Abstract
We defined the genetic landscape of balanced chromosomal rearrangements at nucleotide resolution by sequencing 141 breakpoints from cytogenetically-interpreted translocations and inversions. We confirm that the recently described phenomenon of “chromothripsis” (massive chromosomal shattering and reorganization) is not unique to cancer cells but also occurs in the germline where it can resolve to a karyotypically balanced state with frequent inversions. We detected a high incidence of complex rearrangements (19.2%) and substantially less reliance on microhomology (31%) than previously observed in benign CNVs. We compared these results to experimentally-generated DNA breakage-repair by sequencing seven transgenic animals, and revealed extensive rearrangement of the transgene and host genome with similar complexity to human germline alterations. Inversion is the most common rearrangement, suggesting that a combined mechanism involving template switching and non-homologous repair mediates the formation of balanced complex rearrangements that are viable, stably replicated and transmitted unaltered to subsequent generations.
INTRODUCTION
Our understanding of the genetic architecture of human chromosomal rearrangements has expanded in recent years, as rapid improvements in genomics technology have spawned a growing number of mechanistic hypotheses, many involving some degree of homology between participant sequences1–3. For complex events, replication-based mechanisms have been proposed such as template switching from a stalled or disrupted replication fork (Fork-Stalling and Template Switching, FoSTeS)4 and microhomology-mediated break-induced replication (MMBIR)3,5. While it has been established that chromosomal exchanges which appear to be balanced at lower-resolution can actually involve considerable complexity that may contribute to human disease in unexpected ways6,7, few studies have assessed these events at base-pair resolution. The first massively-parallel sequencing of cancer cells suggested a complex rearrangement landscape8, and more recently an astonishing phenomenon was uncovered in cancer cells that included massive chromosomal shattering and rearrangement, with frequent change in copy number state across the region9. The authors dubbed this “chromothripsis” and hypothesized it to be a unique feature of somatic mutation that might occur in 2–3% of all cancers9, with similar complexity seen in several subsequent sequencing studies10–12.
The mutational mechanism(s) underlying such complex genomic reorganization is unknown, but an intriguing feature of cancer-related chromothripsis was a frequent transition between two copy number states throughout the impacted region. As copy number variants (CNVs) have emerged as a major component of genetic variation in humans13, two recent population-based sequencing studies found a high degree of microhomology at CNV breakpoints and suggested two predominant CNV generating mechanisms: microhomology-mediated end joining (MHEJ) and non-homologous end joining (NHEJ)14,15, similar events postulated to occur by distinct pathways16,17. These mechanistic hypotheses may be somewhat limited as they were restricted to events defined by DNA dosage changes in unphenotyped individuals. The rearrangement landscape of other forms of genomic rearrangements, namely constitutional balanced structural variation (SV) such as reciprocal translocation and inversion, have not been comprehensively assessed at the sequence level. Breakpoint resolution of these SVs is fundamental to the prediction of which rearrangement mechanisms could underlie their formation and to our understanding of the full range of mutational mechanisms involved in human SV.
Here, we provide the first high-throughput, sequence-based assessment of mutational mechanisms associated with breakpoints from 52 subjects with chromosomal abnormalities that were previously defined at cytogenetic resolution as balanced (45 reciprocal translocations and seven inversions) and clinically assessed as likely to be pathogenic (50 arose de novo and two were inherited from an affected parent). We performed a series of next-generation sequencing experiments that included either whole-genome sequencing or targeted capture of breakpoints (see Supplementary Methods and Talkowski et al.18,19) and all breakpoints were confirmed at base-pair resolution by capillary sequencing. Our results show definitively that, as in cancer, remarkably complex genomic reorganization can also occur in the human germline, but that the repair process can resolve to a relatively balanced state rather than yielding extensive gains and losses of DNA. When we surveyed an experimental system of chromosomal rearrangement (transgenic animals), we found that similar genomic reorganization can result from experimentally-generated double-stranded DNA breaks (DSB), in the absence of environmental mutagenic factors and in the presence of an abundance of homologous template. In both human and transgenic animal breakpoints studied, the mechanism(s) that mediated rearrangements did not depend primarily on microhomology, nor were they frequently associated with large DNA dosage changes; both findings in stark contrast to previous studies of chromothripsis in cancer and benign CNV formation. Instead, these results reveal substantial chromosomal reorganization in the germline that can be compatible with viability, stably replicated, and transmitted to subsequent generations.
RESULTS
Complex genomic reorganization in the human germline
We found cytogenetically defined and apparently balanced SVs to be far more complex than originally thought, detecting 141 breakpoints, an average of 2.71 breakpoints per subject. Only two subjects had two derivative chromosomes with no DNA imbalance, but most subjects also did not suffer a substantial loss of genetic material (arbitrarily defined here as >1 kb total genomic imbalance, Table 1, Table S1). Instead, multiple breakpoints were often reassembled in a relatively balanced manner with microdeletions of one to several bases. We found that 19.2% of all karyotypically balanced SVs actually met conventional criterion of a complex chromosomal rearrangement (CCR; three or more breakpoints), a significantly higher frequency than the previous estimate of 2.8% from 246 de novo balanced anomalies assessed from more than 269,000 prenatal diagnoses (p = 2.2 × 10−4)7. Nine of the CCRs involved at least one inverted segment; only two CCRs involved exchanges between more than two chromosomes.
Table 1.
Breakpoint characteristics of karyotypically balanced structural variations
| All | Simple | Complex | Chromothripsis | ||
|---|---|---|---|---|---|
| By Subject | Total N (%)§ | 52 | 42 (80.8%) | 10 (19.2%) | 2 (3.8%) |
| B alanced | 3.8% | 4.8% | 0.0% | 0.0% | |
| Contains inversion | 28.8% | 14.3% | 90.0% | 100.0% | |
| By Breakpoint | Count (%) | 141 | 83 (58.9%) | 58 (41.1%) | 25 (17.7%) |
| Balanced | 18.9% | 20.0% | 17.0% | 20.0% | |
| Inverted | 30.5% | 14.5% | 53.4% | 48.0% | |
| Bases | Deletions | ||||
| < 2 | 49.6% | 51.3% | 46.8% | 40.0% | |
| 2 – 20 | 33.1% | 36.3% | 27.7% | 36.0% | |
| 21 – 100 | 7.9% | 3.8% | 14.9% | 12.0% | |
| 101 – 1,000 | 0.8% | 1.3% | 0.0% | 0.0% | |
| > 1,000 | 8.7% | 7.5% | 10.6% | 12.0% | |
| Insertions | |||||
| < 2 | 80.1% | 77.1% | 84.5% | 88.0% | |
| 2 – 20 | 15.6% | 20.5% | 8.6% | 8.0% | |
| 21 – 100 | 3.5% | 2.4% | 5.2% | 4.0% | |
| 101 – 1,000 | 0.7% | 0.0% | 1.7% | 0.0% | |
| > 1,000 | 0.0% | 0.0% | 0.0% | 0.0% | |
| Microhomology | |||||
| < 2 | 68.8% | 68.7% | 69.0% | 60.0% | |
| 2 – 20 | 30.5% | 31.3% | 29.3% | 40.0% | |
| > 20* | 0.7% | 0.0% | 1.7% | 0.0% | |
Percentages represent percentage of total events (either by subject, n=52, or by breakpoint, n=141) in each category.
Results presented from analyses using EMBOSS needle for direct comparison with Kidd et al. (2010).
Six of the seven karyotypically defined inversions involved just two breakpoints; one involved multiple inversion breakpoints.
A single inverted breakpoint junction contained 654 bp of homology, no other junctions had more than 20 bp of homology.
Deletions restricted to 127 breakpoints for which breakpoint pairs could be unambiguously determined so the precise loss of DNA could be calculated.
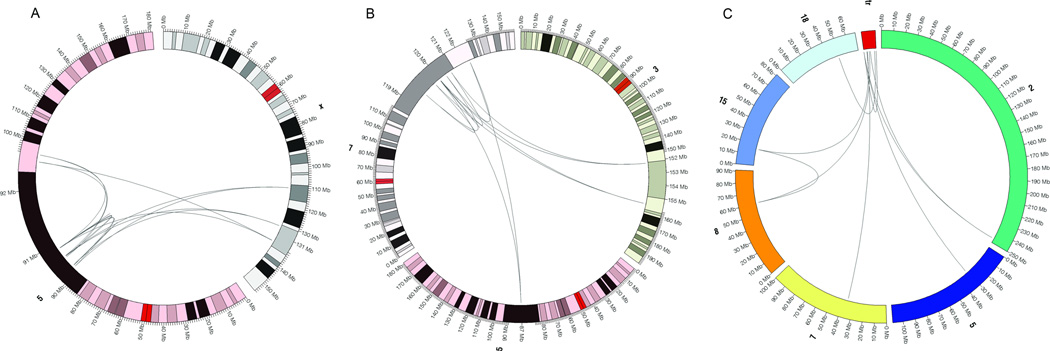
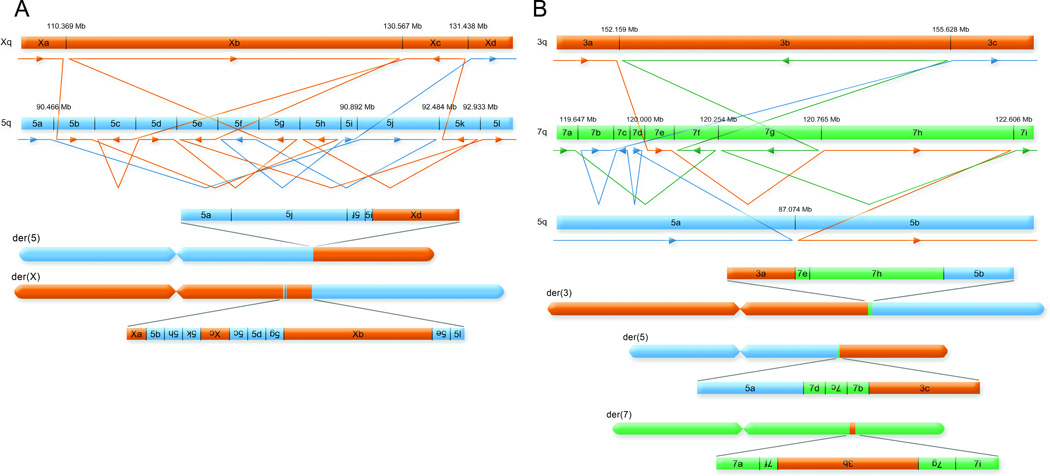
Among the 10 CCR subjects, there was extensive complexity of rearrangement. In the two most intricate cases (BSID42 and BSID43), we observed shattering and reorganization of multiple chromosomal segments similar to chromothripsis seen in cancer, except that these germline events resolved to a largely dosage balanced state rather than exhibiting frequent alterations in copy number9 (Fig. 1, Fig. 2; Supplementary Table 2). For BSID42, we observed 14 different junctions between chromosome (chr) X and chr5, with extensive shattering of localized regions (Fig. 2a, Supplementary Video 1). The 5q14.3 DNA ‘shards’ integrated into chrX with frequent oscillations between inverted and same-strand orientation (Fig. 2a). In both derivative chromosomes, large segments remained fully intact between reassembled DNA fragments. Prior to the most distal telomeric 5q-Xq junction in the der(x), an ~871 kb segment of chrX (designated Xc) was excised, inverted, and inserted into the strand-oscillating chr5 segment approximately 20 Mb from its chrX origin. Similarly, an 8 kb segment of chr5 (designated 5f) was also excised, inverted, and reinserted in der(5). Sequencing thus delineated a fascinating composite wherein shattered exchanges between chromosomes also resulted in extensive intrachromosomal reorganization. Notably, both the clinical karyotype and a 1 million feature aCGH experiment were completely blind to this series of rearrangements that extensively reorganized more than 23.5 Mb of DNA (0.76% of the entire genome for this SV; Table S2), as the sum of DNA imbalance from all junction fragments was just 6,357 bases.
Figure 1. Circos plots of chromothripsis in human germline balanced rearrangements and a transgenic animal.
Three plots are shown for the two most intricately rearranged balanced SV subjects (BSID42 and BSID43) and the multiple fragmented integrations associated a transgenic animal (G0/6). In each plot, lines connect each of the inter and intra-chromosomal junction fragments sequenced and the chromosomes are labeled outside of the circle. 1A. BSID42: Fourteen junctions were confirmed between chromosomes 5 and X. 1B. BSID43: Eleven junction fragments were confirmed between chromosomes 3, 5, and 7. 1C. G0/6: Sequencing in a transgenic sheep revealed that the transgene was apparently ‘shattered’ and integrated into six different chromosomes, including one interchromosomal excision of sheep chromosome 8 (OAR8) and insertion into the junction fragment between OAR7 and the transgene, mimicking a translocation junction. All junction fragments were confirmed by capillary sequencing. Additional validation was performed using FISH to further confirm these results (Supplementary Material). All positions are hg19.
Figure 2. Delineation of two subjects with germline chromothripsis.
Complete sequence resolution of all 25 chromosomal breakpoints in two highly complex chromosomal rearrangements. On the top are diagrams detailing the fully reconciled exchanges of material between chromosomes with lines and arrows connecting the junctions and below are reconstructions of the resultant chromosomal organization. Coordinate distances are not to scale. 2A. Sequencing revealed a similar pattern of the ‘shattering’ and aberrant reorganization of localized genomic regions from karyotypically balanced germline structural variations to those recently reported in cancer cells (i.e., chromothripsis). There were 14 junction fragments from two shattered regions of 5q14.3 and resultant re-organization of both 5q14.3 and the integration sites of Xq26.3 in both derivative chromosomes of the reciprocal translocation. These exchanges were generally balanced (6,357 bp lost from all exchanges combined), with frequent oscillations of strand orientation at junction fragments, inverted insertions of many fragments, and apparent induction of intrachromosomal inverted excision/insertion events in both derivative chromosomes. 2B. Two independent karyotypic analyses indicated a balanced reciprocal translocation between chromosomes 3q and 5q; however, sequencing revealed the shattering of chromatin from 7q and re-integration of 7q DNA shards into the junction fragments of both derivative chromosomes, resulting in no direct 3q–5q junctions. There were four different inverted excision/insertion events, including intrachromosomal excision, inversion, and re-insertion in 7q with a co-occurring inverted insertion of an intact 3.5 Mb segment of 3q, all of which involved only 1,551 bases of total DNA imbalance. All positions are hg19. See also Supplementary Movies 1 and 2.
The second sample, BSID43, had DNA available from two sources, one extracted from whole blood and another from an EBV-transformed lymphoblastoid cell line, both of which were sequenced (214.6X average physical coverage of mapped inserts, Table S2). The fully resolved genomic reorganization involved 11 breakpoints resulting from the apparent shattering of a 3 Mb region of 7q31.31–q31.32 that integrated into the long arms of both chr3 and chr5. The chr3 breakpoints spanned 3.5 Mb (3q25.2–q25.31), whereas chr5 had just a single breakpoint on each derivative (5q14.3) with the loss of only 16 bases between derivatives (Fig 2b, Supplementary Video 2). Despite the karyotypic interpretation of a reciprocal translocation of chr3 and chr5, there were no actual junctions connecting 3q25.2–25.31 to 5q14.3. Rather, all breakpoint junctions detected were joined to shattered fragments of 7q31.31–q31.32, and all events were again cryptic to both karyotype and 1M aCGH diagnostics (a total of only 1,551 bases was lost from all rearrangements) (Fig 2b). All breakpoints were identical between the two DNA sources, indicating that, once established, the rearranged chromosomal organization was stable through EBV transformation and subsequent cell propagation.
Eight additional subjects had CCRs ranging from 3–6 breakpoints each, many of which involved less than 1 kb of total genomic imbalance. All but one had inverted segments associated with a breakpoint, usually directly at the breakpoint junction, with excision, inversion, and insertion events as a common signature (Tables S1 and S2). The only CCR event occurring without an inversion associated with a breakpoint (BSID45) contained a balanced excision of 13.5 Mb of chr4 and reinsertion 5.3 Mb away from the der(13) translocation breakpoint (Tables S1 and S2). Of the seven karyotypically-defined inversions, six were simple and largely balanced events and only 1 event (BSID39) was complex, containing a smaller 4.1 kb inversion nested within the larger 43.7 Mb inversion (Tables S1 and S2).
Complex rearrangements from experimental transgenesis
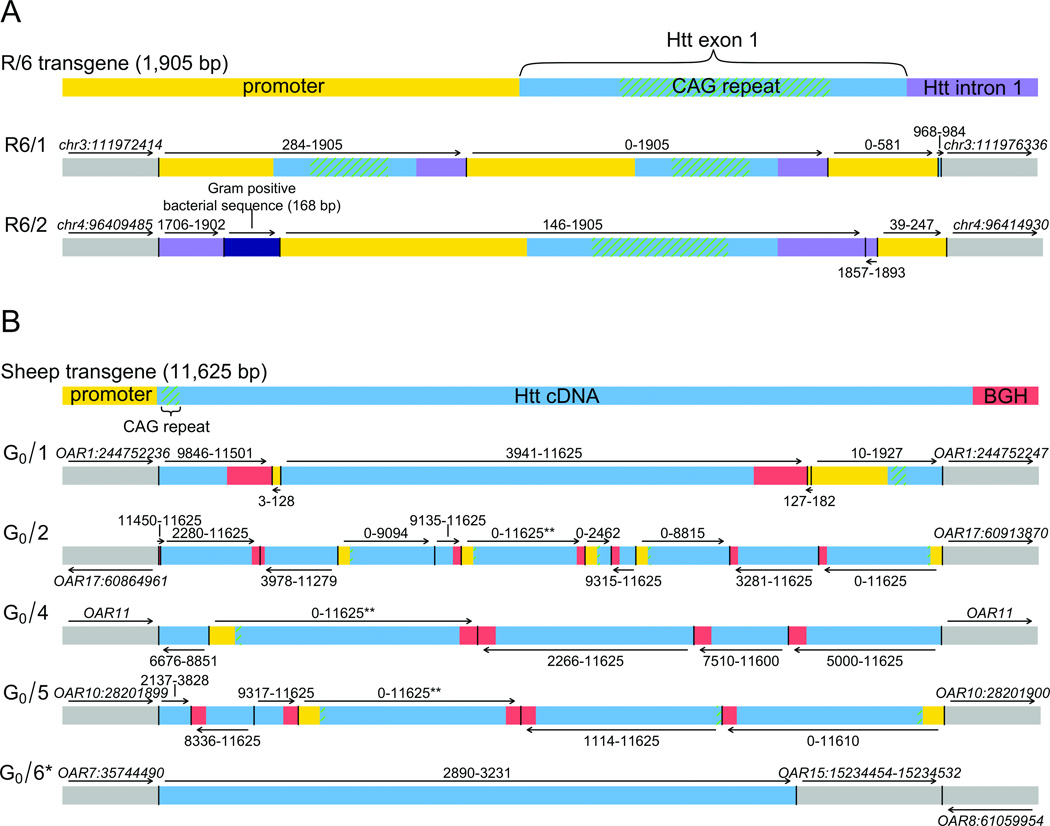
Each of the previous examples of cancer chromothripsis postulated that exogenous factors such as ionizing radiation, reactive oxygen species, or chemotherapy could prime such rearrangements by inducing multiple DSBs (reviewed by Khanna & Jackson20). One previous study assessed DSB repair between germline and experimentally generated, aphidicolin-induced CNV formation, finding non-homologous repair as the driving mechanism in both21. The proposition that the complex reorganization of human and cancer genomes operates through aberrant repair of multiple, apparently simultaneous DNA breaks prompted us to test whether a similar outcome is experimentally induced during transgene integration, where exogenous DNA fragments, which are themselves substrates for DSB repair, are incorporated into the genome. We hypothesized that the process of transgene integration might provide insight into the general mechanisms involved in complex rearrangements. Interestingly, few such transgene integration sites have been characterized at the sequence level22. Using two sequencing approaches (targeted capture of transgenes and whole-genome jumping libraries), we analyzed seven transgenic animals: two well-established lines of mice produced by injection of a 1.9 kb fragment of human 4p16.3 genomic DNA containing exon 1 of the Huntington’s disease gene (HTT) (R6/1 and R6/2)23 and five HTT sheep transgenic lines created by injection of an 11.6 kb full-length HTT cDNA (G0/1, G0/2, G0/5, G0/4 and G0/6)24 (see Methods and Supplemental Information).
Transgene integration sites were identified to base-pair resolution in all seven animals, revealing abundant complex rearrangements in both the internal structure of the final transgene (i.e., deletion, duplication, inversion, and excision/inversion/insertion) and in the host genomic DNA. Inverted segments were a common feature, displaying frequent oscillations in DNA orientation similar to the human CCRs described above (Fig. 3). At least two inverted excision/insertion events were detected in each sheep line with many containing significantly more such events. In the R6/2 line, we also found insertion of a 168 bp segment of foreign DNA with high homology to Corynebacteria. Repair of the DNA breaks commonly resulted in rearrangement of the host genome at the site of integration as well as at remote loci. Genomic deletions were detected in four transgenic animals, the largest of which was a 5.4 kb deletion in R6/2. In G0/2, we also found an 81 kb inversion at the integration site, while in G0/6, an apparent shattering and complex reintegration of the injected DNA into six different chromosomes was observed, including an interchromosomal rearrangement between chromosomes OAR7, OAR8, and OAR15 (Fig. S1). This rearrangement involved a small interchromosomal excision/insertion of OAR15 into an apparent translocation junction between OAR7, transgene sequence, and OAR8, reminiscent of the complex rearrangement seen in subject BSID43.
Figure 3. Complex rearrangements in transgenic animals.
Results from sequencing of two transgenic mice (A) and five transgenic sheep (B). Each transgene prior to pronuclear injection is provided in the first line and resultant transgene integration site and internal structure is shown below for each animal. Genomic integration sites are given by the gray lines flanking the transgene and the most parsimonious transgene structure is shown (R6/1,2 mice and G0/1,2,4,5,6 sheep). The fragment injected into the R6 lines (1,905 bp) comprised the human HTT promotor, exon 1 (~130 CAG repeats), and 264 bp of human intron 1. The fragment injected into the sheep (11,625 bp) included the human HTT promoter and cDNA (69 CAG repeats), followed by exon 4, intron 4, and exon 5 of the bovine growth hormone (BGH) gene. Arrows represent strand orientation. *This transgene was fragmented and inserted into multiple chromosomal locations; one of the small fragments and resultant complex rearrangements of the host genome involving three independent chromosomes is shown here (see also Fig. 1, Supplementary Fig. 1). **Head-to-tail junctions and read depth analyses indicate multiple copies of the segment, however the precise number of duplications could not be determined. Mouse positions are mm9 and sheep positions are OAR2.
These complex rearrangements mirrored the human germline CCRs, suggesting that shared or overlapping mechanisms of DSB repair mediate formation of CCRs. Arguably, the study of transgenic animals has the potential for two independent breakpoint classes: (1) the sites of integration and rearrangements of the host genome, and (2) rearrangements of the injected fragment itself. Having established the gross similarities of rearrangements in the human germline and in transgenic animals, we proceeded to analyze the sequence features of our balanced SVs and to compare these with the breakpoint sequences of human CNVs and the transgenic animals.
Mechanistic signatures differ between balanced structural variation and copy number variants
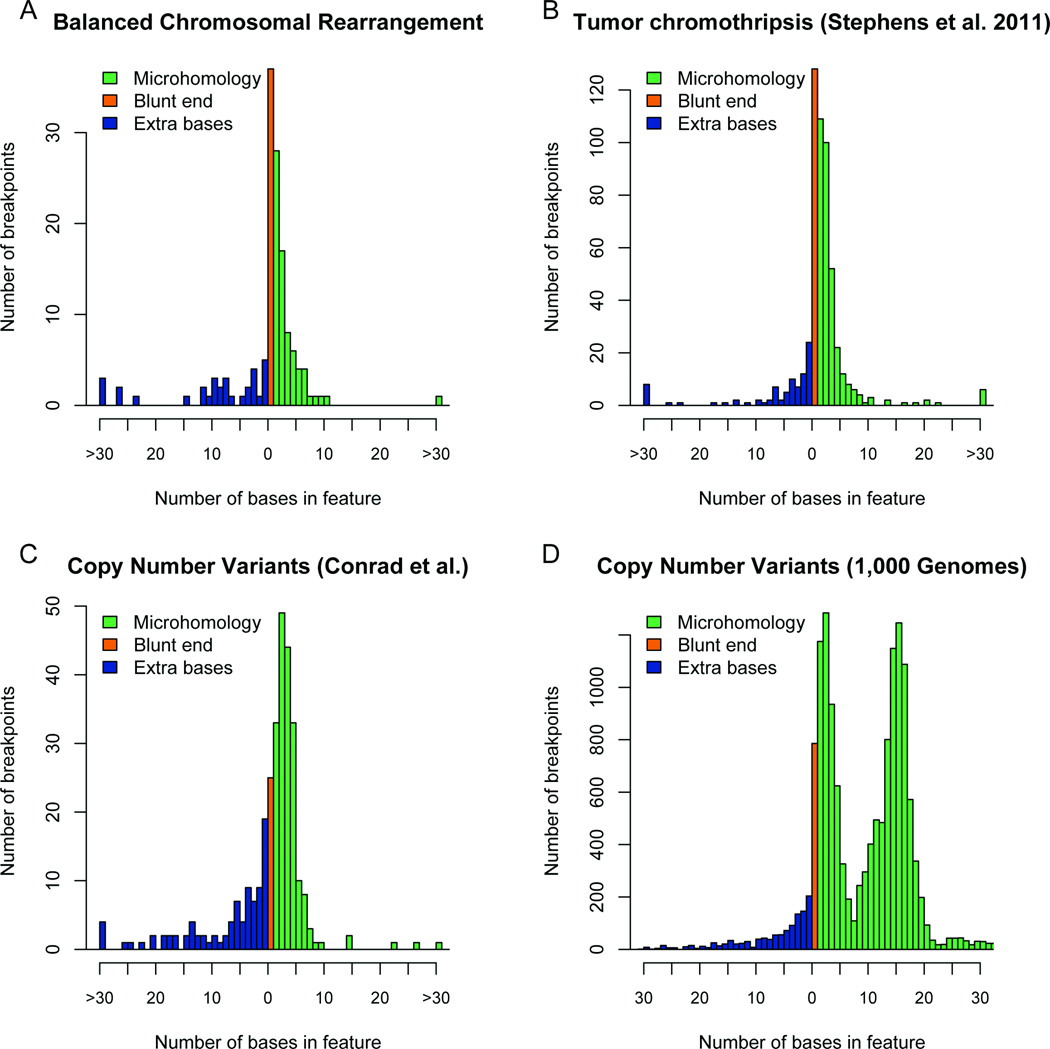
We discovered a dramatic disparity between breakpoint features of the de novo and presumably pathogenic translocations or inversions and the results obtained from population-based CNV studies, with important mechanistic consequences. Only 30.5% of these 141 breakpoints were flanked by microhomology, suggesting that most events were unlikely to have arisen by MHEJ, homology mediated repair, or microhomology-mediated replication-based mechanisms such as MMBIR3,5 (Fig. 4a). This distribution resembled the profile of six tumors sequenced by Stephens et al. (2010) that had undergone chromothripsis (45% of breakpoints contained microhomology; Fig. 4b). By comparison, from recent studies 70%–80% of all CNVs in unphenotyped individuals contained microhomology14,15 (Fig. 4c, 4d).
Figure 4. Breakpoint sequence signatures from balanced structural variations and copy number variation from independent population-based studies.
Histograms of nucleotide distribution for each sample. Breakpoints with no homology or inserted sequences are in orange (blunt ligation), green bars represent sequence microhomology between breakpoints, and blue bars represent the number of inserted nucleotides at the breakpoints. 4A. Breakpoint sequence distribution from karyotypically and presumably pathogenic balanced rearrangements sequenced in this study. 4B. Sequence distribution of 545 breakpoints from six tumors (two CLLs, one colorectal cancer, one thyroid cancer, one renal cancer, and one small cell lung cancer) localized by paired-end sequencing and confirmed with PCR by Stephens et al. (2011). 4C. Sequence distribution of 315 deletions captured and sequenced in normal individuals by Conrad et al. (2010). 4D. Breakpoint signatures of 16,783 deletion breakpoints from the 1,000 Genomes Pilot Project 1 analyzed with the same pipeline as the karyotypically balanced rearrangements in this study (4A) showing an identical distribution of breakpoint sequences to those seen in Mills et al. (2011).
The previous CNV estimates were almost exclusively based upon deletions as these events were most readily detected by the previous CNV capture of Conrad et al. (2010) and the low-depth sequencing of Mills et al. (2011); neither study evaluated balanced events. Kidd et al. (2010) surveyed SVs at fosmid resolution and suggested that NAHR was the dominant mechanism mediating large inversion formation in population-based samples (69% had flanking homologous segments of > 200 bp)25. In our study of potentially pathogenic balanced SVs, we found that just one of the 43 total inverted segments contained greater than 200 bp of homology (654 bp)(Fig. S3). This one instance was not a karyotypically defined large inversion but rather a small 2.3 kb inversion associated with a CCR (Table S1). Collectively, the microhomology profile of the karyotypically-defined inversions mirrored the breakpoints of the translocations, 69% of which contained < 2 bp of microhomology. The disparity between these presumably pathogenic inversions and the variants analyzed by Kidd et al. (2010)25 was not biased by sequencing methodology (NAHR is a notoriously difficult mechanism to delineate by short-read sequencing) as we resolved each of the seven karyotypically-defined de novo inversions to nucleotide resolution, none of which contained long stretches of homology at the breakpoint.
We surveyed breakpoint profiles in the transgenic animals and found that the microhomology associated with this experimentally induced DSB repair provided compelling evidence for mechanisms more similar to de novo balanced SVs than to benign CNVs, as 78% of the host genomic DNA integration sites contained no microhomology. To test sensitivity of the paired-end sequencing results, we performed capillary sequencing validation of the entire transgene sequence in two animals (R6/2 and G0/1, excluding 121 bp of a hairpin formation in G0/1), which confirmed all of the rearrangement junctions detected by paired-end sequencing in Fig. 3. We also found that 76.9% of the internal transgene junctions had less than 2 bp of homology, despite the presence of abundant homologous template for homology mediated mechanisms.
We further explored this striking difference in the frequency of microhomology between de novo, balanced SVs tested here and previous CNV studies. We initially tested for methodological differences using three independent methods to assess microhomology: (1) a high-throughput alignment using BWA Smith-Watherman (BWA-SW; Fig. S2), resulting in 26.2% of breakpoints containing two or more bases of microhomology. (2) The EMBOSS needle program as described by Kidd et al. (2010), which allows concurrent inserted template sequences with microhomology, identifying microhomology in seven additional individuals (31.2%), which was not statistically different from the BWA-SW procedure (p = 0.43). (3) A revised version of BreakSeq26 previously used in Mills et al. (2011), that we customized to analyze balanced SVs with nearly identical results to BWA-SW and EMBOSS (R. Mills, unpublished, see Supplemental Information). We therefore considered both the final breakpoint junctions and the sequence features of the initially intact chromosomes prior to their disruption in our analyses with consistent conclusions between methods. When we compared these data to the breakpoint homology from 16,783 CNV breakpoints assembled for the 1,000 Genomes Pilot 1 study using identical methods, we find a highly significant difference from either the BWA-SW analysis (χ2 = 244.6, 1 d.f., p = 2.6×10−55) or the EMBOSS method (χ2 = 201.6, 1 d.f., p = 1.26×10−45) (Fig. 3). The EMBOSS results are reported in the Tables and Figures as the most conservative interpretation of the distinction between previously published population-based studies and our results in karyotypically-defined balanced rearrangements.
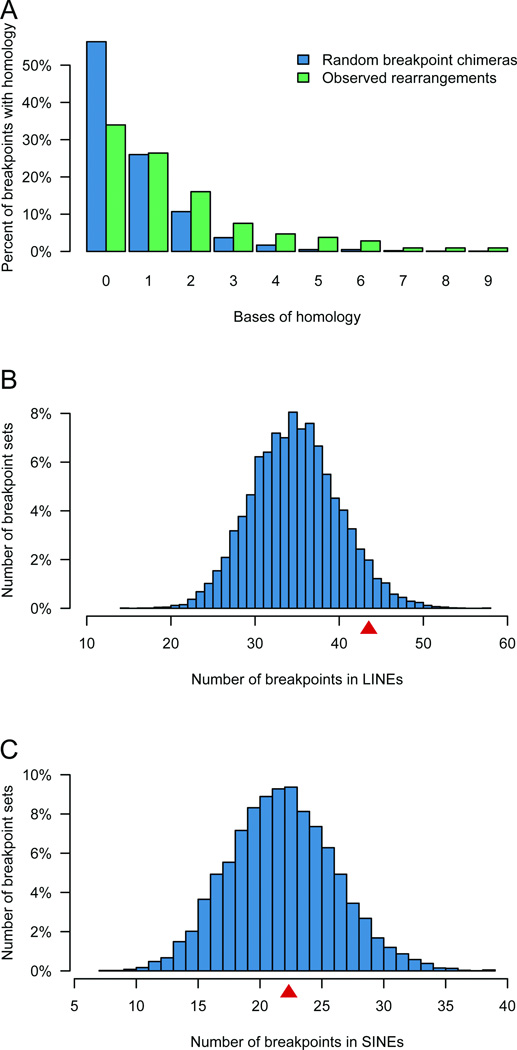
This persistent deficiency in microhomology led us to question the extent to which any mechanisms other than random NHEJ contributed to formation of chromosomal rearrangements in these subjects. We generated 1,000 simulated chimeric sequences of random breakpoints in the genome and analyzed the microhomology at these random junctions relative to the breakpoints in our subjects. The overall distributions of microhomology were significantly different (χ2 = 24.5, 2 d.f., p = 4.88×10−6). When we scrutinized these data we found a close concordance between the experimental set and random breakpoints for exactly two bp of homology (12.1% compared to 10.7%, respectively; p = 0.66). Of the remaining breakpoints (< 2 bp or > 2 bp homology), there was a significant enrichment of microhomology in the experimental set compared to the random breakpoints (experimental vs. simulated for < 2 bp = 68.8% vs. 82.3%, p = 3.6×10−4; > 2 bp = 19.1% vs. 7.0%; p = 1.38×10−5) (Fig. 5a), indicating a non-random reliance on a microhomology-mediated repair mechanism in the formation of these balanced SVs. In comparison with simulations of 10,000 datasets of equal size to our experimental set, we found a marginal enrichment of LINEs at the breakpoints (empirical p = 0.085) but not SINEs (Fig. 5b, 5c), and with 1 million simulations of windows ranging from 2 bp – 500 bp surrounding random genomic breakpoints, we found no significant enrichment in frequency of repetitive DNA motifs that could fold into non-B DNA structure27,28 (Table S3).
Figure 5. Comparison of observed chromosomal rearrangements to random simulations.
5A. Histogram density chart showing homology length of samples with 0 bp or more at the breakpoint. Green bars are observed breakpoints, blue bars are a distribution of 1,000 chimeras of two random genomic sequences. 5B–C. Histogram distribution of breakpoints falling in LINE and SINE elements from 10,000 simulated sets of breakpoints equal in size to the observed set. The arrow indicates the observed dataset (91st and 52nd percentile for SINEs (5B) and LINEs (5C), respectively).
DISCUSSION
This sequence-level evaluation of the genetic architecture of karyotypically balanced, potentially pathogenic chromosomal rearrangements reveals several remarkable and underappreciated features of such events: (1) karyotypically balanced SVs are rarely truly balanced at the primary sequence level; (2) human germline rearrangements can be extraordinarily complex, including localized deconstruction of chromosomal segments into many small fragments that can be rejoined and resolved in an aberrant but largely dosage-balanced manner that is compatible with life; (3) local inversions and what we refer to as ‘inverted translocations’ (inversions associated with translocation breakpoints) are a commonly observed signature of CCRs; and (4) the breakpoint characteristics of potentially pathogenic, karyotypically balanced SVs are markedly different from those of apparently benign CNVs. These studies also provide the first high-throughput sequencing assessment of the complexity resulting from integration of linear exogenous DNA in transgenic animals introduced by pronuclear injection, revealing resolved breakpoints that in many ways mimic those of human germline and cancer chromothripsis9,11,12. These similarities suggest that the mechanisms involved in establishing transgene integrations in model organisms may overlap with those that produce complex genomic DNA rearrangement in humans.
Cytogenetic studies previously established that a small subset of translocations involve multiple chromosomal breakpoints rather than simple reciprocal exchanges between chromosomes6,29. By sequencing karyotypically balanced SVs, we find a substantially greater proportion CCRs than previously estimated. Sequence resolution enabled us to expand the definition of CCR to include local inversions at translocation breakpoints (dubbed ‘inverted translocations’ here) which appear to be an underappreciated feature of CCRs and among the most pervasive characteristics of germline chromothripsis, which displayed frequent switching of strand orientation between integrated fragments. This is consistent with cancer chromothripsis9, as well as the finding that 4.5% of sequenced CNVs contained inserted sequence from the opposite strand in close proximity to the breakpoint in one study14, and small inversions found at the breakpoints of constitutional CNVs in an independent study21. We further observed that inverted excision/insertions in CCRs are frequently accompanied by additional genomic rearrangements, and that these can be interspersed with large, fully intact segments, suggesting an association between local microinversion and aberrant repair of DNA breaks.
Our findings confirm and further illuminate the phenomenon of chromothripsis resulting from highly aberrant DSB repair, however they foster further uncertainty about the cause of the initial event and the mechanism of repair. Our results strengthen the conclusion of Stephens et al. (2011)9 that these events occur in a one-off chromosomal catastrophe rather than by progressive accumulation, as we find the same phenomenon in the human germline and in the first generation transgenic sheep. We also confirm in humans and transgenic animals that, once resolved, the events can be viable, replicate during mitosis with high fidelity, and be stably transmitted to subsequent generations. The cause of chromothripsis thus does not appear to be specific to aberrant repair pathways in cancer, nor is it likely driven by other mechanisms considered by Stephens et al. (2011)9 such as disruption of tumor suppressor genes, formation of double minutes, or telomere disruption from end-to-end chromosome fusion. Although cancer and germline chromothripsis appear to differ in the presence of extensive copy number change, dosage alterations may be favored in cancer cells due to dysregulation of growth pathways via loss of tumor suppressor genes. Frequent dosage alterations likely also occur in the germline, as one recent study found a surprisingly high number of segmental imbalances in cleavage stage embryos (~70%)30. At least a portion of these complex events are viable, as evidenced by Lupski and colleagues in a study published during review of this manuscript that selected for human cases with multiple copy number imbalances on a single chromosome31. It is possible that the co-occurrence of dosage alteration with substantial inter-chromosomal exchanges result in a non-viable embryo or fetus, eliminating such cases before birth. Thus, the relatively balanced dosage of individuals with CCRs may simply reflect a selection for viability. With the increasing application of aCGH in research and clinical diagnostics, additional human cases with extensive copy number alterations will undoubtedly emerge and it will be interesting to determine whether they display the high frequency of concomitant chromosomal reorganization seen here and in cancer chromothripsis.
It is unknown if the NAHR-dominated mechanism of large inversions observed previously by fosmid sequencing25 is a characteristic of recurrent events as inversions were not assessed in other population-based sequencing studies. One recent study of unbalanced, recurrent translocations found predictable microhomology at the recurrent breakpoints32. In contrast, accumulating evidence indicates that formation of both common and rare CNVs is predominantly driven by non-homologous repair mechanisms that are reliant on much smaller degrees of homology between segments than NAHR14,15,25. Our results align with those of Mills et al. (2011) in supporting non-homologous repair as the favored mechanism in formation of deletions and the non-recurrent balanced rearrangements sequenced here; however, the level of microhomology associated with breakpoint signatures of these non-recurrent de novo balanced rearrangements was distinctly lower than those previous studies. There was also a dearth of sequence homology at the sites of integration in the transgenic animals and the chromothripsis seen in cancer cells based on data obtained from Stephens et al. (2011)9. The distinguishing feature of all three groups (balanced SVs, cancer cells, and transgenic animals) is that they presumably all contribute to an abnormal phenotype. In this regard, it may not be surprising that this extreme form of aberrant DNA breakage repair proposes significantly different mechanisms of formation than presumably benign CNVs in the general population. Our data support a version of NHEJ for most exchanges of genetic material between chromosomes in the absence of microhomology and without significant alteration in copy number. However, this mechanism as currently understood is unlikely to account for the frequent strand inversions observed, which instead imply a model of template switching, such as FoSTeS or MMBIR in the formation of CCRs including chromothripsis3,5. Spatially localized DNA breaks due to a single DNA insult could also drive the rearrangements seen here, as the complex CCRs may require multiple simultaneous breaks. These data might also implicate operation of multiple mechanisms, where reorganization of shattered chromosomes to a largely balanced state is driven by blunt end-joining with some degree of template switching and homology mediated repair, or by an alternative and yet undefined mechanism that has the capacity to derive inverted segments frequently, which have been seen in all instances of chromothripsis. Such a model could account for CCR formation in the germline aberrations as well as those seen in cancer cells. Our simulations indicate this possibility as we saw a modest but significant enrichment of microhomology in comparison to randomly selected breakpoints, which increased with the complexity of the SV.
These analyses provide an initial sequence-based survey of karyotypically balanced chromosomal rearrangements that, like the previous findings of chromothripsis in cancer, set the stage for extensive further investigation. Yet, our results already have a potentially wide-ranging impact as they indicate that traditional and current clinical diagnostic methods, including chromosomal microarrays, can be insufficient to understand the true nature of genomic disruption in affected patients. They also raise questions about whether the mechanisms suggested in CNV studies of unphenotyped populations are fully generalizable to other structural genomic alterations, and they suggest that more complex mechanistic hypotheses are required to explain how the genome can dynamically remodel itself in a surprisingly extensive manner to produce a stable, viable, complex constitutional rearrangement that can contribute to altered human development and disease.
Supplementary Material
Acknowledgments
We thank all research participants, their families, and the referring clinicians for their invaluable contributions. We thank Tammy Gillis, Mary Anne Anderson, Whitney Varney, Jayla Ruliera, Tatyanna Henderson, Chengsheng Zhang, Lauren Griffin, The Genomics Core of the MGH CHGR, the Genome Sequencing Platform at the Broad Institute, and the Partners Center for Personalized Genomic Medicine for technical assistance. We thank Mei Sun for performing the MLPA experiments. We also thank the Partners Research Computing at Massachusetts General Hospital. This work was funded by grants from the National Institutes of Health (GM061354, HD065286, NS32765, NS16367), the Simons Foundation Autism Research Initiative, Autism Speaks, and CHDI Foundation, Inc. J. Jacobsen was the recipient of the Philip Wrightson Fellowship of the Neurological Foundation of New Zealand, C. Ernst received a Bisby Fellowship from the CIHR, and M. Talkowski was supported by an NIMH National Research Service Award (F32MH087123) and an MGH ECOR Fund for Medical Discovery Award.
Footnotes
Author Contributions
MET and JFG wrote the manuscript, which was edited by all co-authors, CC, JJ, CE, MJD, MEM, JFG, and MET designed the experiments, JJ, CE, CH, and AL performed the molecular studies, CC, RM, MLB, TKO, MJD, IH, and MET designed the bioinformatic and statistical analyses, CC, AH, AK, IB, TKO, IH, RM, and MET performed the analyses, AL and CCM performed the FISH analyses, JJ, CCM, AL, SRR, CJM, CSB, SJR, RLMF, RGS, YS, MJD, MEM, and JFG provided the subjects and transgenic animals, and CL provided the clinical microarray data for the human chromothripsis samples.
Competing financial interests
The authors have no competing financial interests to report.
REFERENCES
- 1.Iliakis G, et al. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenetic and genome research. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 2.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS genetics. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends in genetics : TIG. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS genetics. 2009;5 doi: 10.1371/journal.pgen.1000327. e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Gregori M, et al. Cryptic deletions are a common finding in "balanced" reciprocal and complex chromosome rearrangements: a study of 59 patients. Journal of medical genetics. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giardino D, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenatal diagnosis. 2009;29:257–265. doi: 10.1002/pd.2215. [DOI] [PubMed] [Google Scholar]
- 8.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011 doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloosterman WP, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Human molecular genetics. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 13.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber MR. The mechanism of human nonhomologous DNA end joining. The Journal of biological chemistry. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 17.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS genetics. 2008;4 doi: 10.1371/journal.pgen.1000110. e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talkowski ME, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talkowski ME, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. American Journal of Human Genetics. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature genetics. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 21.Arlt MF, et al. Comparison of constitutional and replication stress-induced genome structural variation by SNP array and mate-pair sequencing. Genetics. 2011;187:675–683. doi: 10.1534/genetics.110.124776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurtele H, Little KC, Chartrand P. Illegitimate DNA integration in mammalian cells. Gene therapy. 2003;10:1791–1799. doi: 10.1038/sj.gt.3302074. [DOI] [PubMed] [Google Scholar]
- 23.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 24.Jacobsen JC, et al. An ovine transgenic Huntington's disease model. Human molecular genetics. 2010;19:1873–1882. doi: 10.1093/hmg/ddq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nature biotechnology. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends in biochemical sciences. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Molecular carcinogenesis. 2009;48:273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 29.Astbury C, et al. Delineation of complex chromosomal rearrangements: evidence for increased complexity. Human genetics. 2004;114:448–457. doi: 10.1007/s00439-003-1079-1. [DOI] [PubMed] [Google Scholar]
- 30.Vanneste E, et al. What next for preimplantation genetic screening? High mitotic chromosome instability rate provides the biological basis for the low success rate. Human reproduction. 2009;24:2679–2682. doi: 10.1093/humrep/dep266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou Z, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome research. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.