bottom of the tube. Forty microliters of each fraction were subjected to SDS-PAGE/Western blot analyses. Thyroglobulin (19.4 S), catalase (11.3 S), and bovine serum albumin (2.3 S) were used as sedimentation coefficient standards (50).
Labeling of Rat Liver Cytosolic Deubiquitinases (DUBs) with HA-UbVME and Deubiquitination Assays
SEC fractions (40 μl aliquots) were incubated with 2 μm HA-UbVME (see below) for 15 min at 37 °C and subjected to SDS-PAGE/Western blot analyses. For SDS-PAGE/Coomassie Blue analyses, fraction 7 from SEC (derived from 6 mg of cytosolic proteins) was halved, and one-half was incubated with 1 μm of HA-UbVME, as above. The two samples were subjected to precipitation with 10% (w/v) trichloroacetic acid before SDS-PAGE. Deubiquitination assays were performed in 100 μl of buffer B (20 mm MOPS-KOH, pH 7.2, 50 mm KCl, 1 mm EDTA-NaOH, pH 8.0) and contained as substrate either 15 μl of radiolabeled monoubiquitinated PEX5 proteins or 2–3 μg of recombinant ubiquitin-PEX5 fusion proteins, and as a source of DUBs 50 μg of rat liver cytosol/HeLa cell extract or SEC fractions derived from 200 μg of cytosol. For the deubiquitination assays presented in Fig. 1B, rat liver subcellular fractions (50 μg of PNS and the corresponding soluble and organelle fractions) were incubated in import buffer supplemented with 10 mm deoxyglucose (Sigma) and 10 units/ml hexokinase (Sigma) for 5 min at 37 °C before adding radiolabeled Ub-PEX5. The assays were performed at 37 °C for 15 min. Where indicated, HA-Ubal and HA-UbVME were used at 2 μm.
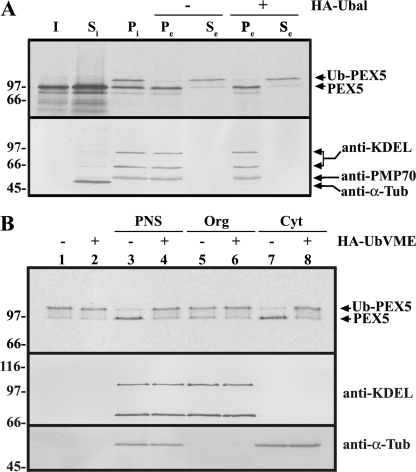
FIGURE 1.
DUB acting on Ub-PEX5 is a cytosolic protein. A, 35S-labeled PEX5 was incubated with a PNS in the presence of AMP-PNP for 20 min at 37 °C. The import reaction was then separated into soluble (Si) and organelle (Pi) components by centrifugation. The organelles were resuspended in ATP-containing buffer, in the absence or presence of HA-Ubal (lanes − and +, respectively), and further incubated for 20 min at 37 °C. Finally, the suspensions were separated into an organelle pellet (Pe) and a supernatant (Se) by centrifugation. Samples were treated with 20 mm N-ethylmaleimide and subjected to SDS-PAGE under nonreducing conditions followed by Western blot. The membrane was first exposed to an x-ray film to detect the 35S-labeled PEX5 (top panel) and afterward was sequentially probed with the following antisera: anti-KDEL (recognizes GRP72 and GRP98, two endoplasmic reticulum proteins); anti-α-tubulin (a cytosolic marker; anti-α-Tub), and anti-PMP70 (an intrinsic protein of the peroxisomal membrane). B, 35S-labeled Ub-PEX5 was incubated alone (lanes 1 and 2), with a PNS (lanes 3 and 4), or with the corresponding organelle (Org; lanes 5 and 6) or cytosolic (Cyt; lanes 7 and 8) fractions, in the absence (−) or presence (+) of HA-UbVME, as indicated. Samples were analyzed as above. The distributions of microsomal (anti-KDEL), and soluble proteins (anti-α-Tub) are also shown. Lane I, 50% of the 35S-labeled PEX5 reticulocyte lysate used in this experiment. Numbers to the left indicate the molecular masses of the reduced protein standards in kDa.
Immunoprecipitations
DUBs present in fraction 7 from SEC (derived from 0.5 mg of cytosol) were incubated with either HA-UbVME or HA-Ub (as a negative control), diluted to 500 μl with 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, and 10% (w/v) glycerol, and subjected to immunoprecipitation using 30 μl of anti-HA antibody agarose beads (Sigma) for 2 h at 4 °C. Beads were washed five times with 500 μl of the same buffer, and the immunoprecipitated proteins were eluted twice with 120 μl of HA peptide (Sigma) at 100 μg/ml. For the preparative immunoprecipitation of rat liver USP9X, 10 mg of cytosolic protein in 1 ml of buffer B were incubated with 30 μl of protein A-Sepharose beads (Sigma) containing 1.5 μg of anti-USP9X IgGs. After 2 h at 4 °C, with gentle shaking, the beads were washed five times with 1 ml of buffer B, and the immunoprecipitated proteins were eluted with 65 μl of Laemmli sample buffer and subjected to SDS-PAGE. The same procedure was used for the immunoprecipitation/immunodepletion experiments performed with rat liver cytosol and HeLa cell extracts. In both cases, 2 mg of protein in 500 μl of buffer B were used as the starting material. Fractions of the antibody-bound and -unbound material in buffer B, derived from 50 μg of the initial extract, were used in the deubiquitination assays. For the immunoprecipitation of V5-tagged USP9X, 5 mg of HEK293T cell protein extract in 500 μl of buffer B and 30 μl of protein A-Sepharose beads (Sigma) containing 1 μg of anti-V5 monoclonal antibody were used. Immunopurified V5-tagged USP9X derived from 200 μg of the initial extract was used in the deubiquitination assays.
Pulldown Assays
GST fusion proteins (120 μg) were bound to 20 μl of glutathione-Sepharose beads (GE Healthcare) in 100 μl of buffer A for 1 h at 4 °C. The beads were washed twice with 500 μl of buffer A, and 1 mg of rat liver cytosol in 250 μl of buffer A was added. After incubation for 2 h at 4 °C with gentle shaking, the beads were washed five times with 250 μl of buffer A, and bound proteins were eluted with Laemmli sample buffer. Aliquots of cytosol, unbound and bound fractions, were subjected to SDS-PAGE/Western blot analyses.
Cell Culture, Transfections, and Fluorescence Microscopy
Cell culture reagents were from Invitrogen, unless otherwise specified. HeLa and HEK293T cells for the immunoprecipitation experiments were cultivated in high glucose DMEM + GlutaMAXTM supplemented with 10% heat-inactivated FBS, 1 mm sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mm nonessential amino acids in 5% CO2 atmosphere at 37 °C. Transfection of HEK293T cells was performed using PolyJetTM (SignaGen® Laboratories) according to the manufacturer's instructions. For siRNA experiments, HeLa cells were grown in DMEM/F-12 (Lonza), supplemented with 10% heat-inactivated FBS, 2 mm GlutaMAXTM, and 0.2% MycoZapTM CL-Plus (Lonza). During and after the transfection with siRNAs the concentration of heat-inactivated FBS in the medium was decreased to 2%. HeLa cells were transfected using DharmaFECT 1 (Dharmacon, ThermoScientific) with a 1:1 mixture of the following custom USP9X-specific siRNA oligonucleotides: GAUGAGGAACCUGCAUUUCtt and GCAGUGAGUGGCUGGAAGUtt (Integrated DNA Technologies). These siRNAs are referred to as FAM1 and FAM2, respectively, in Ref. 51. USP9X knocked down HeLa cells were transiently transfected with the plasmid pMF1706 (52) encoding a peroxisomal matrix reporter protein (roGFP2-PTS1) employing the Neon Transfection System (Invitrogen; pulse voltage 1005 V; 35-ms pulse width; 2 pulses) at different time points after siRNA treatment, as specified. Cells were seeded and imaged in FD-35 Fluorodish Cell culture dishes (World Precision Instruments) at the 480-nm excitation wavelength.
Mass Spectrometry
Protein bands were excised manually from SDS-polyacrylamide gels, subjected to trypsin digestion, and analyzed on a matrix-assisted laser desorption/ionization-time-of-flight MALDI-TOF/TOF mass spectrometer (4800 Proteomics Analyzer, Applied Biosystems, Foster City, CA), exactly as described recently (53). Mascot generic format files were retrieved from each LC-MALDI-TOF/TOF MS/MS run, exporting up to 10 MS/MS spectra per spot, each requiring a minimum signal-to-noise ratio of 50. Recalibration of the peptide masses in MS run was performed using Glu-fib at 15 fmol as internal standard (with a m/z 1570.68). Spectra were processed and analyzed by the Global Protein ServerWorkstation (Applied Biosystems). MS/MS queries were processed using the in-house Mascot database search engine version 2.1.1. (Matrix Science Ltd.). Original MALDI-TOF/TOF MS/MS data were analyzed using the following settings: ≤30 ppm MS precursor mass tolerance and ≤0.3 Da MS/MS mass tolerance. Data received from the protein digest LC-MALDI-TOF/TOF MS/MS runs were searched using the following criteria: database, TrEMBL (release 25012012); taxonomy, rodentia; type of search, MS/MS ion search; enzyme, trypsin; variable modifications, cysteine carbamidomethylation or propionamidation, oxidation on methionine, and the N-terminal loss of ammonia at Gln; number of maximum missed cleavages, 2. Peptides were considered identified if their Mascot individual ion score was higher than 31 (p < 0.05). Proteins with scores ≥71 were confidently assigned. To evaluate the false discovery rate, we performed a decoy database search against a reversed decoy database created by Mascot using identical search parameters and validation criteria yielding below 1% of false discovery rate.
Miscellaneous
The following primary antibodies were used: anti-KDEL (clone 10C3, Abcam), anti-α-tubulin (clone B-5-1-2, Sigma), anti-HA (clone 12CA5, Roche Applied Science), anti-V5 (R96025, Invitrogen), anti-Ataxin-3 (clone 1H9, Millipore), anti-PMP70 (54), and anti-USP9X (A301–351A, Bethyl Laboratories). The commercial anti-USP9X antibody was raised against the C-terminal 51 amino acid residues of human USP9X (GenBankTM accession number CAD18900.2; Ref. 55), which are 100% conserved in the rat USP9X sequence (GenBankTM accession number NM_001135923.1). Whether or not this antibody also recognizes rat USP9Y, a male-specific USP9X-homologous DUB is presently unknown. Therefore, experiments that relied on the use of this antibody were made with female-derived material, unless otherwise indicated. The anti-USP9X antibody recognizes a single 220-kDa protein band in Western blot analyses of total HeLa cell lysates or male or female rat liver cytosolic fractions. Also, when a female rat liver cytosolic fraction was subjected to immunoprecipitation using this antibody, no HA-UbVME-reactive band other than the one corresponding to USP9X can be detected in the immunoprecipitate (see supplemental Fig. S1). Rabbit and mouse antibodies were detected on Western blots using alkaline phosphatase-conjugated anti-rabbit and anti-mouse antibodies (Sigma), respectively. Total IgGs were purified from rabbit sera using protein A-Sepharose beads according to the manufacturer's protocol (Sigma). PEX5 in HeLa cell lysates was detected by blot overlay using 35S-labeled PEX14, as described previously (56). Preparation of samples for SDS-PAGE under nonreducing conditions (necessary to preserve the Ub-PEX5 thioester conjugate) and autoradiography following Western blot were described previously (33, 38). Densitometric analyses of Western blots were performed using the ImageJ software (National Institutes of Health, Bethesda, imagej.nih.gov).
RESULTS
We have previously shown that after being exported from the DTM, a fraction of Ub-PEX5 is deubiquitinated by a ubiquitin-aldehyde (Ubal)-sensitive component present in rat liver post-nuclear supernatant. The sensitivity to Ubal (and to ubiquitin-vinyl methyl ester (UbVME); see below) indicates that this component is a member of the cysteine protease family of DUBs (44). Its identity, however, remained unknown. In this work, we used an unbiased biochemical approach to identify this enzyme. In brief, rat liver proteins were fractionated using standard biochemical techniques, and the fractions obtained were assayed for the following: 1) DUB activity using Ub-PEX5 as the substrate and 2) reactivity with an activity-based probe (HA-UbVME) that covalently labels DUBs of the cysteine protease family (44). The purification procedure was ended when the DUB activity profile could be matched to a specific HA-UbVME-reactive DUB. As described below, this situation was reached after a single chromatography step.
A key reagent required for this approach is Ub-PEX5, the substrate for the DUB activity assays. This species can be easily obtained by incubating 35S-labeled PEX5 with a (peroxisome-containing) rat liver PNS in the presence of ATP and Ubal. Under these conditions, about half of the total PEX5 protein passes through the peroxisomal DTM where it is monoubiquitinated and subsequently exported into the soluble phase (38). However, the presence of Ubal in the soluble phase of these reactions complicates the downstream enzymatic assays. Omission of Ubal from these reactions still allows the production of soluble Ub-PEX5, although in smaller amounts (33, 38). However, Ub-PEX5 produced in this manner is contaminated with cytosolic DUBs, and further incubation of this ubiquitin conjugate in the absence of Ubal leads to its hydrolysis (Ref. 38 and data not shown). We found that reasonable amounts of soluble Ub-PEX5 suitable for these assays can be obtained using a two-step procedure. In the first step, 35S-labeled PEX5 is incubated with a rat liver PNS in the presence of AMP-PNP, a nonhydrolyzable ATP analog. AMP-PNP, like ATPγS, is efficiently used by the ubiquitin-activating enzyme (57) but inhibits the receptor export module, thus leading to the accumulation of Ub-PEX5 at the peroxisomal DTM (this work and Refs. 30, 33). After centrifugation of this suspension and removal of cytosolic proteins, the organelles (Fig. 1A, lane Pi) are then resuspended and incubated in a buffer containing ATP to promote export of the DTM-embedded Ub-PEX5 into the soluble phase of the reaction (lanes Se). The amount of soluble Ub-PEX5 recovered using this strategy does not increase when Ubal is included in the ATP-containing export buffer (lanes Se in the plus and minus HA-Ubal sets) suggesting that the organelle fraction lacks significant DUB activity (see also Fig. 1B, lanes 1 and 2). Besides allowing us to obtain Ub-PEX5 free of contaminating DUBs or Ubal, this experiment also shows that the machinery that extracts Ub-PEX5 from the peroxisomal DTM can be separated from the DUB(s) acting on soluble Ub-PEX5 by a simple centrifugation (see under “Discussion”).
Further data supporting the conclusion that the DUB acting on Ub-PEX5 is not an organelle-associated protein but rather a cytosolic protein were obtained when soluble and organelle proteins from rat liver were tested in DUB activity assays. As shown in Fig. 1B, the vast majority of the DUB acting on Ub-PEX5 is found in the cytosolic fraction (compare lanes 5 and 7).
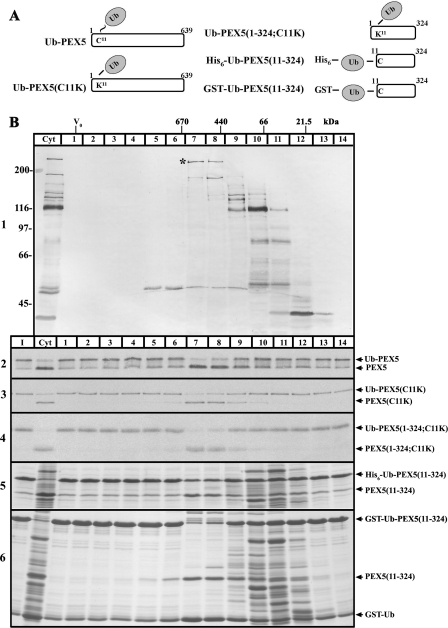
We next subjected a male rat liver cytosol to several fractionation procedures and tested the obtained fractions for the presence of DUBs and DUB activity, as described above. From the several fractionation techniques that were tried, SEC turned out to be the most powerful one. The results of one of these experiments are shown in Fig. 2. Several DUBs were labeled with the HA-UbVME probe (Fig. 2B, panel 1), as expected (44). Their apparent molecular masses upon SDS-PAGE range from about 40 to 230 kDa. When these SEC fractions were incubated with Ub-PEX5 (see Fig. 2A for a schematic representation of this and other PEX5 proteins described below), a robust DUB activity peak was detected in fractions 7 and 8, with the former displaying a slightly higher activity than the latter (Fig. 2B, panel 2). Identical activity profiles were obtained with Ub-PEX5(C11K), a mutant PEX5 molecule monoubiquitinated at lysine 11 (38) and with the truncated protein Ub-PEX5(1–324;C11K), which lacks the C-terminal half of PEX5 (Fig. 2B, panels 3 and 4). These observations suggest, on the one hand, that the relevant DUB does not discriminate between a thioester bond (present in the Ub-PEX5 conjugate) and an isopeptide bond (present in the Ub-PEX5(C11K) and Ub-PEX5(1–324;C11K) conjugates) and, on the other hand, that the C-terminal half PEX5 is not a major determinant in the interaction of Ub-PEX5 with the DUB. Interestingly, identical activity profiles were observed when two (linear) recombinant proteins containing a ubiquitin moiety fused to amino acid residues 11–324 of PEX5, His6-Ub-PEX5(11–324), and GST-Ub-PEX5(11–324) were tested in this assay (Fig. 2B, panels 5 and 6, respectively). Apparently, the active site of the DUB eluting in these fractions is quite flexible with respect to the type of bond that links ubiquitin to PEX5.
FIGURE 2.
Activity profiling of rat liver cytosolic DUBs after SEC. A, schematic representation of the monoubiquitinated PEX5 proteins used in the deubiquitination assays. The thioester bond linking ubiquitin (Ub) to cysteine 11 of PEX5 is indicated with ∼; isopeptide and peptide bonds are indicated with −. B, male rat liver cytosol was subjected to SEC and the collected fractions (lanes 1–14) were incubated with HA-UbVME and analyzed by SDS-PAGE/Western blot with an anti-HA antibody (panel 1). Fractions were also assayed for deubiquitinating activity using the indicated monoubiquitinated PEX5 proteins (panels 2–6). The reactions were terminated with 20 mm N-ethylmaleimide and analyzed by SDS-PAGE. Autoradiographs (panels 2–4) and Coomassie Blue-stained gels (panels 5 and 6) are presented. The asterisk in panel 1 indicates an HA-UbVME-reactive DUB that co-elutes with the hydrolytic activity detected toward monoubiquitinated PEX5 proteins. The elution positions and molecular masses of the protein standards used to calibrate the SEC column, as well as the column void volume (V0), are indicated. Reactions using the initial cytosolic extract (lanes Cyt) and the monoubiquitinated PEX5 proteins alone (lanes I) are also presented. Numbers to the left in panel 1 indicate the molecular masses of protein standards in kDa.
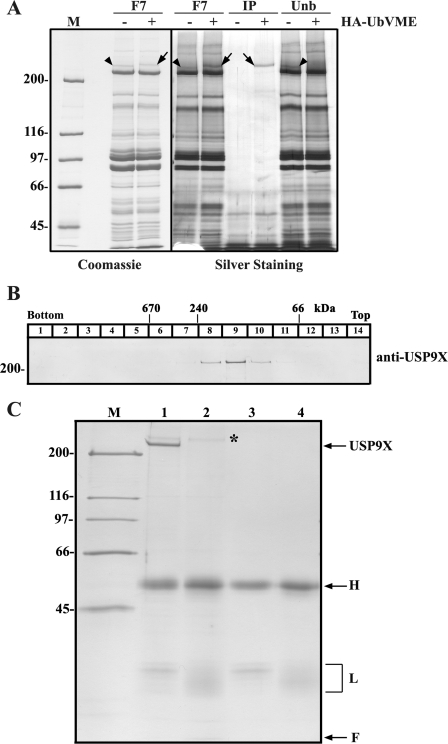
Comparison of the DUB activity profiles obtained with the monoubiquitinated PEX5 proteins (Fig. 2B, panels 2–6) with the elution profiles of the HA-UbVME-labeled protein bands suggested that a protein displaying an apparent molecular mass of about 230 kDa on SDS-PAGE (band marked with an asterisk in panel 1) could be the DUB of interest. To identify this DUB, proteins present in fraction 7 of the SEC column were incubated in the presence or absence HA-UbVME and subjected to SDS-PAGE/Coomassie Blue staining. Side-by-side comparison of these two samples revealed a protein that upon treatment with the HA-UbVME probe was quantitatively shifted to an apparent molecular mass of 230 kDa (Fig. 3A, left panel). That this shift in apparent molecular mass is indeed the result of covalent labeling by the HA-UbVME probe was confirmed by immunoprecipitation experiments using an anti-HA antibody. As shown in Fig. 3A (right panel), the 230-kDa protein was quantitatively immunoprecipitated only in the sample that had been treated with HA-UbVME.
FIGURE 3.
Biochemical characterization of rat liver cytosolic USP9X. A, fraction 7 (F7) from SEC (see Fig. 2B, panel 1) was incubated in the absence (lanes −) or presence (lanes +) of HA-UbVME and subjected to SDS-PAGE. The left panel shows a Coomassie Blue-stained gel and the panel on the right shows the results of an immunoprecipitation experiment using anti-HA agarose beads. Lanes IP, immunoprecipitated proteins. Lanes Unb, unbound protein fractions. Arrows and arrowheads indicate the HA-UbVME-modified and unmodified DUB, respectively, that was subjected to mass spectrometry analysis. Lane M, molecular mass protein standards (1 μg each). B, analyses of female rat liver cytosolic USP9X by sucrose gradient centrifugation. Numbers at the top indicate the molecular masses and the sedimentation positions of protein standards. C, female rat liver cytosol was subjected to immunoprecipitation using either an anti-USP9X antibody (lane 1) or control IgGs (lane 2). The anti-USP9X and control antibody-containing beads were also analyzed (lanes 3 and 4, respectively) to identify antibody-derived bands. A Coomassie Blue-stained gel is shown. The asterisk marks a nonspecific protein recovered in both immunoprecipitates; H and L indicate IgG heavy and light chains, respectively. F indicates front of the gel. Lane M indicates molecular mass protein standards (0.5 μg each). Numbers to the left indicate the molecular masses of protein standards in kDa.
Mass spectrometry analysis of this protein band (Fig. 3A, left panel) identified ubiquitin-specific protease 9X (USP9X) as the protein with the highest score (see supplemental Table S1). USP9X is a 290-kDa DUB encoded in the X chromosome of humans, mice, and rats (58–60). We note that some of the peptides matching USP9X might also be derived from USP9Y (see footnote in supplemental Table S2), a DUB homologous to USP9X but encoded in the Y chromosome (58, 61). Although the presence of USP9Y in rat liver is presently doubtful (USP9Y, in contrast to USP9X, is not expressed in mouse liver; see Ref. 62), it remained a possibility that USP9Y, and not USP9X, was the DUB of interest. If this were the case then a different DUB should be found in cytosolic fractions from female animals because USP9Y is a male-specific enzyme. However, when a female rat liver cytosolic fraction was subjected to SEC, we found that the DUB activity co-eluted in a perfect manner with USP9X (see supplemental Fig. S2). This finding thus suggests that USP9X is the main DUB acting on Ub-PEX5, at least in female animals.
We note that these results do not exclude the possibility that USP9Y may also hydrolyze the Ub-PEX5 conjugate. Actually, such a possibility is quite plausible because USP9X and USP9Y are almost identical proteins (61). However, considering that the complete deletion of the USP9Y-encoding gene in men has no deleterious effects (63), whereas deletion of USP9X in mice is embryonically lethal (64, 65), and that USP9Y is probably expressed at much lower levels than USP9X, as assessed by the number of human/rat/mouse-expressed sequence tags presently available in the GenBankTM database (data not shown), the contribution of USP9Y for the hydrolysis of Ub-PEX5 in male animals, if any, is probably a minor one. For these reasons we focused our efforts in USP9X.
One property of USP9X that called our attention was its behavior upon SEC (see also Ref. 45). Indeed, USP9X presents a Stokes radius of 7.3 nm, eluting from the SEC column as if it were a 550-kDa globular/spherical protein (see supplemental Fig. S2). Considering the theoretical molecular mass of USP9X (290 kDa) (60), this finding might suggest that USP9X is a component of a large protein complex. However, sucrose gradient sedimentation analysis revealed that USP9X is not a spherical protein because it sediments well above catalase (240 kDa, 11.3 S), displaying a sedimentation coefficient of 9.1 S (Fig. 3B). Using the Siegel-Monty equation, which correlates the molecular mass of a protein with its sedimentation coefficient and Stokes radius (66, 67), we estimated that USP9X displays a native molecular mass of 280 kDa. These data suggest that USP9X is a monomeric protein with an elongated shape in solution.
Additional evidence supporting the idea that USP9X is a monomeric protein was obtained when USP9X from a female rat liver cytosol was immunoprecipitated under native conditions. As shown in Fig. 3C, besides USP9X, antibody-derived proteins (bands marked with H and L, respectively) and a high molecular mass protein that was also found in the negative control immunoprecipitate (asterisk), no other proteins in stoichiometric amounts were detected in the USP9X immunoprecipitate.
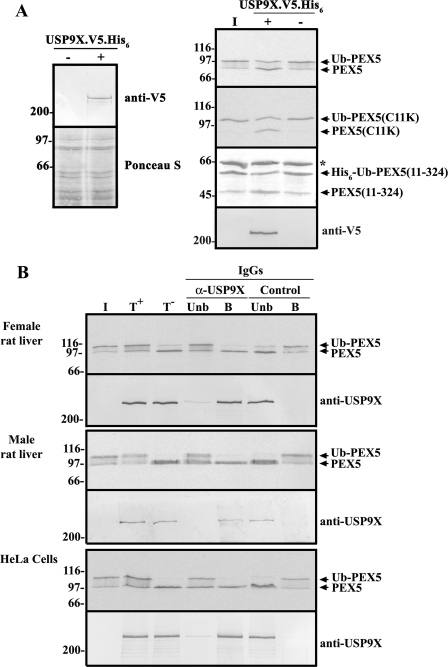
The results described above point to USP9X as the best DUB candidate acting on Ub-PEX5. Several independent approaches were used to validate this possibility. In the first, we expressed V5-tagged USP9X (45) in HEK293T cells (see Fig. 4A, left panel), and after immunopurifying the enzyme using an anti-V5 antibody, we tested its activity in DUB assays using monoubiquitinated PEX5 proteins. Nontransfected HEK293T cells were included in this experiment as a negative control. As shown in Fig. 4A (right panel), the immunoprecipitate obtained from the V5-tagged USP9X-expressing cells was able to cleave Ub-PEX5. Identical results were obtained when Ub-PEX5(C11K) and recombinant Ub-PEX5(11–324) were used in this assay (Fig. 4A).
FIGURE 4.
USP9X is the major DUB acting on monoubiquitinated PEX5 conjugates. A, Western blot analysis using an anti-V5 antibody of nontransfected (lane −) and pDEST51-USP9X-V5-transfected HEK293T cells (lane +) is presented in the left panel. V5-tagged USP9X was immunoprecipitated with an anti-V5 antibody and used in deubiquitination assays (lanes +) with the indicated monoubiquitinated PEX5 proteins. Nontransfected HEK293T cells immunoprecipitates were included in the assays as negative controls (lanes −). The asterisk indicates bovine serum albumin that was added to this assay to minimize protein loss by adsorption. Reactions were stopped with 20 mm N-ethylmaleimide, and the samples were processed for SDS-PAGE/Western blot/autoradiography. Autoradiographs for 35S-labeled PEX5 proteins (Ub-PEX5 and Ub-PEX5(C11K)) and a Ponceau S-stained membrane for His6-Ub-PEX5(11–324) are shown. Immunoprecipitated USP9X was detected with the anti-V5 antibody. B, liver cytosolic fractions from female and male rats (upper and middle panels, respectively) or a HeLa cell extract (lower panel) were subjected to immunoprecipitation/immunodepletion protocol using control (lanes Control) or anti-USP9X IgGs (lanes α-USP9X). The immunoprecipitates (lanes B) and the unbound protein fraction (lanes Unb) were assayed for deubiquitinase activity with 35S-labeled Ub-PEX5. Activity assays with the initial protein extracts were also performed in the presence (T+) or absence (T−) of HA-Ubal. Samples were processed as described above. USP9X was detected with the anti-USP9X antibody. Lanes I,- the 35S-labeled Ub-PEX5 substrate used in the assay.
In another approach, we subjected a cytosolic fraction from female rat liver to immunoprecipitation using the anti-USP9X antibody and the antibody-bound and unbound fractions were then tested in DUB assays using Ub-PEX5 as the substrate. As shown in Fig. 4B (upper panel), USP9X was efficiently retained in the antibody-bound fraction with almost no enzyme being detectable in the unbound fraction. Importantly, Ub-PEX5 was completely hydrolyzed upon incubation with the immunoprecipitated fraction, whereas no significant DUB activity was detected in the unbound fraction. A control experiment using an irrelevant antibody (Fig. 4B, lanes Control) shows that the immunodepletion/immunoprecipitation procedure performed with the anti-USP9X antibody was specific. Identical results were obtained with a male rat liver cytosolic fraction (Fig. 4B, middle panel), although in this case the presence of USP9Y in the immunoprecipitate cannot be formally excluded (see under “Experimental Procedures” for details).
To determine whether human USP9X also acts on Ub-PEX5, we performed a similar immunoprecipitation experiment but this time using a clarified HeLa cell homogenate. The results presented in Fig. 4B (lower panel) are identical to those obtained with rat liver cytosol. Thus, USP9X is the major DUB acting on Ub-PEX5 also in HeLa cells.
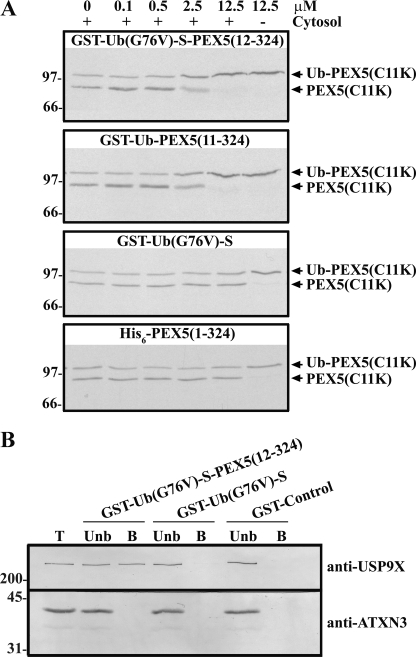
Finally, we asked whether we could detect an interaction between USP9X and ubiquitinated PEX5 using pulldown assays. Because of the fact that chemical amounts of authentically monoubiquitinated PEX5 cannot be easily obtained at present, these assays were performed with (linear) recombinant fusion proteins. One of the proteins produced for this purpose was GST-Ub(G76V)-S-PEX5(12–324), a recombinant protein almost identical to GST-Ub-PEX5(11–324), differing from it in just two amino acid residues; the last glycine of the ubiquitin moiety was changed to a valine, a mutation that no longer allows hydrolysis by DUBs (68); and cysteine 11 of PEX5 was changed to a serine, to avoid the potential oxidation of this residue that might somehow interfere in the assay. A truncated version of this nondeubiquitinatable protein lacking the PEX5 domain (GST-Ub(G76V)-S) and a negative control GST protein (GST-Control; see “Experimental Procedures” for details) were also prepared. To test the functionality of these recombinant proteins, we asked whether they could inhibit cleavage of Ub-PEX5(C11K) by a female rat liver cytosol. The results of this experiment (see Fig. 5A) show that GST-Ub-PEX5(11–324) as well as the noncleavable protein GST-Ub(G76V)-S-PEX5(12–324) inhibit hydrolysis of Ub-PEX5(C11K). The IC50 of these two proteins is ∼2.5 μm. No inhibition was observed when using GST-Ub(G76V)-S or histidine-tagged PEX5(1–324) at the same concentrations. These data suggest that neither ubiquitin nor the N-terminal half of PEX5 alone bind efficiently to the cytosolic DUB that cleaves Ub-PEX5(C11K); only molecules possessing these two moieties seem to have this capacity.
FIGURE 5.
USP9X interacts with a monoubiquitinated PEX5 fusion protein. A, 35S-labeled Ub-PEX5(C11K) was incubated in the presence (lanes +) or absence (lane −) of a female rat liver cytosol, in the absence or presence of increasing concentrations of the indicated recombinant proteins (from 0 to 12.5 μm). The recombinant proteins used in these experiments are indicated at the top of each panel. B, female rat liver cytosol was subjected to GST pulldown assays with the indicated recombinant proteins. The bound (B) and unbound (Unb) fractions derived from 200 and 40 μg of cytosol, respectively, were analyzed by SDS-PAGE/Western blot using anti-USP9X and anti-Ataxin-3 (anti-ATXN3) antibodies. Lane T, 40 μg of female rat liver cytosol. Numbers to the left indicate the molecular masses of protein standards in kDa.
The GST-Ub(G76V)-S-PEX5(12–324), GST-Ub(G76V)-S, and GST-Control recombinant proteins were then used as baits in pulldown assays performed with a female rat liver cytosol. As shown in Fig. 5B, GST-Ub(G76V)-S-PEX5(12–324), but not GST-Ub(G76V)-S, was able to pull down USP9X. This finding strongly suggests that amino acid residues 12–324 of PEX5 contribute for the interaction with USP9X.
The biochemical experiments described above strongly suggest that USP9X is the main DUB acting on soluble Ub-PEX5 both in rat liver and HeLa cells. We aimed at extending these findings using a cell biology approach. Specifically, we knocked down USP9X in HeLa cells and searched for a peroxisomal protein import defect. The assumption behind these experiments was that a decrease in USP9X might lead to an accumulation of soluble Ub-PEX5, which might then become polyubiquitinated and subsequently degraded at the proteasome (see Ref. 38). Unfortunately, although we were able to knock down USP9X by ∼90%, we have been unable to detect a peroxisomal protein import defect or even a decrease in the steady-state levels of PEX5 (see supplemental Fig. S3). Several possibilities could explain this negative result. The most obvious is that the remaining 10% of USP9X is still sufficient to deubiquitinate Ub-PEX5 at a good rate. Another possibility is that in the USP9X-knocked down cells a larger fraction of Ub-PEX5 is deubiquitinated by a nonenzymatic mechanism (e.g. the half-life of Ub-PEX5 in the presence of 5 mm reduced glutathione is 2.3 min; see Ref. 38). Alternatively, other enzyme(s) with a lower affinity/catalytic efficiency for Ub-PEX5 may compensate the partial loss of USP9X. Surely, a different cell biology strategy will be necessary to clarify this issue.
DISCUSSION
Using a cell-free in vitro import system, we have previously shown that the ATP-dependent dislocation of Ub-PEX5 from the peroxisomal DTM into the cytosol and the subsequent deubiquitination of soluble Ub-PEX5 are kinetically distinct events, leading us to propose that these two steps are uncoupled (38). The data presented in this work corroborate this idea. Indeed, we show that the separation of rat liver organelles from cytosolic proteins by a simple centrifugation is sufficient to separate the machinery that extracts Ub-PEX5 from peroxisomes from the DUB that acts on soluble Ub-PEX5. Therefore, it is unlikely that the DUB acting on Ub-PEX5 is a component of the receptor export module, as proposed recently for yeast (69).
The main aim of this work was to identify the mammalian DUB acting on soluble Ub-PEX5. Four independent observations suggest that this enzyme is USP9X. First, SEC analysis of a rat liver cytosol revealed that USP9X co-elutes with the activity that hydrolyzes Ub-PEX5. Second, immunopurified V5-tagged USP9X produced in HEK293T cells was able to hydrolyze Ub-PEX5. Third, immunoprecipitation/immunodepletion assays using an anti-USP9X antibody suggest that USP9X is the major enzyme acting on Ub-PEX5 both in rat liver and HeLa cells. Finally, GST-Ub(G76V)-S-PEX5(12–324), but not GST-Ub(G76V)-S, was able to pull down USP9X from a rat liver cytosol, thus suggesting that USP9X interacts with amino acid residues 12–324 of PEX5.
USP9X has been implicated in many different biological processes, including protein trafficking (45) and tight junction biogenesis in epithelial cells (70), regulation of the transforming growth factor-β pathway (51), and chromosome alignment and segregation (71). In addition, it has been shown that USP9X modulates the levels of proteins such as the following: α-synuclein (72), a protein involved in the pathogenesis of Parkinson disease; MCL1, an anti-apoptotic protein that is overexpressed in some types of cancer (73); some protein kinases involved in stress responses and cellular energy homeostasis (65, 74); and some ubiquitin-ligases, such as MARCH7 and Itch (75, 76). Our work suggests still another function for USP9X, namely the participation in the PEX5-mediated peroxisomal protein import pathway. Considering that (i) USP9X is widely expressed in mammalian tissues (58), (ii) that USP9X, although mostly cytosolic, is also found in the nucleoplasm (51, 71, 73), and (iii) this DUB is capable of removing monoubiquitins as well as polyubiquitin chains of different topologies from quite a number of proteins (71–74), it seems likely that the list of biological pathways in which USP9X is involved will grow in the future.
Understanding the function of the near 100 different mammalian DUBs (77) requires knowledge on their subcellular localization, their interactors, and their substrates. In the last years major advances have been made regarding the first two of these properties (78, 79). In contrast, the list of known substrates for these enzymes remains relatively small. Actually, for many of the mammalian DUBs, not a single substrate has been identified thus far (78–80). The reason behind this lack of knowledge is probably related to the fact that identifying substrates using biochemical approaches centered on the DUBs themselves is not an easy task. The low in vivo steady-state concentrations of many ubiquitinated proteins, the transient nature of the DUB-substrate interaction, and the possibility that at least some DUBs may act on numerous ubiquitin-conjugates make any attempt to isolate DUB-substrate complexes quite difficult (78, 79, 81, 82). Therefore, it is not surprising that the majority of the DUB substrates known presently were identified using substrate-centered approaches. Here, instead of trying to identify substrates that bind a given DUB, the goal is to identify the DUB that acts on a given substrate or biological pathway.
Two main strategies have been used for this purpose. One consists in knocking down all DUBs individually in a given cell line using RNAi technology and to search for a specific protein alteration or cell phenotype (51, 83). The other strategy relies on protein purification techniques to isolate the DUB acting on a particular substrate from a cell/tissue homogenate (84, 85). In this work, we show that this second approach may be quite simple. Indeed, our data indicate that a single chromatography step may be sufficient to resolve at least the most abundant mammalian DUBs to a point where it becomes possible to correlate one of these proteins with an enzymatic activity. In the specific case described here, a large pore size exclusion matrix turned out to be the best option, but naturally, SEC matrices with smaller pores or other types of matrices may provide better results in other situations. Once a correlation between an enzymatic activity and a DUB is established, the DUB can be identified with the help of an electrophilic ubiquitin probe. This strategy should be useful to identify DUBs acting on other substrates.
Supplementary Material
Acknowledgments
We thank Dr. Hidde Ploegh, WhiteHead Institute for Biomedical Research, Cambridge, MA, for the pTYB-HAUb plasmid, and Dr. Wilhelm W. Just (University of Heidelberg, Heidelberg, Germany) for the anti-PMP70 antibody.
This work was supported in part by Fundação para a Ciência e Tecnologia and Fundo Europeu de Desenvolvimento Regional through COMPETE-Programa Operacional Factores de Competitividade in the context of QREN, Portugal, Grants PEst-C/SAU/LA0002/2011, PEst-C/QUI/UI0062/2011, and PTDC/BIA-BCM/64771/2006.

This article contains supplemental Figs. S1–S3 and Tables S1–S2.
- DTM
- docking/translocation machinery
- DUB
- deubiquitinase
- PNS
- postnuclear supernatant
- REM
- receptor export module
- SEC
- size-exclusion chromatography
- Ubal
- ubiquitin-aldehyde
- UbVME
- ubiquitin-vinyl methyl ester
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Lazarow P. B., Fujiki Y. (1985) Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530 [DOI] [PubMed] [Google Scholar]
- 2. Brocard C., Hartig A. (2006) Peroxisome targeting signal 1. Is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 3. Lazarow P. B. (2006) The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604 [DOI] [PubMed] [Google Scholar]
- 4. Braverman N., Dodt G., Gould S. J., Valle D. (1998) An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7, 1195–1205 [DOI] [PubMed] [Google Scholar]
- 5. Galland N., Demeure F., Hannaert V., Verplaetse E., Vertommen D., Van der Smissen P., Courtoy P. J., Michels P. A. (2007) Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei. Biochim. Biophys. Acta 1773, 521–535 [DOI] [PubMed] [Google Scholar]
- 6. Otera H., Okumoto K., Tateishi K., Ikoma Y., Matsuda E., Nishimura M., Tsukamoto T., Osumi T., Ohashi K., Higuchi O., Fujiki Y. (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2. Studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woodward A. W., Bartel B. (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schliebs W., Kunau W. H. (2006) PTS2 co-receptors. Diverse proteins with common features. Biochim. Biophys. Acta 1763, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 9. Stanley W. A., Wilmanns M. (2006) Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim. Biophys. Acta 1763, 1592–1598 [DOI] [PubMed] [Google Scholar]
- 10. Costa-Rodrigues J., Carvalho A. F., Fransen M., Hambruch E., Schliebs W., Sá-Miranda C., Azevedo J. E. (2005) Pex5p, the peroxisomal cycling receptor, is a monomeric nonglobular protein. J. Biol. Chem. 280, 24404–24411 [DOI] [PubMed] [Google Scholar]
- 11. Shiozawa K., Konarev P. V., Neufeld C., Wilmanns M., Svergun D. I. (2009) Solution structure of human Pex5·Pex14·PTS1 protein complexes obtained by small angle x-ray scattering. J. Biol. Chem. 284, 25334–25342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., Gould S. J. (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9, 115–125 [DOI] [PubMed] [Google Scholar]
- 13. Gatto G. J., Jr., Geisbrecht B. V., Gould S. J., Berg J. M. (2000) Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 14. Stanley W. A., Filipp F. V., Kursula P., Schüller N., Erdmann R., Schliebs W., Sattler M., Wilmanns M. (2006) Recognition of a functional peroxisome type 1 target by the dynamic import receptor Pex5p. Mol. Cell 24, 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carvalho A. F., Costa-Rodrigues J., Correia I., Costa Pessoa J., Faria T. Q., Martins C. L., Fransen M., Sá-Miranda C., Azevedo J. E. (2006) The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 356, 864–875 [DOI] [PubMed] [Google Scholar]
- 16. Freitas M. O., Francisco T., Rodrigues T. A., Alencastre I. S., Pinto M. P., Grou C. P., Carvalho A. F., Fransen M., Sá-Miranda C., Azevedo J. E. (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gunkel K., van Dijk R., Veenhuis M., van der Klei I. J. (2004) Routing of Hansenula polymorpha alcohol oxidase. An alternative peroxisomal protein-sorting machinery. Mol. Biol. Cell 15, 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klein A. T., van den Berg M., Bottger G., Tabak H. F., Distel B. (2002) Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277, 25011–25019 [DOI] [PubMed] [Google Scholar]
- 19. Oshima Y., Kamigaki A., Nakamori C., Mano S., Hayashi M., Nishimura M., Esaka M. (2008) Plant catalase is imported into peroxisomes by Pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 49, 671–677 [DOI] [PubMed] [Google Scholar]
- 20. van der Klei I. J., Hilbrands R. E., Swaving G. J., Waterham H. R., Vrieling E. G., Titorenko V. I., Cregg J. M., Harder W., Veenhuis M. (1995) The Hansenula polymorpha PER3 gene is essential for the import of PTS1 proteins into the peroxisomal matrix. J. Biol. Chem. 270, 17229–17236 [DOI] [PubMed] [Google Scholar]
- 21. Grou C. P., Carvalho A. F., Pinto M. P., Alencastre I. S., Rodrigues T. A., Freitas M. O., Francisco T., Sá-Miranda C., Azevedo J. E. (2009) The peroxisomal protein import machinery. A case report of transient ubiquitination with a new flavor. Cell. Mol. Life Sci. 66, 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lanyon-Hogg T., Warriner S. L., Baker A. (2010) Getting a camel through the eye of a needle. The import of folded proteins by peroxisomes. Biol. Cell 102, 245–263 [DOI] [PubMed] [Google Scholar]
- 23. Ma C., Subramani S. (2009) Peroxisome matrix and membrane protein biogenesis. IUBMB Life 61, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf J., Schliebs W., Erdmann R. (2010) Peroxisomes as dynamic organelles. Peroxisomal matrix protein import. FEBS J. 277, 3268–3278 [DOI] [PubMed] [Google Scholar]
- 25. Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H. (2003) Pex8p. An intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646 [DOI] [PubMed] [Google Scholar]
- 26. Reguenga C., Oliveira M. E., Gouveia A. M., Sá-Miranda C., Azevedo J. E. (2001) Characterization of the mammalian peroxisomal import machinery. Pex2p, Pex5p, Pex12p, and Pex14p are subunits of the same protein assembly. J. Biol. Chem. 276, 29935–29942 [DOI] [PubMed] [Google Scholar]
- 27. Gouveia A. M., Guimarães C. P., Oliveira M. E., Sá-Miranda C., Azevedo J. E. (2003) Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J. Biol. Chem. 278, 4389–4392 [DOI] [PubMed] [Google Scholar]
- 28. Gouveia A. M., Reguenga C., Oliveira M. E., Sa-Miranda C., Azevedo J. E. (2000) Characterization of peroxisomal Pex5p from rat liver. Pex5p in the Pex5p-Pex14p membrane complex is a transmembrane protein. J. Biol. Chem. 275, 32444–32451 [DOI] [PubMed] [Google Scholar]
- 29. Alencastre I. S., Rodrigues T. A., Grou C. P., Fransen M., Sá-Miranda C., Azevedo J. E. (2009) Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway. J. Biol. Chem. 284, 27243–27251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oliveira M. E., Gouveia A. M., Pinto R. A., Sá-Miranda C., Azevedo J. E. (2003) The energetics of Pex5p-mediated peroxisomal protein import. J. Biol. Chem. 278, 39483–39488 [DOI] [PubMed] [Google Scholar]
- 31. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., Fujiki Y. (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner. Conserved Pex5p WXXX(F/Y) motifs are critical for matrix protein import. Mol. Cell. Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F. W., Kunau W. H. (1999) Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 274, 5666–5673 [DOI] [PubMed] [Google Scholar]
- 33. Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., Azevedo J. E. (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 [DOI] [PubMed] [Google Scholar]
- 34. Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., Fujiki Y. (2011) Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12, 1067–1083 [DOI] [PubMed] [Google Scholar]
- 35. Williams C., van den Berg M., Sprenger R. R., Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 36. Miyata N., Fujiki Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5. ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 38. Grou C. P., Carvalho A. F., Pinto M. P., Huybrechts S. J., Sá-Miranda C., Fransen M., Azevedo J. E. (2009) Properties of the ubiquitin-Pex5p thiol ester conjugate. J. Biol. Chem. 284, 10504–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fransen M., Brees C., Baumgart E., Vanhooren J. C., Baes M., Mannaerts G. P., Van Veldhoven P. P. (1995) Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J. Biol. Chem. 270, 7731–7736 [DOI] [PubMed] [Google Scholar]
- 40. Gouveia A. M., Guimaraes C. P., Oliveira M. E., Reguenga C., Sa-Miranda C., Azevedo J. E. (2003) Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J. Biol. Chem. 278, 226–232 [DOI] [PubMed] [Google Scholar]
- 41. Ferro A., Carvalho A. L., Teixeira-Castro A., Almeida C., Tomé R. J., Cortes L., Rodrigues A. J., Logarinho E., Sequeiros J., Macedo-Ribeiro S., Maciel P. (2007) NEDD8. A new ataxin-3 interactor. Biochim. Biophys. Acta 1773, 1619–1627 [DOI] [PubMed] [Google Scholar]
- 42. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1. Implications for Lewy body formation. J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grou C. P., Carvalho A. F., Pinto M. P., Wiese S., Piechura H., Meyer H. E., Warscheid B., Sá-Miranda C., Azevedo J. E. (2008) Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283, 14190–14197 [DOI] [PubMed] [Google Scholar]
- 44. Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., Kessler B. M. (2002) Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 45. Murray R. Z., Jolly L. A., Wood S. A. (2004) The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and β-catenin. Mol. Biol. Cell 15, 1591–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinto M. P., Grou C. P., Fransen M., Sá-Miranda C., Azevedo J. E. (2009) The cytosolic domain of PEX3, a protein involved in the biogenesis of peroxisomes, binds membrane lipids. Biochim. Biophys. Acta 1793, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 47. Catic A., Misaghi S., Korbel G. A., Ploegh H. L. (2007) ElaD, a deubiquitinating protease expressed by E. coli. PLoS One 2, e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hershko A., Rose I. A. (1987) Ubiquitin-aldehyde. A general inhibitor of ubiquitin-recycling processes. Proc. Natl. Acad. Sci. U.S.A. 84, 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilkinson K. D., Gan-Erdene T., Kolli N. (2005) Derivatization of the C terminus of ubiquitin and ubiquitin-like proteins using intein chemistry. Methods and uses. Methods Enzymol. 399, 37–51 [DOI] [PubMed] [Google Scholar]
- 50. Oegema K., Wiese C., Martin O. C., Milligan R. A., Iwamatsu A., Mitchison T. J., Zheng Y. (1999) Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., Piccolo S. (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 [DOI] [PubMed] [Google Scholar]
- 52. Ivashchenko O., Van Veldhoven P. P., Brees C., Ho Y. S., Terlecky S. R., Fransen M. (2011) Intraperoxisomal redox balance in mammalian cells. Oxidative stress and interorganellar cross-talk. Mol. Biol. Cell 22, 1440–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carvalho A. F., Pinto M. P., Grou C. P., Vitorino R., Domingues P., Yamao F., Sá-Miranda C., Azevedo J. E. (2012) High yield expression in Escherichia coli and purification of mouse ubiquitin-activating enzyme E1. Mol. Biotechnol., doi: 10.1007/s12033-011-9463-x [DOI] [PubMed] [Google Scholar]
- 54. Köster A., Heisig M., Heinrich P. C., Just W. W. (1986) In vitro synthesis of peroxisomal membrane polypeptides. Biochem. Biophys. Res. Commun. 137, 626–632 [DOI] [PubMed] [Google Scholar]
- 55. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2011) GenBank. Nucleic Acids Res. 39, D32–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oliveira M. E., Reguenga C., Gouveia A. M., Guimarães C. P., Schliebs W., Kunau W. H., Silva M. T., Sá-Miranda C., Azevedo J. E. (2002) Mammalian Pex14p. Membrane topology and characterization of the Pex14p-Pex14p interaction. Biochim. Biophys. Acta 1567, 13–22 [DOI] [PubMed] [Google Scholar]
- 57. Haas A. L., Warms J. V., Rose I. A. (1983) Ubiquitin adenylate. Structure and role in ubiquitin activation. Biochemistry 22, 4388–4394 [DOI] [PubMed] [Google Scholar]
- 58. Jones M. H., Furlong R. A., Burkin H., Chalmers I. J., Brown G. M., Khwaja O., Affara N. A. (1996) The Drosophila developmental gene fat facets has a human homolog in Xp11.4, which escapes X-inactivation and has related sequences on Yq11.2. Hum. Mol. Genet. 5, 1695–1701 [DOI] [PubMed] [Google Scholar]
- 59. Puente X. S., López-Otín C. (2004) A genomic analysis of rat proteases and protease inhibitors. Genome Res. 14, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wood S. A., Pascoe W. S., Ru K., Yamada T., Hirchenhain J., Kemler R., Mattick J. S. (1997) Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech. Dev. 63, 29–38 [DOI] [PubMed] [Google Scholar]
- 61. Brown G. M., Furlong R. A., Sargent C. A., Erickson R. P., Longepied G., Mitchell M., Jones M. H., Hargreave T. B., Cooke H. J., Affara N. A. (1998) Characterization of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum. Mol. Genet. 7, 97–107 [DOI] [PubMed] [Google Scholar]
- 62. Xu J., Burgoyne P. S., Arnold A. P. (2002) Sex differences in sex chromosome gene expression in mouse brain. Hum. Mol. Genet. 11, 1409–1419 [DOI] [PubMed] [Google Scholar]
- 63. Luddi A., Margollicci M., Gambera L., Serafini F., Cioni M., De Leo V., Balestri P., Piomboni P. (2009) Spermatogenesis in a man with complete deletion of USP9Y. N. Engl. J. Med. 360, 881–885 [DOI] [PubMed] [Google Scholar]
- 64. Pantaleon M., Kanai-Azuma M., Mattick J. S., Kaibuchi K., Kaye P. L., Wood S. A. (2001) FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech. Dev. 109, 151–160 [DOI] [PubMed] [Google Scholar]
- 65. Nagai H., Noguchi T., Homma K., Katagiri K., Takeda K., Matsuzawa A., Ichijo H. (2009) Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol. Cell 36, 805–818 [DOI] [PubMed] [Google Scholar]
- 66. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Siegel L. M., Monty K. J. (1966) Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 68. Johnson E. S., Bartel B., Seufert W., Varshavsky A. (1992) Ubiquitin as a degradation signal. EMBO J. 11, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Debelyy M. O., Platta H. W., Saffian D., Hensel A., Thoms S., Meyer H. E., Warscheid B., Girzalsky W., Erdmann R. (2011) Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286, 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Théard D., Labarrade F., Partisani M., Milanini J., Sakagami H., Fon E. A., Wood S. A., Franco M., Luton F. (2010) USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 29, 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vong Q. P., Cao K., Li H. Y., Iglesias P. A., Zheng Y. (2005) Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310, 1499–1504 [DOI] [PubMed] [Google Scholar]
- 72. Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A. J., Shani V., Engelender S. (2011) α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 18666–18671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schwickart M., Huang X., Lill J. R., Liu J., Ferrando R., French D. M., Maecker H., O'Rourke K., Bazan F., Eastham-Anderson J., Yue P., Dornan D., Huang D. C., Dixit V. M. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumor cell survival. Nature 463, 103–107 [DOI] [PubMed] [Google Scholar]
- 74. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. (2008) Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 411, 249–260 [DOI] [PubMed] [Google Scholar]
- 75. Mouchantaf R., Azakir B. A., McPherson P. S., Millard S. M., Wood S. A., Angers A. (2006) The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 281, 38738–38747 [DOI] [PubMed] [Google Scholar]
- 76. Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., Lehner P. J. (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9, 1130–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 78. Kouranti I., McLean J. R., Feoktistova A., Liang P., Johnson A. E., Roberts-Galbraith R. H., Gould K. L. (2010) A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8, e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ventii K. H., Wilkinson K. D. (2008) Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Amerik A. Y., Li S. J., Hochstrasser M. (2000) Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381, 981–992 [DOI] [PubMed] [Google Scholar]
- 82. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Buus R., Faronato M., Hammond D. E., Urbé S., Clague M. J. (2009) Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr. Biol. 19, 1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Joo H. Y., Jones A., Yang C., Zhai L., Smith A. D., 4th, Zhang Z., Chandrasekharan M. B., Sun Z. W., Renfrow M. B., Wang Y., Chang C., Wang H. (2011) Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 286, 7190–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Parsons J. L., Dianova I. I., Khoronenkova S. V., Edelmann M. J., Kessler B. M., Dianov G. L. (2011) USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase β. Mol. Cell 41, 609–615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.