Background: In brain lysates denatured huntingtin is full-length and fragmented.
Results: Blue Native PAGE analysis revealed huntingtin as a soluble full-length monomer and resistant to exogenous protease cleavage. Exposure to denaturants cleaved mutant huntingtin.
Conclusion: Native mutant huntingtin in brain is unstable compared with wild-type huntingtin.
Significance: Native conditions may improve detection of full-length huntingtin in human brain.
Keywords: Brain, Huntington Disease, Mouse, Protein Denaturation, Protein Folding, Human
Abstract
Huntington disease (HD) is caused by polyglutamine expansion in the N terminus of huntingtin (htt). Analysis of human postmortem brain lysates by SDS-PAGE and Western blot reveals htt as full-length and fragmented. Here we used Blue Native PAGE (BNP) and Western blots to study native htt in human postmortem brain. Antisera against htt detected a single band broadly migrating at 575–850 kDa in control brain and at 650–885 kDa in heterozygous and Venezuelan homozygous HD brains. Anti-polyglutamine antisera detected full-length mutant htt in HD brain. There was little htt cleavage even if lysates were pretreated with trypsin, indicating a property of native htt to resist protease cleavage. A soluble mutant htt fragment of about 180 kDa was detected with anti-htt antibody Ab1 (htt-(1–17)) and increased when lysates were treated with denaturants (SDS, 8 m urea, DTT, or trypsin) before BNP. Wild-type htt was more resistant to denaturants. Based on migration of in vitro translated htt fragments, the 180-kDa segment terminated ≈htt 670–880 amino acids. If second dimension SDS-PAGE followed BNP, the 180-kDa mutant htt was absent, and 43–50 kDa htt fragments appeared. Brain lysates from two HD mouse models expressed native full-length htt; a mutant fragment formed if lysates were pretreated with 8 m urea + DTT. Native full-length mutant htt in embryonic HD140Q/140Q mouse primary neurons was intact during cell death and when cell lysates were exposed to denaturants before BNP. Thus, native mutant htt occurs in brain and primary neurons as a soluble full-length monomer.
Introduction
Huntington disease (HD)2 is a neurodegenerative disease caused by an increase in the number of glutamines (>40) in the N terminus of huntingtin (htt). Huntingtin is a large protein containing 3144 amino acids. Multiple pathogenic mechanisms have been proposed to result from polyglutamine expansion in htt. It is speculated that polyglutamine expansion increases the abundance of partially folded intermediates in htt. The unfolded mutant htt forms abnormal protein interactions either as intact protein or as N-terminal fragments that can become insoluble and form aggregates. There is considerable support from studies of cell and animal models that expression of N-terminal mutant htt fragments encoded by exon 1 or exons 1 and 2 is sufficient to cause neuronal dysfunction and death (1–3). Consistent with the idea of a pathogenic fragment, biochemical studies show that endogenous htt from human brain or human cell lines or exogenously expressed htt in cells is cleaved in N-terminal regions by different proteases (4–9). However, in one study of transgenic mice, expression of an N-terminal mutant fragment did not produce motor phenotypes (10). Thus, it is still unproven that a specific N-terminal htt product is critical to pathogenesis in the human disease. Dyer and McMurray (7) studied the human postmortem HD brain. They found lower levels of small N-terminal htt fragments in HD brain compared with control brain and suggested that mutant htt was more resistant to proteolysis than wild-type (WT) htt. The inclusion is a visible sign of mutant htt aggregation detected by immunohistochemistry in the light microscope. Inclusions are seen more easily with expression of N-terminal mutant htt fragments than expression of the full-length protein. However neurodegeneration is not dependent upon the presence of inclusions and is more associated with levels of soluble diffuse mutant htt (11–13). A specific conformation of the expanded polyglutamine region in monomeric full-length as well as truncated htt has been associated with increased cellular toxicity (14–16). Findings in HD animal models favor a pathogenic role for full-length mutant htt. Neuronal dysfunction in Drosophila and neuropathology and motor deficits in BACHD mice occur with expression of human full-length mutant htt in the absence of detectable fragments or aggregate formation (17, 18). BACHD mice expressing full-length mutant htt with modifications at phosphorylation sites Ser-13/Ser-16 show reduced neuropathology and motor deficits (19). These in vitro and in vivo data suggest that soluble full-length mutant htt may be pathogenic in HD.
Most biochemical studies of htt in brain have been performed in denaturing conditions in which the apparent size migrates at about 350 kDa. Studies of recombinant purified htt by Li et al. (8) using native polyacrylamide gel electrophoresis (PAGE), size exclusion chromatography, and dynamic light scattering found that htt was largely intact and monomeric with a mass of about 500–550 kDa. Some proteolysis occurred during purification and could be detected by SDS-PAGE. These authors speculated that full-length htt structure fit the model of an elongated heat repeat-enriched super helical protein with a continuous hydrophobic core; this structure prevented native htt from dissociating despite the presence of “nicks” in the protein that produced cleaved products upon denaturation (8). Seong et al. (20) examined purified WT htt expressed in insect cells. Their highly purified htt preparation showed no N-terminal fragments after denaturing in SDS-PAGE. However, a 10-min exposure of htt to trypsin generated an N-terminal band of 150 kDa, and a 20-min trypsin digest caused the 150-kDa fragment to degrade, resulting in the formation of a 60-kDa N-terminal htt fragment. These data suggested that the N-terminal domains were unstable in the presence of trypsin during SDS-dependent unfolding of the protein.
The propensity for purified recombinant htt to aggregate has made study of its native state a challenge. Huntingtin is known to associate with membranes and phospholipids and inserts into the lipid bilayer (21–23). The association of htt with membranes and its potential structure as a protein enriched in heat repeats with a continuous hydrophobic core (noted above) suggested to us that htt in brain might be amenable to analysis by Blue Native PAGE. This method referred to as BNP has been effective for isolating the components of large molecular complexes and their subdomains in membranes (e.g. mitochondrial membrane proteins, AMPA receptors, and oligomers of β amyloid (24)). The addition of Coomassie Blue dye G250 adds negative charge to protein surfaces significantly reducing aggregation and enhancing protein separation under native conditions (25, 26).
Some of us had studied patients from Venezuela who were homozygous for HD (27). These individuals were part of a larger Venezuelan kindred that was used to isolate the HD gene (28, 29). Our hypothesis was that comparing clinical data and other information obtained from homozygotes for HD with data from heterozygotes and controls would help to elucidate the nature of the defect. Further study revealed that HD is a true dominant disorder. There is no dose effect; the normal allele does not ameliorate the phenotype, and two doses of the pathological allele do not exacerbate it (30). This study marks the first effort to examine postmortem brain tissue of homozygote HD patients by biochemical assay and to study the aberrant protein, undisguised by the normal protein.
Here we report a biochemical analysis using BNP with lysates from postmortem cortex of normal individuals and patients homozygous and heterozygous for the HD gene, from the cortex of WT and HD mice (HD140Q/140Q and BACHD) and from primary neurons of WT and HD140Q/140Q mice. Our findings suggest that most of the htt detected in human brain, mouse brain, and primary neurons using native conditions is full-length monomer. Native mutant htt is more susceptible to cleavage than WT htt if brain lysates are treated with denaturants to stimulate partial protein unfolding.
EXPERIMENTAL PROCEDURES
Human Brain Tissue
Control (n = 4) and HD postmortem brain tissue (n = 7) was obtained from the New York Brain Bank at Columbia University and the Massachusetts General Hospital Neuropharmacology Laboratory Brain Bank and Harvard Brain Tissue Resource Center. This tissue was received frozen and was stored at −80 °C until use. The two homozygotes analyzed in this study were cousins. One had 43/48 CAG repeats, and the other had 42/46 CAG repeats. Age of disease onset was 23 years in one individual and 37 years in the other. Duration of illness was 9 years in one case and 21 years in the other. The clinical features of these patients did not differ from those of heterozygotes. The heterozygote HD individuals had CAG repeats of 27Q/42Q, 17Q/42Q, 25Q/40Q, 17Q/43Q, and 15Q/69Q in the huntingtin gene. CAG repeats for two of the four controls were known (17Q/17Q and 9Q/17Q). The interval between death and brain dissection ranged from 4 to 48 h. With the exception of tissue from two homozygote HD brains and two controls, all of the human brain tissue samples had been studied previously and had coded designations that are also used in this paper (31–34). In these previous studies using SDS-PAGE and Western blot, we found that a postmortem delay of up to 48 h did not influence the level of detection of huntingtin in control or HD brain.
Mouse Brain Tissue
WT and HD140Q/140Q mice (C57BL/6 strain background) are maintained at the Massachusetts General Hospital (MGH) animal facility in building 114. The HD mice are “knock-in” mice and have human exon 1 with 140 CAG repeats inserted into the mouse huntingtin gene (35). Mice are bred to be homozygous for the HD mutation. WT and BACHD mice (18) are maintained at the University of Massachusetts Medical School (UMMS). The BACHD mice are transgenic mice that express the gene encoding human full-length htt with 97 glutamines from mixed CAG/CAA repeats under control of human htt gene promoter (18). The animal protocols were reviewed and approved by the Subcommittee on Research Animal Care (SRAC)-OLAW Assurance at MGH #A3596-01 and at UMMS #A-978. The protocol conforms to the USD Animal Welfare Act, PHS Policy on Humane Care and Use of Laboratory Animals, the “ILAR Guide for the Care and Use of Laboratory Animals,” and other applicable laws and regulations. Brains were removed from the animals and frozen at −80 °C until use in biochemical assays. Fresh brain tissue from embryonic mice was used as a source of primary neurons (see below).
Sample Preparation for Native PAGE
Approximately 20 mg of frozen cortical brain tissue was homogenized in 500 μl of 1× NativePAGETM sample buffer (Invitrogen) + 1% n-dodecyl-β-d-maltoside + protease inhibitors (Roche Applied Science) + 1 mm NaF + 1 mm Na3VO4 then centrifuged at 100,000 × g for 15 min at 4 °C. The supernatant was placed in a fresh tube and used for most of the analysis in this study. The insoluble pellet was resuspended in 300 μl of the same 1× Native PAGETM sample buffer as above. 1 unit/μl of benzonase (Sigma) and 2 mm MgCl2 (final concentration) were added to the soluble and resuspended pellet fraction and incubated at room temperature for 30 min. The soluble fraction was centrifuged at 100,000 × g for 15 min at 4 °C, and the resuspended pellet fraction was sonicated for 2 s. Protein content was determined using the Bradford method (Bio-Rad), and samples were frozen at −80 °C until use. In some experiments 1 μg/ml pepstatin A (Sigma) or 20 μm N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (Calbiochem) was added to the lysis buffer before homogenization. In some experiments SDS (2%) ± 100 mm DTT was added to the lysates and incubated at room temperature for 40 min. Some lysates were treated with 8 m urea + 100 mm DTT for 30 min at room temperature or 0.1 mg/ml trypsin for 20–60 min at 37 °C.
Preparation of Subcellular Fractions; S2 and P2 Membrane Fractions
Small pieces of frozen cortex were homogenized in 5 volumes of 20 mm Tris, pH 7.4, 250 mm sucrose, 1 mm EDTA + protease inhibitor tablet (Roche Applied Science) and centrifuged at 2000 × g for 10 min at 4 °C. The supernatant (S1) was removed to a fresh tube, and the pellet (P1) was resuspended in the same buffer and sonicated for 10 s. The S1 fraction was then further centrifuged at 100,000 × g for 1 h at 4 °C. The supernatant (S2) was placed in a fresh tube, and the pellet (P2) was resuspended in the same buffer. Protein content was determined using the Bradford method (Bio-Rad). Immediately before electrophoresis, samples were diluted in 1× Native PAGETM sample buffer and incubated in 2 mm MgCl2 and 1 unit/μl benzonase (Sigma) for 15 min at room temperature.
Preparation of Primary Neurons
Embryonic cortex of WT and HD140Q/140Q mice were used as the source of primary neurons and prepared according to our previously published method (23). The protocol resulted in >99% neurons in the cultures. Cultures were harvested into 50 mm Tris, pH 7.4, 250 mm sucrose, 1% Nonidet P-40, 5 mm EDTA, protease inhibitors (Roche Applied Science), 1 mm NaF, and 1 mm Na3VO4 on ice for 15 min, scraped into an Eppendorf tube, and centrifuged at 10,000 × g for 3 min. Protein content in the supernatant was determined using the Bradford method (Bio-Rad). Before electrophoresis, samples were diluted in 1× Native PAGETM sample buffer and incubated in 2 mm MgCl2 and 1 unit/μl benzonase (Sigma) for 15 min at room temperature. Parallel cultures prepared from the same embryos were used to detect viability by MTT assay as previously described (23, 36).
Blue Native PAGE and Western Blot
The Native PAGETM Novex Bis-Tris gel system (Invitrogen) was used to perform electrophoresis under native conditions. Coomassie G250 was added to samples at a concentration of 0.25% as well as to the cathode buffer at concentrations of 0.02–0.002%, and 10–50 μg of sample/lane was loaded. Proteins were separated in NativePAGETM Novex 3–12 or 4–16% Bis-Tris gels and transferred to PVDF at 100 V for 1 h using the Mini Trans-Blot Cell (Bio-Rad). Blots were treated with 8% acetic acid for 15 min, air-dried, then soaked in methanol to remove excess Coomassie Blue as well as to visualize the molecular mass markers. Native protein standards from GE Healthcare were thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and albumin (66 kDa). Native protein standards from Invitrogen were IgM hexamer (1236 kDa), IgM pentamer (1048 kDa), apoferritin band 1 (720 kDa), apoferritin band 2 (480 kDa), B phycoerythrin (242 kDa), lactate dehydrogenase (146 kDa), bovine serum albumin (66 kDa), and soybean trypsin inhibitor (20 kDa). After washing in TBS + 0.1% Tween 20 (TBST), blots were processed for Western blot by blocking in 5% nonfat dry milk (Bio-Rad) in TBST then incubated in primary antibody diluted in blocking solution overnight at 4 °C. Blots were incubated in peroxidase-labeled secondary antibody (Jackson Immunoresearch) in blocking solution, and bands were visualized using the SuperSignal West-Pico Chemiluminescent Substrate (Pierce) and Hyperfilm ECL (GE Healthcare).
For SDS-PAGE, proteins were loaded 20–50 μg/lane in NuPAGE Novex 3–8% Tris acetate or Novex 4–12% Tris-glycine gels (Invitrogen). Samples were treated with 100 mm DTT and boiled for 5 min before loading. Proteins were transferred to nitrocellulose using the iblot system (Invitrogen); blots were washed in TBST and processed immediately for Western blot as described above.
In Vitro Translation in Reticulocytes
Protein was generated using rabbit reticulocyte lysates (TnT Quick Coupled Transcription/Translation Systems, Promega) following the protocol provided and using 1 μg of pcDNA plasmids encoding huntingtin cDNAs as templates. The construction of cDNAs encoding huntingtin fragments have been described previously (21, 22).
Sources of Antibodies Used in This Study
Antibodies used in this study along with their antigenic sites and sources are Ab1 (htt-(1–17)) and 585 (htt-(585–725)) (37), PW0595 (htt1–17, Enzo Life Sciences), MAB1574 (1C2, Chemicon), 3B5H10 (Sigma, anti-polyQ, made to htt1–171), MAB2166 (htt-(443–457)), MAB5490 (htt-(115–129)), MAB2168 (htt-(2146–2541)) (Chemicon), Ab1173 (htt-(1173–1196) (38)), and Ab2527 (htt-(2527–2547) (39)), GAPDH (Millipore), and βIII-tubulin (Sigma). MW1 (anti-polyQ) and MW8 (htt83–90), developed by Paul H. Patterson, were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
RESULTS
BNP and Western Blot Analysis of Huntingtin in Human Brain
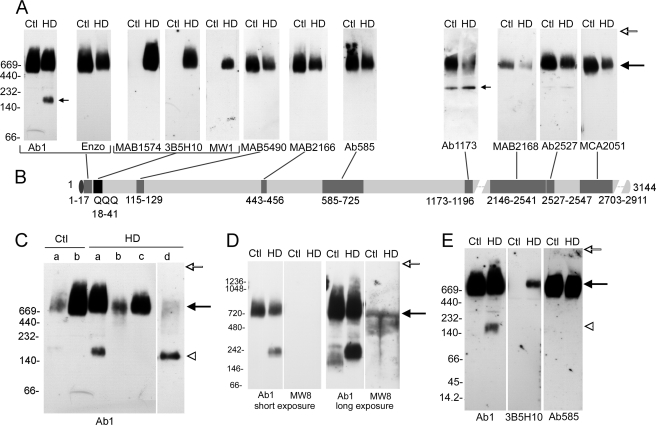
We examined postmortem brain lysates from the cortex of four control subjects and seven HD patients. Proteins were separated using NativePAGETM Novex 3–12% Bis-Tris gels, transferred to PVDF, and Western blot analysis was performed with different anti-htt and anti-polyglutamine antibodies. Each antibody was tested in two to seven independent assays. Fig. 1A shows results for postmortem cortex of control and age matched homozygote HD individuals for one set of results. Anti-htt antisera that recognized different regions in htt (htt-(1–17), -115–129, -443–456, -585–725, -1173–1196, -2146–2541, -2527–2547, and -2703–2911) (Fig. 1B) detected WT and mutant htt typically as broad bands migrating around the 669-kDa marker, which was the highest molecular mass in the set of standards used for these experiments. Antisera that recognize expanded polyglutamine regions (MAB1574 (1C2), 3B5H10, and MW1) detected mutant htt but not WT htt (Fig. 1A). Other postmortem samples from normal and HD individuals also showed a broad band for native htt migrating around the 669-kDa standard when detected by Western blot with antibody Ab1 (htt-(1–17)) (Fig. 1C and supplemental Fig. S1). To estimate the molecular mass range of htt by BNP, we used another set of standards that included proteins with molecular masses of 720, 1048, and 1236 kDa and performed Western blot analysis with antibody Ab1. Results showed an apparent mass for native WT htt to be 575–850 kDa and for mutant htt to be 650–885 kDa. At longer exposures of the Western blot films, WT htt migrated between 540 and 1120 kDa, and mutant htt migrated between 615 and 1270 kDa (Fig. 1D). There was no htt signal detected at the top of the blots (open arrows in Fig. 1) that would indicate insoluble protein had failed to migrate into the gel during BNP. Thus the predominant signal for native WT and mutant htt in the human brain was soluble full-length protein. Migration of htt occurred over a wide molecular mass range and was slowed by polyglutamine expansion.
FIGURE 1.
Biochemical analysis of native huntingtin in human brain. A, lysates were prepared from postmortem control (Ctl) and homozygous HD patient cortex. Proteins were separated with BNP using NativePAGETM Novex 3–12% Bis-Tris gels and transferred to PVDF. Each Western blot was probed with a different anti-htt- or anti-expanded polyglutamine-sensitive antibody as indicated at the bottom of the blots. The levels of the native molecular mass markers are shown on the left and apply to all blots. Note that the predominant forms of WT and mutant htt migrate near the 669-kDa marker with all antisera (large arrow). Some antibodies (Ab1, Ab1173) also detect smaller complexes (small arrows). Each antibody was used in at least two experiments. B, the schematic shows the epitopes recognized by different anti-htt and anti-polyglutamine antisera. C, lysates from two controls and four HD brains were prepared from frozen postmortem brain as described under “Experimental Procedures.” HDa and HDd are from adult homozygotes (43Q/48Q and 42Q/46Q), HDb is presymptomatic grade 1 heterozygote (17Q/42Q, A4), and HDc is an adult heterozygote, grade 3 (27Q/42Q, A12). Note that in this blot only the two homozygous HD cases show evidence of a prominent N-terminal fragment (open arrowhead). D, shown is detection of native WT htt and mutant htt with Ab1 antibody and mutant htt with MW8 antibody at short and longer exposures of the same Western blot film. The gel was run using different molecular mass standards than in A. The sizes of WT and mutant htt encompass a large size range, and polyglutamine expansion slows htt migration. Detection of mutant htt by MW8 is seen only at the long exposure (arrow). E, BNP was performed using NativePAGETM Novex 4–16% Bis-Tris gels and Western blot with different antisera as indicated. Migration of molecular mass markers from 14.2 to 669 kDa is shown on the left. Ab1 detects the 180-kDa band but not smaller fragments. No fragments are seen with antibodies 3B5H10 or Ab585. 10 μg/lane were loaded for all lysates. The open arrow on the right in A, C, D, and E indicates the top of the gel.
BNP and Western Blot Analysis of Human HD Brain Shows Presence of N-terminal Fragment
In brain lysates of two homozygote HD brains, antibody Ab1 (htt-(1–17)) detected a mutant htt fragment (Fig. 1, A and C). The fragment migrated at about 180 kDa. The control brain lysate examined in the same assay did not show a fragment. However, a htt fragment of about 160 kDa was seen in long exposures of the Western blot films, suggesting that the 160-kDa WT htt band was much less abundant than the 180-kDa mutant htt fragment (Fig. 1D). The difference in size of the bands in control and homozygous HD brain is likely due to the difference in polyglutamine length and would support the interpretation that the bands are fragments of N-terminal htt. In two of five human heterozygote HD brains examined there was a fragment detected with antibody Ab1 (supplemental Fig. S1, HD brains A11 and A3). However, unlike the band in the homozygote HD brain, we cannot be sure if the fragment in the heterozygotes belongs to mutant htt or WT htt. Antisera Ab1173 was the only other antibody to robustly detect native htt fragments. The bands migrated similarly (about 250 kDa) in control and HD brain, suggesting that they were C-terminal cleavage products.
To increase detection of N-terminal mutant htt fragments, we used antibody MW8. Previous studies show that MW8 detects a small mutant htt fragment in denaturing conditions (40). BNP and Western blot of control and homozygous HD brain with MW8 showed that the antibody detected a band at around the 669-kDa marker and a broader band between 440 and 669 kDa in both control and HD brain (Fig. 1D). No small fragments were seen. We also separated proteins using NativePAGETM Novex 4–16% Bis-Tris gels in which the molecular mass standards of 45 and 14.2 kDa migrated within the gel. Western blot analysis with antibodies Ab1, 3B5H10, and Ab585 showed no mutant htt bands other than the 180-kDa band detected with Ab1 antibody (Fig. 1E). Thus, the 180-kDa band detected by Ab1 was the only N-terminal mutant htt fragment observed in the homozygous HD brain using native conditions.
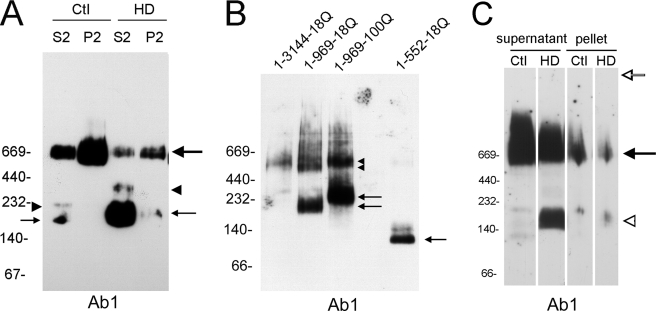
Htt is detected in soluble and membrane fractions when examined under denaturing conditions (41). To determine the subcellular distribution of the 180-kDa native mutant htt band, we examined soluble (S2) and membrane (P2) fractions by BNP and Western blot with Ab1 antibody (htt-(1–17)) (Fig. 2A). Native full-length htt was more enriched in the P2 fraction than the S2 fraction of control and HD brain. In contrast, the 180-kDa native htt band was enriched in the soluble (S2) fraction compared with the membrane (P2) fraction. The 160-kDa band in control brain was detected in the S2 fraction but at much lower levels than the 180-kDa band seen in the HD S2 fraction. Additional bands migrating at around 220 kDa in control brain and at about 300 kDa in HD brain were also detected in the S2 fractions (Fig. 2A, arrowheads).
FIGURE 2.
Native htt in human brain and in vitro translated in reticulocytes. Anti-htt1–17 Ab1 was used for Western blots, 10 μg of protein was loaded per lane, and molecular mass markers are shown on the left. A, distribution of 180 kDa mutant htt in soluble and membrane compartments by BNP is shown. The soluble (S2) and membrane (P2) fractions were prepared as described under “Experimental Procedures.” Control and HD brain have more native full-length htt in the P2 fraction than the S2 fraction (large arrow, right). The 180-kDa mutant htt fragment (small arrow, right) is more enriched in the S2 fraction than in the P2 fraction. A 340-kDa fragment was also detected (arrowhead, right). Control brain shows 160 kDa (arrow, left) and 230-kDa fragments (arrowhead, left). B, BNP analysis of recombinant full-length htt and htt fragments is shown. cDNAs encoding htt and htt fragments as indicated were translated in vitro using a reticulocyte assay, and the proteins were separated by BNP and probed by Western blot with Ab1 antibody. Arrows identify the positions of htt-(1–552)-18Q (two bands) and htt-(1–969)-18Q and -(1–969)-100Q,and arrowheads identify the dimeric forms of htt-(1–969). Most of htt-(1–552)-18Q may undergo processing to the smaller fragment in vitro or during processing. C, shown is a BNP analysis of detergent-insoluble pellet. Detergent-soluble supernatant and resuspended pellet were prepared as described under “Experimental Procedures.” The signal for full-length htt (arrow) is an average of 4.4× greater in the soluble fraction than in the resuspended pellet, and the 180-kDa band was detected in both fractions (open arrowhead). There is no signal at the top of the gel, suggesting that aggregated protein has entered the gel (open arrow).
To approximate the N-terminal domain that formed the 180-kDa mutant htt, we examined the migration of recombinant full-length htt and truncated N-htt fragments of different sizes. Htt cDNAs encoding full-length htt-(1–3144)-18Q, htt-(1–969)-18Q, htt-(1–969)-100Q, and htt-(1–552)-18Q were expressed by in vitro translation in reticulocytes and examined by BNP and Western blot (Fig. 2B). The in vitro expressed full-length WT htt migrated at about 550 kDa. htt-(1–552)-18Q migrated at about 120–140 kDa. htt-(1–969)-18Q and htt-(1–969)-100Q migrated as monomers at about 220 and 270 kDa, respectively, and as dimers at about 500 and 600 kDa, respectively. Based on these data, the 180-kDa native mutant htt fragment seen in human homozygous HD brain would be expected to terminate between amino acids 552 and 969 if it is a monomer.
BNP Analysis of Detergent-insoluble Pellet Fraction
To determine if mutant htt aggregates were missed by analysis of total lysates and S2 and P2 fractions, we also examined the resuspended detergent insoluble pellet using BNP and Western blot with Ab1 antibody. The pellet fraction would be expected to contain any insoluble mutant htt. The results showed that full-length mutant htt migrated at the same size in the soluble and resuspended pellet fractions (Fig. 2C). No signal appeared at the top of the blot in the lanes with the resuspended pellet to suggest a retarded migration of aggregated proteins into the gel. The signal intensity for native full-length WT or mutant htt was much greater in the soluble fraction than in the resuspended pellet fraction (on average about 4.4× more). The 180-kDa N-terminal mutant huntingtin fragment was also more prevalent in the soluble fraction than the resuspended pellet fraction. However the resuspended pellet fraction had a higher ratio of N-terminal band to full-length htt (1.49) than did the soluble fraction (0.43). The greater representation of the 180-kDa fragment relative to full-length in the pellet fraction suggests that the 180-kDa fragment may be more prone to accumulate in a detergent resistant compartment. However, there was no change in mobility of the 180-kDa band in the resuspended pellet compared with the soluble fraction to suggest increased oligomerization. Thus, using the tissue sample preparation and BNP method performed in this study, most of the native mutant htt detected by Western blot in human HD brain is soluble.
Full-length Mutant htt Is Degraded by Partial Denaturing
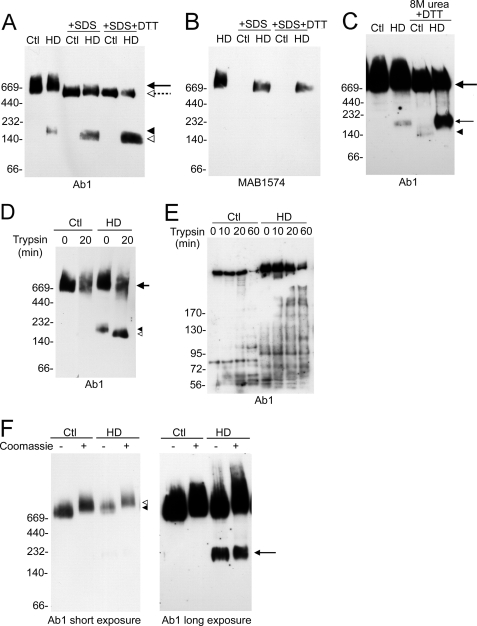
Our findings revealed the presence of a 180-kDa mutant htt fragment in the HD brains using BNP and Western blot analysis with Ab1 (htt-(1–17)) antibody. The 180-kDa fragment was detected at low levels compared with full-length htt. To increase the levels of the 180-kDa fragment, we pretreated HD brain lysates with denaturants before BNP. SDS alone (Fig. 3A), DTT alone (not shown), or SDS + DTT (Fig. 3A) increased the levels of the 180-kDa band compared with untreated lysates and reduced levels of full-length htt when detected with Ab1 antibody. Western blot with MAB1574 (1C2) antibody did not show evidence of the 180-kDa fragment even in the presence of SDS or SDS + DTT (Fig. 3B). Treating control brain lysates with these denaturants did not generate a WT htt band, although migration of intact WT htt was increased. The addition of 8 m urea alone did not change the levels of 180-kDa band (not shown). However, the combination of 8 m urea + DTT pretreatment (30 min) before BNP significantly increased levels of 180-kDa mutant htt band (Fig. 3C). Treatment of control brain lysates with 8 m urea + DTT induced formation of a 160-kDa band at much lower levels compared with the 180-kDa mutant htt band (Fig. 3C). 8 m urea + DTT treatment reduced the levels of full-length WT htt and mutant htt. Thus, pretreatment with 8 m urea + DTT was the most effective means of increasing the levels of the 180-kDa mutant htt band detected by BNP.
FIGURE 3.
Biochemical analysis of native htt in human brain; effects of partial denaturing. BNP and Western blot analysis of human control (Ctl) and HD homozygous brain lysates are shown in A–D and F. Anti-htt1–17 Ab1 was used in all Western blots except B, and 10 μg of protein was loaded per lane. Molecular mass markers are indicated on the left. A, effects of partial unfolding by SDS or SDS + DTT on native htt and formation of 180-kDa mutant htt fragment are shown. Lysates were treated with denaturant SDS (2%) or SDS + DTT (100 mm) for 40 min before BNP. Htt complexes migrate faster in the presence of SDS or SDS + DTT (the large arrow and dashed arrow indicate change in migration), and there is an increase in the levels of 180-kDa mutant htt band (the filled arrowhead and open arrowhead indicate shift in migration). B, Western blot shows that MAB1574 (1C2) detects mutant htt but not WT htt. SDS or SDS + DTT treatment increased the mobility and reduces the levels of full-length mutant htt, but no N-terminal fragment was detected. C, effects of denaturing in 8 m urea on native htt and formation of 180-kDa mutant htt fragment are shown. Lysates were treated with 8 m urea + DTT (100 mm) for 30 min before loading for BNP. The 180-kDa mutant htt (small arrow) increases in the presence of 8 m urea + DTT. Htt fragments of about 140 and160 kDa (arrowhead) are present in control brain lysates in the presence of 8 m urea + DTT. D, effects of trypsin digestion on native htt are shown. Brain lysates were pretreated with 0.1 mg/ml trypsin for 20 min before BNP. The Western blot shows increased migration and loss of full-length WT and mutant htt (arrow) and a marked increase in the levels of the N-terminal mutant htt domain (arrowheads). E, shown are the effects of trypsin digestion and analysis by SDS-PAGE and Western blot with anti-htt Ab1 antibody. Control and HD brain lysates were incubated in trypsin for time periods indicated. Increasing the incubation time in trypsin increases the levels of N-terminal fragments of WT and mutant htt. F, Coomassie dye was omitted or added to control and homozygous HD brain lysates before BNP. Two exposures of the film are shown. Short exposure, the presence of Coomassie dye slows migration of full-length htt (arrowhead and open arrowhead). Long exposure, the absence of dye increases the migration of htt and raises the levels of the N-terminal mutant htt band at 180 kDa (arrow).
We also addressed if trypsin pretreatment of brain lysates affected mutant htt degradation. Trypsin digestion of HD brain lysates (0.1 mg/ml trypsin for 20 min) before BNP increased the levels of the 180-kDa N-terminal domain compared with untreated lysates (Fig. 3D); no other htt fragments were detected. Native full-length mutant htt migrated faster and with reduced signal intensity in trypsin-treated lysates. Control brain lysates exposed to the same trypsin treatment did not generate a htt fragment, but the signal for full-length htt was reduced compared with untreated lysates. It is noteworthy that when the trypsin-treated brain lysates were separated by SDS-PAGE, multiple N-htt fragments were evident by Western blot in the control and HD brain lysates. Notably, mutant htt generated more large fragments than WT htt (Fig. 3E). Thus, except for increasing the levels of the 180-kDa mutant htt band, trypsin-treated mutant htt and WT htt remained intact under native conditions and degraded under denaturing SDS-PAGE.
Coomassie dye is usually added to the brain lysates before BNP. To determine if the addition of Coomassie dye affected mutant htt proteolysis, we examined brain lysates in the presence (standard method) or absence of Coomassie dye. The absence of Coomassie dye increased the migration of full-length htt but had little effect on the levels of the 180-kDa mutant htt band detected by BNP (Fig. 3F, long exposure).
Studies in vitro show that htt is processed near its N terminus by a β secretase-like aspartyl protease and in vivo by a calpain-like protease after ischemic injury (42, 43). To determine if native mutant htt was degraded by the increased activity of one of these proteases despite the addition of a protease inhibitor mixture, we pretreated brain lysates with pepstatin A (1 μg/ml) or N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (20 μm) before BNP. Results showed that the signal intensity of the 180-kDa band was not affected by the presence of these inhibitors (results not shown).
Analysis of Human htt Migration Using Two Dimension Native-denaturing PAGE
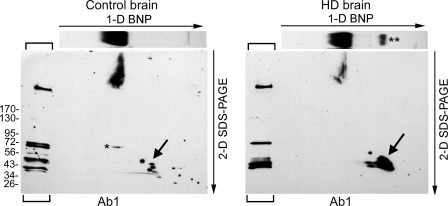
Native htt migrated at 575–850 kDa in control brain and at 650–885 kDa in homozygote HD brain. We wondered if the high molecular mass of htt was due to co-migration of full-length htt with N-terminal htt fragments. To test this idea we combined first dimension BNP with a second dimension denaturing gel (26). First, proteins from control and HD brain lysates were separated using BNP (1-D-BNP), then the lane for each sample was isolated and overlaid on top of a wide lane gel to serve as the source of protein for the SDS-PAGE (2-D SDS-PAGE) (Fig. 4). Lysates from the control and HD brains were run in the first lane of the two-dimension SDS-PAGE instead of molecular mass standards (Fig. 4, brackets). Western blot analysis with anti-htt1–17 showed that denatured WT htt and mutant htt detected in the two-dimension SDS-PAGE migrated as discrete bands at about 350 kDa and also formed multiple N-terminal htt fragments. These features have been described previously for htt in human brain when examined by SDS-PAGE and Western blot with Ab1 antibody (32, 44). The WT and mutant htt that entered the two-dimension SDS-PAGE from the one-dimension BNP gel piece migrated as broad bands barely reaching the position of htt seen using SDS-PAGE (first lane, brackets) (Fig. 4). In the two-dimension SDS-PAGE, the signal for mutant htt was much reduced compared with that of WT htt. There were no N-terminal htt fragments migrating in two-dimension SDS-PAGE below the level of the full-length protein, suggesting that the native WT and mutant htt signals >575 and 650 kDa detected using BNP contained only intact full-length WT and mutant htt. At the position where the 180-kDa band migrated in the one-dimension BNP, the two-dimension SDS-PAGE showed abundant mutant htt bands migrating at about 43–50 kDa (Fig. 4). Low levels of WT htt fragments migrated at around 40 kDa. Thus, the 180-kDa mutant htt band was replaced by smaller htt fragments after SDS-PAGE.
FIGURE 4.
Biochemical analysis of human brain htt using two-dimension BNP and SDS-PAGE. Western blots were probed with Ab1 antibody, and 10 μg of protein was loaded per lane. Approximate molecular mass is indicated on the left. Brain lysates were separated in BNP (the first dimension (1-D)), and the gel strip containing the separated proteins was used as the source of protein for the SDS-PAGE (second dimension (2-D)). The Western blot at the top shows a one-dimension BNP as an illustration to show the migration of htt complexes in BNP. The lanes on the left of each panel (brackets) were loaded with the same brain lysates and were separated only by SDS-PAGE. Full-length htt from one dimension migrates to the levels of monomeric htt in the second dimension at around 350 kDa. There was a marked loss of full-length mutant htt compared with WT htt in the transfer from one to two dimensions. Some WT htt was detected at the 72-kDa marker (small single asterisk). The 180-kDa mutant htt band seen by BNP (small double asterisk) was not detected in the two-dimension blot. Instead, there is a large signal present at 43–50 kDa (large arrow).
Analysis of Native Mutant htt in HD Mouse Models
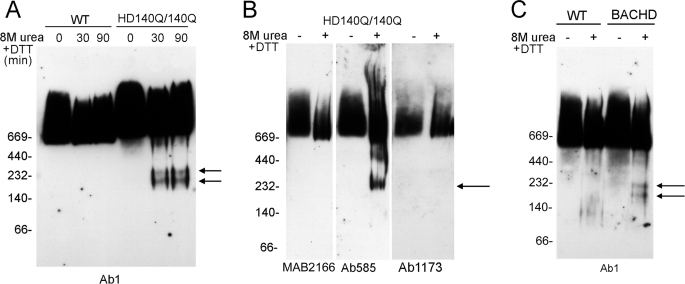
BNP analysis of human homozygote HD brain revealed a 180-kDa N-terminal domain in homozygous HD brain when examined by Western blot with Ab1 antibody (htt-(1–17)). To determine if a comparable N-terminal domain could be detected in the brain of HD mice, we examined native htt complexes in brain lysates from HD140Q/140Q mice and BACHD mice. The HD homozygous mice have a human exon 1 with 140Q inserted into mouse huntingtin gene. The BACHD transgenic mice express human htt with 97 glutamines and the endogenous mouse htt. Similar to htt in human brain, full-length htt in the HD mice migrated as broad bands of high molecular mass when detected with Ab1 antibody (Fig. 5). Mutant htt bands migrating at 180 and 240 kDa in the HD140Q/140Q mice and at 180 and 200 kDa in the BACHD mice were detected only after brain lysates were treated with 8 m urea + DTT (Fig. 5, A and C). Antibody Ab585 (htt-(585–725)) also recognized an N-terminal htt band after treatment with 8 m urea + DTT, but antibodies MAB2166 (htt-(443–457)) and Ab1173 (htt-(1173–1196)) did not (Fig. 5B). These data suggested that 8 m urea + DTT was effective in unfolding native mutant htt and generating cleavage of an N-terminal domain. Results also suggest that the N-terminal domain includes the epitope recognized by Ab585 antibody (amino acids 585–725); a modification in htt may prevent detection of the N-terminal domain by monoclonal MAB2166.
FIGURE 5.
Western blot analysis of native htt from brain of two HD mouse models expressing full-length mutant htt-effects of denaturing with 8 m urea + DTT. Brain lysates from cortex were prepared from WT and HD knock-in 140Q/140Q mice (A and B) and from WT and BACHD mice (C) and examined by BNP and Western blot. Lysates (10 μg) were untreated or pretreated with 8 m urea + DTT for 30 min (A–C) or 90 min (A) before BNP. Western blots in A and C were probed with Ab1 (anti-htt1–17) and with MAB2166, Ab585, and Ab1173 in B (see Fig. 1 for the epitopes for these antisera). A, in the presence of 8 m urea + DTT, N-terminal htt bands at 180 and 240 kDa (arrows) appear in homozygote HD brain but not WT brain. Levels of the bands in HD brain lysates were not different if incubations in 8 m urea + DTT were 30 or 90 min. In B antibody Ab585 detected an N-terminal band (arrow) in the presence of 8 m urea + DTT, but Ab2166 and Ab1173 do not. In C, two N-terminal htt bands migrating at 180 and 200 kDa (arrows) were detected in BACHD lysates treated with 8 m urea + DTT.
Analysis of Native Mutant htt in HD140Q/140Q Primary Neurons
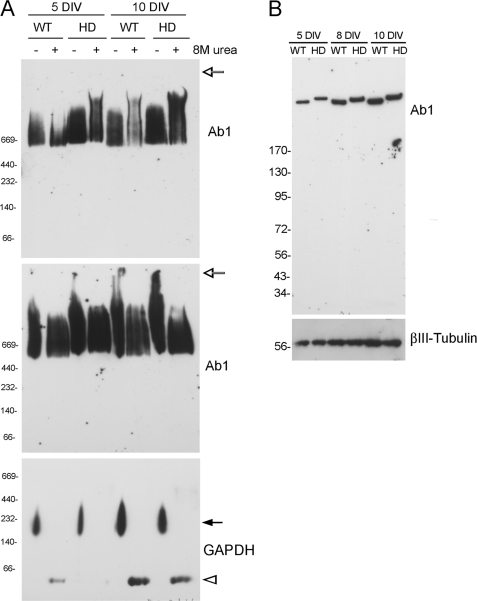
In recent studies we showed that primary embryonic cortical neurons from homozygous HD140Q/140Q mice develop elevated levels of reactive oxygen species at day 8 in vitro and increased cell death at day 10 in vitro compared with WT primary cortical neurons (23, 36). The HD140Q/140Q neurons in culture do not show visible signs of aggregates using anti-htt antibody.3 We examined lysates of WT and HD140Q/140Q primary neurons using BNP and SDS-PAGE followed by Western blot analysis for detection of htt with antisera Ab1. At days 5, 8, and 10 in vitro only full-length WT and mutant htt were detected using native and denatured conditions (Fig. 6, A and B). Even if higher than normal protein concentrations were used for BNP (50 μg instead of 10 μg), no N-terminal mutant htt bands were detected in the cultures (Fig. 6A, top and middle panels). Moreover, the addition of 8 m urea + DTT did not generate a detectable cleavage product of native mutant htt in the HD140Q/140Q primary neurons (Fig. 6A) even though the denaturing treatment was effective in reducing GAPDH from an oligomer to a monomer (Fig. 6A, bottom panel). In parallel cultures, we confirmed using the MTT assay that the viability of the HD140Q/140Q neurons was reduced compared with WT neurons (results not shown). Thus native full-length mutant htt in embryonic primary HD140Q/140Q neurons remains intact if cell lysates are exposed to denaturants before BNP or if cell death has occurred in the cultures.
FIGURE 6.
Western blot analysis of primary cortical neurons from WT and HD140Q/140Q embryonic mice. Recent studies from our laboratory show that neurodegeneration occurs in HD140Q/140Q primary neurons compared with WT neurons at 10 days in vitro (DIV) (23, 36). Cultures were prepared from embryos as described under “Experimental Procedures” and harvested at 5, 8, and 10 days in culture. Lysates were prepared for BNP (A) and SDS-PAGE (B), transferred to membranes, and probed by Western blot with antibody Ab1 (htt-117), then re-probed with antibody against GAPDH (A) or βIII-tubulin (B). Parallel cultures from the same embryos were used for MTT assay as described in our recent studies (23, 36). Reduced survival of the HD140Q/140Q neurons compared with WT neurons was confirmed in the 10-day old HD cultures. A, shown is BNP and Western blot analysis of neuronal lysates that were untreated or treated with 8 m urea + DTT and loaded at 10 μg/lane (top panel) or 50 μg/lane (middle panel). The open arrow indicates the top of gel. Lysates examined at low or high concentrations and pretreated with 8 m urea + DTT showed no N-terminal mutant htt bands even at 10 days in vitro when neurodegeneration is detected. GAPDH was reduced to a monomer (open arrowhead, at about 37 kDa) with treatment of 8 m urea + DTT (bottom panel). Native wild-type and mutant htt migrated into the gel in top panel. In the middle panel, although there is 5-fold more protein loaded, most of the htt migrated to its usual position. The location of native molecular mass markers from 66 to 669 kDa is shown on the left. B, SDS-PAGE and Western blot analysis of cell lysates (10 μg/lane) shows the presence of full-length WT and mutant htt and the absence of N-terminal htt fragments in the neurons. Mutant htt migrates more slowly than WT due to polyglutamine expansion. Some of the increase in signal for htt and βIII-tubulin with days in culture is due to the growth of the neurons. The location of molecular mass markers from 34 to 170 kDa is shown on the left.
DISCUSSION
Biochemical analysis of huntingtin in human postmortem brain has been evaluated previously using denatured conditions. These findings show intact protein and htt fragments containing the polyglutamine tract (44). Here, we sought to detect htt in human and mouse brain under native conditions. Because huntingtin partly associates with membranes, we used the method of Blue Native PAGE (25, 45). This method has been found to improve the resolution of native membrane protein complexes. Adding Coomassie Brilliant Blue to protein lysates and to the running buffer creates a net negative charge to protein surfaces and to the cathode buffer, which is at neutral pH. Proteins migrate to the anode independent of intrinsic pI. Hydrophobic proteins such as those in membranes avoid each other, thereby reducing protein aggregation and improving the resolution of native complexes.
Native full-length htt in human brain was detected by Western blot with nine anti-htt antisera made to regions spanning the entire protein. Native htt migrated over a broad molecular mass range of 575–850 kDa for WT htt and 650–885 kDa for mutant htt. As also observed using SDS-PAGE, the migration of mutant htt using BNP was slowed by the presence of the polyglutamine tract. Recombinant WT htt migrates in native PAGE as a monomer at about 550 kDa and as a dimer at an undetermined size well above the 669-kDa marker (8). Htt purified from brain was reported to be dimeric (8, 46). In our study of brain lysates using BNP, we did not observe discrete native htt bands corresponding to monomer and dimer, and the molecular mass is more consistent with a monomer. Pretreatment with denaturants did not markedly affect native htt migration. However, the molecular masses for full-length WT htt (540–1120 kDa) and for full-length mutant htt seen with long exposures of the Western blot films (615–1270 kDa) spanned a broad range and, therefore, might include monomeric and dimeric forms. Moreover, BNP analysis of in vitro translated htt-(1–969) resolved monomeric and dimeric forms, which suggested that a dimer interface in htt may occur between amino acids 552 and 969. Alternatively, the broad range in molecular mass of native full-length monomeric htt in brain may be due to the presence of modifications in the monomer such as phosphorylation and acetylation that modify htt migration. We confirmed by a second-dimension SDS-PAGE after BNP that the signals for native full-length WT and mutant htt did not form complexes with smaller N-terminal htt fragments. We were surprised that few native htt fragments were detected in brain. This was not a limit of the BNP/Western blot methods as we were able to detect in vitro translated products of htt-(1–552)-18Q, htt-(1–969)-18Q, and htt-(1–969)-100Q as well as htt1–3144 (full-length). Remarkably, exposure of brain lysates to trypsin had relatively little effect on native htt even though there was marked htt proteolysis detected in denatured conditions.
Native full-length mutant htt but not WT htt was detected with three antisera sensitive to the expanded polyglutamine region. Antibody 3B5H10, unlike the other two antisera (MW1 and MAB1574), does not detect mutant htt aggregates in cells but does recognize a soluble mutant htt monomer that is associated with cell death in vitro (15, 16). Antibody MW8 recognizes an epitope outside of the polyglutamine tract (amino acids 83–90). By SDS-PAGE and Western blot antibody, MW8 detects insoluble mutant htt in the stacking gel and a neoepitope of a mutant htt fragment (40). MW8 also labels fibrillar aggregates by immunostaining methods (47). In our analysis of human HD brain, MW8 recognized native full-length WT and mutant htt monomer weakly compared with the other antisera and did not detect a fragment of native mutant htt. Antibody Ab1 robustly recognized native WT and mutant htt in our study. This antibody is known to recognize soluble diffuse mutant htt and globular oligomers but not fibrillar forms of mutant htt (32, 48, 49).
Despite their distinct affinities for various forms of mutant htt, all of the antisera mentioned above detected a signal on Western blot for the native full-length mutant htt at the size reported for monomeric htt. None of the antisera detected signal at the top of the blot that would be evidence of protein failing to migrate into the gel during native PAGE. Native mutant htt also migrated to its usual position when examined in a resuspended pellet fraction of HD brain lysates. The absence of SDS in PAGE would be expected to diminish mutant htt solubility compared with WT htt and reveal slowly migrating insoluble protein. Instead, it appears that the methods for BNP minimize the aggregation of mutant htt compared with SDS-PAGE.
BNP analysis revealed a prominent mutant htt band at about 180 kDa in soluble fractions of human HD brain and a corresponding fragment of 160 kDa at much lower levels in control brain lysates. The 180-kDa mutant htt band in human HD brain increased markedly in soluble fractions compared with membrane fractions and also when denaturants (SDS, DTT, or urea) were added to the lysates. In contrast, the WT band was weakly detected in soluble fractions and also after treatment with 8 m urea + DTT. These results suggest that an unstable form of soluble full-length mutant htt is present in the HD brain. Membrane association may stabilize mutant htt and reduce the levels of unstable conformers compared with soluble htt. Study of Na+K+-ATPase unfolding by urea showed that the cytoplasmic domain was more sensitive to urea than the transmembrane domain (50). It is possible that dissociation of the N-terminal domain in mutant htt occurred during the process of separation in BNP. Even if this is the case, the tendency of full-length mutant htt to undergo cleavage of its N-terminal domain more than WT htt is a measure of instability.
Analysis of human brain lysates using a first dimension BNP and second dimension SDS-PAGE showed that WT and mutant htt were degraded to small fragments during SDS-PAGE. The levels of the small fragments were much greater for mutant htt than WT htt, suggesting that mutant htt is less stable than is WT htt during SDS-PAGE. Seong et al. (20) examined purified WT htt by SDS-PAGE and found only intact full-length htt. However, briefly exposing WT htt to trypsin produced an N-terminal segment of about 150 kDa; longer exposure yielded a fragment of 60 kDa. Seong et al. (20) proposed that a flexible domain in WT htt is released by trypsin digestion at amino acids 1184–1250. In another study, recombinant purified WT htt was found to be sensitive during preparation to cleavage at about amino acids 632, resulting in the presence of an N-terminal 100-kDa fragment (8). These findings suggest that native WT htt is relatively stable unless exposed to exogenous proteases. In our study of human brain, BNP and Western blot analysis with antibody Ab1 (htt-(1–17)) revealed more N-terminal mutant htt fragment (180 kDa) compared with WT htt fragment (160 kDa). Native mutant htt, being in a partially unfolded state compared with WT htt, may be prone to expose protease-sensitive sites and thereby have an increased susceptibility to cleavage by an endogenous protease. The addition of an aspartyl protease inhibitor or a calpain inhibitor to HD brain lysates before BNP did not affect htt proteolysis. However, we found that treating HD brain lysates with trypsin before BNP increased the abundance of the 180-kDa mutant htt band seen by Western blot. We suggest that a trypsin cleavage in htt generating the 180-kDa band occurs between amino acids 670 and 880. This is based on our analysis of the migration of in vitro translated htt fragments and a report that this region in htt has trypsin-sensitive sites (51). It is possible that the protease cleaving mutant htt during BNP is released from an internal cellular compartment during the preparation of brain lysates for BNP. Another possibility is that the cleavage is by autoproteolysis through a conformation-dependent event as described for some other proteins (52).
The native mutant htt fragment detected in human HD brain was more robust in tissue from homozygotes than heterozygotes possibly due to higher levels of mutant htt in homozygotes. Htt fragments were also observed in brain lysates of HD140Q/140Q mice and BACHD mice but only after treatment with 8 m urea + DTT. In addition to antibody Ab1, antibody Ab585 also detected an N-terminal mutant htt band in HD140Q/140Q mouse brain lysates treated with 8 m urea + DTT. However, a fragment was not detected in primary HD140Q/140Q neurons with antibody Ab1 even after denaturing with 8 m urea + DTT. These results suggest that the ability to detect a cleavage product of unstable mutant htt may depend on the antibody and the levels and source of full-length mutant htt.
In summary, our biochemical analysis provides support that soluble native full-length mutant htt can be detected as a monomer in human HD brain and that compared with WT htt it is less stable and more prone to release its N-terminal domain by an as yet unknown process. The results from human homozygotes confirm the identity of the expanded huntingtin as the unstable species in the HD brain as no WT htt is present. Because full-length mutant htt from brain lysates of two HD mouse models showed the same characteristic of instability as that seen in human brain lysates, it should be possible to address experimentally if partially unfolded mutant htt has a pathogenic role. Although WT htt was more stable than mutant htt under the conditions we examined, a more extensive analysis of human brain samples would be required to know if there are other factors independent of polyglutamine expansion in htt such as other disease states that can alter the stability of WT htt. It is unclear if the N-terminal domain released from partially unfolded mutant htt is functionally important or harmful. However, our analysis of embryonic primary neurons from HD140Q/140Q mice suggests that the presence of the full-length mutant protein is sufficient to cause the death of HD140Q/140Q neurons in vitro. BNP appears to be a sensitive method for detecting native monomeric full-length mutant htt in human HD brain with different antisera and may prove useful for tracking the presence of conformations of mutant htt that are pathogenic.
Supplementary Material
Acknowledgments
We thank Dr. Carl Johnson of the Hereditary Disease Foundation and Dr. Robert Matthews of the University of Massachusetts School of Medicine for advice during this work. We are very grateful to the Venezuelan families and the U. S.-Venezuela Collaborative Research Project for helpful participation over the years.
This work was supported, in whole or in part, by National Institutes of Health Grants 38194 (NINDS; to N. A. and M. D.) and P01-AG07232, R37-AG15473, and P50-AG08702 (NIA; to J. P. V.). This work was also supported by the Huntington Disease Society of America Coalition for the Cure (to M. D.), the UMass DERC (DIC 32520) (to N. A.), The Iseman Fund, The Louis and Rachel Rudin Foundation (to J. P. V.), Hereditary Disease Foundation (to J. P. V. and N. W.), and the W. M. Keck Foundation (to A. B. Y. and N. W.).

This article contains supplemental Fig. S1.
E. Sapp, A. Valencia, X. Li, N. Aronin, K. B. Kegel, J.-P. Vonsattel, A. B. Young, N. Wexler, and M. DiFiglia, unpublished data.
- HD
- Huntington disease
- BNP
- Blue Native PAGE
- htt
- huntingtin
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BAC
- Bacterial Artificial Chromosome
- BACHD
- BAC-mediated mouse model of HD.
REFERENCES
- 1. Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 2. Schilling G., Becher M. W., Sharp A. H., Jinnah H. A., Duan K., Kotzuk J. A., Slunt H. H., Ratovitski T., Cooper J. K., Jenkins N. A., Copeland N. G., Price D. L., Ross C. A., Borchelt D. R. (1999) Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 8, 397–407 [DOI] [PubMed] [Google Scholar]
- 3. Schilling G., Klevytska A., Tebbenkamp A. T., Juenemann K., Cooper J., Gonzales V., Slunt H., Poirer M., Ross C. A., Borchelt D. R. (2007) Characterization of huntingtin pathologic fragments in human Huntington disease, transgenic mice, and cell models. J. Neuropathol. Exp. Neurol. 66, 313–320 [DOI] [PubMed] [Google Scholar]
- 4. Wellington C. L., Singaraja R., Ellerby L., Savill J., Roy S., Leavitt B., Cattaneo E., Hackam A., Sharp A., Thornberry N., Nicholson D. W., Bredesen D. E., Hayden M. R. (2000) Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- 5. Kim Y. J., Sapp E., Cuiffo B. G., Sobin L., Yoder J., Kegel K. B., Qin Z. H., Detloff P., Aronin N., DiFiglia M. (2006) Lysosomal proteases are involved in generation of N-terminal huntingtin fragments. Neurobiol. Dis. 22, 346–356 [DOI] [PubMed] [Google Scholar]
- 6. Mende-Mueller L. M., Toneff T., Hwang S. R., Chesselet M. F., Hook V. Y. (2001) Tissue-specific proteolysis of Huntingtin (htt) in human brain. Evidence of enhanced levels of N- and C-terminal htt fragments in Huntington disease striatum. J. Neurosci. 21, 1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dyer R. B., McMurray C. T. (2001) Mutant protein in Huntington disease is resistant to proteolysis in affected brain. Nat. Genet. 29, 270–278 [DOI] [PubMed] [Google Scholar]
- 8. Li W., Serpell L. C., Carter W. J., Rubinsztein D. C., Huntington J. A. (2006) Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 281, 15916–15922 [DOI] [PubMed] [Google Scholar]
- 9. Toneff T., Mende-Mueller L., Wu Y., Hwang S. R., Bundey R., Thompson L. M., Chesselet M. F., Hook V. (2002) Comparison of huntingtin proteolytic fragments in human lymphoblast cell lines and human brain. J. Neurochem. 82, 84–92 [DOI] [PubMed] [Google Scholar]
- 10. Slow E. J., Graham R. K., Osmand A. P., Devon R. S., Lu G., Deng Y., Pearson J., Vaid K., Bissada N., Wetzel R., Leavitt B. R., Hayden M. R. (2005) Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc. Natl. Acad. Sci. U.S.A. 102, 11402–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 12. Saudou F., Finkbeiner S., Devys D., Greenberg M. E. (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 13. Kim M., Lee H. S., LaForet G., McIntyre C., Martin E. J., Chang P., Kim T. W., Williams M., Reddy P. H., Tagle D., Boyce F. M., Won L., Heller A., Aronin N., DiFiglia M. (1999) Mutant huntingtin expression in clonal striatal cells. Dissociation of inclusion formation and neuronal survival by caspase inhibition. J. Neurosci. 19, 964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finkbeiner S. (2011) Huntington Disease. Cold Spring Harb. Perspect. Biol. 3, a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller J., Arrasate M., Brooks E., Libeu C. P., Legleiter J., Hatters D., Curtis J., Cheung K., Krishnan P., Mitra S., Widjaja K., Shaby B. A., Lotz G. P., Newhouse Y., Mitchell E. J., Osmand A., Gray M., Thulasiramin V., Saudou F., Segal M., Yang X. W., Masliah E., Thompson L. M., Muchowski P. J., Weisgraber K. H., Finkbeiner S. (2011) Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 7, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Q. C., Yeh T. L., Leyva A., Frank L. G., Miller J., Kim Y. E., Langen R., Finkbeiner S., Amzel M. L., Ross C. A., Poirier M. A. (2011) A compact β model of huntingtin toxicity. J. Biol. Chem. 286, 8188–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romero E., Cha G. H., Verstreken P., Ly C. V., Hughes R. E., Bellen H. J., Botas J. (2008) Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron 57, 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray M., Shirasaki D. I., Cepeda C., André V. M., Wilburn B., Lu X. H., Tao J., Yamazaki I., Li S. H., Sun Y. E., Li X. J., Levine M. S., Yang X. W. (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 28, 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu X., Greiner E. R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J. S., Thompson L. M., Wetzel R., Yang X. W. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin-induced disease pathogenesis in HD mice. Neuron 64, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seong I. S., Woda J. M., Song J. J., Lloret A., Abeyrathne P. D., Woo C. J., Gregory G., Lee J. M., Wheeler V. C., Walz T., Kingston R. E., Gusella J. F., Conlon R. A., MacDonald M. E. (2010) Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 19, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kegel K. B., Sapp E., Alexander J., Valencia A., Reeves P., Li X., Masso N., Sobin L., Aronin N., DiFiglia M. (2009) Polyglutamine expansion in huntingtin alters its interaction with phospholipids. J. Neurochem. 110, 1585–1597 [DOI] [PubMed] [Google Scholar]
- 22. Kegel K. B., Sapp E., Yoder J., Cuiffo B., Sobin L., Kim Y. J., Qin Z. H., Hayden M. R., Aronin N., Scott D. L., Isenberg G., Goldmann W. H., DiFiglia M. (2005) Huntingtin associates with acidic phospholipids at the plasma membrane. J. Biol. Chem. 280, 36464–36473 [DOI] [PubMed] [Google Scholar]
- 23. Valencia A., Reeves P. B., Sapp E., Li X., Alexander J., Kegel K. B., Chase K., Aronin N., DiFiglia M. (2010) Mutant huntingtin and glycogen synthase kinase 3β accumulates in neuronal lipid rafts of a presymptomatic knock-in mouse model of Huntington disease. J. Neurosci. Res. 88, 179–190 [DOI] [PubMed] [Google Scholar]
- 24. Westmeyer G. G., Willem M., Lichtenthaler S. F., Lurman G., Multhaup G., Assfalg-Machleidt I., Reiss K., Saftig P., Haass C. (2004) Dimerization of β-site β-amyloid precursor protein-cleaving enzyme. J. Biol. Chem. 279, 53205–53212 [DOI] [PubMed] [Google Scholar]
- 25. Wittig I., Schägger H. (2008) Features and applications of blue native and clear native electrophoresis. Proteomics 8, 3974–3990 [DOI] [PubMed] [Google Scholar]
- 26. Burré J., Wittig I., Schägger H. (2009) Non-classical two-dimensional electrophoresis. Methods Mol. Biol. 564, 33–57 [DOI] [PubMed] [Google Scholar]
- 27. Wexler N. S. (2012) Huntington disease. Advocacy driving science. Annu. Rev. Med. 63, 1–22 [DOI] [PubMed] [Google Scholar]
- 28. The Huntington Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 29. Gusella J. F., Wexler N. S., Conneally P. M., Naylor S. L., Anderson M. A., Tanzi R. E., Watkins P. C., Ottina K., Wallace M. R., Sakaguchi A. Y. (1983) A polymorphic DNA marker genetically linked to Huntington disease. Nature 306, 234–238 [DOI] [PubMed] [Google Scholar]
- 30. Wexler N. S., Young A. B., Tanzi R. E., Travers H., Starosta-Rubinstein S., Penney J. B., Snodgrass S. R., Shoulson I., Gomez F., Ramos Arroyo M. A. (1987) Homozygotes for Huntington disease. Nature 326, 194–197 [DOI] [PubMed] [Google Scholar]
- 31. Aronin N., Chase K., Young C., Sapp E., Schwarz C., Matta N., Kornreich R., Landwehrmeyer B., Bird E., Beal M. F., (1995) CAG expansion affects the expression of mutant Huntingtin in the Huntington disease brain. Neuron 15, 1193–1201 [DOI] [PubMed] [Google Scholar]
- 32. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 33. Sapp E., Penney J., Young A., Aronin N., Vonsattel J. P., DiFiglia M. (1999) Axonal transport of N-terminal huntingtin suggests early pathology of corticostriatal projections in Huntington disease. J. Neuropathol. Exp. Neurol. 58, 165–173 [DOI] [PubMed] [Google Scholar]
- 34. Sapp E., Schwarz C., Chase K., Bhide P. G., Young A. B., Penney J., Vonsattel J. P., Aronin N., DiFiglia M. (1997) Huntingtin localization in brains of normal and Huntington disease patients. Ann. Neurol. 42, 604–612 [DOI] [PubMed] [Google Scholar]
- 35. Menalled L. B., Sison J. D., Dragatsis I., Zeitlin S., Chesselet M. F. (2003) Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington disease with 140 CAG repeats. J. Comp. Neurol. 465, 11–26 [DOI] [PubMed] [Google Scholar]
- 36. Li X., Valencia A., Sapp E., Masso N., Alexander J., Reeves P., Kegel K. B., Aronin N., Difiglia M. (2010) Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington disease. J. Neurosci. 30, 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J. P., Carraway R., Reeves S. A., (1995) Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 38. Kegel K. B., Meloni A. R., Yi Y., Kim Y. J., Doyle E., Cuiffo B. G., Sapp E., Wang Y., Qin Z. H., Chen J. D., Nevins J. R., Aronin N., DiFiglia M. (2002) Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem. 277, 7466–7476 [DOI] [PubMed] [Google Scholar]
- 39. Velier J., Kim M., Schwarz C., Kim T. W., Sapp E., Chase K., Aronin N., DiFiglia M. (1998) Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp. Neurol. 152, 34–40 [DOI] [PubMed] [Google Scholar]
- 40. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L. M., Trottier Y., Richards W. G., Osmand A., Paganetti P., Bates G. P. (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 285, 8808–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kegel K. B., Kim M., Sapp E., McIntyre C., Castaño J. G., Aronin N., DiFiglia M. (2000) Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 20, 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim M., Roh J. K., Yoon B. W., Kang L., Kim Y. J., Aronin N., DiFiglia M. (2003) Huntingtin is degraded to small fragments by calpain after ischemic injury. Exp. Neurol. 183, 109–115 [DOI] [PubMed] [Google Scholar]
- 43. Kegel K. B., Sapp E., Alexander J., Reeves P., Bleckmann D., Sobin L., Masso N., Valencia A., Jeong H., Krainc D., Palacino J., Curtis D., Kuhn R., Betschart C., Sena-Esteves M., Aronin N., Paganetti P., Difiglia M. (2010) Huntingtin cleavage product A forms in neurons and is reduced by γ-secretase inhibitors. Mol. Neurodegener. 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reisinger V., Eichacker L. A. (2008) Solubilization of membrane protein complexes for blue native PAGE. J. Proteomics 71, 277–283 [DOI] [PubMed] [Google Scholar]
- 46. Bao J., Sharp A. H., Wagster M. V., Becher M., Schilling G., Ross C. A., Dawson V. L., Dawson T. M. (1996) Expansion of polyglutamine repeat in huntingtin leads to abnormal protein interactions involving calmodulin. Proc. Natl. Acad. Sci. U.S.A. 93, 5037–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., Paganetti P., Muchowski P. J., Wilson S., Bates G. P. (2010) Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington disease. Hum. Mol. Genet. 19, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bhide P. G., Day M., Sapp E., Schwarz C., Sheth A., Kim J., Young A. B., Penney J., Golden J., Aronin N., DiFiglia M. (1996) Expression of normal and mutant huntingtin in the developing brain. J. Neurosci. 16, 5523–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qin Z. H., Wang Y., Sapp E., Cuiffo B., Wanker E., Hayden M. R., Kegel K. B., Aronin N., DiFiglia M. (2004) Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J. Neurosci. 24, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Babavali M., Esmann M., Fedosova N. U., Marsh D. (2009) Urea-induced unfolding of Na,K-ATPase as evaluated by electron paramagnetic resonance spectroscopy. Biochemistry 48, 9022–9030 [DOI] [PubMed] [Google Scholar]
- 51. Schilling B., Gafni J., Torcassi C., Cong X., Row R. H., LaFevre-Bernt M. A., Cusack M. P., Ratovitski T., Hirschhorn R., Ross C. A., Gibson B. W., Ellerby L. M. (2006) Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J. Biol. Chem. 281, 23686–23697 [DOI] [PubMed] [Google Scholar]
- 52. Sandberg A., Johansson D. G., Macao B., Härd T. (2008) SEA domain autoproteolysis accelerated by conformational strain. Energetic aspects. J. Mol. Biol. 377, 1117–1129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.