Background: CLP-1 heterozygous mice exhibit enhanced susceptibility to cardiac stress.
Results: Angiotensin-II-induced left ventricular hypertrophy and fibrosis were enhanced, and the Smad3 and Stat3 signaling was stimulated in CLP-1+/− mice.
Conclusion: CLP-1 controls the hypertrophy and fibrotic response during cardiac remodeling.
Significance: Our results offer the potential of targeting CLP-1 for therapeutic intervention in cardiac disease.
Keywords: Angiotensin II, Cardiac Hypertrophy, Cardiovascular Disease, Fibroblast, Fibrosis, Heart, Matrix Metalloproteinase (MMP), Myofibroblast, Natriuretic Peptides
Abstract
It is well known that the renin-angiotensin system contributes to left ventricular hypertrophy and fibrosis, a major determinant of myocardial stiffness. TGF-β1 and renin-angiotensin system signaling alters the fibroblast phenotype by promoting its differentiation into morphologically distinct pathological myofibroblasts, which potentiates collagen synthesis and fibrosis and causes enhanced extracellular matrix deposition. However, the atrial natriuretic peptide, which is induced during left ventricular hypertrophy, plays an anti-fibrogenic and anti-hypertrophic role by blocking, among others, the TGF-β-induced nuclear localization of Smads. It is not clear how the hypertrophic and fibrotic responses are transcriptionally regulated. CLP-1, the mouse homolog of human hexamethylene bis-acetamide inducible-1 (HEXIM-1), regulates the pTEFb activity via direct association with pTEFb causing inhibition of the Cdk9-mediated serine 2 phosphorylation in the carboxyl-terminal domain of RNA polymerase II. It was recently reported that the serine kinase activity of Cdk9 not only targets RNA polymerase II but also the conserved serine residues of the polylinker region in Smad3, suggesting that CLP-1-mediated changes in pTEFb activity may trigger Cdk9-dependent Smad3 signaling that can modulate collagen expression and fibrosis. In this study, we evaluated the role of CLP-1 in vivo in induction of left ventricular hypertrophy in angiotensinogen-overexpressing transgenic mice harboring CLP-1 heterozygosity. We observed that introduction of CLP-1 haplodeficiency in the transgenic α-myosin heavy chain-angiotensinogen mice causes prominent changes in hypertrophic and fibrotic responses accompanied by augmentation of Smad3/Stat3 signaling. Together, our findings underscore the critical role of CLP-1 in remodeling of the genetic response during hypertrophy and fibrosis.
Introduction
Cardiac hypertrophy, manifested by an increase in the size of cardiomyocytes and proliferation of cardiac fibroblasts, is initially a compensatory response to cardiac insults designed to maintain the myocardial output and wall stress. When the workload becomes chronic, the compensatory remodeling of the heart becomes decompensatory and is accompanied by defined structural alterations due to the increase in the extracellular matrix (ECM)2 proteins, which culminate into excessive fibrosis, loss of compliance, and heart failure. It is known that the renin-angiotensin system (RAS) contributes to left ventricular hypertrophy (LVH) and fibrosis (1, 2), which is characterized by a disproportionate accumulation of collagen type I, a major determinant of myocardial stiffness. It is also known that the transforming growth factor β (TGF-β)/Mothers against decapentaplegic homolog 3 (Smad3) signaling is activated in LVH and fibrotic response (3). Mechanistically, the TGF-β/Smad3 signaling induces a genetic program that leads to a disproportionate increase in collagen, excessive deposition of ECM, and remodeling of the heart (4, 5). TGF-β1 and RAS signaling alters the fibroblast phenotype by promoting its differentiation into morphologically distinct pathological myofibroblasts that, in turn, promotes collagen synthesis and causes enhanced ECM deposition. Recent reports suggest the participation of atrial natriuretic peptides (ANP) as an anti-fibrogenic molecule in hypertrophic response in the heart via modulation of ECM gene expression (6). ANP, whose expression is increased in various deleterious conditions to the heart, such as pressure and volume overload (6, 7) and heart failure (8), is known to block TGF-β-induced nuclear localization of the causes of Smads and the inhibition of TGF-β-induced ECM proteins (9). Thus, it appears that Smad signaling plays a prominent role in AngII-moderated hypertrophic response. In addition to Smad, the signal transduction and activator of transcription 3 (STAT3) signaling also plays a role in cardiac hypertrophy (10, 11). We have previously shown that activation of Janus kinase (Jak)/STAT signaling up-regulates angiotensinogen (ANG) gene transcription promoting the autocrine RAS loop in cardiomyocytes (10–13). Despite the wealth of information, it is not clear how the hypertrophic and fibrotic responses are transcriptionally regulated.
Regulation of transcription at the level of chain elongation is now considered as the primary site for eukaryotic gene control and is mediated by the positive transcription elongation factor (pTEFb) complex that consists of cyclin-dependent kinase 9 (Cdk9) and its partner protein CyclinT1. CLP-1, the mouse homolog of human hexamethylene bis-acetamide inducible 1 (HEXIM-1), regulates pTEFb activity via its dynamic association/dissociation with pTEFb causing inhibition/activation of Cdk9-mediated serine 2 phosphorylation in the carboxyl-terminal domain of RNA polymerase II. We previously reported that the “on” and “off” association of CLP-1 with pTEFb is Jak2-dependent (14), a phenomenon that is paramount to control of hypertrophic growth (14–17). More recently, it was reported that the serine kinase activity of Cdk9 not only targets RNA polymerase II but also the conserved serine residues of the polylinker region in Smad3 (18), leading to speculation that CLP-1-mediated changes in pTEFb activity may trigger Cdk9-dependent Smad3 signaling that can modulate collagen expression and fibrosis. We have also reported previously that CLP-1 null mice die during late embryonic development (19), but the heterozygote CLP-1+/− mice survive and harbor enhanced susceptibility to hypertrophic agonists providing a useful animal model for investigation of the mechanistic aspects of compensated hypertrophy (19). In this study, we evaluate the role of CLP-1 in vivo in induction of LVH in ANG-overexpressing transgenic mice harboring CLP-1 heterozygosity. We observed that introduction of CLP-1 haplodeficiency in the transgenic α-myosin heavy chain (αMHC)-ANG mice causes prominent changes in hypertrophic and fibrotic responses. Our findings underscore the critical role of CLP-1 in remodeling of the genetic response during hypertrophy and fibrosis.
EXPERIMENTAL PROCEDURES
Transgenic Lines
Production of transgenic mice with cardiac-restricted expression of αMHC-ANG, a kind gift of Dr. Thierry Pedrazzini, has been described previously (20, 21). Male αMHC-ANG (TG1306)/CLP-1+/− mice were produced by crossing C57BL6 CLP-1+/− mice generated in our laboratory with αMHC-ANG mice. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH Publication 85-23, revised 1996) and the approval of the institutional Animal Care and Use Committee at State University of New York-Downstate Medical School (protocol 03-209-10).
Immunoprecipitation and Western Blot Analysis
The hearts were homogenized with a glass tissue grinder in ice-chilled buffer A (10 mmol/liter HEPES (pH 7.9). 1.5 mmol/liter MgCl2, 10 mmol/liter KCl, 200 mmol/liter NaCl, 0.2 mmol/liter EDTA) supplemented with 1 mmol/liter DTT, protease inhibitor mixture (P-8340, Sigma), 40 units/ml RNasin (Promega), phosphatase inhibitor mixtures (P2850 and P5726, Sigma), and 0.1% Nonidet P-40. The extracts were subjected to sequential centrifugations for 5 min each at 500 and 9000 × g at 4 °C. The final supernatant was used in all experiments. For immunoprecipitation, the supernatant was incubated overnight with rabbit antibody against Jak2 (Santa Cruz Biotechnology), followed by 3 h of incubation with protein G-agarose. Proteins were separated by PAGE (10% Tris-HCl) and electrotransferred onto nitrocellulose membrane. The blots were blocked in Tris-buffered saline, 0.1% Tween 20 with 5% bovine serum albumin powder for 1 h at room temperature. After incubation with the primary antibody, detection was performed using secondary horseradish peroxide-coupled antibody and ECL-enhanced chemiluminescence according to the recommendations of the supplier (Amersham Biosciences). The following antibodies were used: rabbit anti-CLP-1 (Proteintech Group Inc.); rabbit anti-GAPDH (Abcam); rabbit anti-cyclin T1 mouse monoclonal anti-cdk9, rabbit anti-Stat3, and rabbit anti-Nox4 (Santa Cruz Biotechnology); rabbit anti-phosphotyrosine-Stat3, rabbit anti-phosphotyrosine-Jak2, rabbit anti-Jak2 (Cell Signaling); anti-smooth muscle α-actin (Sigma); and anti-rabbit cysteine-rich 61-connective tissue growth factor-nephroblastoma overexpressed (CCN) 1 from Biovision Inc. (Mountain View, CA). Monoclonal antibodies against matrix metalloproteases 3 and 9 antibodies were from Calbiochem, and anti-phosphoserine 208 Smad3 was from Dr. Fang Liu (Center for Advanced Biotechnology and Medicine, Rutgers).
Quantitative Real Time-PCR (qPCR)
The following oligonucleotide primers specific for mouse cardiac genes were used in this study: ANF, sense 5′-gaacctgctagaccacct-3′ and antisense 5′-cctagtccactctgggct-3′; brain natriuretic peptide (BNP), sense 5′-aagctgctggagctgataaga-3′ and antisense 5′-gttacagcccaaacgactgac-3′; and GAPDH, 5′-tgaccacagtccatgccatc-3′ and antisense 5′-gacggacacattgggggtag-3′. Total RNA was extracted from frozen cardiac tissues of mice group using TRIzol reagent (Invitrogen), and cDNA was generated from 1 μg of RNA using a random hexamer and the Omniscript RT (Qiagen) kit. qPCR was performed as described in the qPCR core kit from SBYR Green (Eurogentec). The data were analyzed using the ΔΔCT method and presented as arbitrary units.
Morphological Analysis
Hearts from 6-month-old male mice of wild type, CLP-1+/−, αMHC-ANG, and αMHC-ANG/CLP-1+/− genotypes were fixed in 10% buffered formalin for 24 h, followed by washing in phosphate-buffered saline (PBS) and processed for paraffin wax tissue sectioning. Ten-μm tissue sections were prepared and stained with hematoxylin and eosin and Masson Trichrome staining performed in accordance with the instructions of the manufacturer (HT15-KT, Sigma HT15 Trichrome stain (Masson) kit). For myocyte cross-sectional area, the outlines of 100 myocytes were determined in each group and measured with an image quantitative digital analysis system (National Institutes of Health Image 1.6). Collagen content was determined in Trichrome-stained sections and measured by using the software Image-Pro Plus.
Angiotensin II Measurement
The intra-cardiac AngII peptide levels were determined by enzyme immunoassay as suggested by the manufacturer RayBiotech, Inc. Briefly, left ventricle tissues were harvested from each mouse group and frozen in liquid nitrogen. Heart tissue were homogenized in lysis buffer: 50 mm HEPES (pH 8.0), 10 mm MgCl2, 120 mm NaCl, 0.5% Nonidet P-40, 2.5 mm MnCl2, and protease inhibitor mixture (P-8340, Sigma). The assay minimum detectable concentration of angiotensin II is 2.62 pg/ml.
Echocardiography
For echocardiography, the mouse chest was shaved, and echocardiography was done on conscious mice to avoid any possible cardiac effects of anesthesia. Mice were imaged using the optimal two-dimensionally guided M-mode parasternal view. Echocardiography was performed using the Phillips SONOS 5500 with a 15 MHz linear probe. Machine and gain settings were optimized for best image quality using maximal sweep speed. Images were stored digitally on optical disk for analysis. Left ventricular systolic (LVESD), diastolic (LVEDD), septal (SW), and posterior wall (PW) thicknesses were measured. Left ventricular fractional shortening (FS) was calculated from the following formula: FS (%) = ((LVEDD − LVESD)/LVEDD) × 100. The ejection fraction (EF) is calculated from the following formula: EF = ((LVEDD2 − LVESD2)/LVEDD2) × 100. Left ventricular mass is calculated from the following formula: LV mass = 1.05 ((LVEDD + SW + PW)3 − (LVEDD3)).
Statistical Analysis
Data are presented as mean ± S.E. The Graph Pad Prism5 software was used for statistical analysis. Differences were compared using analysis of variance; p < 0.05 was considered statistically significant.
RESULTS
Hypertrophy in αMHC-ANG/CLP-1+/− Hearts
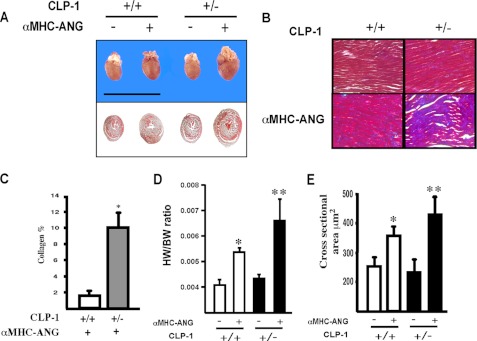
To establish a direct cause/effect relationship between CLP-1-mediated changes in P-TEFb activity and cardiac hypertrophy, we crossed the αMHC-ANG transgenic mice with CLP-1+/− mice to obtain the bigenic αMHC-ANG/CLP-1+/− mice. The bigenic mice have characteristically an enlarged heart with significant thickening of the ventricular wall compared with the hearts of age-matched ANG/CLP-1+/+ and CLP-1+/− mice (Fig. 1A). Transverse sections of the hearts of each genotype, cut at identical landmarks and stained with Masson Trichrome, showed an increase in accumulation of collagen in the left ventricle of the αMHC-ANG/CLP-1+/− hearts (Fig. 1B), significantly more than in ANG/CLP-1+/+ (Fig. 1, B and C) and consistent with previous reports (20). Likewise, there was an increase in the heart weight to body weight ratio (Fig. 1D) accompanied with an increase in muscle fiber diameter in αMHC-ANG/CLP-1+/− hearts (Fig. 1E). The morphological and functional changes of all four genotypes were also evaluated by M-mode echocardiography (see under “Experimental Procedures”) (Table 1), which shows an increase in LV mass, ED-SWT, and ES-SWT in the αMHC-ANG/CLP-1+/− mice. It also appears that αMHC-ANG/CLP-1+/− hearts have better systolic function as revealed by fractional shortening. Further characterization of the αMHC-ANG/CLP-1+/− transgenic mice was obtained by determining the levels of intracardiac AngII peptide. The data in Table 2 show that the levels of intracardiac AngII in αMHC-ANG and αMHC-ANG/CLP-1+/− transgenic mice are similar and support the findings of Mazzolai et al. (21).
FIGURE 1.
Enhanced left ventricular hypertrophy in αMHC-ANG/CLP-1+/− mice. A, representative cross-section at the level of the left ventricle of adult mouse hearts of wild type heart, transgenic wild type hearts, nontransgenic heterozygous heart, and transgenic heterozygous heart. B, panel shows collagen deposition in a section of the left ventricle of αMHC-ANG/CLP-1+/+ and αMHC-ANG/CLP-1+/− mice obtained by Masson Trichrome stain in magnification of ×200. C, bar graph shows the distribution of collagen deposition between αMHC-ANG/CLP-1+/+ and αMHC-ANG/CLP-1+/− hearts sections (n = 3 per group). *, p < 0.05 between αMHC-ANG/CLP-1+/− versus αMHC-ANG/CLP-1+/+. D, heart/body weight ratios for nontransgenic wild type heart, nontransgenic heterozygous heart, and transgenic heterozygous heart. *, p values < 0.05. For wild type versus αMHC-ANG, the values are means ± S.E.; **, p values are <0.01 αMHC-ANG/CLP-1+/− versus αMHC-ANG. E, bar graph shows the quantification of cross-sectional area. Values are mean ± S.E.; *, p < 0.05 αMHC-ANG versus wild type; **, p values are <0.01 αMHC-ANG/CLP-1+/− versus αMHC-ANG.
TABLE 1.
Functional analysis of wild-type (WT), heterozygous (CLP-1+/−), transgenic wild type-αMHC-ANG (TG+/+), and transgenic heterozygous-αMHC-ANG (TG+/−) mice hearts obtained by M-mode echocardiograms
LVESD indicates left ventricle end-systolic diameter; LVEDD, LV end diastolic diameter; ED-SWT, end-diastolic septal wall thickness; ES-SWT, end-systolic septal wall thickness; LVmass left ventricular mass and % FS, fractional shortening. Values are means ± S.E.
| αMHC-ANG CLP-1 | WT+/+n = 8 | TG+/+n = 10 | WT+/−n = 10 | TG+/−n = 12 |
|---|---|---|---|---|
| Heart rate (beats/min) | 610 ± 13 | 590 ± 33 | 650 ± 17 | 630 ± 11 |
| LVESD (mm) | 1.08 ± 0.006 | 1.29 ± 0.004 | 0.93 ± 0.005 | 1.06 ± 0.003 |
| LVEDD (mm) | 2.18 ± 0.010 | 2.37 ± 0.011 | 2.03 ± 0.024 | 2.35 ± 0.009 |
| ED-SWT (mm) | 0.97 ± 0.020 | 1.26 ± 0.010a | 0.88 ± 0.060 | 1.25 ± 0.040a |
| ES-SWT (mm) | 1.20 ± 0.030 | 1.60 ± 0.070a | 1.40 ± 0.030 | 1.98 ± 0.050a |
| LV mass (mg) | 60 ± 3 | 80 ± 4a | 62 ± 2 | 93 ± 3b |
| % FS | 50 ± 2 | 45 ± 4 | 54 ± 1 | 54 ± 3b |
a p < 0.05 versus corresponding control group.
b p < 0.05 αMHC-ANG/CLP-1+/− versus αMHC-ANG.
TABLE 2.
Intracardiac of angiotensin II peptide
Tissue extracts from the left ventricle of 6-month-old wild-type (WT), heterozygous (CLP-1+/−), transgenic wild type-αMHC-ANG (TG+/+), and transgenic heterozygous-αMHC-ANG (TG+/−) mice were used to determined by enzyme immunoassay of the AngII peptide levels. Data are expressed as means ± S.E. of six mice per group.
| Angiotensin II (heart) | |||
|---|---|---|---|
| CLP-1+/+ without αMHC-ANG | CLP-1+/− without αMHC-ANG | CLP-1+/+ with αMHC-ANG | CLP-1+/− with αMHC-ANG |
| fmol/g | |||
| 128.3 ± 7.4 | 125.8 ± 9.5 | 332.8 ± 33.4a | 360.2 ± 70.5b |
a p < 0.05 αMHC-ANG versus wild type.
b p < 0.01 CLP-1+/− versus CLP-1 heterozygous mice.
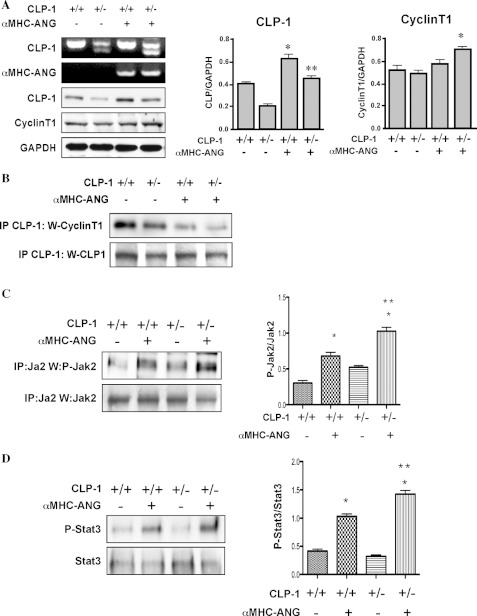
Overexpression of CyclinT1, the Cdk9 kinase partner protein in the P-TEFb complex, has been shown to potentiate cardiac hypertrophy suggesting that CyclinT1 is limiting for activation of P-TEFb activity (15, 16). We examined the protein levels of CyclinT1 in Western blot that show an increase in αMHC-ANG/CLP-1+/− transgene as compared with the wild type hearts without overexpression of ANG (Fig. 2A), suggesting that a synergistic relationship exists between heterozygosity of CLP-1 and expression of CyclinT1. The protein level of CLP-1 was low in the heterozygous control mice compared with wild type mice, as expected. Because CLP-1 is known to be associated with CyclinT1 (14), the reduction in its level in heterozygous hearts is expected to cause an increase of CLP-1-free CyclinT1 in the bigenic hearts, which can be ascribed to enhancement of transcriptional response in hypertrophy. We therefore examined the association of CLP-1 with P-TEF-b in extracts from αMHC-ANG/CLP-1+/− hypertrophied hearts by immunoprecipitation with anti-CLP-1 antibody and Western blotting with anti-CyclinT1 antibody. Fig. 2B shows that there was a reduction in association of CLP-1 with the P-TEFb complex in AngII-induced hypertrophied hearts compared with mice with wild type CLP-1 levels. We then examined whether introduction of CLP-1 heterozygosity in the ANG transgenes would influence the Jak2/Stat3 growth signaling by immunoprecipitation with anti-Jak2 antibody and Western blotting with anti-phosphotyrosine Jak2 and Stat3 antibodies. Fig. 2, C and D, shows that both Jak2 and Stat3 tyrosine phosphorylation levels were increased significantly in the bigenic hearts, suggesting the RAS loop activation is functionally involved in enhancement of transcriptional activity in which CLP-1 apparently plays a regulatory role.
FIGURE 2.
A, two upper panels represent the CLP-1 and αMHC-ANG genotypes of each genetic background Western-blotted with specific antibodies. GAPDH was used as loading control (n = 4). Statistical analysis on the expression levels of CLP-1 and cyclin T1 is depicted in the graphs. CLP-1 expression, *, p < 0.05 CLP-1+/+ versus αMHC-ANG/CLP-1+/+; **, p < 0.05 CLP-1+/− versus αMHC-ANG/CLP-1+/−. p value was not significant between CLP-1+/+ versus αMHC-ANG/CLP-1+/−. CyclinT1 expression, *, p < 0.05 CLP-1+/− versus αMHC-ANG/CLP-1+/−. B, representative Western blot showing CLP-1/CyclinT1 interaction by immunoprecipitation (IP) of total protein heart extracts from mice of each genetic background with anti-CLP-1 followed by blotting with anti-CyclinT1 antibody. Loading control was performed using a chicken anti-CLP-1 antibody (bottom panel). C, AngII triggers activation of the Jak/Stat pathway. The upper panel is a Western blot showing tyrosine phosphorylation of Jak2. Total heart tissue extracts were immunoprecipitated with polyclonal anti-Jak2 antibody and probed with anti-phosphotyrosine-Jak2 antibody. A direct Western blot using anti-Jak2 antibody serves as loading control. D, lower panel shows the tyrosine phosphorylation of Stat3. As above, total heart tissue extracts were immunoprecipitated with rabbit anti-Stat3 followed by Western blotting with anti-phosphotyrosine Stat3 antibodies. Data are expressed as means ± S.E. of four independent experiments. *, p < 0.05 control mice versus transgenic mice. **, p < 0.05 αMHC-ANG/CLP-1+/− versus αMHC-ANG.
Extracellular Matrix Deposition in αMHC-ANG/CLP-1+/− Hearts
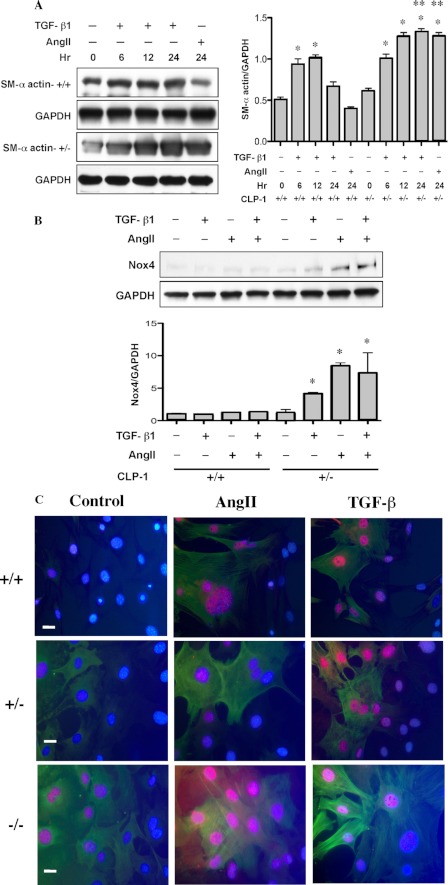
Evidence suggests that TGF-β induction in remodeling of the heart is mediated via AngII, which also stimulates fibroblast proliferation and ECM protein production. The finding that TGF-β1−/− mice chronically treated with AngII failed to develop LVH (3) supports the notion that AngII-mediated LVH is intrinsically linked to TGF-β1 signaling. Collagen turnover is triggered by fibroblasts undergoing TGF-β-induced transition to myofibroblasts representing the pathological feature of the failing heart. This prompted us to examine whether the fibroblast transition is controlled by CLP-1/pTEFb activity. We measured the expression of smooth muscle α-actin (SM-α-actin), a known marker for myofibroblasts (22, 23). Fibroblasts were isolated from CLP-1+/− heterozygous mouse hearts and treated with either AngII or TGF-β. Fig. 3A shows that there was a time-dependent increase in SM-α-actin in fibroblasts isolated from CLP-1+/− hearts compared with those from the wild type hearts. It is also known that fibrosis triggered by either AngII or TGF-β involves the utilization of reactive oxygen species (ROS), derived from NAD(P)H oxidases (23) and Nox4, and plays a role in ECM deposition. We show here that the expression of Nox4 is augmented in CLP-1+/− fibroblasts treated with TGF-β or AngII (Fig. 3B). To evaluate in situ whether decreased expression of CLP-1 correlates with enhanced Smad3 activation and smooth muscle expression, we performed immunofluorescence studies in mouse embryonic wild type (+/+), heterozygous (CLP-1+/−), and null (CLP-1−/−) fibroblasts subjected to AngII or TGF-β1 induction. As seen in Fig. 3C, a distinct nuclear localization of serine 208-Smad3 was observed that correlated with α-smooth muscle actin expression.
FIGURE 3.
Enhanced expression of smooth muscle α-actin in CLP-1+/− fibroblasts. A, Western blots show the expression of smooth muscle α-actin in wild type (SM-α-actin+/+) and CLP-1+/− (SM-α-actin+/+) fibroblasts. GAPDH expression was used as loading control. Data are expressed as means ± S.E. of four independent experiments. SM-α-actin, *, p < 0.05 wild type versus wild type treated with agonist; *, p < 0.05 control untreated versus treated with TGF-β, and **, p < 0.05 CLP-1+/− versus wild type at 24 h treated with agonists. B, Western blot of wild type (CLP-1+/+) and heterozygous (CLP-1+/−) cardiac fibroblasts treated for 24 h with TGF-β (10 ng/ml) and/or AngII peptide (10−7 m). The Western blot shows that the expression of Nox4 paralleled the increase expression of smooth muscle α-actin in CLP-1+/− fibroblast. Data are expressed as means ± S.E. of four independent experiments. Nox4, *, p < 0.05 wild type versus CLP-1+/− treated with agonists. C, immunofluorescence assay performed in mouse embryonic in wild type (+/+), heterozygous (CLP-1+/−) and null (CLP-1−/−) fibroblasts, showing the AngII or TGF-β1 induction of nuclear translocation of serine-phosphorylated Smad3 (red) and expression of smooth muscle α-actin (green). Nuclear staining was obtained with DAPI (blue).
Modulation of Smad3 Signaling in Haplodeficient CLP+/− Hearts
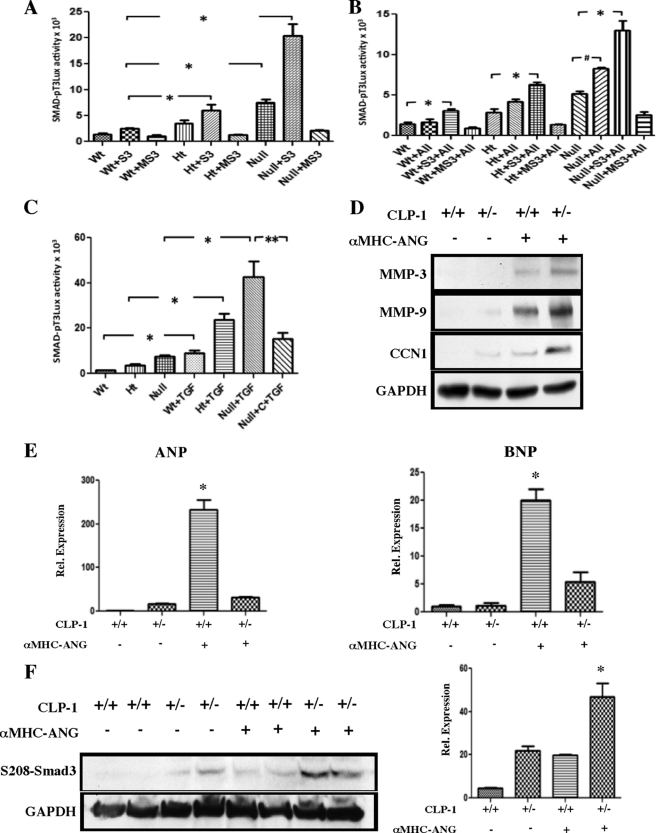
It is known that there is a link between AngII and TGF-β signaling, which through activation of Smad3 induces fibrotic remodeling and contributes to diastolic dysfunction. A previous report (18) indicated that the linker region of Smad3 can be targeted for phosphorylation by Cdk9, which results in enhanced Smad3 activity (18). Despite this, there is no clear evidence on the mechanism by which Smad3 signaling influences the fibrotic process. To establish that Smad3 activity is influenced by CLP-1/pTEFb control of transcription, we performed co-transfection experiments using a Smad3-luciferase reporter (p3Tx-Luc), FLAG-Smad3 expression vector, and a FLAG-MSmad3 expression vector carrying a substitution mutation of the serine residues in the linker region (18). As observed in Fig. 4A, haplodeficiency of CLP-1 correlated inversely with the Smad-luciferase reporter activity driven by FLAG-Smad3. Under the same experimental conditions, a substitution mutation in the serine residue of the polylinker (FLAG-MSmad3) expression vector failed to activate the reporter Smad-luciferase. We then examined the Smad-luciferase reporter activity after induction with AngII (Fig. 4B) or TGF-β1 treatment (Fig. 4C). The increase observed in CLP-1−/− embryonic fibroblast treated with TGF-β1 was significantly reversed by reintroducing CLP-1 via co-transfection with the expression vector pcDNA-CLP-1 (Null + C + TGF). Further support for the role of CLP-1 in AngII-induced remodeling was obtained by expression of connective tissue remodeling markers, such as metalloprotease 9 and cysteine-rich 61-connective tissue growth factor-nephroblastoma. As seen in Fig. 4D, there was a significant increase in the expression of MMP9 and CCN1 markers in extracts from the ANG/CLP-1+/− hearts.
FIGURE 4.
CLP-1 gene dosage modulates serine-Smad3 transcriptional activity. A, transient transfections using the Smad3-luciferase reporter (pT3×Luc), the FLAG-Smad3, or the mutant FLAG-MSmad3 were examined in mouse embryonic fibroblasts wild type, heterozygous, and mice null for the CLP-1 gene. In the absence of the ligands AngII or TGF-β1, there was a significant increase in the luciferase reporter activity inversely proportional to the CLP-1 gene dosage. However, substitution mutations of the conserved serine residues in the polylinker region of Smad3 (MSmad3) completely abolished luciferase activity. B, AngII mediates Smad3 transcriptional activity in mouse embryonic fibroblasts, and a similar response as above using AngII as inducer of Smad activity was observed. C, even a more significant transcriptional induction was observed with TGF-β1 treatment that was reversed by co-transfection with pcDNA CLP-1 (Null + C (CLP-1) + TGF). Data are expressed as means ± S.E. of four independent experiments. *, p < 0.05 control versus TGF-β treatment; **, p < 0.05 Null TGF-β-treated versus Null TGF-β treated and CLP-1 added. D, upper panels shows a representative Western blot with specific antibodies against MMP3, MMP9, and CCN1. GAPDH was used as loading control (n = 4). E, bar graphs represent the quantitative determination of atrial natriuretic peptide and brain natriuretic peptide by qPCR. Data are expressed as means ± S.E. of three independent experiments. ANP and BNP expression shows a *, p < 0.05 αMHC-ANG/CLP-1+/+ versus αMHC-ANG/CLP-1+/−. F, Western blots show the expression of serine 208-Smad3 in heart protein extracts from wild type (WT), heterozygous (CLP-1+/−), transgenic wild type-αMHC-ANG (TG+/+), and transgenic heterozygous-αMHC-ANG (TG+/−) mice hearts. GAPDH expression was used as loading control. Data are expressed as means ± S.E. of three independent experiments. Serine 203-Smad3 expression shows the following: *, p < 0.05 αMHC-ANG/CLP-1+/− versus αMHC-ANG/CLP-1+/+.
It was reported (24, 25) recently that ANP exerts an anti-fibrogenic effect on pro-fibrogenic TGF-β and the intracardiac RAS signaling, suggesting that the expression pattern of the antifibrogenic ANP may be distinct in the wild type CLP-1+/+ and CLP-1+/− harboring the MHC-AngII expression in hearts. We therefore evaluated by quantitative PCR the expression of ANP and BNP peptides. cDNA obtained from the heart tissues of each mouse group was used to perform qPCR using primers for ANP and BNP (see “Experimental Procedures”). We observed a marked increase in expression of ANP and BNP in the transgenic ANG/CLP-1+/+ mice. However, the expression was notably reduced in the ANG/CLP-1+/− mice (Fig. 4E). We conclude that CLP-1/pTEFb transcriptional control is involved in diverse aspects of LVH and fibrosis remodeling, including the negative transcriptional control of ANP and BNP expression. Finally, we performed a Western blot of heart extracts to evaluate activation of the Cdk9/TGF-β1/Smad3 axis in wild type (WT), heterozygous (CLP-1+/−), transgenic wild type-αMHC-ANG (TG+/+), and transgenic heterozygous-αMHC-ANG (TG+/−) mouse hearts. As seen, in Fig. 4F, there was a significant increase in serine 208-Smad3 phosphorylation in extracts from transgenic heterozygous αMHC-ANG (TG+/−), supporting our in vitro findings (Fig. 3C) that a decrease in levels of CLP-1 enhances Smad3-mediated ECM deposition.
DISCUSSION
This report provides evidence that CLP-1 plays a significant role in modulation of the disease processes in left ventricular remodeling and fibrosis during cardiac hypertrophy due to its regulatory effects on pTEFb activity controlling transcription elongation, considered now to be the primary site for eukaryotic gene control. The CLP-1 haplodeficiency caused exacerbation of the various aspects of LVH development and fibrogenesis. Because CLP-1-mediated control of pTEFb is universal, targeting genes associated with LVH and fibrogenic responses perhaps arise from the interplay among TGF-β/Smad3/Jak2 signaling for the context-specific control of pTEFb activity.
The Jak/Stat signaling has been documented to play a prominent role in cardiac hypertrophy (10–12). Jak2 is also known to promote ROS production and ECM deposition (26). Although the precise nature of the cross-talk between these signaling events is not known, the regulation of the P-TEFb complex by Jak2 signaling was previously described (14), suggesting perhaps that the modulation of the hypertrophic features observed in αMHC-ANG/CLP-1+/− hearts is the consequence of enhanced Jak2 function. Precedent also exists for Jak2/Stat3 signaling to undergo functional association with a number of regulatory proteins (27, 28). For example, Stat3-mediated expression of Nox4 transcription factor (26), which influences the trans-differentiation of fibroblast into myofibroblasts, is an ROS known to be essential in myofibroblast formation in AngII-induced cardiac hypertrophy (23). We speculate that a functional linkage between TGF-β1/Smad3 and Jak/STAT signaling might be essential in eliciting the regulatory response triggered by AngII in cardiomyocytes. AngII-mediated LVH thus appears to be intrinsically linked to TGF-β1/Smad3/Jak2/Stat3 signaling, which is a contributing factor in ECM deposition and apoptotic cell death (29).
It is known that AngII contributes significantly to left ventricular remodeling and fibrosis and plays an important role in mediating TGF-β up-regulation in the heart undergoing remodeling. However, the precise nature of the molecular components activated by TGF-β leading to both fibrosis and hypertrophic growth is not known and needs further analysis. Indeed, it was recently reported that Smad4 gene knock-out mice developed cardiac hypertrophy, collagen deposition, and impaired fractional shortening (30, 31). Similarly, Smad3 knock-out mice revealed an increase in cardiac hypertrophy but a decrease in ECM deposition (32). These observations suggest that Smad4 and/or Smad3 may not be the components of TGF-β1 hypertrophy response. However, the enhanced left ventricular remodeling in the Smad4 knock-out was recently shown to be due to an increase in the levels of miR27b, a microRNA normally inhibited by Smad4 (31). Whether a similar explanation applies to the findings of Divakaran et al. (32) has yet to be seen. Alternatively, TGF-β may promote hypertrophy in a Smad-independent pathway.
Regardless, our objective here was to explore whether CLP-1 mediates the activation of the TGF-β-Smad3 signaling axis. The prevailing view is that activation of fibroblast and its transformation into myofibroblasts, which is integrally involved as the key regulator of ECM, are the primary events associated with fibrotic disease. TGF-β expression is directly correlated with the transition of compensated hypertrophy to failure. Activation of TGF-β and, by extension, of fibrosis is in part dependent on Smad3 signaling. The Smad3 null mice exhibit an attenuation of fibrosis and ventricular remodeling. Other signaling events, such as NADH oxidase-dependent ROS production, are also involved in myofibroblast transition and fibrosis (23).
Another complexity in the transcriptional control of hypertrophy is the finding that ANP has an antifibrogenic effect via interrupting TGF-β-stimulated signaling, inhibiting myofibroblast transformation and ECM deposition, and consequently playing an anti-fibrogenic role in the pathogenesis of LVH and fibrosis (9). Nuclear translocation of Smad3 is fundamental to its action in positive or negative regulation of target genes. Mechanistically, this is achieved by phosphorylation of Smad3 at a site different from that required for its TGF-β-induced nuclear translocation and consequently disrupting the nuclear entry of Smad3. We observed that deficiency of CLP-1 in CLP-1+/− reduced effectively the expression of ANP. Whether CLP-1 deficiency extends its control to phosphorylation of the alternate site in Smad3 is not known. Evidence is, however, available in support of the role of CLP-1 in modulation of expression of a defined gene in the controlled expression of vascular endothelial growth factor (VEGF) (33).
Previous reports described that establishment of AngII-mediated LVH was not accompanied by collagen deposition (3, 20, 34). We show here that reduction in CLP-1 levels triggers enhancement of collagen deposition in vivo linking collagen synthesis to the CLP-1-mediated modulation of P-TEFb activity. The unique role of CLP-1 in augmenting the AngII-TGF-β1-Smad3 signaling network is supported by the evidence that reduction of CLP-1 gene dosage resulted in an increase in Cdk9 activity followed by nuclear translocation of phosphoserine 208-Smad3 and increased expression of α-smooth muscle actin in mouse embryonic fibroblast. The consequences of phosphoserine 208-Smad3 signaling in transcriptional modulation are accompanied by trans-differentiation of fibroblasts subjected to administration of AngII or TGF-β. We also demonstrate that CLP-1 has a role in modulation of the transcriptional activity of Smad3 in fibroblasts co-transfected with a Smad-luciferase reporter, a finding that contributes to the role of Smad-Stat signaling in fibrotic remodeling. Taken together, our data underscore the subtle role of CLP-1 not only in the transcription elongation machinery but also in the complex control of AngII-TGF-β1-CLP-1-Smad3 signaling axis and natriuretic peptide expression.
This work was supported, in whole or in part, by National Institutes of Health Grant HL073399 (to M. A. Q. S.).
- ECM
- extracellular matrix
- RAS
- renin-angiotensin system
- LVH
- left ventricular hypertrophy
- ANP
- atrial natriuretic peptide
- qPCR
- quantitative PCR
- BNP
- brain natriuretic peptide
- ANG
- angiotensinogen
- SM-α-actin
- smooth muscle α-actin
- ROS
- reactive oxygen species.
REFERENCES
- 1. Schnee J. M., Hsueh W. A. (2000) Angiotensin II, adhesion, and cardiac fibrosis. Cardiovasc. Res. 46, 264–268 [DOI] [PubMed] [Google Scholar]
- 2. Re R. N., Cook J. L. (2007) Mechanisms of disease. Intracrine physiology in the cardiovascular system. Nat. Clin. Pract. Cardiovasc. Med. 4, 549–557 [DOI] [PubMed] [Google Scholar]
- 3. Schultz Jel J., Witt S. A., Glascock B. J., Nieman M. L., Reiser P. J., Nix S. L., Kimball T. R., Doetschman T. (2002) TGF-β1 mediates the hypertrophic cardiomyocyte growth induced by angiotensin II. J. Clin. Invest. 109, 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmierer B., Hill C. S. (2007) TGF-β-SMAD signal transduction. Molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970–982 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y., Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 6. Wang D., Oparil S., Feng J. A., Li P., Perry G., Chen L. B., Dai M., John S. W., Chen Y. F. (2003) Effects of pressure overload on extracellular matrix expression in the heart of the atrial natriuretic peptide-null mouse. Hypertension 42, 88–95 [DOI] [PubMed] [Google Scholar]
- 7. Mori T., Chen Y. F., Feng J. A., Hayashi T., Oparil S., Perry G. J. (2004) Volume overload results in exaggerated cardiac hypertrophy in the atrial natriuretic peptide knock-out mouse. Cardiovasc. Res. 61, 771–779 [DOI] [PubMed] [Google Scholar]
- 8. McGrath M. F., de Bold M. L., de Bold A. J. (2005) The endocrine function of the heart. Trends Endocrinol. Metab. 16, 469–477 [DOI] [PubMed] [Google Scholar]
- 9. Li Y., Kishimoto I., Saito Y., Harada M., Kuwahara K., Izumi T., Takahashi N., Kawakami R., Tanimoto K., Nakagawa Y., Nakanishi M., Adachi Y., Garbers D. L., Fukamizu A., Nakao K. (2002) Guanylyl cyclase-A inhibits angiotensin II type 1A receptor-mediated cardiac remodeling, an endogenous protective mechanism in the heart. Circulation 106, 1722–1728 [DOI] [PubMed] [Google Scholar]
- 10. Mascareno E., Dhar M., Siddiqui M. A. (1998) Signal transduction and activator of transcription (STAT) protein-dependent activation of angiotensinogen promoter. A cellular signal for hypertrophy in cardiac muscle. Proc. Natl. Acad. Sci. U.S.A. 95, 5590–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beckles D. L., Mascareno E., Siddiqui M. A. (2006) Inhibition of Jak2 phosphorylation attenuates pressure overload cardiac hypertrophy. Vascul. Pharmacol. 45, 350–357 [DOI] [PubMed] [Google Scholar]
- 12. Guo Y., Mascareno E., Siddiqui M. A. (2004) Distinct components of Janus kinase/signal transducer and activator of transcription signaling pathway mediate the regulation of systemic and tissue localized renin-angiotensin system. Mol. Endocrinol. 18, 1033–1041 [DOI] [PubMed] [Google Scholar]
- 13. Fukuzawa J., Booz G. W., Hunt R. A., Shimizu N., Karoor V., Baker K. M., Dostal D. E. (2000) Cardiotrophin-1 increases angiotensinogen mRNA in rat cardiac myocytes through STAT3. An autocrine loop for hypertrophy. Hypertension 35, 1191–1196 [DOI] [PubMed] [Google Scholar]
- 14. Espinoza-Derout J., Wagner M., Shahmiri K., Mascareno E., Chaqour B., Siddiqui M. A. (2007) Pivotal role of cardiac lineage protein-1 (CLP-1) in transcriptional elongation factor P-TEFb complex formation in cardiac hypertrophy. Cardiovasc. Res. 75, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Espinoza-Derout J., Wagner M., Salciccioli L., Lazar J. M., Bhaduri S., Mascareno E., Chaqour B., Siddiqui M. A. (2009) Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circ. Res. 104, 1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sano M., Abdellatif M., Oh H., Xie M., Bagella L., Giordano A., Michael L. H., DeMayo F. J., Schneider M. D. (2002) Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8, 1310–1317 [DOI] [PubMed] [Google Scholar]
- 17. Sano M., Wang S. C., Shirai M., Scaglia F., Xie M., Sakai S., Tanaka T., Kulkarni P. A., Barger P. M., Youker K. A., Taffet G. E., Hamamori Y., Michael L. H., Craigen W. J., Schneider M. D. (2004) Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 23, 3559–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alarcón C., Zaromytidou A. I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A. N., Manova-Todorova K., Macias M. J., Sapkota G., Pan D., Massagué J. (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang F., Wagner M., Siddiqui M. A. (2004) Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech. Dev. 121, 559–572 [DOI] [PubMed] [Google Scholar]
- 20. Mazzolai L., Nussberger J., Aubert J. F., Brunner D. B., Gabbiani G., Brunner H. R., Pedrazzini T. (1998) Blood pressure-independent cardiac hypertrophy induced by locally activated renin-angiotensin system. Hypertension 31, 1324–1330 [DOI] [PubMed] [Google Scholar]
- 21. Mazzolai L., Pedrazzini T., Nicoud F., Gabbiani G., Brunner H. R., Nussberger J. (2000) Increased cardiac angiotensin II levels induce right and left ventricular hypertrophy in normotensive mice. Hypertension 35, 985–991 [DOI] [PubMed] [Google Scholar]
- 22. Swaney J. S., Roth D. M., Olson E. R., Naugle J. E., Meszaros J. G., Insel P. A. (2005) Inhibition of cardiac myofibroblast formation and collagen synthesis by activation and overexpression of adenylyl cyclase. Proc. Natl. Acad. Sci. U.S.A. 102, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cucoranu I., Clempus R., Dikalova A., Phelan P. J., Ariyan S., Dikalov S., Sorescu D. (2005) NAD(P)H oxidase 4 mediates transforming growth factor-β1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ. Res. 97, 900–907 [DOI] [PubMed] [Google Scholar]
- 24. Oliver P. M., Fox J. E., Kim R., Rockman H. A., Kim H. S., Reddick R. L., Pandey K. N., Milgram S. L., Smithies O., Maeda N. (1997) Hypertension, cardiac hypertrophy, and sudden death in mice lacking natriuretic peptide receptor A. Proc. Natl. Acad. Sci. U.S.A. 94, 14730–14735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franco V., Chen Y. F., Oparil S., Feng J. A., Wang D., Hage F., Perry G. (2004) Atrial natriuretic peptide dose-dependently inhibits pressure overload-induced cardiac remodeling. Hypertension 44, 746–750 [DOI] [PubMed] [Google Scholar]
- 26. Manea A., Tanase L. I., Raicu M., Simionescu M. (2010) Jak/STAT signaling pathway regulates nox1 and nox4-based NADPH oxidase in human aortic smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 30, 105–112 [DOI] [PubMed] [Google Scholar]
- 27. Mitsuyama K., Sata M., Rose-John S. (2006) Interleukin-6 trans-signaling in inflammatory bowel disease. Cytokine Growth Factor Rev. 17, 451–461 [DOI] [PubMed] [Google Scholar]
- 28. Ho M. K., Su Y., Yeung W. W., Wong Y. H. (2009) Regulation of transcription factors by heterotrimeric G proteins. Curr. Mol. Pharmacol. 2, 19–31 [DOI] [PubMed] [Google Scholar]
- 29. Rosenkranz S. (2004) TGF-β1 and angiotensin networking in cardiac remodeling. Cardiovasc. Res. 63, 423–432 [DOI] [PubMed] [Google Scholar]
- 30. Wang J., Xu N., Feng X., Hou N., Zhang J., Cheng X., Chen Y., Zhang Y., Yang X. (2005) Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ. Res. 97, 821–828 [DOI] [PubMed] [Google Scholar]
- 31. Wang J., Song Y., Zhang Y., Xiao H., Sun Q., Hou N., Guo S., Wang Y., Fan K., Zhan D., Zha L., Cao Y., Li Z., Cheng X., Yang X. (2011) Cardiomyocyte overexpression of miR-27b induces cardiac hypertrophy and dysfunction in mice. Cell Res. doi: 10.1038/cr.2011.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Divakaran V., Adrogue J., Ishiyama M., Entman M. L., Haudek S., Sivasubramanian N., Mann D. L. (2009) Adaptive and maladaptive effects of SMAD3 signaling in the adult heart after hemodynamic pressure overloading. Circ. Heart Fail. 2, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montano M. M., Doughman Y. Q., Deng H., Chaplin L., Yang J., Wang N., Zhou Q., Ward N. L., Watanabe M. (2008) Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through down-regulation of vascular endothelial growth factor. Circ. Res. 102, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Domenighetti A. A., Wang Q., Egger M., Richards S. M., Pedrazzini T., Delbridge L. M. (2005) Angiotensin II-mediated phenotypic cardiomyocyte remodeling leads to age-dependent cardiac dysfunction and failure. Hypertension 46, 426–432 [DOI] [PubMed] [Google Scholar]