Background: NHE3 is regulated by a signaling complex on its C terminus, only some of the components of which are known.
Results: CaMKIIγ binds to the NHE3 C terminus and phosphorylates and inhibits basal NHE3 activity by altering turnover number.
Conclusion: CaMKIIγ is part of an NHE3 signaling complex.
Significance: Signaling complexes that form on transport proteins take part in regulation of the transporter
Keywords: Epithelial Cell, Protein Kinases, Protein-Protein Interactions, Signal Transduction, Sodium Proton Exchange, Calmodulin Kinase II
Abstract
The epithelial brush border (BB) Na+/H+ exchanger 3 (NHE3) accounts for most renal and intestinal Na+ absorption. Ca2+/calmodulin-dependent protein kinase II (CaMKII) inhibits NHE3 activity under basal conditions in intact intestine, acting in the BB, but the mechanism is unclear. We now demonstrate that in both PS120 fibroblasts and polarized Caco-2BBe cells expressing NHE3, CaMKII inhibits basal NHE3 activity, because the CaMKII-specific inhibitors KN-93 and KN-62 stimulate NHE3 activity. This inhibition requires NHERF2. CaMKIIγ associates with NHE3 between aa 586 and 605 in the NHE3 C terminus in a Ca2+-dependent manner, with less association when Ca2+ is increased. CaMKII inhibits NHE3 by an effect on its turnover number, not changing surface expression. Back phosphorylation demonstrated that NHE3 is phosphorylated by CaMKII under basal conditions. This overall phosphorylation of NHE3 is not affected by the presence of NHERF2. Amino acids downstream of NHE3 aa 690 are required for CaMKII to inhibit basal NHE3 activity, and mutations of the three putative CaMKII phosphorylation sites downstream of aa 690 each prevented KN-93 stimulation of NHE3 activity. These studies demonstrate that CaMKIIγ is a novel NHE3-binding protein, and this association is reduced by elevated Ca2+. CaMKII inhibits basal NHE3 activity associated with phosphorylation of NHE3 by effects requiring aa downstream of NHE3 aa 690 and of the CaMKII-binding site on NHE3. CaMKII binding to and phosphorylation of the NHE3 C terminus are parts of the physiologic regulation of NHE3 that occurs in fibroblasts as well as in the BB of an intestinal Na+-absorptive cell.
Introduction
There are nine Na+/H+ exchanger (NHE)3 isoforms that can be divided into two groups as follows: isoforms 1–5 (NHE1–5) function predominantly in the plasma membrane, and isoforms 6–9 (NHE6–9) reside in the membrane of subcellular organelles, although NHE8 also occurs in the BB of several epithelia. NHE3 is present in the BB of intestinal Na+-absorptive cells and the renal proximal tubule where it accounts for the majority of gastrointestinal and renal Na+ absorption (1, 2). NHE3 also influences other BB transport processes. NHE3 is functionally linked to the intestinal BB Cl−/HCO3− exchangers DRA (down-regulated in adenoma) and PAT-1 (putative anion transporter 1) (3, 4), Cl− secretion mediated by cystic fibrosis transmembrane regulator (5), and peptide absorption mediated by PEPT1 (6).
NHE3 shares a common organizational structure with other mammalian NHE isoforms, being made up of an N-terminal transmembrane domain that mediates Na+ and H+ exchange followed by a long C-terminal cytoplasmic domain that regulates Na+/H+ exchange activity and interacts with the cytoskeleton and other proteins. NHE3 is acutely regulated predominantly by changes in its Vmax by processes that involve changes in its continual trafficking to/from the plasma membrane by endocytosis and exocytosis; however, some of its regulation predominantly involves changes in the NHE3 turnover number (7).
Regulation of Na+/H+ exchange activity involves several distinct domains in the NHE3 C terminus (7–14). NHE3 exists as a member of large multiprotein complexes with sizes of up to 1–2 MDa rather than its predicted size of 87 kDa. Its C-terminal binding partners include the multi-PDZ domain containing Na+/H+ exchanger regulatory factor gene family members (NHERF1–4), CK2, phospholipase Cγ, calmodulin, megalin, dipeptidyl peptidase IV, calcineurin homologous protein, and ezrin (7–14). At least six of these regulatory proteins (NHERF1–4, CK2, and phospholipase Cγ) directly associate with NHE3 via a small, putative α-helical domain of NHE3, between amino acids 586 and 605 (14). These interactions modulate acute stimulation and inhibition of NHE3 activity, which mimic changes that occur in digestive physiology.
On a molecular and cellular level, there are multiple examples in which NHE3 regulation is dependent on changes in its protein phosphorylation (15, 16), and NHE3 phosphorylation by different protein kinases is part of the signal transduction that modulates NHE3 activity. For example, cAMP phosphorylates NHE3 at aa Ser552 and Ser605, although inhibition of NHE3 by cAMP requires an additional but unknown event (17). Oppositely, basal NHE3 activity requires phosphorylation at Ser719 by CK2, and glucocorticoid stimulation of NHE3 requires phosphorylation at Ser663 (18, 19).
Ca2+/calmodulin-dependent protein kinase II (CaMKII) is a multifunctional kinase that has been implicated in a multitude of Ca2+-regulated biologic processes, including synaptic transmission, gene transcription, and channel regulation (20). There are four isoforms of CaMKII encoded by separate genes. Two of them, the γ and δ isoforms, are ubiquitously expressed, whereas the α and β isoforms are expressed mostly in the neural system. Involvement of CaMKII in the basal regulation of small intestinal BB Na+/H+ exchange was demonstrated because addition of Ca2+/CaM, ATP, and Mg2+ to the inside of right-side out rabbit ileal BB membrane vesicles inhibited Na+/H+ exchange (21). These membranes contain CaMKII, and a specific inhibitory peptide of CaMKII reversed the inhibition and stimulated BB Na+/H+ exchange activity (21). Thus, CaMKII inhibits basal BB NHE activity (21). Moreover, the CaMKII inhibition of BB Na+/H+ exchange was consistent with phosphorylation, because it required Mg2+/ATP, and the Ca2+ concentration dependence of the effects on Na+/H+ exchange and on Ca2+/CaM-dependent phosphorylation in the same vesicles was similar (21). However, direct NHE3 phosphorylation by CaMKII was not demonstrated, and how CaMKII regulated NHE3 activity was not determined.
Further mechanistic studies of CaMKII regulation of NHE3 were carried out in endogenous NHE3 null PS120 fibroblasts stably expressing rabbit NHE3 and in a polarized intestinal epithelial cell model, Caco-2 cells (22). We now show that NHE3 activity is stimulated by CaMKII inhibitors under basal Ca2+ conditions, further suggesting that CaMKII inhibits basal NHE3 activity. This CaMKII inhibition of basal NHE3 activity was shown to be associated with NHE3 phosphorylation by CaMKII, and the mechanism of this regulation is the further subject of this study.
MATERIALS AND METHODS
Reagents
Reagents were from Sigma unless otherwise stated. Protein A-Sepharose was from Amersham Biosciences. Protein G-agarose beads were from Millipore. KN-62, KN-93, and KN-92 were from Calbiochem (catalog nos. 422706, 422708, 422709, respectively). KN-04 was from Seikagaku America Inc (catalog no. 71208321, Ijamsville, MD). HaltTM phosphatase inhibitor mixture was from Thermo Scientific. Nigericin was from Invitrogen. 2′,7′-Bis(carboxyethyl)-5–6-carboxyfluorescein-AM was from Molecular Probes. EZ-Link Sulfo-NHS-SS-biotin was from Pierce. Rat recombinant CaMKIIα was from Calbiochem (catalog no. 208706), and CaMKIIγ recombinant protein was from Novus Biologicals (catalog no. H00000818, Littleton, CO). [γ-32P]ATP was from Amersham Biosciences (catalog no. PB10168). ECL reagent was from Amersham Biosciences. Monoclonal anti-hemagglutinin (HA) affinity matrix was from Roche Diagnostics (catalog no. 11815016001).
Cell Culture
Studies were performed using either PS120 fibroblasts stably transfected with rabbit NHE3 (rNHE3) or Caco-2BBe cells transiently infected (adenovirus) with rNHE3. PS120 cells, which lack all endogenous plasma membrane NHEs, were stably transfected with rNHE3 tagged either at the C terminus with a vesicular stomatitis virus-glycoprotein epitope tag (NHE3V) or on the N terminus with a triple HA epitope tag (HA-NHE3), as described previously (23). They were transfected as described previously (23, 24). Where indicated, PS120/HA-NHE3 or PS120/NHE3V cells were stably co-transfected with human NHERF1 or NHERF2, as described previously (25). All stable PS120 cell lines were maintained at 37 °C in a humidified atmosphere with 5% CO2 and 95% O2 in Dulbecco's modified Eagle's medium with sodium pyruvate (catalog no. 10-013-CV, Mediatech Inc.) supplemented with 10% (v/v) fetal calf serum and G418 (400 μg/ml) (Invitrogen). The cells co-transfected with NHERF1/2 were additionally supplemented with hygromycin (600 μg/ml). To maintain high levels of NHE3 expression, the stably transfected cells were “acid-loaded” weekly, as described previously (26).
The Caco-2BBe cell line, originally derived from a human adenocarcinoma, was obtained from M. Mooseker (Yale University) via J. Turner (University of Chicago) and grown at 37 °C in a humidified atmosphere with 5% CO2 and 95% O2 in Dulbecco's modified Eagle's medium without sodium pyruvate (10-017-CM, Mediatech Inc.) supplemented with 15 mm HEPES and 10% fetal bovine serum (referred to as “Caco-2 medium”).
Adenoviral Constructs/Infection
Caco-2BBe cells, which endogenously express the four members of the NHERF gene family and small amounts of NHE3, were transiently infected with triple HA-tagged rNHE3 previously engineered into replication-deficient adenoviral shuttle vector ADLOX.HTM under a cytomegalovirus promoter. Caco-2BBe cells were first grown on Transwell filters (Corning Glass) until 12 days post-confluence in Caco-2 medium. Cells were then treated with serum-free media containing 6 mm EGTA for 2 h at 37 °C to allow the tight junctions to open, further exposing apical and basolateral surfaces to the virus. Cells were then infected by appropriate amounts of viral particles diluted (109–1010 particles/ml) in serum-free Caco-2 medium at 37 °C for 6 h, and then cells were allowed to recover in Caco-2 medium over the next 40 h before transport assays or Western analyses.
Antibodies
Rabbit polyclonal CaMKII antibodies (G-301), raised against synthetic peptide corresponding to residue 281–302 of the α subunit of rat brain CaMKII, a sequence that is highly conserved among isoforms, was generously provided by F. S. Gorelick/A. Czernig (Yale University). Affinity-purified rabbit polyclonal antibodies against human NHERF2 (Ab2570) have been described previously (27). Mouse monoclonal CaMKIIα (catalog no. sc-13141), rabbit polyclonal p-CaMKIIα (catalog no. sc-12886-R), and goat polyclonal CaMKIIδ and CaMKIIγ antibodies (catalog nos. sc-5392 and sc-1541, respectively) were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-vesicular stomatitis virus (VSVG) antibodies were derived from the P5D4 hybridoma from T. Kreiss via D. Louvard (Curie Institute, Paris, France). Monoclonal mouse antibodies to the hemagglutinin (HA) epitope were from Covance Research Products (Princeton, NJ).
Construction and Expression of NHE3 Truncation Mutants
DNA fragments of NHE3/585V, NHE3/605V, NHE3/640V, NHE3/660V, and NHE3/690V (the final number in the name of each mutant indicates the C-terminal amino acid number following the truncation) were amplified from pcDNA 3.1 containing the full-length NHE3V (pcDNA 3.1/NHE3V) by PCR to generate HindIII-XhoI fragments. Sense primer was engineered to contain the HindIII restriction site on the 5′ end of NHE3. The XhoI restriction site at the junction between NHE3 and the VSVG sequence allowed the design of antisense primers with the internal sequence at the desired sites of truncation and an XhoI site inserted at the 3′ end. Amplicons were restricted with HindIII and XhoI and electrophoresed on 1% agarose gels. The DNA fragments were ligated into the pcDNA 3.1/NHE3V expression vector and selected with G418. Plasmids expressing these C-terminal truncation mutants were verified by restriction analysis and sequencing. The NHE-deficient PS120 cells were transfected with each plasmid construct using Lipofectamine 2000 (Invitrogen). Transfected cell lines resistant to G418 (400 μg/ml) and/or to hygromycin (600 μg/ml), where indicated, were selected for measurement of Na+/H+ exchange activity by exposing cells to repetitive cycles of acid loading, as described previously (26).
Measurement of Na+/H+ Exchange Activity
Na+/H+ exchange activity in PS120/NHE3 and Caco-2BBe/HA-NHE3 cells was determined fluorometrically using the intracellular pH-sensitive fluorescent dye 2′,7′-bis(carboxyethyl)-5–6-carboxyfluorescein-AM (5 μm) as described previously (26, 28). Briefly, stably transfected PS120 cells were grown on glass coverslips up to ∼70% confluence and studied after serum starvation for 3–6 h, and polarized Caco-2BBe cells were grown to confluence on small pieces of Transwell filter membranes glued to plastic coverslips (0.4-μm pore size, Corning Glass), called “filter slips,” and polarized cells were studied ∼14 days after reaching confluence. To achieve an initial pHi of 6.0–6.2, the cells were pulsed with NH4Cl solution, containing 98 mm NaCl, 1 mm NaPO4, 25 mm dextrose, and 40 mm NH4Cl, pH 7.4, during 30 min (PS120 cells) or 60 min (Caco-2BBe cells) of dye loading. During the final 20 min of the dye loading, cells were incubated with CaMKII inhibitors, KN-62 and KN-93, or with their nonactive analogs KN-04 and KN-92, respectively. Glass coverslips and filter slips were then mounted in a cuvette and placed in the fluorometer (Photon Technology International, Lawrenceville, NJ). In polarized Caco-2BBe cells, the apical and basolateral monolayer surfaces were perfused separately, with sequential apical perfusion with TMA and Na+ solutions and basal only with TMA solution throughout. Removal of NH4Cl and perfusion with TMA+ solution (130 mm TMA chloride, 1 mm TMAPO4, 25 mm dextrose, pH 7.4) resulted in acidification of the cells. Perfusion of the cells with Na+ solution (130 mm NaCl, 1 mm NaPO4, 25 mm dextrose, pH 7.4) was used for determining Na+/H+ exchange activity. In Caco-2BBe cells, the endogenous NHE1 and NHE2 activity was inhibited by 50 μm HOE694 present in TMA and Na+ solutions. At the end of each experiment, the fluorescence ratio was calibrated to pHi, using the high potassium/nigericin method (22); the cells were equilibrated in external pH clamp media containing 20 mm HEPES, 20 mm MES, 110 mm KCl, 14 mm NaCl, 1 mm MgSO4, 1 mm CaCl2, 1 mm TMA, 25 mm glucose, and 10 mm nigericin at pH 6.1, 6.6, and 7.4.
For PS120 cells, Na+/H+ exchange activity, calculated as H+ efflux rate (in μm/s), was determined by multiplying the rate of change in intracellular pH (pHi) by the cellular buffering capacity at the corresponding pHi. Intracellular buffering capacity for the transfected cells was determined as described previously (26, 29). All measurements in control and experimental cells were made on cells from the same passage and assayed on the same day. The nonlinear regression data analysis computer program (Microcal Origin), which allows fitting of data to a general allosteric model described by the Hill equation, was used to analyze the data. Estimates were determined for Vmax and K′(H+)i by comparing means ± S.E. of the Vmax and K′(H+) of Na+/H+ exchange rates of multiple separate experiments (daily average was considered n = 1) using Student's t test or ANOVA.
For Caco-2BBe cells, Na+/H+ exchange activity for a given pHi was calculated as initial rates of sodium-dependent intracellular alkalinization (efflux of H+, in μm/s); ∼1 min of the initial rate of intracellular alkalinization was analyzed as ΔpH/ΔT. Means ±S.E. were determined from at least three experiments.
Co-immunoprecipitation Experiments
Both Caco-2BBe cells grown on 10-cm2 Transwell filters and infected as described above and PS120 cells stably expressing either VSVG or HA-tagged full-length NHE3 or NHE3 truncation mutants grown in 10-cm2 Petri dishes were collected and lysed in 500 μl of ice-cold 50 mm Tris-Cl, 150 mm NaCl, 1% Triton X-100, pH 7.4, plus protease inhibitor mixture using a 23-gauge needle. Cells were then solubilized with gentle rotation for 30 min at 4 °C, and cellular debris was cleared by centrifugation at 14,000 × g for 30 min at 4 °C. The supernatants (1–1.5 mg of total protein per ml of cell lysate) were incubated with 30 μl of monoclonal anti-HA affinity agarose beads or with polyclonal anti-CaMKII antibody in the lysis buffer for 2 h or overnight at 4 °C. Where indicated, the lysis buffer contained 1 mm EGTA, 100 μm Ca2+, or 0.1 and 3.0 μm free Ca2+ (set with 1 mm EGTA and calculated with a computer program) (30). Samples incubated with polyclonal antibody were then incubated with prewashed protein beads for either 2 h or overnight at 4 °C with gentle rotation. In the last step, beads with immunocomplexes were precipitated by brief centrifugation at 3000 rpm and then washed five times with ice-cold lysis buffer containing Ca2+ in concentrations corresponding to the immunoprecipitation conditions.
After washing, the immunoprecipitated proteins were eluted from the beads with 2× Laemmli buffer, separated by 10% SDS-PAGE, and transferred to nitrocellulose. Proteins were detected with anti-VSVG, anti-HA, or anti-CaMKII antibodies where indicated and visualized by the enhanced chemiluminescence method or by an Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE). Results were obtained from at least three individual experiments.
“Pulldown” Assays Using His6-tagged Fusion Proteins
Pulldown assays were used to identify which fragment of the NHE3 C terminus binds to CaMKII. For these purposes, we used the NHE3 C terminus divided into four different His6-tagged fragments as follows: F1 (aa 475–581), F2 (aa 582–667), F3 (aa 668–744), and F4 (aa 745–832) (31). His6-tagged fragments (3 μg each) were first linked to Ni-NTA beads and then incubated with CaMKII recombinant protein (∼2 μg) for an hour at 30 °C in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40 plus protease inhibitor mixture, in the presence of either 2 mm Ca2+ or 1 mm EGTA. Ni-NTA beads alone were used as a negative control, and the amounts of the samples loaded were identified with monoclonal anti-His6 antibody. Beads were spun down at 3000 rpm for 5 min, and the supernatant was discarded, and beads were washed three times with lysis buffer (containing either Ca2+ or EGTA). Nonbound material was washed off the beads with ice-cold lysis buffer, and subsequently the binding complexes were eluted by sample loading buffer. The samples were resolved by SDS-PAGE and analyzed by Western blot with monoclonal anti-His6 antibody or anti-CaMKII antibody.
In Vitro Back Phosphorylation of NHE3
An in vitro back phosphorylation assay was used to assess CaMKII-mediated phosphorylation of NHE3, as described previously (32). They assay was performed in two steps. In step 1, PS120/NHE3V and PS120/NHE3V/NHERF2 cells were first exposed in vivo to KN-62 or KN-93 (30 μm for 30 min) under conditions allowing in vivo phosphorylation, and then NHE3 was immunoprecipitated. In step 2, immunoprecipitated NHE3 was phosphorylated, in vitro, with recombinant CaMKII in the presence of Ca2+/CaM, Mg2+, and [γ-32P]ATP. By this method, all sites that were not occupied by endogenous phosphate residues in vivo should be available for in vitro phosphorylation by recombinant CaMKII. Briefly, cells grown in 10-cm2 Petri dishes were first incubated (where indicated) with KN-62 or KN-93 (30 μm for 30 min), and then NHE3 was lysed in 50 mm HEPES, 150 mm NaCl, 5 mm Na3EDTA, 10 mm sodium orthovanadate, 50 μm NaF, pH 7.4, and 1% Triton X-100, plus protease and phosphatase inhibitor mixtures. The supernatants from cell lysate were incubated with protein A-Sepharose beads for 2 h at 4 °C with gentle rotation, and then the beads were precipitated by brief centrifugation at 2400 rpm and washed three times with ice-cold lysis buffer without Triton X-100. In the second step, immunoprecipitated NHE3 was first washed three times with CaMKII buffer containing 20 mm Tris, 0.5 mm DTT, 0.1 mm Na2EDTA, 2.0 mm CaCl2, pH 7.4, and then immunoprecipitated NHE3 was phosphorylated with recombinant CaMKII (1.2 μg/ml) for 20 min at 30 °C in 10 mm MgCl2, 100 μm ATP, 10 μCi of [γ-32P]ATP, 1.2 μm CaM, 2 mm CaCl2. The reaction was terminated by boiling in Laemmli buffer and separated on 10% SDS-polyacrylamide gels. The proteins were transferred to nitrocellulose membranes, and the phosphoproteins labeled during in vitro phosphorylation were visualized by autoradiography. After autoradiography, Western immunoblotting using polyclonal anti-VSVG antibodies was performed to assess the amount of immunoprecipitated NHE3V and sample loading of the gels.
Dephosphorylation of CaMKII with Alkaline Phosphatase
Calf intestinal alkaline phosphatase was used to release phosphate groups from phosphorylated CaMKII in Caco-2BBe cells. Cells were lysed in 100 mm NaCl, 50 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol, pH 7.9, with 1% Triton X-100 and incubated (100 μg of total lysate protein) with 1.0 unit of intestinal alkaline phosphatase/μg of protein. The mixture was incubated for 60 min at 37 °C. Then the reaction was terminated by boiling in Laemmli buffer, and the proteins were separated on 10% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. CaMKIIγ bands were visualized by anti-CaMKIIγ antibody.
Immunoblot Analyses
The immunoprecipitates solubilized in 50 μl of the sample buffer were resolved on 10% SDS-PAGE and transferred to nitrocellulose membranes. Unbound sites were blocked with 5% (w/v) fat free milk powder in PBS (10 mm potassium phosphate buffer, 0.15 m NaCl, pH 7.4) for 1 h at room temperature. The blots were probed with primary antibodies (where indicated: polyclonal anti-CaMKIIγ (1:200); monoclonal anti-VSVG (1:100); anti-HA (1:1000); and anti-His (1:5000)) in 5% milk/PBST (PBS with 0.1% Tween 20) at room temperature for 1 h and then washed three times for 10 min each with PBST. Where indicated, IRDye 800- or 680-conjugated anti-mouse or anti-rabbit IgG (1:15000; Rockland Immunochemicals, Gilbertsville, PA) or horseradish peroxidase (HRP)-conjugated IgG (2 μg/ml) were used as secondary antibodies for 1 h followed by three washes. Protein bands were detected by using either the Odyssey infrared system at 700 and 800 nm wavelengths (LI-COR, Lincoln, NE) or standard ECL. Signal intensities from each Western blot were quantified by either the LI-COR/Odyssey Image Studio software or for ECL by scanning the developed films, followed by analysis with MetaMorph software.
Cell Surface Biotinylation and Immunoblotting
Caco-2BBe cells transiently infected with triple HA-tagged rabbit NHE3 as described above were grown on 10-cm Transwell filters until 14 days post-confluence. The cells were first serum-starved for ∼4 h and then exposed in vivo to CamKII inhibitors/controls for 20 min. All subsequent manipulations were performed at 4 °C as described previously (33). Briefly, cells were first rinsed three times with ice-cold PBS and once in borate buffer (154 mm NaCl, 10 mm boric acid, 7.2 mm KCl, and 1.8 mm CaCl2, pH 9.0). For surface labeling of NHE3, cells were incubated with 0.5 mg/ml NHS-SS biotin (Pierce) in ice-cold borate buffer by gently shaking for 20 min and repeated once. The cells were then washed with the quenching buffer (20 mm Tris and 120 mm NaCl, pH 7.4) to scavenge the unreacted biotin and subsequently washed twice with PBS. Cells were scraped at 4 °C and solubilized with the lysis buffer (60 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm KCl, 5 mm EDTA trisodium, 3 mm EGTA, 1 mm Na3VO4, and 1% Triton X-100), sonicated, and then centrifuged to remove insoluble cellular debris. Lysates, containing biotinylated surface proteins, were incubated with streptavidin-agarose beads for 16 h (overnight) to determine the percentage of cell surface NHE3. After precipitation of avidin-agarose beads, the supernatant was retained as the intracellular fraction, and the beads were washed five times with the lysis buffer. Biotinylated proteins bound to the beads were eluted by Laemmli sample buffer, and bound proteins were resolved by SDS-PAGE (10% gel) and transferred to nitrocellulose membranes. After blocking, the blots were probed with monoclonal anti-HA polyclonal antibodies and anti-β-actin antibodies. Western analysis and the quantification of the total and surface NHE3 fractions and total actin were performed using the Odyssey system (LI-COR). Multiple volumes for each total, surface, and intracellular sample were used with linear regression of intensity of signal to obtain a single value for each sample. Percentage of surface NHE3 was calculated from the ratio ((surface NHE3 signal/total NHE3 signal/total actin) × dilution factor of surface and total NHE3 samples) of the cells treated with KN-93 or KN-92 normalized to the ratio of control cells from three separate experiments.
Mass Spectrometry Analyses
HA-NHE3 was first immunoprecipitated from a 6-mg lysate of PS120/NHERF2/HA-NHE3 fibroblasts prepared in lysis buffer (50 mm Tris, 150 mm NaCl, 10 mm EDTA, 1% Triton X-100, 0.1% SDS, 20 mm N-ethylmaleimide (Sigma), pH 7.4) and incubated with 100 μl of monoclonal anti-HA affinity matrix. Cell lysates incubated with anti-VSVG antibody and immobilized to agarose were used as a negative control. Immunoprecipitated NHE3 was resolved by 12% SDS-PAGE, and then protein bands were visualized by antigen staining. The bands present in the anti-HA affinity matrix but not in the negative control were cut out from the gel and trypsinized, and proteins were identified by liquid chromatography-Tandem mass spectrometry (LC-MS/MS) in the Mass Spectrometry Core of The Johns Hopkins University School of Medicine, as described previously (19). Briefly, samples were destained, in gel digested, extracted, and run on an LTQ ion trap mass spectrometer. The data were analyzed with the search program Mascot (Matrix Science, London, UK) to search the NCBI database with a wide open species designation (mammals and all species).
RESULTS
CaMKII Inhibits NHE3 under Basal Conditions by an NHERF2-dependent Process
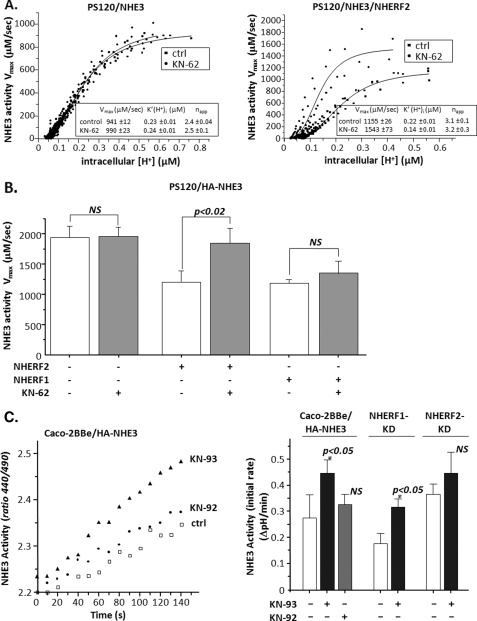
We previously demonstrated that CaMKII inhibits ileal neutral NaCl absorption and its constituent BB Na+/H+ exchanger under basal conditions (21, 34). In examining the mechanism for this effect, we reported that pretreatment of PS120 cells stably expressing NHE3 with the CaMKII inhibitor KN-62 increased Na+/H+ exchange activity (22). However, in subsequent studies, these results were variable and seemed to vary from clone to clone of PS120/NHE3 cells. Given the recent appreciation that many aspects of NHE3 regulation depend on the presence of members of the NHERF family of multi-PDZ domain containing regulatory proteins, CaMKII inhibitors were studied in cells stably expressing NHERF1 or NHERF2. PS120 cells express small amounts of NHERF1 and do not express NHERF2 (5, 13). Therefore, to determine whether NHERF1 or -2 was involved in the KN-62-dependent stimulation of NHE3 activity, clones of PS120/NHE3 were stably co-transfected with NHERF2 or NHERF1.
As shown in single experiments in Fig. 1A and in a series of experiments in Fig. 1B, NHERF2 reconstituted KN-62 stimulation of NHE3 in PS120 cells. KN-62 caused ∼45% (p < 0.05) higher Vmax values compared with untreated control cells and increased the K′(H+)i (p < 0.05) without affecting the napp (Hill coefficient). In contrast, NHERF1 expression did not lead to a reproducible KN-62 effect. Pretreatment of the cells with the KN-62 control, KN-04 (30 μm), did not affect NHE3 activity (data not shown).
FIGURE 1.
CaMKII inhibitors KN-62 and KN-93 increase NHE3 activity by a mechanism requiring NHERF2. A, representative experiment of the effect of KN-62 on NHE3 activity in PS120 cells with or without stable expression of NHERF2. Left panel, in PS120/NHE3 cells, KN-62 had no effect on Na+/H+ exchange compared with untreated control cells. Right panel, in PS120/NHE3 cells stably expressing NHERF2, KN-62 increased Na+/H+ exchange activity Vmax by ∼35% versus untreated controls with a simultaneous decrease in K′(H+)i. Kinetic parameters from the single experiment are shown in the box, with S.E. indicating the variance among the multiple coverslips studied (3–4 per condition). B, KN-62 stimulation of NHE3 in PS120 cells is NHERF2-dependent. Cells were acidified by pulsing with NH4Cl (with or without KN-62) followed by perfusion with TMA media. Cells were then allowed to recover to steady state pHi in Na+ medium. H+ efflux rates equivalent to Na+/H+ exchange were plotted against intracellular [H+], and Na+/H+ exchange activity was determined. Results shown are mean ± S.E.; n = at least 3. p values in comparison with untreated control (paired t test). NS, not significant. C, KN-93 stimulates NHE3 in Caco-2BBe cells by an NHERF2-dependent but not NHERF1-dependent process. Na+/H+ exchange activity was measured in wild type Caco-2BBe/HA-NHE3 or with NHERF2 or NHERF1 stably knocked down via lentivirus shRNA ∼14 days after reaching confluency. Cells were exposed to 50 mm NH4Cl either alone or with KN-93 or KN-92 during the final 20 min of a 50-min dye-loading period. Endogenous NHE1 and NHE2 activity were inhibited by 50 μm HOE694. In the wild type cells, KN-93 treatment significantly increased activity of NHE3, although KN-92 had no effect. In cells with NHERF1 knocked down, KN-93 treatment significantly increased activity of NHE3. However, KN-93 did not increase NHE3 activity in the Caco-2-BBe cells with NHERF2 knocked down. Left panel shows results from a single experiment. Right panel shows mean ± S.E. of n ≥ 3, with p values in comparison with basal NHE3 activity in each untreated control (paired t test).
Similar studies were performed using Caco-2BBe cells adenovirally infected with HA-rNHE3. In Fig. 1C, left panel, a single example is shown in which 30 μm KN-93 (but not KN-92, a control) stimulates basal NHE3 activity, which is consistent with the results in PS120 fibroblasts. These cells endogenously express NHERF1 and NHERF2; thus, the roles of NHERF1 and -2 were determined using shRNA. We have previously reported the use of shRNA to knock down NHERF1 and NHERF2 separately in these Caco-2BBe cells, using lentivirus-shRNA for NHERF1 and NHERF2 compared with shRNA for GFP (does not occur endogenously in Caco-2-BBe cells) as the negative control (35). As shown in Fig. 1C, right panel, initial rates of NHE3 activity in Caco-2BBe cells, as reported previously (35), were affected by KD NHERF1 (inhibits basal NHE3 activity) and NHERF2 (increases basal NHE3 activity). Moreover, KN-93 stimulation of NHE3 activity occurred similarly to wild type in NHERF1 KD, whereas this KN-93 effect was abolished in the NHERF2 KD (Fig. 1C, right panel). Note that KN-92 did not affect basal NHE3 activity in wild type Caco-2BBe cells. Taken together, these results suggest that CaMKII inhibits NHE3 under basal conditions in PS120/NHERF2/NHE3 and Caco-2BBe/HA-NHE3 cells and that NHERF2 but not NHERF1 is necessary for this CaMKII inhibition.
CaMKII Inhibitor KN-62 Has No Additive Effect with the EGF Stimulation of NHE3
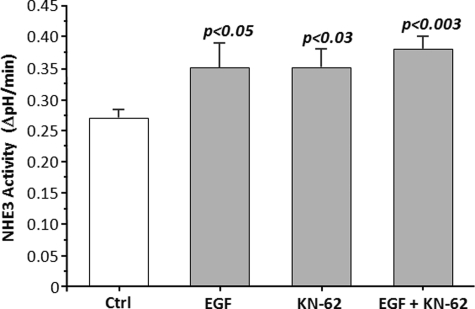
We next determined whether the stimulatory pathway unmasked by CaMKII inhibition overlapped with other NHE3 stimulatory processes. EGF stimulates NHE3 via basolateral membrane EGF receptors (36) and via apical EGF receptor transactivation (37). Thus, Caco-2BBe/HA-NHE3 cells had NHE3 activity determined in the presence/absence of 30 μm KN-62, and the effects compared with a maximally effective concentration of EGF added both apically and basolaterally with determination of whether both stimulatory processes were additive. As shown in Fig. 2, EGF and KN-62 both stimulated NHE3 activity in Caco-2BBe/HA-NHE3 cells, with effects that were similar in magnitude. These effects were not additive. This suggests the involvement of at least some overlap in the signaling pathways used in NHE3 regulation by CaMKII and EGF.
FIGURE 2.
KN-62 (30 μm) and EGF (100 ng/ml) both acutely stimulate NHE3 activity in Caco-2BBe/HA-NHE3 cells in a nonadditive manner. Caco-2BBe cells expressing HA-NHE3 were exposed to KN-62 and EGF for 30 min, and the apical NHE3 activity was determined (initial rates). Both stimulated NHE3 activity similarly with the effects not being additive. Results are from 3 to 4 experiments, with p values representing comparison to untreated controls (Ctrl) (paired t tests).
CaMKII Does Not Alter Cell Surface Expression of NHE3
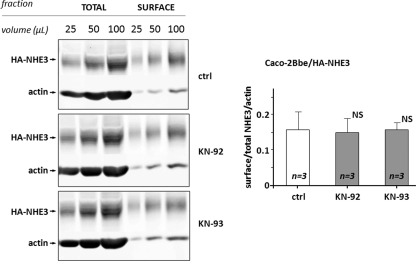
NHE3 activity is regulated by mechanisms involving either changes in trafficking and/or changes in turnover number. Regulation by changes in trafficking alters the percent of total NHE3 present on the cell surface, which can be determined by cell surface biotinylation studies. Therefore, to determine the mechanism by which CaMKII inhibits NHE3 activity under basal conditions, the amount of the total NHE3 and plasma membrane-expressed NHE3 was determined in polarized Caco-2-BBe/HA-NHE3 exposed in vivo to the CaMKII inhibitor KN-93 or to its inactive analog KN-92 with biotin added apically. Western immunoblotting of the total amount of NHE3 and apical surface-expressed NHE3 protein was used for quantification and comparison between control cells and cells exposed to KN-93 or KN-92. A Western blot of total and surface-biotinylated NHE3 in a single experiment is shown in Fig. 3, left panel, showing that the ratios between total NHE3/total actin and surface-expressed NHE3 signal were similar in control cells and cells treated with KN-93 or KN-92. Quantification and analysis of the data from three independent experiments (Fig. 3, right) showed that neither KN-93 nor KN-92 altered the percentage of NHE3 expressed on the plasma membrane. Similar results were obtained when cell surface biotinylation studies were performed in PS120/NHERF2/NHE3V (VSV-G epitope-tagged cells) (data not shown). Thus, these results indicate that CaMKII regulation of basal NHE3 activity is via changes in turnover number and not by changes in trafficking.
FIGURE 3.
CaMKII inhibitor KN-93 does not alter surface expression of NHE3 in Caco-2BBe/HA-NHE3 cells. The amounts of the total NHE3 with actin as a loading control and apical membrane-expressed NHE3 were determined in polarized Caco-2BBe/HA-NHE3 cells exposed in vivo to CaMKII inhibitor KN-93 (30 μm), its inactive analog KN-92 (30 μm) for 20 min, or untreated control. NHS-SS-biotin was added only apically. Left panel, a single example of Western blot of three dilutions of total and avidin-precipitated (surface-biotinylated) NHE3 with actin staining on the total sample used as a loading control for control; KN-92- and KN-93-exposed cells demonstrate no difference in ratio between total and surface-expressed NHE3 signal. Right panel, quantification of the total amount of NHE3/actin and apical surface NHE3 protein performed using Western immunoblotting from three experiments such as shown in the left panel. Results are presented as mean ± S.E. of the ratio of surface/total NHE3/actin. Neither KN-93 nor KN-92 altered the percent of NHE3 expressed on the apical plasma membrane. p values in comparison with untreated control (Ctrl) (paired t test) are shown. NS, not significant.
CaMKII Increases Total NHE3 Phosphorylation under Basal Conditions, and NHERF2 Is Not Required for This Effect
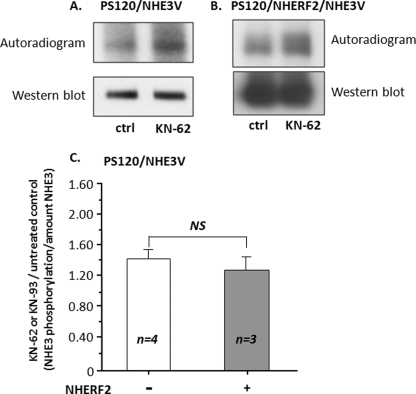
Another mechanism that is likely to be involved in NHE3 regulation by CaMKII is phosphorylation, especially because NHE3 contains multiple CaMKII consensus sites in its C terminus. Therefore, whether CaMKII caused phosphorylation of NHE3 under basal conditions was examined by studying the effect of the CaMKII inhibitors KN-62 and KN-93 on NHE3 phosphorylation. Evidence that CaMKII increased NHE3 phosphorylation under basal conditions was provided by an in vitro back phosphorylation assay. NHE3 was studied in control conditions and after the PS120 cells were exposed in vivo to KN-62 or KN-93 and before in vitro phosphorylation with recombinant CaMKII in the presence of Ca2+/CaM, Mg2+, and [γ-32P]ATP. In step 1, PS120/NHE3 (Fig. 4A) and PS120/NHE3V/NHERF2 (Fig. 4B) cells were first exposed in vivo to KN-62 or KN-93 under conditions allowing in vivo phosphorylation, and then NHE3 was immunoprecipitated. In step 2, immunoprecipitated NHE3 was phosphorylated in vitro with recombinant CaMKII in the presence of Ca2+/CaM, Mg2+, and [γ-32P]ATP. In this technique, all sites on NHE3 that are not occupied by endogenous phosphate residues during the in vivo treatment with CaMKII inhibitor should have been available for γ-32P labeling of phosphate residues during the in vitro phosphorylation by recombinant CaMKII. By this method, an increase in in vitro phosphorylation indicates a decrease in vivo phosphorylation. Shown in Fig. 4A (upper panel), in vitro phosphorylation of immunoprecipitated NHE3 was increased in PS120/NHE3V cells treated by KN-62 or KN-93, and this increase in phosphorylation was not attributed to changes in the amount of NHE3 as shown by Western blotting (lower panel). Because NHERF2 is necessary for CaMKII inhibition of basal NHE3 activity, we examined whether NHERF2 played a role in CaMKII-mediated phosphorylation of NHE3 under basal conditions. As shown in Fig. 4B (upper panel), in vitro phosphorylation of immunoprecipitated NHE3 was increased in the cells treated by KN-62 or KN-93 during in vivo treatment of PS120/NHERF2/NHE3V cells. Moreover, the magnitude of the increase in NHE3 phosphorylation caused by KN-62 or KN-93 was similar in the presence and absence of NHERF2 (Fig. 4C). These data indicate that CaMKII phosphorylates NHE3 under basal conditions and the presence of NHERF2 does not alter the extent of total NHE3 phosphorylation.
FIGURE 4.
CaMKII phosphorylates NHE3 under basal conditions, and NHERF2 does not alter the magnitude of this phosphorylation. A back phosphorylation assay was used to assess the in vivo change of NHE3 phosphorylation level after treatment with KN-62 or KN-93. This approach is based on the fact that all potential CaMKII phosphorylation sites on NHE3 that are not occupied by endogenous phosphate residues during the in vivo treatment should have been available to γ-32P during the in vitro phosphorylation. Therefore, by this method, increased in vitro phosphorylation indicates less in vivo phosphorylation. A, PS130/NHE3V. B, PS130/NHE3V cells stably expressing NHERF2 were studied in the absence of KN-62 or KN-93 (control (Ctrl)) or following 20 min of in vivo treatment with 30 μm KN-62 or KN-93. Immunoprecipitated NHE3 was then phosphorylated in vitro by recombinant CaMKII in the presence of Ca2+/CaM, Mg2+, and [γ-32P]ATP. Representative autoradiographs (upper panel) and Western immunoblots (lower panel) of four (no NHERF2) or three (NHERF2 containing) similar experiments are shown. C, quantitative analysis of KN-62 or KN-93 effects on NHE3 back phosphorylation in the presence or absence of stable expression of NHERF2 in PS120 cells. Quantitation was by MetaMorph of autoradiograph/immunoblots of NHE3. Results for KN-62- or KN-93-treated cells were normalized to untreated controls in each experiment. These results were similar in KN-62 or KN-93 treatments, and these results were combined. Results are mean ± S.E. of 3–4 experiments. NS, not significant.
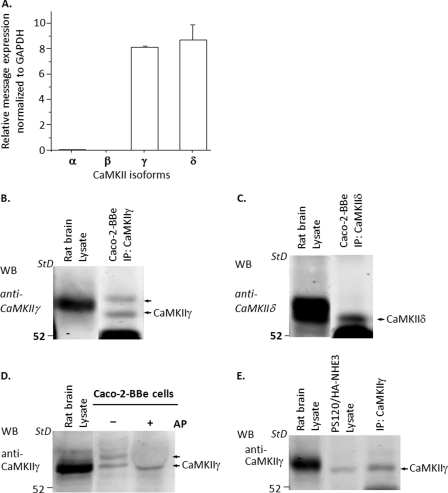
Identification of CaMKII Isoforms in Caco-2BBe and PS120 Cells
There are four isoforms of CaMKII with the γ and δ isoforms ubiquitously expressed, although α and β isoforms are mostly expressed in the neural system (38). To identify which isoforms of CaMKII were expressed in Caco-2BBe cells, we first used quantitative RT-PCR. Shown in Fig. 5A, quantitative RT-PCR revealed expression of only γ and δ isoforms of CaMKII, and these were expressed in similar amounts. This expression was confirmed using immunoprecipitation and Western blotting with antibodies directed against γ and δ isoforms. Both CaMKII γ and δ proteins were expressed in Caco-2BBe cells (Fig. 5, B and C). In addition, immunoprecipitation and Western blotting with anti-CaMKIIα antibody did not identify the corresponding CaMKII protein in Caco-2BBe cells (data not shown). Using brain as a standard, the γ isoform present appeared to be the 54-kDa splice variant (39). As is shown in Fig. 5, B and D, in Caco-2BBe cells, CaMKIIγ was present as two bands. These bands appear to be different states of phosphorylation of CaMKIIγ, as determined by exposure to alkaline phosphatase. After exposing the Caco-2BBe cell lysates to alkaline phosphatase, the upper band was no longer detected, although the large amount of alkaline phosphatase depressed all bands in this area of the gel (Fig. 5D).
FIGURE 5.
Expression of CaMKII isoforms γ and δ in Caco-2BBe cells and PS120 cells. A, Caco-2BBe cells. Quantitative RT-PCR revealed that messages for only two CaMKII isoforms, CaMKIIγ and CaMKIIδ, were expressed in the Caco-2BBe cells, and they were expressed in similar quantity. B and C, an immunoprecipitation (IP)/Western blot (WB) approach was used with specific anti-CaMKIIγ and anti-CaMKIIδ antibodies to confirm the presence of these two CaMKII isoforms. B, CaMKIIγ (n = 7); C, CaMKIIδ (n = 2) in Caco-2BBe cells. Rat brain was used as a positive control. D, Western blotting of Caco-2BBe cells with CaMKIIγ antibodies reveals two bands. The upper band disappeared after the Caco-2 cell lysate was treated with alkaline phosphatase at 37 °C for 60 min, but there was no change in the lower band (n = 2). E, immunoprecipitation/Western blot was used to identify isoforms of CaMKII proteins expressed in PS120 cells. Only CaMKIIγ was present (n = 5). Specific CaMKIIα and CaMKIIδ antibodies failed to identify these isoforms (data not shown).
Immunoprecipitation/Western blotting was also used to identify which isoforms of CaMKII were expressed in PS120 cells. As shown in Fig. 5E, only CaMKIIγ was expressed; negative immunoblots studies for CaMKIIα and CaMKIIδ are not shown.
Identification of CaMKIIγ as an NHE3 Co-precipitating Protein in PS120/NHERF2/HA-NHE3 Cells
To further understand how CaMKII affects basal NHE3 activity, NHE3 was immunoprecipitated from PS120/NHERF2/HA-NHE3 cell lysates. Immunoprecipitation with anti-HA affinity matrix was compared with anti-VSVG affinity matrix as a negative control, followed by separation of the immunoprecipitates on 12% SDS-PAGE and then antigen staining. The bands immunoprecipitated by the anti-HA affinity matrix but not the negative control were cut from the gel and trypsinized, and proteins were identified by LC-MS/MS (data not shown). One protein identified was CaMKIIγ (95% certainty) based on identification of a single peptide (FTDDYQLFEELGK). Blast search failed to identify any other proteins with an identical sequence to this peptide.
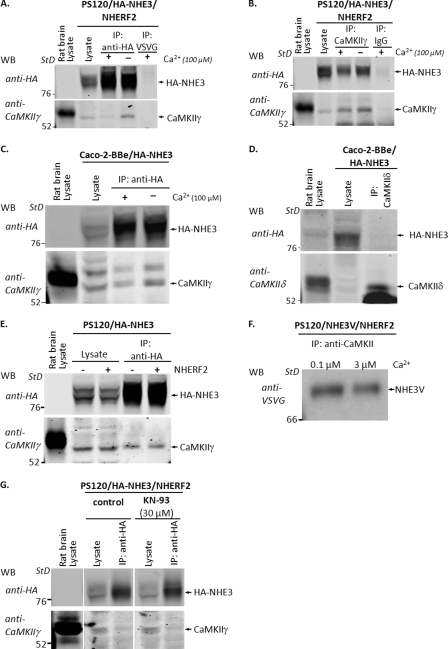
CaMKII Associates with NHE3 in Vivo by a Ca2+-inhibitable Process
CaMKII binds to some of its target proteins in addition to phosphorylating them (40, 41). To understand more about CaMKII association with NHE3 under basal conditions, NHE3 and CaMKII were co-immunoprecipitated from PS120/NHERF2/HA-NHE3 cells (Fig. 6), using either anti-HA antibodies (Fig. 6A) or anti-CaMKII antibodies (Fig. 6B). As negative controls, the cell lysate was incubated with anti-VSVG or IgG. As shown in Fig. 6A, CaMKIIγ was precipitated by anti-HA antibody but not by anti-VSVG antibody. The association between NHE3 and CaMKIIγ was confirmed by the reciprocal co-precipitation (Fig. 6B) in which immunoprecipitated CaMKIIγ co-precipitated HA-NHE3. IgG did not co-precipitate HA-NHE3. A similar association between NHE3 and CaMKIIγ was demonstrated in Caco-2BBe/HA-NHE3 cells. Co-immunoprecipitation with anti-HA antibodies showed that NHE3 co-precipitated CaMKII (Fig. 6C). In contrast, CaMKIIδ did not co-precipitate NHE3 in Caco-2BBe cells (Fig. 6D).
FIGURE 6.
CaMKII associates with NHE3 in vitro and in vivo by a Ca2+-inhibitable process. A, PS120/NHERF2/HA-NHE3 cells lysates were immunoprecipitated (IP) with anti-HA affinity beads in the presence (100 μm) or absence (1 mm EGTA) of Ca2+ and Western blotted (WB) with anti-CaMKIIγ antibodies. Immunoprecipitation with anti-VSVG antibodies was used as a negative control. Immunoprecipitated HA-NHE3 co-precipitated CaMKIIγ in the presence and absence of Ca2+; however, the co-precipitation decreased with elevation of Ca2+. Rat brain lysate was used as a positive control for CaMKIIγ. B, reciprocal experiment in which the immunoprecipitated CaMKII-co-precipitated HA-NHE3 confirmed the association between NHE3 and CaMKIIγ in the presence and absence of Ca2+. The co-precipitation also decreased with elevation of Ca2+. C, Caco-2BBe/HA-NHE3 cells lysates were immunoprecipitated with anti-HA affinity beads and then Western blotted with anti-CaMKIIγ antibodies. Immunoprecipitated HA-NHE3 co-precipitated CaMKIIγ in the presence (100 μm) or absence (1 mm EGTA) of Ca2+, but the association between CaMKIIγ and NHE3 decreased with elevation of Ca2+. Note, blotting with anti-CaMKIIγ antibodies revealed two bands with the upper band representing the phosphorylated form of CaMKIIδ. D, Caco-2BBe/HA-NHE3 cells lysates were immunoprecipitated with anti-CaMKIIδ and then Western-blotted with anti-HA antibodies. Immunoprecipitated CaMKIIγ did not co-precipitate HA-NHE3. Rat brain lysate was used as a positive control for CaMKIIδ. E, PS120/HA-NHE3 (2nd and 4th lanes) and PS120/NHE3/NHERF2 (3rd and 5th lanes) cell lysates were immunoprecipitated with anti-HA affinity beads and then Western-blotted with anti-CaMKIIγ antibodies. HA-NHE3 co-precipitated CaMKIIγ in the presence and absence of NHERF2, indicating that NHERF2 is not required for the binding of CaMKIIγ with NHE3. F, PS120/NHERF2/NHE3V cell lysates were immunoprecipitated with anti-CaMKIIγ antibody in the presence of 0.1 and 3.0 μm free Ca2+ (set with EGTA), and co-precipitated NHE3V was visualized by Western blotting with monoclonal anti-VSVG antibodies. Co-precipitation occurred in the presence of both 0.1 and 3.0 μm free Ca2+ but was decreased with 3 μm Ca2+. G, PS120/NHERF2/HA-NHE3 cells were pretreated with 30 μm KN-93 for 30 min, and cell lysates were incubated with anti-HA affinity beads, and co-immunoprecipitated CaMKIIγ was detected by immunoblotting using anti-CaMKIIγ antibodies. KN-93 did not affect the co-immunoprecipitation between CaMKII and NHE3, compared with untreated controls. All experiments were at least n = 3.
The relationship between NHE3 and CaMKII was further examined, asking whether NHERF2 affected their co-precipitation. HA-NHE3 was immunoprecipitated from total lysates of PS120/HA-NHE3 and PS120/NHERF2/HA-NHE3 cells, respectively. As shown in Fig. 6E, CaMKIIγ was co-immunoprecipitated equally with HA-NHE3 in the absence or presence of NHERF2, indicating that NHERF2 is not required for the CaMKIIγ binding with NHE3.
Because the increase of intracellular Ca2+ is known to induce binding of CaMKII to some of its binding partners (42), we examined whether Ca2+ affects the association between NHE3 and CaMKII. For these purposes we initially used co-immunoprecipitation assays with cell lysate in the presence (100 μm) and absence (1 mm EGTA) of Ca2+, with all studies performed by changing Ca2+ concentration in vitro. Fig. 6, A and B, shows that CaMKII and NHE3 co-precipitated in the presence and absence of Ca2+, but the association between CaMKIIγ and NHE3 decreased with elevation of Ca2+. Similar in vitro studies in Caco-2BBe/HA-NHE3 cells (Fig. 6C) demonstrated that increased Ca2+ decreased CaMKIIγ binding with NHE3.
To determine the effect of physiologic levels of [Ca2+] on CaMKIIγ binding to NHE3 in vivo, immunoprecipitation of PS120/NHE3V cell lysates by anti-CaMKII antibody was performed in the presence of either 0.1 or 3.0 μm free [Ca2+] (set with EGTA (30)). As shown in Fig. 6F, CaMKII co-precipitated NHE3V in the presence of both 0.1 and 3.0 μm free [Ca2+], but this co-precipitation decreased with 3.0 μm [Ca2+]. These results indicate that in vivo changes of intracellular [Ca2+] are likely to be involved in CaMKII association with NHE3, with increases of intracellular [Ca2+] causing decreased CaMKII binding to NHE3.
The effect of CaMKII inhibitor KN-93 on CaMKII binding with NHE3 was examined to determine whether the increase in NHE3 activity after exposing cells to KN-93 is associated with a change in the CaMKII association with NHE3. For these purposes PS120/NHERF2/HA-NHE3 cells were incubated with and without KN-93 for 20 min prior to co-immunoprecipitation with anti-HA affinity beads. As shown in Fig. 6G, KN-93 did not affect the co-immunoprecipitation between CaMKII and NHE3, suggesting that CaMKII activity was not necessary for CaMKII binding to NHE3.
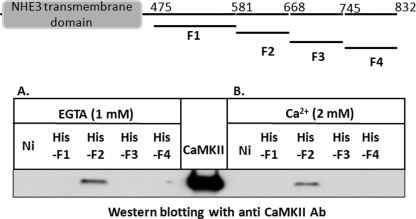
CaMKII Directly Binds NHE3
CaMKII pulldown assays were used to identify the following: (i) whether CaMKII interacts with the NHE3 C terminus directly or through another protein and (ii) whether the binding sites for CaMKII were within the NHE3 C terminus. The NHE3 C-terminal region was divided into four contiguous fragments and produced as His6-tagged fusion proteins as follows: His-F1 (aa 475–581), His-F2 (aa 582–667), His-F3 (aa 668–744), and His-F4 (aa 745–832) (Fig. 7). These fusion proteins (∼3 μg each) were attached to Ni-NTA beads and incubated with recombinant CaMKII protein (∼2 μg) in the absence of Ca2+ (1 mm EGTA) or presence of 2 mm Ca2+. All pulldown assays were done in the absence of CaM and ATP. Ni-NTA beads alone or incubated with CaMKII recombinant protein without the NHE3 fusion proteins were used as negative controls. Sample loading was examined with monoclonal anti-His antibodies (data not shown). Fig. 7 shows a representative Western blot probed with anti-CaMKII antibody. Recombinant CaMKII was bound to His6-F2 fusion protein but not to the His6-F1, -F3, and -F4. This binding to the F2 domain of NHE3 occurred both in the presence (Fig. 7A) and absence (Fig. 7B) of Ca2+, although there was less binding in the presence of Ca2+. This binding was specific because recombinant CaMKII was not pulled down using Ni-NTA beads alone either in the presence or absence of Ca2+. These results indicate that CaMKII binds to NHE3 directly, involving NHE3 aa 582–667, without involvement of another protein. In addition, this direct binding of CaMKII to the NHE3 C terminus does not require the presence of Ca2+, calmodulin, or ATP.
FIGURE 7.
CaMKII directly binds to NHE3 C terminus. Schematic showing four His6-tagged NHE3 C-terminal fusion proteins, His-F1 (aa 475–581), His-F2 (aa 582–667), His-F3 (aa 668–744), and His-F4 (aa 745–832), is at top of figure. Pulldown of CaMKII recombinant protein with these four His6-tagged fusion proteins of the NHE3 C terminus (∼3 μg each) bound to Ni-NTA beads was performed in the absence of Ca2+ (1 mm EGTA) (A) or in the presence 2 mm Ca2+ (B). Western blotting with polyclonal anti-CaMKII antibody visualized CaMKII binding only to His6-F2 fusion protein. Probing of the blots with anti-His antibodies indicated that comparable amounts of the Ni-NTA beads were present in all samples (data not shown). Note reduction of binding with Ca2+. CaMKII loading control is in the middle. Experiment shown was performed twice with similar results.
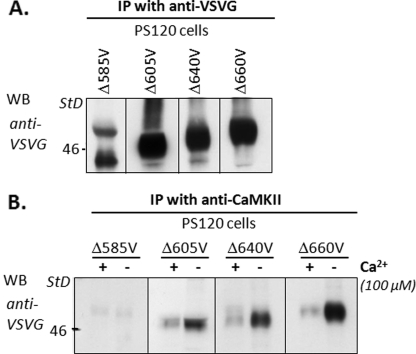
Further Localization of the CaMKIIγ Binding Domain within the NHE3 C Terminus
To further localize the site of the CaMKIIγ binding domain within the NHE3 C terminus, cell lysates of PS120 cells expressing NHE3 C-terminal truncation mutants epitope-tagged with VSVG were immunoprecipitated by anti-VSVG antibodies (Fig. 8A) or co-precipitated with anti-CaMKIIγ antibody (Fig. 8B). The NHE3 C-terminal mutants were truncated at aa 585, 605, 640, 660, and 690 (NHE3/Δ585V, NHE3/Δ605V, NHE3/Δ640V, NHE3/Δ660V, and NHE3/Δ690V, respectively). As shown in Fig. 8B (for all except NHE3/Δ690V), CaMKII co-immunoprecipitated full-length NHE3 and all NHE3 C-terminal truncation mutants except NHE/585V. These co-precipitations studies were done in the presence of 100 μm Ca2+ or EGTA. There was less binding when Ca2+ was elevated for wild type NHE3 and each truncation. These data demonstrate that CaMKIIγ binding to the NHE3 C terminus requires NHE3 aa 586–605.
FIGURE 8.
CaMKII binds in vivo to NHE3 between aa 586 and 605. A, immunoprecipitation (IP) of a series of NHE3 C-terminal truncation mutants truncated at aa 585, 605, 640, 660, and 690 (NHE3/Δ585V, NHE3/Δ605V, NHE3/Δ640V, NHE3/Δ660V, and NHE3/Δ690V, respectively) were identified with anti-VSVG polyclonal antibodies (data not shown for Δ690). B, cell lysates with NHE3 C-terminal truncation mutants were immunoprecipitated with anti-CaMKIIγ antibody in the presence (100 μm) and absence (1 mm EGTA) of Ca2+, and coprecipitated NHE3 truncation mutants were visualized with anti-VSVG antibodies. CaMKII co-immunoprecipitated all tested NHE3 C-terminal truncation mutants except NHE3/Δ585V. NHE3 mutants were all co-precipitated in the presence and absence of Ca2+ with less binding when Ca2+ was elevated.
CaMKII Requires a Domain C-terminal to Its Binding Site to Regulate NHE3 Activity
Having determined that CaMKII binds to the NHE3 C terminus using aa 586–605, we studied NHE3 truncation mutations to determine whether the part of NHE3 required for CaMKII regulation of basal NHE3 activity was the same as that required for CaMKII binding. As shown in Fig. 9, unlike KN-62 stimulation of wild type NHE3 activity, KN-62 had no effect on NHE3 activity truncated to aa 690 (NHE3/Δ690V). Similar results were obtained using KN-93 (data not shown). This identified the C-terminal 143 aa of NHE3 as necessary for the CaMKII inhibition of NHE3 activity.
FIGURE 9.
CaMKII requires a domain C-terminal of its binding site to regulate NHE3 activity. PS120/NHERF2 cells stably transfected with NHE3 truncated to aa 690 (NHE3/Δ690V) were exposed to 30 μm KN-62 as in Fig. 1, and the effect on NHE3 activity was determined. KN-62 failed to affect NHE3 activity of the truncation mutant suggesting that the C-terminal 143 aa of NHE3 are required for the CaMKII inhibition of NHE3 activity. Note that basal NHE3 activity in NHE3/Δ690V cells is much faster than full-length NHE3, as reported (33). N refers to the number of separate experiments. NS, not significant.
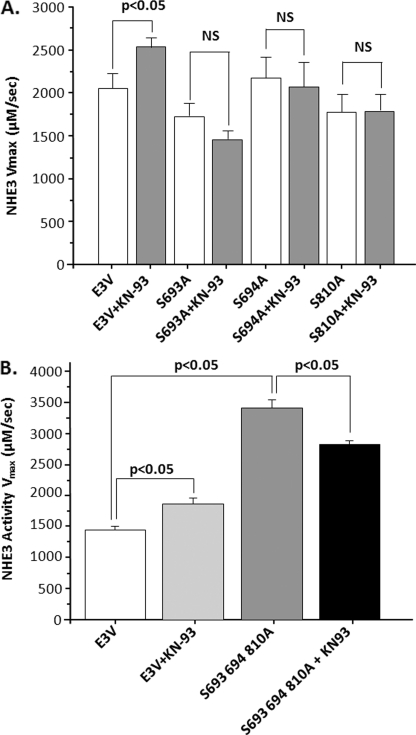
Using software from several bioinformatics/kinase consensus sequence identification programs (43, 44), three CaMKII phosphorylation consensus sequences were identified in NHE3, which are conserved across species (rabbit Ser693, Ser694, and Ser810). Each of these were mutated Ser to Ala individually and together, confirmed by sequencing, and stably expressed in PS120/NHERF2 cells. The effect of KN-93 (30 μm) on each mutant was compared with wild type NHE3. As shown in Fig 10A, mutation of each putative CaMKII consensus abolished KN-93 stimulation of NHE3. Basal activity was not altered in any of the three mutants. In contrast, as shown in Fig 10B, in which all three Ser were mutated to Ala, basal NHE3 activity was increased, and KN-93 failed to further increase basal NHE3 activity. Together, these results demonstrate that these three Ser are all needed for CaMKII to inhibit basal NHE3 activity.
FIGURE 10.
Serines in putative CaMKII phosphorylation consensus sequences C-terminal to amino acid 690 are all necessary to increase basal NHE3 activity and for KN-93 stimulation of NHE3 activity. PS120/NHERF2 cells were stably transfected separately with three NHE3 mutants, S693A, S694A, and S810A (A). These are the serines of the three putative CaMKII phosphorylation consensus sequences that are present in NHE3 C-terminal to aa 690. After multiple rounds of acid loading to increase NHE3 expression, basal NHE3 activity and activity after KN-93 (30 μm) were determined. KN-93 increased basal NHE3 activity in wild type NHE3-expressing cells studied at the same time as the three mutants but failed to increase activity in any of the three mutants. Note basal NHE3 activity in the three mutants was similar to that of wild type. B, single construct containing all three mutants, S693A, S694A, S810A. Basal NHE3 activity was significantly higher in the triple mutant than in wild type, but this mutant failed to increase NHE3 activity with KN-93. Results shown are NHE3 Vmax (mean ± S.E.) from three separate experiments. NS, not significant.
DISCUSSION
This study demonstrated that under basal conditions, CaMKII binds to the NHE3 C terminus, phosphorylates NHE3, and reduces NHE3 activity, and these effects help to set the overall basal NHE3 activity. One of the consistently identified characteristics of NHE3 is that it is active under basal conditions (45). This allows the large swings in its activity that occur as part of digestion in which it is initially inhibited and then is sequentially stimulated by release of multiple neurohumoral ligands. Being active under fasting conditions allows NHE3 to carry out its major physiologic function during digestion in the intestine in which it is inhibited in the early post-prandial state, which, via water transport coupling, helps spread the digestive enzymes over the digestive and absorptive surface of the intestine, following which there is increased NHE3 activity, which helps prevent dehydration occurring as part of digestion.
The characteristics of the effects of CaMKII in regulation of NHE3 in PS120 and Caco-2 cells demonstrated here are consistent with and extend mechanistic understanding of our previous studies of Ca2+ regulation of NHE3 in intact mammalian ileum. We previously showed that CaMKII is present in small intestinal BB where it inhibits neutral NaCl absorption under basal conditions, and studied in isolated ileal villus cell BB, it inhibits NHE3 activity by a process consistent with phosphorylation, because it was Mg2+-, ATP-, Ca2+-, and calmodulin-dependent (21, 34). The similarity of the CaMKII effects in intact ileal Na+-absorptive cells, isolated absorptive cell BB, PS120 fibroblasts, and Caco-2-BBe cells, as shown here, supports that this inhibition of basal NHE3 activity by CaMKII is intrinsic to NHE3 rather than being tissue- or cell-specific. This study provides some mechanistic understanding of how this occurs, which is by altering the NHE3 turnover number without affecting surface expression. In addition, it is important to note that this study concentrated on the regulation of NHE3 under basal conditions. In intact intestine, elevation of Ca2+ inhibits basal NaCl absorption, acting independently of CaMKII via activation of PKC (46). The role of CaMKII in additional regulation (stimulation and inhibition as part of digestion physiologically) of NHE3 remains to be defined.
Acute regulation of NHE3, as occurs in the post-prandial state, involves signaling complexes that form on the NHE3 C terminus. We have estimated that at any time up to 20 proteins of an average size of 50 kDa associate with NHE3, only some of which have been identified (14). The NHE3 C-terminal domain, which is necessary for CaMKII binding is aa 586–605, which is predicted by multiple modeling programs to be α-helical. Please note that we do not know whether there are additional sequences in the NHE3 C terminus or in its N-terminal transport domain that are involved in the binding of CaMKII to NHE3. However, other proteins that associate with NHE3 also require this NHE3 domain, including NHERF1–4, phospholipase Cγ, and CK2α (10, 19). Based on point mutations of this part of NHE3, this signaling complex is necessary for part of basal NHE3 activity (the phosphatidylinositol 3-kinase-dependent component) and is necessary for all cAMP, cGMP, and elevated Ca2+ inhibition of NHE3 activity.4 CaMKII appears to bind to NHE3 at one site (586–605) but requires a domain C-terminal to aa 690 to inhibit basal NHE3 activity. This part of NHE3 contains three putative CaMKII phosphorylation consensus sequences conserved across species (in rabbit NHE3, Ser693, Ser694, and Ser810), all of which are necessary for CaMKII to inhibit basal NHE3 activity as indicated by increased basal NHE3 activity only when all three were removed simultaneously, whereas removal of any one eliminated the KN-93 stimulation of NHE3 (Fig 10). Although we have not shown that any of these sites are phosphorylated by CaMKII under basal conditions, there is another example in which three sites in NHE3 are phosphorylated by a single protein kinase, all of which are required for the kinase to regulate NHE3 function. In starfish NHE3, the Mos-MEK-MAPK-p90RSK pathway stimulates NHE3 via Rsk phosphorylation of NHE3 at three distal C-terminal sites, all of which are necessary for NHE3 phosphorylation and stimulation of NHE3 activity (47). CaMKII is not the only kinase that binds to this domain of NHE3 and is part of the large NHE3 signaling complex that forms there. CK2α, an active subunit, also associates with NHE3 in this domain (also aa 586–605) and like CaMKII affects basal NHE3 activity (stimulates) by phosphorylating a site C-terminal to the kinase-binding site (Ser719) (19). It is important to note that although mutation of the three Ser in putative CaMKII phosphorylation sites C-terminal to NHE3 aa 690 prevented a CaMKII inhibitor from stimulating basal NHE3 activity, they did not increase basal NHE3 activity by themselves. We speculate that this might indicate a more complex regulation of NHE3 by CaMKII in addition to possible interactions among these three Ser phosphorylation sites in basal regulation.
Thus, this NHE3 signaling complex, which sets basal NHE3 activity, includes two active kinases that stimulate and inhibit NHE3 and almost certainly contribute to the “footprint of NHE3,” which is its ability to be both stimulated and inhibited as part of digestion (14). It is not known how the CK2 stimulation and CaMKII inhibition of NHE3 are coordinated, but it is of interest that two different mechanisms of regulation of NHE3 activity are used (CK2 trafficking; CaMKII change in turnover number without changes in trafficking), suggesting potential coordination of their effects. Also, how such a small α-helical domain can be necessary for interactions with so many large proteins is not understood structurally. NHE3 exists as a dimer, and even if the two NHE3 monomers making up a dimer bind to somewhat different components of the signaling complex, such a large complex suggests that multiple areas in the NHE3 C terminus are likely to interact, as has been suggested previously (14, 48).
The dynamic aspect of the CaMKII-NHE3 association in the plasma membrane with elevated Ca2+ is consistent with the NHE3 signaling complex changing with signaling; we and others have demonstrated that the NHE3 signaling complexes change in size not only with elevated Ca2+ but also with other signaling that alters NHE3 activity, including d-glucose, lysophosphatidic acid, and parathyroid hormone (14, 49, 50). The role of changes in the NHE3-CaMKII association or in changes in other NHE3 associating proteins (elevated Ca2+ reduces binding to NHE3 of NHERFs 2 and 3 (but not NHERF1), phospholipase Cγ, and CK2, in addition to CaMKII as demonstrated here) in mediating signaling related changes in NHE3 activity has not been defined (10, 51, 52).
Binding of the specific isoform CaMKIIγ to NHE3 was initially identified by immunoprecipitation of NHE3 followed by LC-MS/MS, which was undertaken to identify further components of the NHE3 signaling complexes. Intestinal cells, including Caco-2 cells, contain only the γ and δ isoforms of CaMKII, which are ubiquitously expressed, with the 54-kDa γ splice variant being the form expressed in Caco-2 and PS120 cells; and it is only the γ isoform that associates with NHE3. As shown in Fig. 5D, CaMKIIγ is phosphorylated under basal conditions, which, along with the transport and NHE3 phosphorylation studies reported here, demonstrate that at least a pool of this isoform is active in Caco-2 cells under basal conditions. We have assumed that it is the CaMKIIγ that binds NHE3, which is the pool involved in basal NHE3 inhibition. This is an interpretation consistent with our previous demonstration that BB vesicular CaMKII inhibits NHE3 (21). Moreover, there is no evidence that CaMKIIδ is involved in any aspect of NHE3 regulation.
What explains the unusual circumstances of having an active CaMKII binding to NHE3 at basal Ca2+ where it inhibits NHE3 activity? CaMKII is generally inactive at basal Ca2+, and an active CaMKII generally is created by autophosphorylation and release of the kinase sequence from binding to the CaMKII autoinhibitory domain. There are at least two possible mechanisms by which CaMKII can retain activity at basal Ca2+, both of which include initial activation of CaMKII by Ca2+/CaM and subsequent release of the kinase catalytic domain from its autoinhibitory domain. This is followed by autophosphorylation at Thr286/287 (based on species) or oxidation of Met281 or Met282 (53), which cause a conformational change in CaMKII, allowing high affinity interactions with the target proteins, and prevent inactivation of the kinase by re-association of the catalytic domain with the autoinhibitory domain after Ca2+ returns to basal levels. In the first of these mechanisms, exemplified by CaMKII binding and stimulating the NMDA receptor, CaMKII activated by Ca2+/CaM binds the NMDA receptor, which blocks access of the catalytic domain to the autoinhibitory domain despite Ca2+/CaM no longer being present. In the second mechanism, the catalytic subunit of the activated CaMKII (freed from its autoinhibitory domain) binds to a domain in its substrate that resembles the kinase autoinhibitory domain but fails to inactivate the catalytic domain, while preventing access to the kinase autoinhibitory domain (54, 55). Similarity in the sequence alignments of the CaMKII binding domain of NHE3 with the CaMKII autoinhibitory domain (Fig 11) suggests involvement of the second mechanism, although testing this hypothesis by mutagenesis has not been completed.
FIGURE 11.
Alignment of CaMKII autoinhibitory domain and related sequences in NHE3. CaMKII autoinhibitory domain is underlined. * represents amino acids that when mutated convert CaMKII inhibition to activation (54). # represents high homology; +, represents identity.
Multiple aspects of NHE3 regulation depend on the presence of the NHERF family of multi-PDZ domain proteins. Consequently, we evaluated the role of CaMKII in basal NHE3 regulation in cells containing either NHERF1 or NHERF2. We demonstrate here that CaMKII inhibits NHE3 basal activity by a mechanism that depends on the presence of NHERF2 but not NHERF1. This was shown both in cells in which NHERF1 and NHERF2, which are absent or present in low amounts, were stably transfected (PS120 fibroblasts) and in Caco-2BBe cells that express both NHERF1 and NHERF2, in which they were separately knocked down (35). The mechanism of the dependence of the CaMKII inhibition of NHE3 under basal conditions has not been identified, although several possible mechanisms have been eliminated. Despite both CaMKII and the NHERF family members binding to the same 20 aa of the NHE3 C terminus (56), the NHE3-CaMKIIγ co-precipitation is not NHERF2-dependent (Fig. 6E). In addition, although CaMKII phosphorylates NHE3 under basal conditions, the total amount of phosphorylation is not influenced by the presence of NHERF2 (Fig. 4); although it is not known whether NHERF2 is necessary for the specific CaMKII phosphorylation of NHE3 that regulates NHE3 activity.
In summary, our results reveal several layers of complexity for CaMKII regulation and binding to NHE3. We demonstrate here that NHE3 is a novel CaMKIIγ-binding protein, which directly binds NHE3 under basal conditions between aa 586–605, and this binding is rapidly reduced by physiologic levels of elevated Ca2+. CaMKII inhibition of basal NHE3 activity is NHERF2-dependent, occurs via changes in the NHE3 turnover number, and is associated with phosphorylation of NHE3, with the regulatory effect requiring amino acids C-terminal to the CaMKII binding domain and downstream of NHE3 aa 690 (Ser693, Ser694, and Ser810), which are part of putative CaMKII phosphorylation consensus sequences. It appears that CaMKII binding to the NHE3 C terminus and its effect on NHE3 activity are part of the physiologic regulation of NHE3 that occurs in multiple cell types, including fibroblasts as well as in the brush border of intestinal Na+-absorptive cells, and in this function, CaMKII acts as part of a large multiprotein- and multikinase-containing signaling complex that forms on the NHE3 C terminus.
This work was supported, in whole or in part, by National Institutes of Health NIDDK Grants R01DK26523, R01DK61765, P01DK072084, and R24DK64388 (to Hopkins Basic Research Digestive Diseases Development Core Center), P30DK089502 (to Hopkins Digestive Diseases Basic and Translational Core Center), and K08DK088950 and T32DK2007632 (to Hopkins Center for Epithelial Biology).
B. Cha and M. Donowitz, unpublished data.
- NHE
- Na+/H+ exchanger
- BB
- brush border
- aa
- amino acid
- VSVG
- vesicular stomatitis virus G protein
- Ni-NTA
- nickel-nitrilotriacetic acid
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- rNHE3
- rabbit NHE3
- TMA
- tetramethylammonium.
REFERENCES
- 1. Tse C. M., Brant S. R., Walker M. S., Pouyssegur J., Donowitz M. (1992) Cloning and sequencing of a rabbit cDNA encoding an intestinal and kidney-specific Na+/H+ exchanger isoform (NHE-3). J. Biol. Chem. 267, 9340–9346 [PubMed] [Google Scholar]
- 2. Orlowski J., Kandasamy R. A., Shull G. E. (1992) Molecular cloning of putative members of the Na+/H+ exchanger gene family. cDNA cloning, deduced amino acid sequence, and mRNA tissue expression of the rat Na+/H+ exchanger NHE-1 and two structurally related proteins. J. Biol. Chem. 267, 9331–9339 [PubMed] [Google Scholar]
- 3. Melvin J. E., Park K., Richardson L., Schultheis P. J., Shull G. E. (1999) Mouse down-regulated in adenoma (DRA) is an intestinal Cl−/HCO3− exchanger and is up-regulated in colon of mice lacking the NHE3 Na+/H+ exchanger. J. Biol. Chem. 274, 22855–22861 [DOI] [PubMed] [Google Scholar]
- 4. Lamprecht G., Heil A., Baisch S., Lin-Wu E., Yun C. C., Kalbacher H., Gregor M., Seidler U. (2002) The down-regulated in adenoma (dra) gene product binds to the second PDZ domain of the NHE3 kinase A regulatory protein (E3KARP), potentially linking intestinal Cl−/HCO3− exchange to Na+/H+ exchange. Biochemistry 41, 12336–12342 [DOI] [PubMed] [Google Scholar]
- 5. Ahn W., Kim K. H., Lee J. A., Kim J. Y., Choi J. Y., Moe O. W., Milgram S. L., Muallem S., Lee M. G. (2001) Regulatory interaction between the cystic fibrosis transmembrane conductance regulator and HCO3− salvage mechanisms in model systems and the mouse pancreatic duct. J. Biol. Chem. 276, 17236–17243 [DOI] [PubMed] [Google Scholar]
- 6. Thwaites D. T., Kennedy D. J., Raldua D., Anderson C. M., Mendoza M. E., Bladen C. L., Simmons N. L. (2002) H/dipeptide absorption across the human intestinal epithelium is controlled indirectly via a functional Na+/H+ exchanger. Gastroenterology 122, 1322–1333 [DOI] [PubMed] [Google Scholar]
- 7. Donowitz M., Li X. (2007) Regulatory binding partners and complexes of NHE3. Physiol. Rev. 87, 825–872 [DOI] [PubMed] [Google Scholar]
- 8. Pang T., Su X., Wakabayashi S., Shigekawa M. (2001) Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J. Biol. Chem. 276, 17367–17372 [DOI] [PubMed] [Google Scholar]
- 9. Cha B., Tse M., Yun C., Kovbasnjuk O., Mohan S., Hubbard A., Arpin M., Donowitz M. (2006) The NHE3 juxtamembrane cytoplasmic domain directly binds ezrin. Dual role in NHE3 trafficking and mobility in the brush border. Mol. Biol. Cell 17, 2661–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zachos N. C., van Rossum D. B., Li X., Caraveo G., Sarker R., Cha B., Mohan S., Desiderio S., Patterson R. L., Donowitz M. (2009) Phospholipase C-γ binds directly to the Na+/H+ exchanger 3 and is required for calcium regulation of exchange activity. J. Biol. Chem. 284, 19437–19444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biemesderfer D., Nagy T., DeGray B., Aronson P. S. (1999) Phospholipase C-γ binds directly to the Na+/H+ exchanger 3 and is required for calcium regulation of exchange activity. J. Biol. Chem. 274, 17518–17524 [DOI] [PubMed] [Google Scholar]
- 12. Girardi A. C., Degray B. C., Nagy T., Biemesderfer D., Aronson P. S. (2001) Association of Na+-H+ exchanger isoform NHE3 and dipeptidyl peptidase IV in the renal proximal tubule. J. Biol. Chem. 276, 46671–46677 [DOI] [PubMed] [Google Scholar]
- 13. Yun C. H., Oh S., Zizak M., Steplock D., Tsao S., Tse C. M., Weinman E. J., Donowitz M. (1997) cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 94, 3010–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donowitz M., Mohan S., Zhu C. X., Chen T. E., Lin R., Cha B., Zachos N. C., Murtazina R., Sarker R., Li X. (2009) NHE3 regulatory complexes. J. Exp. Biol. 212, 1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao H., Wiederkehr M. R., Fan L., Collazo R. L., Crowder L. A., Moe O. W. (1999) Acute inhibition of Na+/H+ exchanger NHE-3 by cAMP. Role of protein kinase A and NHE-3 phosphoserines 552 and 605. J. Biol. Chem. 274, 3978–3987 [DOI] [PubMed] [Google Scholar]
- 16. Zizak M., Lamprecht G., Steplock D., Tariq N., Shenolikar S., Donowitz M., Yun C. H., Weinman E. J. (1999) cAMP-induced phosphorylation and inhibition of Na+/H+ exchanger 3 (NHE3) are dependent on the presence but not the phosphorylation of NHE regulatory factor. J. Biol. Chem. 274, 24753–24758 [DOI] [PubMed] [Google Scholar]
- 17. Kocinsky H. S., Dynia D. W., Wang T., Aronson P. S. (2007) NHE3 phosphorylation at serines 552 and 605 does not directly affect NHE3 activity. Am. J. Physiol. Renal Physiol 293, F212–F218 [DOI] [PubMed] [Google Scholar]
- 18. Wang D., Sun H., Lang F., Yun C. C. (2005) Activation of NHE3 by dexamethasone requires phosphorylation of NHE3 at Ser663 by SGK1. Am. J. Physiol. Cell Physiol. 289, C802–C810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarker R., Grønborg M., Cha B., Mohan S., Chen Y., Pandey A., Litchfield D., Donowitz M., Li X. (2008) Casein kinase 2 binds to the C terminus of Na+/H+ exchanger 3 (NHE3) and stimulates NHE3 basal activity by phosphorylating a separate site in NHE3. Mol. Biol. Cell 19, 3859–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hudmon A., Schulman H. (2002) Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 364, 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen M. E., Reinlib L., Watson A. J., Gorelick F., Rys-Sikora K., Tse M., Rood R. P., Czernik A. J., Sharp G. W., Donowitz M. (1990) Rabbit ileal villus cell brush border Na+/H+ exchange is regulated by Ca2+/calmodulin-dependent protein kinase II, a brush border membrane protein. Proc. Natl. Acad. Sci. U.S.A. 87, 8990–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levine S. A., Nath S. K., Yun C. H., Yip J. W., Montrose M., Donowitz M., Tse C. M. (1995) Separate C-terminal domains of the epithelial specific brush border Na+/H+ exchanger isoform NHE3 are involved in stimulation and inhibition by protein kinases/growth factors. J. Biol. Chem. 270, 13716–13725 [DOI] [PubMed] [Google Scholar]
- 23. Murtazina R., Kovbasnjuk O., Donowitz M., Li X. (2006) Na+/H+ exchanger NHE3 activity and trafficking are lipid Raft-dependent. J. Biol. Chem. 281, 17845–17855 [DOI] [PubMed] [Google Scholar]
- 24. Hoogerwerf W. A., Tsao S. C., Devuyst O., Levine S. A., Yun C. H., Yip J. W., Cohen M. E., Wilson P. D., Lazenby A. J., Tse C. M., Donowitz M. (1996) NHE2 and NHE3 are human and rabbit intestinal brush-border proteins. Am. J. Physiol. 270, G29–G41 [DOI] [PubMed] [Google Scholar]
- 25. Lamprecht G., Weinman E. J., Yun C. H. (1998) The role of NHERF and E3KARP in the cAMP-mediated inhibition of NHE3. J. Biol. Chem. 273, 29972–29978 [DOI] [PubMed] [Google Scholar]
- 26. Levine S. A., Montrose M. H., Tse C. M., Donowitz M. (1993) Kinetics and regulation of three cloned mammalian Na+/H+ exchangers stably expressed in a fibroblast cell line. J. Biol. Chem. 268, 25527–25535 [PubMed] [Google Scholar]
- 27. Yun C. H., Lamprecht G., Forster D. V., Sidor A. (1998) NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J. Biol. Chem. 273, 25856–25863 [DOI] [PubMed] [Google Scholar]
- 28. Janecki A. J., Montrose M. H., Zimniak P., Zweibaum A., Tse C. M., Khurana S., Donowitz M. (1998) Subcellular redistribution is involved in acute regulation of the brush border Na+/H+ exchanger isoform 3 in human colon adenocarcinoma cell line Caco-2. Protein kinase C-mediated inhibition of the exchanger. J. Biol. Chem. 273, 8790–8798 [DOI] [PubMed] [Google Scholar]
- 29. Tse C. M., Levine S. A., Yun C. H., Brant S. R., Pouyssegur J., Montrose M. H., Donowitz M. (1993) Functional characteristics of a cloned epithelial Na+/H+ exchanger (NHE3):resistance to amiloride and inhibition by protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 90, 9110–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bartfai T. (1979) Preparation of metal-chelate complexes and the design of steady state kinetic experiments involving metal nucleotide complexes. Adv. Cyclic Nucleotide Res. 10, 219–242 [PubMed] [Google Scholar]
- 31. Cha B., Kim J. H., Hut H., Hogema B. M., Nadarja J., Zizak M., Cavet M., Lee-Kwon W., Lohmann S. M., Smolenski A., Tse C. M., Yun C., de Jonge H. R., Donowitz M. (2005) cGMP inhibition of Na+/H+ antiporter 3 (NHE3) requires PDZ domain adapter NHERF2, a broad specificity protein kinase G-anchoring protein. J. Biol. Chem. 280, 16642–16650 [DOI] [PubMed] [Google Scholar]
- 32. Weinman E. J., Steplock D., Donowitz M., Shenolikar S. (2000) NHERF associations with sodium-hydrogen exchanger isoform 3 (NHE3) and ezrin are essential for cAMP-mediated phosphorylation and inhibition of NHE3. Biochemistry 39, 6123–6129 [DOI] [PubMed] [Google Scholar]
- 33. Akhter S., Kovbasnjuk O., Li X., Cavet M., Noel J., Arpin M., Hubbard A. L., Donowitz M. (2002) Na+/H+ exchanger 3 is in large complexes in the center of the apical surface of proximal tubule-derived OK cells. Am. J. Physiol. Cell Physiol. 283, C927–C940 [DOI] [PubMed] [Google Scholar]
- 34. Emmer E., Rood R. P., Wesolek J. H., Cohen M. E., Braithwaite R. S., Sharp G. W., Murer H., Donowitz M. (1989) Role of calcium and calmodulin in the regulation of the rabbit ileal brush-border membrane Na+/H+ antiporter. J. Membr. Biol. 108, 207–215 [DOI] [PubMed] [Google Scholar]
- 35. Sarker R., Valkhoff V. E., Zachos N. C., Lin R., Cha B., Chen T. E., Guggino S., Zizak M., de Jonge H., Hogema B., Donowitz M. (2011) NHERF1 and NHERF2 are necessary for multiple but usually separate aspects of basal and acute regulation of NHE3 activity. Am. J. Physiol. Cell Physiol. 300, C771–C782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yanaka A., Suzuki H., Shibahara T., Matsui H., Nakahara A., Tanaka N. (2002) EGF promotes gastric mucosal restitution by activating Na+/H+ exchange of epithelial cells. Am. J. Physiol. Gastrointest Liver Physiol. 282, G866–G876 [DOI] [PubMed] [Google Scholar]
- 37. Yoo B. K., He P., Lee S. J., Yun C. C. (2011) Lysophosphatidic acid 5 receptor induces activation of Na+/H+ exchanger 3 via apical epidermal growth factor receptor in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 301, C1008–C1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bayer K. U., Löhler J., Schulman H., Harbers K. (1999) Developmental expression of the CaM kinase II isoforms: ubiquitous γ- and δ-CaM kinase II are the early isoforms and most abundant in the developing nervous system. Brain Res. Mol. Brain Res. 70, 147–154 [DOI] [PubMed] [Google Scholar]
- 39. Gangopadhyay S. S., Barber A. L., Gallant C., Grabarek Z., Smith J. L., Morgan K. G. (2003) Differential functional properties of calmodulin-dependent protein kinase IIγ variants isolated from smooth muscle. Biochem. J. 372, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barria A., Muller D., Derkach V., Griffith L. C., Soderling T. R. (1997) Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long term potentiation. Science 276, 2042–2045 [DOI] [PubMed] [Google Scholar]
- 41. Leonard A. S., Lim I. A., Hemsworth D. E., Horne M. C., Hell J. W. (1999) Calcium/calmodulin-dependent protein kinase II is associated with the N-methyl-d-aspartate receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 3239–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bayer K. U., Schulman H. (2001) Regulation of signal transduction by protein targeting: the case for CaMKII. Biochem. Biophys. Res. Commun. 289, 917–923 [DOI] [PubMed] [Google Scholar]
- 43. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0. Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goel R., Harsha H. C., Pandey A., Prasad T. S. (2012) Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol. Biosyst. 8, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zachos N. C., Tse M., Donowitz M. (2005) Molecular physiology of intestinal Na+/H+ exchange. Annu. Rev. Physiol. 67, 411–443 [DOI] [PubMed] [Google Scholar]
- 46. Cohen M. E., Wesolek J., McCullen J., Rys-Sikora K., Pandol S., Rood R. P., Sharp G. W., Donowitz M. (1991) Carbachol- and elevated Ca2+-induced translocation of functionally active protein kinase C to the brush border of rabbit ileal Na+ absorbing cells. J. Clin. Invest. 88, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harada K., Fukuda E., Hirohashi N., Chiba K. (2010) Regulation of intracellular pH by p90Rsk-dependent activation of an Na+/H+ exchanger in starfish oocytes. J. Biol. Chem. 285, 24044–24054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ikeda T., Schmitt B., Pouysségur J., Wakabayashi S., Shigekawa M. (1997) Identification of cytoplasmic subdomains that control pH-sensing of the Na+/H+ exchanger (NHE1). pH maintenance, ATP-sensitive, and flexible loop domains. J. Biochem. 121, 295–303 [DOI] [PubMed] [Google Scholar]
- 49. Lin R., Murtazina R., Cha B., Chakraborty M., Sarker R., Chen T. E., Lin Z., Hogema B. M., de Jonge H. R., Seidler U., Turner J. R., Li X., Kovbasnjuk O., Donowitz M. (2011) d-Glucose acts via sodium/glucose cotransporter 1 to increase NHE3 in mouse jejunal brush border by a Na+/H+ exchange regulatory factor 2-dependent process. Gastroenterology 140, 560–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mahon M. J., Donowitz M., Yun C. C., Segre G. V. (2002) Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signaling. Nature 417, 858–861 [DOI] [PubMed] [Google Scholar]
- 51. Zachos N.C., Li X., Kovbasnjuk O., Hogema B., Sarker R., Lee L.J., Li M., de Jonge H., Donowitz M. (2009) NHERF3 (PDZK1) contributes to basal and calcium inhibition of NHE3 activity in Caco-2BBe cells. J. Biol. Chem. 284, 23708–23818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu X., Cha B., Zachos N. C., Sarker R., Chakraborty M., Chen T. E., Kovbasnjuk O., Donowitz M. (2011) Elevated calcium acutely regulates dynamic interactions of NHERF2 and NHE3 proteins in opossum kidney (OK) cell microvilli. J. Biol. Chem. 286, 34486–34496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erickson J. R., Joiner M. L., Guan X., Kutschke W., Yang J., Oddis C. V., Bartlett R. K., Lowe J. S., O'Donnell S. E., Aykin-Burns N., Zimmerman M. C., Zimmerman K., Ham A. J., Weiss R. M., Spitz D. R., Shea M. A., Colbran R. J., Mohler P. J., Anderson M. E. (2008) A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang E., Schulman H. (1999) Structural examination of autoregulation of multifunctional calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 274, 26199–26208 [DOI] [PubMed] [Google Scholar]
- 55. Griffith L. C. (2004) Regulation of calcium/calmodulin-dependent protein kinase II activation by intramolecular and intermolecular interactions. J. Neurosci. 24, 8394–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cha B., Chen Y., Sharma A., Murtizana R., Sharker R., Tse M., Donowitz M. (2007) LPA/LPA5 receptor stimulation of NHE3 involves two separate events: stimulation of NHE3 exocytosis vis EGFR and ERK and dynamic dissociation of NHE3 from the microvillar cytoskeleton/NHERF2 via PLC and a novel PKC. Gastroenterology 132, 575 [Google Scholar]