Background: Intracellular myo-inositol homeostasis involves both de novo synthesis and uptake of myo-inositol from the environment.
Results: Down-regulation of the myo-inositol transporter in Trypanosoma brucei causes depletion of bulk inositol lipids, but not glycosylphosphatidylinositols, and leads to parasite death.
Conclusion: De novo synthesis of myo-inositol is not sufficient to ensure bulk inositol lipid production.
Significance: myo-Inositol metabolism in T. brucei is compartmentalized.
Keywords: Glycosylphosphatidylinositol Anchors, Golgi, Inositol Phospholipid, Membrane Lipids, Phosphatidylinositol, Phospholipid Metabolism, RNA Interference (RNAi), Transporters, Trypanosoma brucei, Xenopus
Abstract
myo-Inositol is an essential precursor for the production of inositol phosphates and inositol phospholipids in all eukaryotes. Intracellular myo-inositol is generated by de novo synthesis from glucose 6-phosphate or is provided from the environment via myo-inositol symporters. We show that in Trypanosoma brucei, the causative pathogen of human African sleeping sickness and nagana in domestic animals, myo-inositol is taken up via a specific proton-coupled electrogenic symport and that this transport is essential for parasite survival in culture. Down-regulation of the myo-inositol transporter using RNA interference inhibited uptake of myo-inositol and blocked the synthesis of the myo-inositol-containing phospholipids, phosphatidylinositol and inositol phosphorylceramide; in contrast, it had no effect on glycosylphosphatidylinositol production. This together with the unexpected localization of the myo-inositol transporter in both the plasma membrane and the Golgi demonstrate that metabolism of endogenous and exogenous myo-inositol in T. brucei is strictly segregated.
Introduction
The polyol, myo-inositol, is the most common isomeric form of cyclohexane hexols, a group of molecules having diverse cellular functions (reviewed in Ref.1). In eukaryotes, myo-inositol is used to synthesize the following: (i) phosphatidylinositol (PI),2 a structural membrane component and precursor for other inositol-containing membrane lipids such as inositol phosphorylceramide (IPC); and (ii) glycosylphosphatidylinositol (GPI) for membrane anchoring of proteins (reviewed in Refs. 2–4). In addition, PI is a precursor for a large and complex group of membrane-bound signaling molecules and soluble second messengers that are involved in the regulation of cellular processes, such as growth, membrane trafficking, cell migration, differentiation, and apoptosis. Furthermore, myo-inositol and its derivatives are considered compatible solutes for cytoprotection in various eukaryotic cells (reviewed in Refs. 1, 5, 6).
Intracellular myo-inositol can be acquired by two major pathways. First, it can be generated de novo in a two-step reaction by a mechanism that is conserved from bacteria to eukaryotes (1). Biosynthesis of myo-inositol has been well documented in eukaryotic organisms, including plants, yeast, protozoa, and mammals (1, 7–9), as well as in many archaea (reviewed in Ref. 10) and certain bacteria (reviewed in Refs. 11, 12).
Second, myo-inositol can be imported from the environment via Na+- or H+-linked myo-inositol symporters, known as SMITs or HMITs, respectively. Whereas SMITs of mammals belong to the Na+/glucose transporter family, SGLT (TC 2.A.21.3.1 (13)), HMITs of eukaryotes and prokaryotes are members of the major facilitator superfamily, MFS (TC 2.A.1 (13, 14)), which includes proteins capable of transporting a wide range of substrates in response to chemiosmotic ion gradients (15). Although many myo-inositol symporters have been characterized biochemically, the essentiality of SMITs and HMITs for cell viability has only been addressed in a few cases. In Saccharomyces cerevisiae (16) and Candida albicans (17), myo-inositol uptake from the environment was not essential for growth, demonstrating that endogenous production was able to fully compensate for the lack of exogenous myo-inositol. In contrast, in Schizosaccharomyces pombe (18) and Arabidopsis thaliana (19), myo-inositol uptake was shown to be essential for normal development. Furthermore, in mammals, homozygous SMIT1 knock-out mice showing severe depletion of intracellular brain myo-inositol levels, but normal PI content, died shortly after birth (20). However, neonatal lethality could be prevented by prenatal maternal myo-inositol supplementation (21, 22). Taken together, although both pathways (de novo myo-inositol synthesis and uptake of myo-inositol from the extracellular medium) contribute to intracellular myo-inositol homeostasis, it is unclear in most organisms whether endogenous production and exogenous supply can complement each other or whether both pathways are essential, possibly providing myo-inositol for different cellular processes.
A first indication that myo-inositol from the two pathways may be used for different cellular events came from studies in Trypanosoma brucei, a protozoan parasite causing sleeping sickness in humans and nagana in animals. Deletion of the gene for inositol-3-phosphate synthase in T. brucei bloodstream forms to block endogenous production of myo-inositol resulted in complete inhibition of GPI synthesis. Because the levels of bulk PI and IPC were unaffected, the report suggested that de novo synthesized myo-inositol is primarily used to generate PI for GPI synthesis and hypothesized that myo-inositol for membrane lipid formation might be taken up from the medium (23, 24). In support of this hypothesis, the study found that T. brucei bloodstream forms contain two pools of PI synthase as follows: one pool localizing to the ER and possibly being involved in the formation of PI for GPI synthesis using de novo synthesized myo-inositol, and the other pool associating with the Golgi and possibly using myo-inositol taken up from the environment for inositol lipid production (25). However, it was not clear how the two pools of PI synthase recruit their respective substrates, endogenously produced and exogenous myo-inositol. In addition, the requirement for exogenous myo-inositol for lipid synthesis and parasite survival was not tested experimentally (reviewed in Refs. 9, 26).
In T. brucei, myo-inositol uptake has not been studied before. In contrast, in the related trypanosomatid, Leishmania donovani, myo-inositol import was characterized biochemically and is mediated by H+-linked electrogenic symport (27, 28). Disruption of the L. donovani myo-inositol transporter gene resulted in reduced parasite growth in culture, but the cells remained viable (29). In addition, in Trypanosoma cruzi, transport activity studies suggested that two distinct myo-inositol transporters (one Na+-coupled and the other H+-coupled) may mediate myo-inositol uptake (30), yet neither transporter was cloned nor experimentally characterized. Only one candidate gene has been identified in the T. cruzi genome (31). In this study, we describe for the first time the identification and functional characterization of an essential myo-inositol transporter in T. brucei. Its down-regulation revealed compartmentalization of myo-inositol metabolism.
EXPERIMENTAL PROCEDURES
Unless indicated, all reagents were of analytical grade and purchased from Merck, Sigma, or ICN Biomedicals (Tägerig, Switzerland). Antibiotics, DNA polymerase, and fetal bovine serum (FBS) were obtained from Invitrogen. Primers and sequencing services were from Microsynth AG (Balgach, Switzerland). All restriction enzymes were purchased from Fermentas (Nunningen, Switzerland). myo-[2-3H]Inositol (15–20 Ci/mmol) (myo-[3H]inositol) and [1-3H]ethanolamine hydrochloride (40–60 Ci/mmol) ([3H]Etn) were from American Radiolabeled Chemicals (St. Louis), and [α-32P]dCTP (3000 Ci/mmol) was from PerkinElmer Life Sciences. Kodak MXB and BioMax MS films were from Kodak SA (Lausanne, Switzerland).
Trypanosomes and Culture Conditions
T. brucei strain 29-13 procyclic forms co-expressing a tetracycline repressor and T7 polymerase (obtained from Paul Englund, John Hopkins University School of Medicine) (32) were cultured at 27 °C in SDM-79 (33) containing 15% heat-inactivated FBS, 25 μg ml−1 hygromycin, and 15 μg ml−1 G418. T. brucei strain 427 procyclic forms were cultured at 27 °C in SDM-79 (33) containing 5% heat-inactivated FBS.
RNAi-mediated Gene Silencing
Expression of Tb11.02.3020 (annotated in GeneDB/TriTrypDB as putative sugar transporter) was down-regulated by RNAi using a stem loop construct containing a puromycin resistance gene. Selection of the gene sequences for RNAi was done with RNAit, a prediction algorithm designed to prevent potential cross-talk and hence off-target effects (34). Cloning of the gene fragments into the tetracycline-inducible vector, pALC14 (a kind gift of André Schneider, University of Bern), was performed as described previously (35), using two separate PCR products obtained with primers Tb3020-F (5′-GCCCAAGCTTGGATCCCGCTGCAATCAACACACTCT-3′) and Tb3020-R (5′-GCTCTAGACTCGAGTGGGAACACCTGTGAAACAA-3′) (spanning nucleotides 842–1181 of Tb11.02.3020), resulting in plasmid pAG3020. Plasmid extraction was performed using the Qiagen plasmid midi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Before transfection of T. brucei procyclic forms, plasmid DNA was linearized with NotI and precipitated with phenol and chloroform.
Stable Transfection of Trypanosomes and Selection of Clones
T. brucei 29-13 or 427 procyclic forms (4–5 × 107 cells) were harvested at mid-log phase (0.5–0.8 × 107 cells/ml) by centrifugation at 1250 × g for 10 min, washed once in buffer (132 mm NaCl, 8 mm KCl, 8 mm Na2HPO4, 1.5 mm KH2PO4, 0.5 mm magnesium acetate, 0.09 mm calcium acetate, pH 7.0), resuspended in 450 μl of the same buffer, and mixed with 15 μg of linearized plasmids pAG3020 or pAG3020-3. Electroporation was performed with a BTX Electroporation 600 System (Axon Lab, Baden, Switzerland) with one pulse (1.5 kV charging voltage, 2.5 kV resistance, 25-microfarad capacitance timing, and 186 ohm resistance timing) and using a 0.2-cm pulse cuvette (Bio-Rad). Electroporated cells were immediately inoculated in 10 ml of SDM-79, containing 15% heat-inactivated FBS, and, if required for selection, 25 μg ml−1 hygromycin and 15 μg ml−1 G418. Clones were obtained by limiting dilutions in 24-well plates in SDM-79, containing 20% conditioned medium, in the presence of 2 μg ml−1 puromycin (RNAi selection) or 25 μg ml−1 hygromycin (in situ tagging). Antibiotic-resistant clones were tested for the presence of the introduced genes by PCR. Expression of HA-tagged TbHMIT or induction of RNAi was started by addition of 1 μg ml−1 tetracycline to parasite cultures.
RNA Isolation and Northern Blot Analysis
Total RNA for Northern blotting was isolated using the SV Total RNA Isolation System (Protégé, Madison, WI), following the manufacturer's instructions. RNA (10 μg) was separated on formaldehyde-agarose gels (1% agarose, 2% formaldehyde in MOPS) and transferred to GeneScreen Plus nylon membranes (PerkinElmer Life Sciences). 32P-Labeled probes were made by random priming the same PCR products used as inserts in the stem-loop vector using the Prime-a-Gene labeling system (Promega). Hybridization was performed overnight at 60 °C in hybridization buffer containing 7% (w/v) SDS, 1% (w/v) bovine serum albumin, 0.9 mm EDTA, 0.5 m Na2HPO4, pH 7.2, and the membrane was analyzed by autoradiography using BioMax MS film and a TransScreen-HE intensifying screen. Ribosomal RNA was visualized on the same formaldehyde-agarose gel by ethidium bromide staining to control for equal loading.
myo-inositol Uptake Assays
T. brucei procyclic forms (1 × 108 cells) at mid-log phase (0.9–1.1 × 107 cells/ml) were harvested by centrifugation at 1250 × g for 10 min and resuspended in phosphate-buffered saline (PBS; 135 mm NaCl, 1.3 mm KCl, 3.2 mm Na2HPO4, 0.5 mm KH2PO4, pH 7.4) at 27 °C. Uptake of myo-[3H]inositol was measured by adding 50 nm myo-[3H]inositol to cells at 27 °C. At various time points, uptake of label was terminated by pelleting 1.5 × 107 parasites by centrifugation (1500 × g, 5 min, 4 °C) and washing three times in ice-cold PBS. After resuspension of the pellet in 100 μl of PBS, radioactivity was measured by scintillation counting using a Packard Tri-Carb 2100TR liquid scintillation analyzer (PerkinElmer Life Sciences). Aliquots of the parasite suspensions before centrifugation were used to determine the total amount of radioactivity in the assay. All measurements were made in triplicates. The uptake of myo-[3H]inositol at each time point was calculated and plotted as a function of incubation time. Uptake of label was linear for 90 min, with regression coefficients of 0.977 or higher.
Substrate Specificity Studies
To study the substrate specificity of the putative myo-inositol transporter, d-glucose, d-mannose, d-sorbitol, d-mannitol, xylitol, scyllo-inositol, and myo-inositol 2-monophosphate-bis-cyclohexylammonium salt (myo-inositol 2-phosphate) were added to T. brucei procyclic forms together with myo-[3H]inositol. Uptake was terminated after 60 min of incubation at 27 °C. All measurements were made in triplicates. Statistical analysis of the data was performed by the paired sample t test with two-tailed p values.
Metabolic Labeling of Trypanosomes and Extraction Protocols
Metabolic labeling of trypanosomes was performed essentially as described before (36). Briefly, myo-[3H]inositol or [3H]Etn was added to procyclic form trypanosomes at a density of 0.7–1.0 × 107 cells/ml, and incubation was continued for 16 h. Cells were harvested by centrifugation at 1750 × g for 10 min and washed with ice-cold Tris-buffered saline (10 mm Tris, 144 mm NaCl, pH 7.4) to remove unincorporated label, and bulk phospholipids were extracted with 2× 10 ml of chloroform/methanol (CM; 2:1, by volume). CM fractions were pooled, dried under nitrogen, and resuspended in a small volume of CM. For isolation of GPI precursors and free GPIs, the remaining pellet was extracted three times with 5 ml of chloroform/methanol/water (CMW; 10:10:3, by volume). The delipidated pellet was extracted two times with 0.5 ml of 9% (v/v) butan-1-ol in water (butanol fraction), followed by 0.1% (w/v) Triton X-100 in 20 mm Tris-HCl, pH 7.4 (TX fraction), to solubilize GPI-anchored procyclins. The remaining material was resuspended in 1% (w/v) SDS (SDS fraction). CMW fractions were pooled, dried under nitrogen, and partitioned between butan-1-ol (CMWbut) and water (CMWaqu) to separate GPI precursors and free GPIs, respectively (37).
Lipid Analysis by Thin Layer Chromatography (TLC)
TLC was performed on Silica Gel 60 plates (Merck). To analyze the major inositol phospholipid classes, CM extracts were separated by one-dimensional TLC using solvent system 1 composed of chloroform/methanol/acetone/acetic acid/water (40:15:15:12:8, by volume). Complete separation of all phospholipid classes was achieved by two-dimensional TLC using solvent system 2 (chloroform/methanol/ammonia/water, 90:74:12:9; by volume) for the first and solvent system 1 for the second dimension (38). On each plate, appropriate lipid standards were run alongside the samples to be analyzed. Lipid spots on TLC plates were visualized by exposure to iodine vapor. 3H-Labeled GPI precursors were analyzed by one-dimensional TLC using solvent system 3 (chloroform:methanol:water, 10:10:3; by volume). Radioactivity was detected by scanning the air-dried plate with a radioisotope detector (Berthold Technologies, Regensdorf, Switzerland) and quantified using the Rita Star software provided by the manufacturer. Alternatively, plates were sprayed with EN3HANCE (PerkinElmer Life Sciences) and exposed to MXB films at −70 °C.
Lipid Phosphorus Determination
Phospholipids in chromatographic fractions were scraped from TLC plates and digested by boiling in 70–72% perchloric acid for 30–45 min. The released inorganic phosphate was reacted with ammonium molybdate and quantified photometrically (39). Each determination was accompanied by a series of inorganic phosphate standards. The assay was linear between 0 and 200 nmol of phosphate per sample (linear correlation coefficients were >0.99 in all experiments). Statistical analysis of the data were performed as mentioned above.
SDS-PAGE and Fluorography
Extracted proteins were separated on 12% polyacrylamide gels under reducing conditions (40). For detection of 3H-labeled proteins, gels were fixed, stained with Coomassie Brilliant Blue, immersed in Amplify (GE Healthcare), dried, and exposed to MBX films at −70 °C.
Expression of Tb11.02.3020 in Xenopus Oocytes
The target gene (Tb11.02.3020) was amplified using primers Tb3020-2-F (5′-CCGCTCGAGATGAAGTGGCGCGTGAAGAT-3′) and Tb3020–2-R (5′-CCCAAGCTTCTAAATGGGCGCACGGGC-3′), resulting in plasmid pAG3020-2, and inserted between the XhoI and HindIII sites of pCDNA3.1(−) vector (Invitrogen). From the linearized vector, the capped cRNA was synthesized (Ambion, Applied Biosystems, Rotkreuz, Switzerland), and a poly(A) tail of about 300 residues was added to the transcript using yeast poly(A) polymerase (United States Biologicals, Cleveland, OH). The concentration of the cRNA was quantified on a formaldehyde gel using Radiant Red stain (Bio-Rad) for visualization of the RNA. Known concentrations of RNA ladder (Invitrogen) were loaded as standard on the same gel. The cRNA was precipitated in ethanol/isoamyl alcohol (19:1; v/v), and the dried pellet was dissolved in water and stored at −80 °C. Aliquots containing 100 nm RNA were prepared from this stock solution and stored at −80 °C. Xenopus laevis oocytes were prepared, injected, and defollicated as described previously (41, 42). Each oocyte was injected with 50 nl of cRNA solution (5 fmol of RNA/oocyte) followed by incubation in modified Barth's solution (10 mm HEPES, pH 7.5, 88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 0.82 mm MgSO4, 0.34 mm Ca(NO3)2, 0.41 mm CaCl2, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin) at 18 °C for 3 days before measurements.
Functional Characterization in Xenopus Oocytes
Electrophysiological experiments were performed using an OC-725C two-electrode voltage clamp amplifier (Warner Instruments Corp., Hamden, CT) in combination with an XY-recorder, or the signal was digitized at 100 Hz with MacLab/200 (AD Instruments, Spechbach, Germany). Experiments were carried out at a holding potential of −80 mV. The perfusion medium contained 90 mm NaCl, 1 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 5 mm HEPES-NaOH, pH 7.4. The perfusion solution (6 ml/min) was applied through a glass capillary with an inner diameter of 1.35 mm, the mouth of which was placed about 0.5 mm from the surface of the oocyte (43). Concentration-response curves were fitted with the equation I(c) = Imax/(1 + (EC50/c)), where c is the concentration of myo-inositol; EC50 is the concentration of myo-inositol eliciting the half-maximal current amplitude; Imax is the maximal current amplitude, and I is the current amplitude. myo-Inositol was applied for 20 s for each measurement.
Generation of HA-tagged TbHMIT
To generate in situ HA-tagged TbHMIT, one allele of Tb11.02.3020 was C-terminally tagged with 3× HA using the one-step PCR strategy described elsewhere (44). Alternatively, to overexpress HA-tagged TbHMIT using the inducible vector pALC14, genomic DNA was amplified using primers Tb3020-3-F (5′-GCGCCCAAGCTTATGAAGTGGCGCGTGAAGATA-3′) and Tb3020-3-R (5′-CCCCGCGGATCCCTAAATGGGCGCACGGGCGGT-3′), containing a 3× HA tag and a stop codon at the 5′-end. The PCR product was ligated between HindIII and BamHI sites of pALC14, resulting in plasmid pAG3020-3. Before transfection of T. brucei procyclic forms, plasmid DNA was linearized with NotI and precipitated with phenol and chloroform.
Microscopy
For immunolocalization of in situ HA-tagged TbHMIT, trypanosomes were collected by centrifugation at 1250 × g for 10 min, washed once with PBS, and allowed to settle onto Superfrost Plus Microscope Slides (Thermo Scientific, Braunschweig, Germany). Parasites were fixed with 4% paraformaldehyde in PBS for 10 min, washed three times with PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and blocked with 2% (w/v) bovine serum albumin in PBS. Trypanosomes harboring the inducible vector for HA-tagged TbHMIT were cultured in the presence of tetracycline for 24 h to induce protein expression and processed for immunolocalization as described above. HA-tagged proteins were detected using monoclonal mouse anti-HA antiserum (Covance, Munich, Germany) at a dilution of 1:250 in PBS for 1 h at room temperature. Golgi was labeled by incubating fixed parasites for 1 h at room temperature with rabbit anti-TbGRASP antibody (45) (kindly provided by G. Warren, University of Vienna; used at a dilution of 1:1000). Subsequently, the slides were washed three times with PBS and incubated with the corresponding secondary antibodies, Alexa Fluor 594 goat anti-mouse IgG or Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) at a dilution of 1:1000 for 1 h at room temperature. Slides were washed three times with PBS and mounted using Vectashield containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories). Fluorescence microscopy was performed on a Leica AF6000 microscope (Leica Microsystems, Heerbrugg, Switzerland), using the software provided by the manufacturer.
In Silico Methods
Trypanosomatid myo-inositol transporter orthologs were retrieved from UniProt Knowledgebase (UniProtKB release 2011_04) by a blastp search using the L. donovani H+-myo-inositol transporter LdMIT (UniProtKB E9BGT1) as query. The obtained hits were supplemented with all annotated polyol and inositol transporters of the manually curated Swiss-Prot section of UniProtKB. The sequences were aligned with ClustalW2 (46), and an unrooted tree was constructed with Dendroscope (47).
RESULTS
Identification of an Essential Gene in T. brucei Encoding a Putative myo-Inositol Transporter
Using the translated protein sequence of the experimentally confirmed H+-myo-inositol transporter from L. donovani (UniProtKB E9BGT1 (48)), we searched the predicted proteome of T. brucei and identified a gene encoding a putative T. brucei myo-inositol transporter (GeneDB Tb11.02.3020). The gene had been annotated as putative sugar transporter and showed 54 and 64% identity at the protein level to the L. donovani transporter and to a putative T. cruzi myo-inositol transporter (TriTrypDB Tc00.1047053505183.130 (31)), respectively. Using Phobius (49) and TMHMM (50) algorithmic predictions, the translated T. brucei protein, named TbHMIT, consists of 12 transmembrane regions. Homologous proteins could also be found in other trypanosomatids, such as Trypanosoma congolense, Trypanosoma vivax, Leishmania braziliensis, Leishmania major, Leishmania mexicana, and Leishmania infantum. Amino acid sequence comparison shows that the highest degree of identity between the homologous proteins of the different species resides in transmembrane domains 1, 4, and 11 and in a short loop between transmembrane domains 11 and 12 (data not shown).
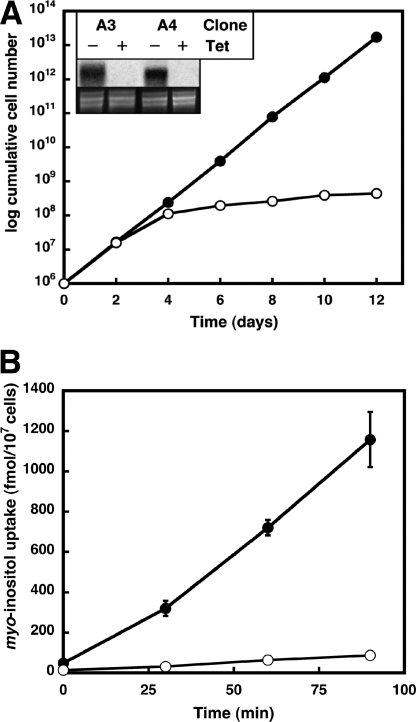
To study if TbHMIT is essential in T. brucei procyclic forms in culture, we generated tetracycline-inducible RNAi cell lines against Tb11.02.3020. Transfection of T. brucei procyclic forms with plasmid pAG3020 and selection by resistance to puromycin resulted in several mutant clones, two of which (A3 and A4) were selected for all subsequent experiments. After induction of RNAi by addition of tetracycline, both clones showed a clear growth reduction compared with uninduced (control) cells, i.e. parasite growth stopped after 4 days of culture (Fig. 1A). Northern blot analysis showed that after 2 days of RNAi, Tb11.02.3020 mRNA could no longer be detected (Fig. 1A, inset). Together, these results demonstrate that expression of Tb11.02.3020 is essential for normal growth of T. brucei procyclic forms in culture.
FIGURE 1.
A, growth phenotype of T. brucei procyclic form RNAi parasites. Expression of putative TbHMIT in T. brucei procyclic forms was down-regulated by tetracycline-inducible RNAi, and growth of trypanosomes was monitored for 12 days. Data points represent cumulative cell numbers of RNAi cells incubated in the absence (filled symbols) or presence (open symbols) of tetracycline and correspond to mean values from two experiments using clones A3 and A4. The inset shows Northern blot analyses of total RNA extracted from trypanosomes after 2 days of incubation in the absence (−) or presence (+) of tetracycline (Tet) and probed with 32P-labeled oligonucleotides used as inserts for the respective stem-loop vectors (top panel); rRNA was stained with ethidium bromide and used as loading control (bottom panel). B, myo-inositol uptake into T. brucei procyclic forms. After 2 days of incubation in the absence (filled symbols) or presence (open symbols) of tetracycline, RNAi cells (clone A3) were washed and subsequently incubated in PBS with trace amounts of myo-[3H]inositol (50 nm, final concentration). After the indicated times, parasites were washed, and the amount of radioactivity in the cell pellet was determined. Data points represent means ± S.D. from three independent experiments.
RNAi against TbHMIT Inhibits myo-Inositol Uptake into T. brucei
To investigate if TbHMIT is involved in uptake of myo-inositol into T. brucei, trace amounts of myo-[3H]inositol were added to RNAi clone A3 before and after down-regulation of TbHMIT, and time-dependent incorporation of label into parasites was determined by measuring radioactivity in the cell pellet after centrifugation. The results in Fig. 1B show that uptake of myo-[3H]inositol into control trypanosomes increased linearly over 90 min. In contrast, in the RNAi clones after down-regulation of TbHMIT, uptake of myo-[3H]inositol was reduced by >90% compared with control cells, demonstrating that TbHMIT is involved in myo-inositol uptake into T. brucei procyclic forms.
TbHMIT Is a myo-Inositol Transporter
To study if TbHMIT itself mediates myo-inositol transport, a cRNA coding for TbHMIT was microinjected into Xenopus oocytes. After 3 days, perfusion of oocytes with 10 mm myo-inositol resulted in an inward current of 55 ± 15 nA (mean value ± S.D. using 13 oocytes from four independent batches). In control oocytes injected with water instead of cRNA, no current was induced upon perfusion with 10 mm myo-inositol (n = 9). To determine the apparent affinity of the transport system, oocytes were exposed to increasing concentrations of myo-inositol (Fig. 2A). The data from four experiments were averaged and indicated an EC50 value of the current elicited by myo-inositol of 61 ± 5 μm (Fig. 2B). When the measurements were repeated in a medium, in which all Na+ had been replaced by K+, the current amplitudes induced by myo-inositol were not changed significantly, indicating that uptake of myo-inositol was independent of the extracellular Na+ concentration. In contrast, a change in pH of the assay medium from 7.4 to 6.5 increased the inward current in response to 10 mm myo-inositol to 152 ± 11%, whereas a change in pH to 8.5 decreased it to 42 ± 3% of the value observed at pH 7.4 (means ± S.D. using nine oocytes from three independent batches; Fig. 2C). These results indicate that uptake of myo-inositol is coupled to H+ transport.
FIGURE 2.
Functional expression of TbHMIT in Xenopus oocytes. A, concentration dependence of the current responses elicited by myo-inositol in an oocyte expressing TbHMIT. Holding potential was −80 mV. The substrate was applied for 20 s in each measurement (indicated by the bars); myo-inositol concentrations are indicated above each bar. B, currents elicited by myo-inositol from four concentration response curves using four different oocytes. Current amplitudes obtained at a given myo-inositol concentration were averaged. The data points represent mean values ± S.D. and were fitted as indicated under “Experimental Procedures.” C, currents elicited in an oocyte by 10 mm myo-inositol were measured as in B. During the application of myo-inositol, the pH of the perfusion medium was changed from pH 7.4 to 6.5 or pH 8.5 and back to pH 7.4, as indicated below the bars. Acidification resulted in an increase and alkalinization in a decrease in current amplitude.
Transport Specificity of TbHMIT
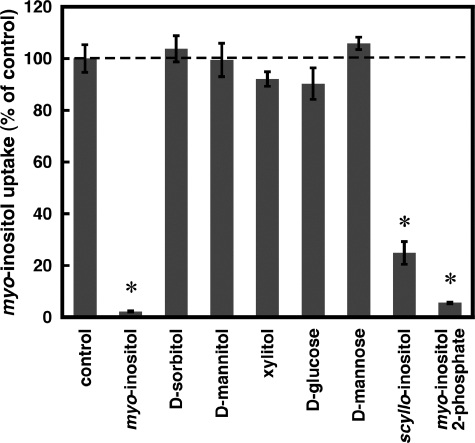
The substrate specificity of TbHMIT was analyzed in T. brucei procyclic forms by measuring uptake of myo-[3H]inositol (50 nm final concentration) in the absence or presence of high concentrations of unlabeled potential substrates (Fig. 3). The results in Fig. 3 show that uptake of myo-[3H]inositol was not affected by the presence of the polyols d-sorbitol, d-mannitol, or xylitol (5 mm, final concentrations). Similarly, no effect on myo-[3H]inositol uptake was observed in the presence of the two sugars, d-glucose and d-mannose (5 mm, final concentrations). In contrast, uptake of myo-[3H]inositol was blocked by the presence of 5 mm unlabeled myo-inositol (>98% inhibition; positive control), whereas scyllo-inositol and myo-inositol 2-phosphate (5 mm, final concentrations) inhibited uptake of myo-[3H]inositol by 75 and 95%, respectively. This, together with the observation that scyllo-inositol and myo-inositol-2-phosphate also elicited small current responses in TbHMIT-expressing oocytes, indicates that the two inositol isomers are also substrates of TbHMIT. When uptake of myo-[3H]inositol in T. brucei procyclic forms was measured in the presence of increasing concentrations of unlabeled myo-inositol (0–5 mm, final concentrations), an EC50 value similar to that determined in TbHMIT-expressing Xenopus oocytes was obtained (data not shown).
FIGURE 3.
Inhibition of myo-[3H]inositol uptake into T. brucei procyclic forms. RNAi clone A3 was incubated with trace amounts of myo-[3H]inositol (50 nm, final concentration) in the presence of unlabeled myo-inositol or potential substrates as indicated (5 mm, final concentrations). After 60 min, parasites were washed, and the amount of radioactivity in the cell pellet was determined. The dashed line indicates myo-[3H]inositol uptake in the absence of unlabeled substrates (control uninhibited uptake). The bars represent means ± S.D. from three independent experiments. *, p < 0.05.
Effect of TbHMIT RNAi on PI and IPC Synthesis in T. brucei Procyclic Forms
De novo synthesis of inositol-containing lipids in T. brucei RNAi cells before and after down-regulation of TbHMIT was studied by labeling parasites with myo-[3H]inositol for 18 h. We found that lipid extracts from RNAi cells after 3 days of induction with tetracycline incorporated <11% of radioactivity of control uninduced cells (data from three experiments using clones A3 and A4). Subsequent analysis of 3H-labeled lipids by one-dimensional TLC and fluorography demonstrated that most radioactivity co-migrated with PI and IPC (Fig. 4A). Quantification of the spots after scraping and liquid scintillation counting showed that the amounts of radioactivity in PI and IPC from induced cells were <10% (values from two independent TLC analyses) of those in the respective lipids from uninduced cells (Fig. 4A, compare lanes A3+ and A4+ versus lane A3−). The identification of the labeled lipids as PI and IPC was confirmed by two-dimensional TLC and mass spectrometry. Together, these results demonstrate that down-regulation of TbHMIT inhibits incorporation of exogenous myo-[3H]inositol into PI and IPC.
FIGURE 4.
A, analysis of myo-[3H]inositol-labeled lipids. RNAi against TbHMIT was induced in T. brucei procyclic clones A3 and A4 for 72 h by the addition of tetracycline to the culture medium (Tet +). Uninduced cells of clone A3 served as a control (Tet −). During the last 16 h of incubation, parasites (2 × 108 cells) were labeled with 25 μCi of myo-[3H]inositol. Subsequently, phospholipids were extracted and separated by one-dimensional TLC using solvent system 1. 3H-Labeled lipids were visualized by fluorography. The lanes contain extracts from equal cell equivalents. The migration of PI and IPC is indicated. B, GPI synthesis in TbHMIT RNAi cells. T. brucei TbHMIT RNAi cells were incubated in the absence or presence of tetracycline for 2 days. Subsequently, parasites were labeled for 16 h with [3H]Etn, and GPI precursors, free GPIs, and GPI-anchored proteins were extracted from the delipidated cell pellets as described under “Experimental Procedures.” 3H-Labeled GPI precursors from uninduced (top panel) and induced (bottom panel) cells were analyzed by one-dimensional TLC in solvent system 3, and radioactive lipids were visualized by scanning the plate. Extracts from equal cell equivalents were applied. PP1 refers to the major GPI precursor lipid in T. brucei procyclic forms (see Ref. 51 for characterization and identification of GPI lipids). The vertical lines indicate the site of sample application (left line) and the solvent front (right line), respectively. Inset, 3H-labeled GPI-anchored proteins from uninduced (−) and induced (+) cells were analyzed by SDS-PAGE and fluorography. The lanes contain protein from equal cell equivalents. EP refers to EP procyclin, the major GPI-anchored protein in T. brucei 29-13 wild-type cells. Molecular mass markers are indicated in kDa in the margin.
Down-regulation of TbHMIT Changes Parasite Phospholipid Composition
Possible changes in steady-state levels of T. brucei phospholipids after RNAi-mediated down-regulation of TbHMIT were analyzed by lipid quantification after two-dimensional TLC. Total lipid extracts from TbHMIT clones A3 and A4 grown in the absence or presence of tetracycline for 4 days revealed a >92 and >83% reduction in IPC and PI, respectively, in TbHMIT knockdown parasites compared with untreated cells (Table 1). In contrast, RNAi against TbHMIT resulted in an increase in the sphingomyelin content by >2.3-fold as compared with uninduced cells. In addition, we observed a small increase in phosphatidylcholine (PC) and phosphatidylserine levels and a small decrease in phosphatidylethanolamine (PE). The cardiolipin content was not significantly changed in RNAi cells compared with control parasites (Table 1).
TABLE 1.
Phospholipid composition of TbHMIT RNAi cells
RNAi against TbHMIT was induced in T. brucei procyclic form clones A3 and A4 for 4 days by addition of tetracycline to the culture medium (+Tet). Uninduced cells served as controls (− Tet). Phospholipid classes were extracted by CM, separated by two-dimensional TLC, and quantified by lipid phosphorus determination. Numbers represent means ±S.D. from six independent experiments using both clones. Statistical significance of induced cells compared with control cells is indicated, with p < 0.05. CL is cardiolipin; PS is phosphatidylserine.
| Phospholipid class | −Tet | +Tet |
|---|---|---|
| PC | 58.0 ± 1.6 | 65.4 ± 3.0a |
| PE | 17.2 ± 1.6 | 13.4 ± 2.6a |
| IPC | 8.9 ± 1.8 | 0.8 ± 0.6b |
| PI | 7.8 ± 0.9 | 1.4 ± 0.8b |
| SM | 4.3 ± 1.3 | 13.4 ± 2.3b |
| CL | 1.7 ± 0.8 | 1.9 ± 0.6 |
| PS | 1.7 ± 0.5 | 2.8 ± 0.7a |
a p < 0.05.
b p < 0.001.
Effect of TbHMIT Down-regulation on GPI Synthesis
Because myo-inositol is a component of the GPI core structure and thus is essential for GPI synthesis, we studied a possible effect of TbHMIT down-regulation on de novo GPI formation in T. brucei procyclic forms. RNAi cells were cultured in the absence or presence of tetracycline for 2 days and subsequently incubated with [3H]Etn for 16 h to label GPI precursors, free GPIs and GPI-anchored proteins (36). Sequential extraction of GPI molecules revealed that the amounts of radioactivity recovered in the fractions containing the GPI precursors and free GPIs were similar in RNAi cells incubated in the absence or presence of tetracycline (data not shown). Analysis of the GPI precursor fraction by one-dimensional TLC followed by radioactivity scanning showed no major differences between extracts from induced and noninduced cells (Fig. 4B). In both samples, >65% of radioactivity associated with the major GPI precursor lipid, designated PP1 (51), with the rest being in GPI precursors migrating between the origin and PP1. In addition, each lane showed a small amount of residual [3H]PE that was not extracted together with bulk phospholipids. The radioactivity scans were similar to those from a previous study, in which the individual GPI lipids had been characterized in detail (51). In addition, similar amounts of [3H]Etn were incorporated into the major GPI-anchored protein of T. brucei procyclic forms, EP procyclin, in uninduced and induced cells (Fig. 4B, inset), indicating that de novo GPI protein synthesis was not affected by RNAi against TbHMIT. In control experiments, phospholipid analysis of these [3H]Etn-labeled parasites confirmed that TbHMIT expression was down-regulated in all samples used for GPI lipid and protein analysis, i.e. the phospholipid composition in [3H]Etn-labeled TbHMIT-depleted cells showed the same alterations compared with control cells (see Table 1). Together, these results demonstrate that down-regulation of TbHMIT by RNAi has no effect on the biosynthesis of GPI molecules in T. brucei procyclic forms.
Localization of TbHMIT in T. brucei Procyclic Forms
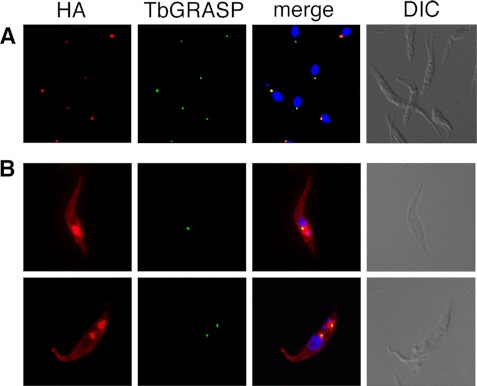
Localization of in situ HA-tagged TbHMIT in T. brucei procyclic forms showed staining of an intracellular structure located between the nucleus and the kinetoplast (Fig. 5A). Similar staining using immunofluorescence microscopy and immunolabeling at the electron microscope level has been reported before in T. brucei procyclic forms labeled with an antibody against the Golgi marker TbGRASP (45). Co-staining of parasites expressing in situ HA-tagged TbHMIT with anti-HA and anti-TbGRASP showed that the two signals completely co-localized (Fig. 5A). Similar results were obtained in parasites in which HA-tagged TbHMIT was overexpressed (Fig. 5B); although in these cells the area stained by anti-HA was larger than that stained by anti-TbGRASP, the two signals always co-localized, even in cells undergoing Golgi duplication (Fig. 5B, 2nd row (45)). In addition, anti-HA clearly revealed surface staining in parasites overexpressing HA-TbHMIT (Fig. 5B); similar staining could also be seen in cells expressing in situ HA-tagged TbHMIT; however, the signal only became apparent when the slides were overexposed (data not shown). The localization of in situ tagged HA-TbHMIT did not change when parasites were incubated in low myo-inositol-containing medium for 24 h (data not shown).
FIGURE 5.
Immunofluorescence microscopy of T. brucei procyclic forms. A, trypanosomes expressing in situ HA-tagged TbHMIT were washed, allowed to settle onto microscope slides, fixed with paraformaldehyde, and permeabilized with Triton X-100. B, trypanosomes were cultured in the presence of tetracycline for 24 h to induce overexpression of HA-tagged TbHMIT and treated as described in A. Proteins were detected using antibodies against HA and TbGRASP, in combination with the corresponding fluorescent secondary antibodies. DNA (shown in the merged pictures) was stained with DAPI. DIC, differential interference contrast.
Phylogenetic Positioning of TbHMIT
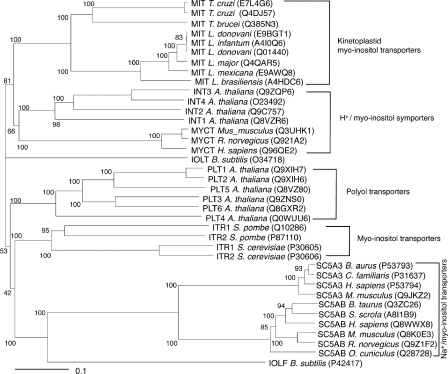
To determine the phylogenetic position of TbHMIT relative to other myo-inositol/polyol transporters, we generated an unrooted tree based on the multiple alignment of all trypanosomatid myo-inositol transporter orthologs with various known or predicted myo-inositol/polyol transporters (Fig. 6). In this phylogram, the trypanosomatid myo-inositol transporters group as a separate clade clearly distinct from their nearest neighbors, the HMITs from plants and mammals (e.g. 38% amino acid identity between TbHMIT and Homo sapiens HMIT Q96QE2), although they share considerably less similarity with mammalian SMITs (e.g. 13% amino acid identity between TbHMIT and H. sapiens SMIT P53794). Interestingly, the T. cruzi Brener Esmeraldo haplotype carries an myo-inositol transporter pseudogene (TriTrypDB Tc00.1047053504125.100) containing multiple in-frame stop codons at the syntenic position, whereas the T. cruzi Brener non-Esmeraldo-like haplotype possesses a full-length myo-inositol transporter ortholog (Tc00.1047053505183.130).
FIGURE 6.
Trypanosomatid myo-inositol transporter orthologs and their relation to other polyol transporter families. Protein names and UniProt accession numbers are given for each transporter. Numbers indicate the percentage of positives of 1000 rounds of bootstrap analysis. The scale bar indicates the number of amino acid substitutions per site.
DISCUSSION
Most organisms able to synthesize myo-inositol de novo from glucose 6-phosphate also express transporters for uptake of myo-inositol from the environment, suggesting that both processes may be required to fulfill the cellular demand for myo-inositol or that de novo synthesized myo-inositol is used for different cellular processes than myo-inositol imported from the surrounding medium.
In trypanosomatids, myo-inositol transport has been documented in L. donovani (27, 29, 48) and T. cruzi (30, 31). However, although an H+-myo-inositol transporter from L. donovani has been functionally characterized, the dependence of trypanosomatids on uptake of extracellular myo-inositol for growth has not been studied in detail. By using RNAi in combination with biochemical transport assays and phospholipid analyses in T. brucei procyclic forms and functional expression studies in Xenopus oocytes, we describe the first identification of an essential H+-coupled myo-inositol symporter in T. brucei.
Recombinant TbHMIT expressed in Xenopus oocytes mediated an electrogenic transport of myo-inositol with micromolar affinity. Although the transport was clearly not dependent on the presence of Na+ ions, the pH sensitivity indicated that the charge is contributed by H+, strongly suggesting that TbHMIT represents a H+-coupled myo-inositol symporter. This classification is consistent with the close phylogenetic relationship of TbHMIT to plant and mammalian HMITs and more distant relationship to SMITs. In addition, the observed specificity of TbHMIT for myo-inositol, and certain inositol derivatives, is in line with previous studies on HMITs from L. donovani, T. cruzi, A. thaliana, and C. albicans (29, 30, 52, 53), showing that other polyols and hexoses are not transported by HMITs, in contrast to SMITs, which readily transport glucose (54–56). Our observation that scyllo-inositol and myo-inositol phosphate inhibited myo-[3H]inositol uptake in T. brucei procyclic forms and elicited small currents in Xenopus oocytes indicates that these derivatives are also substrates for TbHMIT. To our knowledge, this is the first study in a protozoan parasite showing that myo-inositol uptake is essential. The specificity of TbHMIT (and likewise other trypanosomatid HMITs) could potentially be exploited to design toxic myo-inositol analogs as trypanocidal drugs. In addition, the strict dependence of T. brucei from exogenous myo-inositol validates TbHMIT as a potential drug target.
It has been shown in T. brucei bloodstream forms that de novo synthesis of myo-inositol is essential for parasite growth, providing myo-inositol for GPI synthesis, and that this endogenously produced pool of myo-inositol cannot be substituted by uptake of myo-inositol from the extracellular environment (23, 25). We now show that the converse is true as well, i.e. that uptake of myo-inositol from the medium is essential for T. brucei growth and that exogenous myo-inositol is used for the synthesis of membrane phospholipids but not for GPI formation. RNAi-mediated ablation of TbHMIT expression resulted in inhibition of myo-inositol uptake into T. brucei procyclic forms and caused a growth arrest. In addition, in vivo labeling experiments using myo-[3H]inositol and quantitative analysis of the phospholipid composition demonstrated that down-regulation of TbHMIT inhibited de novo formation of the myo-inositol-containing phospholipids, PI and IPC, resulting in a grossly altered trypanosome membrane lipid composition. Based on previous studies showing that inhibition of de novo phospholipid synthesis in T. brucei is lethal, by affecting cell cycle progression and organelle integrity (23, 25, 57–59), it is likely that the reduced levels of inositol-containing phospholipids in TbHMIT-depleted parasites are the major cause for the observed growth arrest. In contrast, by analyzing GPI formation using [3H]Etn labeling, we found that de novo synthesis of GPI lipids and GPI-anchored proteins is not affected by down-regulation of TbHMIT expression, i.e. GPI formation is independent of the supply of myo-inositol from the extracellular medium.
Besides depletion of PI and IPC, we noticed an increase in the SM content in T. brucei procyclic forms after down-regulation of TbHMIT. Because the decrease in IPC matched the increase in SM, it is likely that the lack of myo-inositol prompted a shift in ceramide usage from IPC production to SM synthesis, perhaps in an attempt to maintain the levels of ceramide-based membrane lipids. In addition, TbHMIT-depleted parasites showed an increase in both PC and phosphatidylserine and a decrease in PE. Although the higher relative amounts of PC and phosphatidylserine can be considered compensatory increases due to the reduction in PI, the relative decrease in PE is difficult to explain and may indicate a connection between PI and PE turnover.
The localization of TbHMIT in the Golgi was unexpected. It is generally believed that PI synthesis from exogenous inositol, mediated by PI synthase, occurs on the cytosolic side of the ER. Experiments using purified PI synthase from S. cerevisiae showed that the enzyme was asymmetrically incorporated into artificial vesicles with the active site facing outward (60). However, to our knowledge, no detailed studies have been done to determine the in vivo topology of PI synthase in any organism. Based on our results, we propose that, at least in T. brucei, the active site of PI synthase may be exposed to the lumen of the ER, catalyzing the synthesis of PI for GPI synthesis from endogenously produced inositol. In contrast, PI synthase in the Golgi exclusively uses inositol taken up from the environment into the Golgi via TbHMIT for PI production. Such a model is consistent with the following experimental observations: (i) PI in T. brucei shows a dual localization in the ER and Golgi (25); (ii) endogenously produced inositol from glucose 6-phosphate is used for PI synthesis in the ER for GPI anchor production (24); (iii) inositol taken up from the environment is used for bulk PI production but not GPI anchor synthesis (this study); (iv) extracellular inositol is taken up via TbHMIT localized in both the plasma membrane and the Golgi (this study); and (v) down-regulation of TbHMIT blocks bulk PI and IPC production (this study). Because IPC is produced from PI via transfer of the inositol head group, mediated by sphingolipid synthase 1 (TbSLS1) (61), our results predict that TbSLS1 would also localize to the Golgi. Interestingly, the related enzyme, TbSLS4, catalyzing the production of sphingomyelin and ethanolamine phosphorylceramide from PC and PE, respectively, has been shown to be in the Golgi in T. brucei bloodstream forms (62). Furthermore, the proposed compartmentalization of myo-inositol and PI synthesis in T. brucei is consistent with the previous observation that only a subset of PI molecular species, presumably the subset produced from endogenous myo-inositol, is used for GPI anchor synthesis in T. brucei procyclic forms (63).
Interestingly, in a recent report, HMIT has been found to co-localize with a Golgi marker in primary cultured rat neurons (64). Although the study did not address the metabolism of myo-inositol, the observation suggests that membrane PI synthesis in mammalian cells may also occur in the Golgi. This in turn would explain why in mammalian cells myo-[3H]inositol is often a poor precursor to label (and identify) GPI-anchored proteins; in analogy to T. brucei, exogenous inositol transported into the Golgi would not be available for GPI anchor synthesis in the ER. In contrast, myo-[3H]inositol is the preferred substrate to label GPI anchors in S. cerevisiae (reviewed in Ref. 65), suggesting that yeast may be able to transport myo-inositol into the ER or that the active site of yeast PI synthase faces the cytosol.
Acknowledgments
We thank G. Warren for the anti-TbGRASP antibody. P. B. thanks R. Tedder for valuable input.
This work was supported by Sinergia Grant CRSII3-127300 and in part by Grant 31003A-130815 from the Swiss National Science Foundation.
- PI
- phosphatidylinositol
- IPC
- inositol phosphorylceramide
- GPI
- glycosylphosphatidylinositol
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- SM
- sphingomyelin
- SMIT
- sodium-linked myo-inositol transporter
- HMIT
- proton-linked myo-inositol transporter
- TbHMIT
- T. brucei proton-linked myo-inositol transporter
- CM
- chloroform/methanol
- CMW
- chloroform/methanol/water
- Etn
- ethanolamine
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Michell R. H. (2008) Inositol derivatives. Evolution and functions. Nat. Rev. Mol. Cell Biol. 9, 151–161 [DOI] [PubMed] [Google Scholar]
- 2. Reynolds T. B. (2009) Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi, and protozoa. Making it and taking it. Microbiology 155, 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orlean P., Menon A. K. (2007) Thematic review series. Lipid post-translational modifications. GPI anchoring of protein in yeast and mammalian cells, or how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 4. Guan X., Wenk M. R. (2008) Biochemistry of inositol lipids. Front. Biosci. 13, 3239–3251 [DOI] [PubMed] [Google Scholar]
- 5. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 6. Yancey P. H. (2005) Organic osmolytes as compatible, metabolic, and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 7. Loewus F. A. (2006) Inositol and plant cell wall polysaccharide biogenesis. Subcell. Biochem. 39, 21–45 [DOI] [PubMed] [Google Scholar]
- 8. Carman G. M., Han G. S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith T. K., Bütikofer P. (2010) Lipid metabolism in Trypanosoma brucei. Mol. Biochem. Parasitol. 172, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koga Y., Morii H. (2007) Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 71, 97–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Majumder A. L., Chatterjee A., Ghosh Dastidar K., Majee M. (2003) Diversification and evolution of l-myo-inositol-1-phosphate synthase. FEBS Lett. 553, 3–10 [DOI] [PubMed] [Google Scholar]
- 12. Morita Y. S., Fukuda T., Sena C. B., Yamaryo-Botte Y., McConville M. J., Kinoshita T. (2011) Inositol lipid metabolism in mycobacteria. Biosynthesis and regulatory mechanisms. Biochim. Biophys. Acta 1810, 630–641 [DOI] [PubMed] [Google Scholar]
- 13. Saier M. H., Jr., Yen M. R., Noto K., Tamang D. G., Elkan C. (2009) The transporter classification database. Recent advances. Nucleic Acids Res. 37, D274–D278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marger M. D., Saier M. H., Jr. (1993) A major superfamily of transmembrane facilitators that catalyze uniport, symport, and antiport. Trends Biochem. Sci. 18, 13–20 [DOI] [PubMed] [Google Scholar]
- 15. Law C. J., Maloney P. C., Wang D. N. (2008) Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nikawa J., Tsukagoshi Y., Yamashita S. (1991) Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266, 11184–11191 [PubMed] [Google Scholar]
- 17. Chen Y. L., Kauffman S., Reynolds T. B. (2008) Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 76, 2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niederberger C., Gräub R., Schweingruber A. M., Fankhauser H., Rusu M., Poitelea M., Edenharter L., Schweingruber M. E. (1998) Exogenous inositol and genes responsible for inositol transport are required for mating and sporulation in Shizosaccharomyces pombe. Curr. Genet. 33, 255–261 [DOI] [PubMed] [Google Scholar]
- 19. Schneider S., Beyhl D., Hedrich R., Sauer N. (2008) Functional and physiological characterization of Arabidopsis inositol transporter 1, a novel tonoplast-localized transporter for myo-inositol. Plant Cell 20, 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berry G. T., Wu S., Buccafusca R., Ren J., Gonzales L. W., Ballard P. L., Golden J. A., Stevens M. J., Greer J. J. (2003) Loss of murine Na+/myo-inositol cotransporter leads to brain myo-inositol depletion and central apnea. J. Biol. Chem. 278, 18297–18302 [DOI] [PubMed] [Google Scholar]
- 21. Chau J. F., Lee M. K., Law J. W., Chung S. K., Chung S. S. (2005) Sodium/myo-inositol cotransporter-1 is essential for the development and function of the peripheral nerves. FASEB J. 19, 1887–1889 [DOI] [PubMed] [Google Scholar]
- 22. Bersudsky Y., Shaldubina A., Agam G., Berry G. T., Belmaker R. H. (2008) Homozygote inositol transporter knockout mice show a lithium-like phenotype. Bipolar Disord. 10, 453–459 [DOI] [PubMed] [Google Scholar]
- 23. Martin K. L., Smith T. K. (2006) The glycosylphosphatidylinositol (GPI) biosynthetic pathway of bloodstream-form Trypanosoma brucei is dependent on the de novo synthesis of inositol. Mol. Microbiol. 61, 89–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin K. L., Smith T. K. (2005) The myo-inositol-1-phosphate synthase gene is essential in Trypanosoma brucei. Biochem. Soc. Trans. 33, 983–985 [DOI] [PubMed] [Google Scholar]
- 25. Martin K. L., Smith T. K. (2006) Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem. J. 396, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Serricchio M., Bütikofer P. (2011) Trypanosoma brucei. A model micro-organism to study eukaryotic phospholipid biosynthesis. FEBS J. 278, 1035–1046 [DOI] [PubMed] [Google Scholar]
- 27. Seyfang A., Landfear S. M. (2000) Four conserved cytoplasmic sequence motifs are important for transport function of the Leishmania inositol/H+ symporter. J. Biol. Chem. 275, 5687–5693 [DOI] [PubMed] [Google Scholar]
- 28. Seyfang A., Kavanaugh M. P., Landfear S. M. (1997) Aspartate 19 and glutamate 121 are critical for transport function of the myo-inositol/H+ symporter from Leishmania donovani. J. Biol. Chem. 272, 24210–24215 [DOI] [PubMed] [Google Scholar]
- 29. Mongan T. P., Ganapasam S., Hobbs S. B., Seyfang A. (2004) Substrate specificity of the Leishmania donovani myo-inositol transporter. Critical role of inositol C-2, C-3, and C-5 hydroxyl groups. Mol. Biochem. Parasitol. 135, 133–141 [DOI] [PubMed] [Google Scholar]
- 30. Einicker-Lamas M., Almeida A. C., Todorov A. G., de Castro S. L., Caruso-Neves C., Oliveira M. M. (2000) Characterization of the myo-inositol transport system in Trypanosoma cruzi. Eur. J. Biochem. 267, 2533–2537 [DOI] [PubMed] [Google Scholar]
- 31. Einicker-Lamas M., Nascimento M. T., Masuda C. A., Oliveira M. M., Caruso-Neves C. (2007) Trypanosoma cruzi epimastigotes. Regulation of myo-inositol transport by effectors of protein kinases A and C. Exp. Parasitol. 117, 171–177 [DOI] [PubMed] [Google Scholar]
- 32. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 33. Brun R., Schönenberger M. (1979) Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36, 289–292 [PubMed] [Google Scholar]
- 34. Redmond S., Vadivelu J., Field M. C. (2003) RNAit. An automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128, 115–118 [DOI] [PubMed] [Google Scholar]
- 35. Bochud-Allemann N., Schneider A. (2002) Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849–32854 [DOI] [PubMed] [Google Scholar]
- 36. Bütikofer P., Ruepp S., Boschung M., Roditi I. (1997) “GPEET” procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem. J. 326, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Field M. C., Menon A. K., Cross G. A. (1992) Developmental variation of glycosylphosphatidylinositol membrane anchors in Trypanosoma brucei. In vitro biosynthesis of intermediates in the construction of the GPI anchor of the major procyclic surface glycoprotein. J. Biol. Chem. 267, 5324–5329 [PubMed] [Google Scholar]
- 38. Bütikofer P., Lin Z. W., Kuypers F. A., Scott M. D., Xu C. M., Wagner G. M., Chiu D. T., Lubin B. (1989) Chlorpromazine inhibits vesiculation, alters phosphoinositide turnover, and changes deformability of ATP-depleted RBCs. Blood 73, 1699–1704 [PubMed] [Google Scholar]
- 39. Rouser G., Fkeischer S., Yamamoto A. (1970) Two-dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 40. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 41. Sigel E. (1987) Properties of single sodium channels translated by Xenopus oocytes after injection with messenger ribonucleic acid. J. Physiol. 386, 73–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sigel E., Minier F. (2005) The Xenopus oocyte. System for the study of functional expression and modulation of proteins. Mol. Nutr. Food Res. 49, 228–234 [DOI] [PubMed] [Google Scholar]
- 43. Baur R., Sigel E. (2007) Replacement of histidine in position 105 in the α5 subunit by cysteine stimulates zolpidem sensitivity of α5β2γ2 GABA(A) receptors. J. Neurochem. 103, 2556–2564 [DOI] [PubMed] [Google Scholar]
- 44. Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 45. He C. Y., Ho H. H., Malsam J., Chalouni C., West C. M., Ullu E., Toomre D., Warren G. (2004) Golgi duplication in Trypanosoma brucei. J. Cell Biol. 165, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X, Version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 47. Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Dendroscope. An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drew M. E., Langford C. K., Klamo E. M., Russell D. G., Kavanaugh M. P., Landfear S. M. (1995) Functional expression of a myo-inositol/H+ symporter from Leishmania donovani. Mol. Cell. Biol. 15, 5508–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Käll L., Krogh A., Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 50. Sonnhammer E. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 51. Field M. C., Menon A. K., Cross G. A. (1991) Developmental variation of glycosylphosphatidylinositol membrane anchors in Trypanosoma brucei. Identification of a candidate biosynthetic precursor of the glycosylphosphatidylinositol anchor of the major procyclic stage surface glycoprotein. J. Biol. Chem. 266, 8392–8400 [PubMed] [Google Scholar]
- 52. Jin J. H., Seyfang A. (2003) High affinity myo-inositol transport in Candida albicans. Substrate specificity and pharmacology. Microbiology 149, 3371–3381 [DOI] [PubMed] [Google Scholar]
- 53. Schneider S., Schneidereit A., Konrad K. R., Hajirezaei M. R., Gramann M., Hedrich R., Sauer N. (2006) Arabidopsis inositol transporter 4 mediates high affinity H+ symport of myo-inositol across the plasma membrane. Plant Physiol. 141, 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hager K., Hazama A., Kwon H. M., Loo D. D., Handler J. S., Wright E. M. (1995) Kinetics and specificity of the renal Na+/myo-inositol cotransporter expressed in Xenopus oocytes. J. Membr. Biol. 143, 103–113 [DOI] [PubMed] [Google Scholar]
- 55. Wright E. M., Hirayama B. A., Loo D. F. (2007) Active sugar transport in health and disease. J. Intern. Med. 261, 32–43 [DOI] [PubMed] [Google Scholar]
- 56. Coady M. J., Wallendorff B., Gagnon D. G., Lapointe J. Y. (2002) Identification of a novel Na+/myo-inositol cotransporter. J. Biol. Chem. 277, 35219–35224 [DOI] [PubMed] [Google Scholar]
- 57. Signorell A., Rauch M., Jelk J., Ferguson M. A., Bütikofer P. (2008) Phosphatidylethanolamine in Trypanosoma brucei is organized in two separate pools and is synthesized exclusively by the Kennedy pathway. J. Biol. Chem. 283, 23636–23644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Signorell A., Gluenz E., Rettig J., Schneider A., Shaw M. K., Gull K., Bütikofer P. (2009) Perturbation of phosphatidylethanolamine synthesis affects mitochondrial morphology and cell cycle progression in procyclic form Trypanosoma brucei. Mol. Microbiol. 72, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 59. Gibellini F., Hunter W. N., Smith T. K. (2009) The ethanolamine branch of the Kennedy pathway is essential in the bloodstream form of Trypanosoma brucei. Mol. Microbiol. 73, 826–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fischl A. S., Homann M. J., Poole M. A., Carman G. M. (1986) Phosphatidylinositol synthase from Saccharomyces cerevisiae. Reconstitution, characterization, and regulation of activity. J. Biol. Chem. 261, 3178–3183 [PubMed] [Google Scholar]
- 61. Sevova E. S., Goren M. A., Schwartz K. J., Hsu F. F., Turk J., Fox B. G., Bangs J. D. (2010) Cell-free synthesis and functional characterization of sphingolipid synthases from parasitic trypanosomatid protozoa. J. Biol. Chem. 285, 20580–20587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sutterwala S. S., Hsu F. F., Sevova E. S., Schwartz K. J., Zhang K., Key P., Turk J., Beverley S. M., Bangs J. D. (2008) Developmentally regulated sphingolipid synthesis in African trypanosomes. Mol. Microbiol. 70, 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Güther M. L., Lee S., Tetley L., Acosta-Serrano A., Ferguson M. A. (2006) GPI-anchored proteins and free GPI glycolipids of procyclic form Trypanosoma brucei are nonessential for growth, are required for colonization of the tsetse fly, and are not the only components of the surface coat. Mol. Biol. Cell 17, 5265–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Di Daniel E., Mok M. H., Mead E., Mutinelli C., Zambello E., Caberlotto L. L., Pell T. J., Langmead C. J., Shah A. J., Duddy G., Kew J. N., Maycox P. R. (2009) Evaluation of expression and function of the H+/myo-inositol transporter HMIT. BMC Cell Biol. 10, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pittet M., Conzelmann A. (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 405–420 [DOI] [PubMed] [Google Scholar]