Abstract
As vertebrate embryos develop to adulthood, their organs dramatically increase size and change tissue architecture. Here, we used a multicolor clonal analysis to define contributions of many individual cardiomyocytes as the zebrafish heart undergoes morphogenesis from a primitive embryonic structure into its complex adult form. We find that the single cardiomyocyte-thick wall of the juvenile ventricle forms by lateral expansion of several dozen cardiomyocytes into muscle patches of variable sizes and shapes. As juveniles mature into adults, this structure becomes fully enveloped by a new lineage of cortical muscle. Adult cortical muscle originates from a small number (~8) of cardiomyocytes that display clonal dominance reminiscent of stem cell populations. Cortical cardiomyocytes initially emerge from internal myofibers that in rare events breach the juvenile ventricular wall and expand over the surface. Our study illuminates dynamic proliferative behaviors that generate adult cardiac structure, revealing clonal dominance as a key mechanism that shapes a vertebrate organ.
Vertebrate organ development is a complex process that begins in the early embryo and continues until the functional capacity of the organ meets adult requirements. Organ growth and maturation occupy a significant period of life and involve major anatomic changes, yet relatively little is known about the cellular dynamics that underlie these changes.
New technologies can illuminate cellular mechanisms that drive organ morphogenesis. Recently, Livet and colleagues introduced a system that allowed labeling designations of ~90 colors to murine neurons1. With this technology, termed ‘Brainbow’, they could visualize adjacent neurons and their connections in the brain with high resolution. The ability to assign many colors to different cells in a population can also be applied to understand cell proliferation and lineage decisions.
The heart is a set of chambers comprised predominantly of the contractile units, cardiomyocytes. Genetic fate-mapping has been performed to determine how separate lineages contribute to developing cardiac structures in mice and zebrafish2–6. Additionally, single-marker clonal analysis has traced the activity of individual cells during embryonic heart patterning7–10. These studies have enhanced our understanding of cardiogenic mechanisms in early embryos. Nevertheless, a large gap remains in comprehending how the size, shape, and structure of an adult heart are finalized through the individual and population behaviors of many cardiac cells.
Here, we employed a multicolor clonal analysis to map the proliferative histories of many individual cardiomyocytes as the zebrafish cardiac ventricle transitions from a simple tube of single-cardiomyocyte thickness into a complex adult structure. Our experiments yielded several unexpected discoveries relevant to the number, nature, and mechanisms of cardiomyocyte contributions during heart morphogenesis.
Multicolor labeling of cardiomyocytes
To study cell clones in zebrafish, we adapted the Brainbow 1.0L construct for combinatorial expression of 3 spectrally different fluorescent reporter proteins1. Multiple copy integration of this transgene at a single genetic locus is a common outcome of transgenesis. Thus, combinatorial Cre recombinase-mediated excision events at paired loxP recognition sites can generate many possible permanent colors. (Fig. 1a, b). We generated several transgenic lines containing a β-actin2 promoter-driven multicolor construct, and assessed them in combination with a strain harboring a tamoxifen-inducible, cardiomyocyte-restricted Cre recombinase, cmlc2:CreER11. We identified one line, Tg(β-act2:Brainbow1.0L)pd49 (referred to subsequently as priZm), showing limited mosaic recombination only in the presence of 4-hydroxytamoxifen (4-HT; Supplementary Fig. 1).
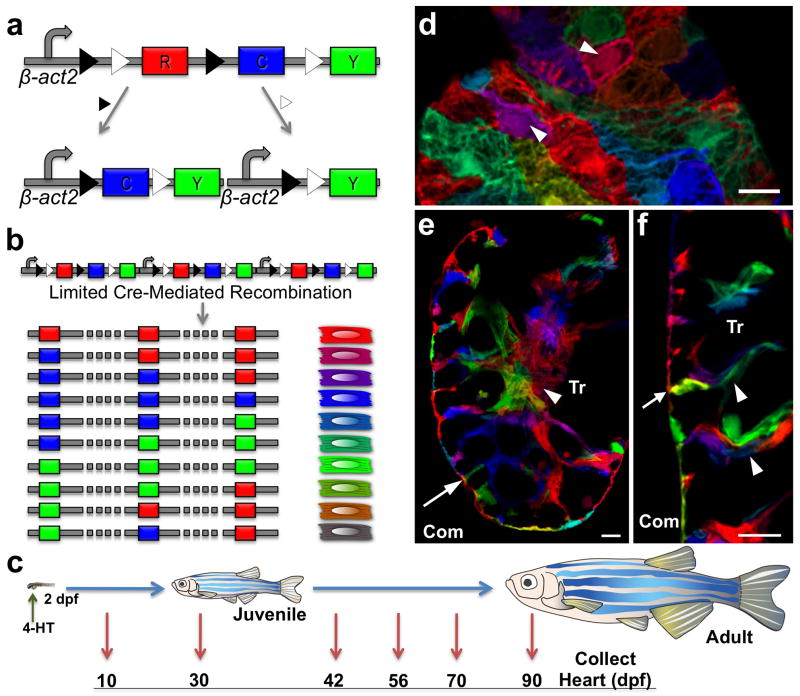
Figure 1. Multicolor clonal labeling of embryonic zebrafish cardiomyocytes.
a, Recombination at paired lox2272 (black triangles) or loxP (white triangles) sites leads to expression of CFP or YFP, respectively. b, Limited Cre-mediated recombination of tandem cassette insertions results in combinatorial expression of fluorescent proteins. c, Cartoon of lineage-tracing experiments. d, 10 dpf ventricular surface myocardium. Single cardiomyocytes are predominantly labeled with unique colors (arrowheads). e, f, 10 dpf ventricular confocal slice, indicating trabecular cardiomyocytes connected with clonally unrelated cardiomyocytes at the wall (e, f, arrow) and within trabeculae (f, arrowheads). Com, compact muscle. Tr, trabecular muscle. Scale bars, 10 μm.
The size and structure of the zebrafish heart is conducive to clonal analysis of cardiomyocytes. The 2–3 dpf zebrafish heart is looped, has a wall of single-cardiomyocyte thickness, and consists of 250–300 muscle cells; by our measurements ~115 of these are contained within the ventricular wall (Supplementary Fig. 2). Cardiomyocyte proliferation is detectable at 2 dpf and thought to account for most or all subsequent cardiogenesis2,12–15. To trace the fates of individual embryonic cardiomyocytes, we briefly incubated 2 dpf cmlc2:CreER; priZm embryos in 4-HT and raised them to different ages (Fig. 1c).
We first assessed cardiac fluorescence at 10 dpf, a stage comparable in organismal size to 2 dpf. Our optimized 4-HT regimen generated >20 unique colors in cardiomyocytes, as viewed at 10 dpf and subsequent stages. Red fluorescent protein (RFP) is the initial reporter cassette and default expression marker, and 4-HT treatment induced recombination (non-red colors) in ~50% of ventricular cardiomyocytes. Importantly, different colors were consistently assigned to adjacent cardiomyocytes on the surface of the 10 dpf ventricular wall (Fig. 1d–f; Supplementary Fig. 3), a prerequisite for multicolor clonal analysis.
The zebrafish ventricle is recognized to contain two types of cardiac muscle16. These include a peripheral wall of compact muscle, and inner myofibers organized into trabeculae that initiate formation at 3 dpf17. Histological examination of 10 dpf hearts revealed three notable observations. First, the ventricular wall remained at single-cardiomyocyte thickness, as at 2 dpf. Second, trabecular myocytes connected to the wall were most often clonally unrelated to adjoining wall myocytes (58 of 63 observations, n = 5 ventricles; Fig. 1e, f). This observation supported a mechanism for trabecularization proposed recently based on different lines of evidence, in which myocytes delaminate from the ventricular wall, seed elsewhere in the chamber, and initiate trabecular growth from a second site17. Third, we saw that the trabecular myofibers themselves were comprised of cardiomyocytes arising from different clonal origins within the 2 dpf wall (Fig. 1f).
Juvenile ventricular wall formation
We next examined the ventricles of juvenile zebrafish at 30 dpf, at which time major organismal and cardiac growth have occurred since cardiomyocyte labeling with 4-HT. By whole-mount imaging of surface myocardium, we readily identified multicellular, single-color regions of myocardium, indicating that progeny of embryonic cardiomyocytes generally remained connected with one another after division (Fig. 2a–c; Supplementary Fig. 4a–c). Although less common (9.8% of clones), we also observed instances suggesting complete separation of a cardiomyocyte from its clonal partners (Supplementary Fig. 4d).
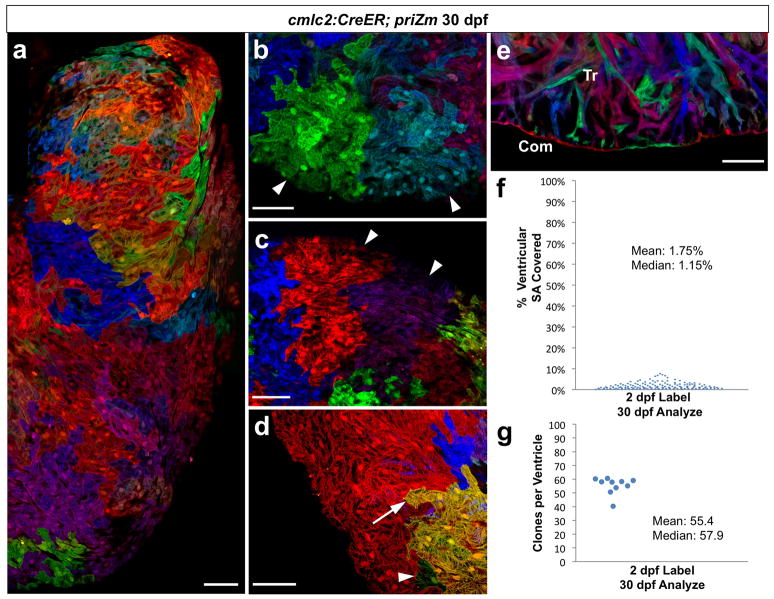
Figure 2. Several dozen embryonic cardiomyocytes build the juvenile ventricular wall.
a, Surface myocardium of half of a 30 dpf ventricular side, displaying clonal patches of varied shapes and sizes. b, c, Cardiomyocyte clones near the apex or chamber midpoint forming wedge/stripe shapes (arrowheads). d, Single-cell clone (green, arrowhead) positioned near a large clone (yellow, arrow). e, 30 dpf ventricular confocal slice, depicting a wall of single-cardiomyocyte thickness (Com) surrounding trabecular muscle (Tr). f, %Surface area occupied by 30 dpf clones (146 clones, 5 ventricles). g, Surface clones per ventricle (n=10). Scale bars, 50 μm.
We expected some uniformity to the clonal patches comprising the ventricular wall, with cardiomyocyte clones of similar shape and size. By contrast, ventricles from different animals presented unique patterns of surface color clones, and the shapes of clones within each ventricle were highly diverse (Fig. 2a; Supplementary Fig. 5). Clone size also varied. Some clones displayed evidence of many cell divisions, while others contained a single cardiomyocyte (Fig. 2d). Histological analysis showed retention of a wall of single-cardiomyocyte thickness, indicating that the expansion of wall clones was limited to lateral directions along the surface. They also revealed that substantial expansion of trabecular myocardium had occurred since 10 dpf (Fig. 2e).
To estimate the number of embryonic cardiomyocytes that create the juvenile ventricular wall, we calculated the surface areas occupied by individual clones and divided each by the total ventricular surface area. Most clones (99/146; n = 5) each occupied less than 2% of the ventricular surface area, while a small number (15/146) of larger clones each represented ~4–8% of the surface (Fig. 2f). Extrapolating for unrecombined myocardium, we determined that the 30 dpf ventricular surface was represented by 55.4 ± 1.9 color clones (n = 10; mean ± SEM; Fig. 2g). Thus, our data indicate that the juvenile zebrafish ventricular wall is built by lateral expansion of ~55 embryonic cardiomyocytes. These cardiogenic events create a patchwork of diverse clonal shapes and sizes that varies from animal to animal, indicating that juvenile cardiac form can be acquired with significant developmental plasticity.
Emergence of a new adult muscle lineage
Zebrafish are typically recognized as adults at 3 months post-fertilization. We examined ventricles of 6, 8, and 10 week-old animals that had undergone cardiomyocyte labeling at 2 dpf. At 6 weeks post-fertilization (wpf), most clonal patches comprising the surface myocardium appeared similar to those of 30 dpf ventricles. Additionally, we detected a population of clonally related cardiomyocytes layered upon these patches near the chamber base (Fig. 3a, b). By 8 wpf, large single-color swaths containing several hundreds of such cardiomyocytes extended from the ventricular base, wrapping around both sides of the chamber and often reaching its midpoint (Fig. 3d–f). At 10 wpf, we began to see evidence of these surface clones converging with each other. These external cardiomyocytes were typically more rod-shaped with more distinct striation than underlying cardiomyocytes (Supplementary Fig. 6).
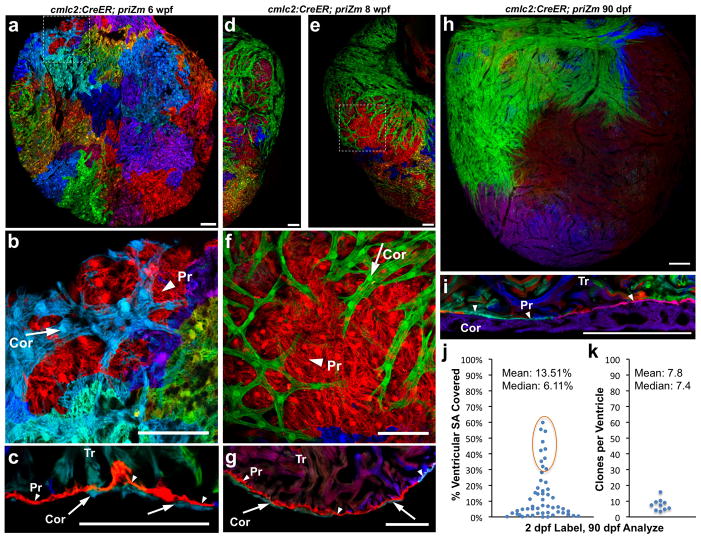
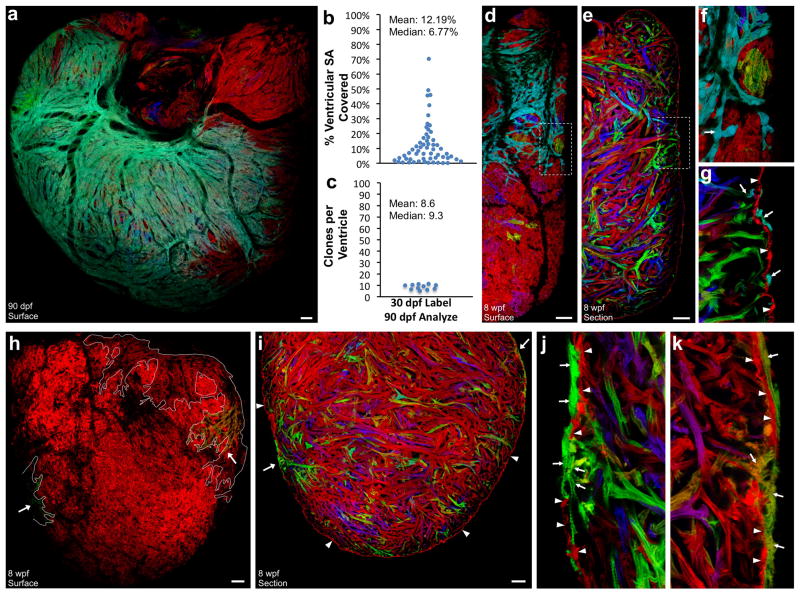
Figure 3. Clonally dominant cortical cardiomyocytes.
a–c, 6 weeks post-fertilization (wpf) ventricular surface (a, b) and confocal slice (c), indicating cortical (Cor, arrow), primordial (Pr, arrowhead), and trabecular (Tr) muscle. d–g, 8 wpf ventricular surface indicating a large green basal clone (d–f), and section indicating 3 muscle types (g). h, 90 dpf ventricular surface, showing only a few large cortical clones. i, 90 dpf ventricular section. j, %Surface area occupied by 90 dpf clones (56 clones, 10 ventricles); basal clones representing each ventricle are circled. k, Surface clones per ventricle (n=10). Scale bars, 50 μm (a–g); 100 μm (h, i)
We examined histological sections and confocal slices through ventricles from 6–10 wpf animals, which confirmed that a new layer of ventricular muscle had emerged externally to the wall of single-cardiomyocyte thickness present at earlier stages (Fig. 3c, g). As indicated by whole-mount imaging, this external layer typically displayed substantial regions of clonally related cardiomyocytes. We will refer subsequently to the inner wall muscle as the ‘primordial layer’, as it retains the same single-cardiomyocyte thickness and characteristics of the embryonic ventricle throughout subsequent life stages. We will refer to the late-emerging, outermost muscle of the ventricular wall as the ‘cortical layer’. Thus, by multicolor clonal analysis, we revealed two developmentally distinct and previously unrecognized forms of ventricular wall myocardium.
Dominant clones build adult wall muscle
We next examined ventricles from mature, 90 dpf animals that had undergone cardiomyocyte labeling at 2 dpf. At this stage, the entire surface of the ventricle in each animal was covered by cortical cardiomyocytes (Fig. 3h; Supplementary Fig. 7). Similar to the clonal representation of the primordial layer at earlier stages, cortical clone patterns appeared different in each animal. However, a common theme for each ventricle was the presence of one or two large cortical clones at the base of the heart extending across the ventricular surface toward the apex. We infer from our timepoint analysis that these areas of cortex formed by a base-to-apex wave of expansion over the primordial layer. A number of smaller surface clones were apparent at other locations. Clones converged by weaving or fitting with some overlay into each other, and were penetrated by coronary vessels (Supplementary Fig. 8).
Histological analysis of adult ventricular tissue indicated that the primordial layer remained at single-cardiomyocyte thickness and was comprised of multiple color clones. This appearance contrasted with the overlying cortical myocardium, which was predominantly single-color and could be several cells thick (Fig. 3i). We calculated the areas of surface clones from whole-mount images of ventricles that displayed basal clones of a recombined color. Ten of 20 ventricles met this criterion, each of which had a large cortical clone covering 30–60% of the total ventricular surface (Fig. 3j). An average of 7.8 ± 1.2 clones contributed the entire cortical muscle of each ventricle (n = 10; Fig. 3k), a number several times lower than that clonal surface representation of the much smaller juvenile ventricle. Thus, a rare group of ~8 clonally dominant cardiomyocytes in the embryonic ventricle ultimately contribute to building the adult cortical myocardium.
Muscle lineage regeneration after injury
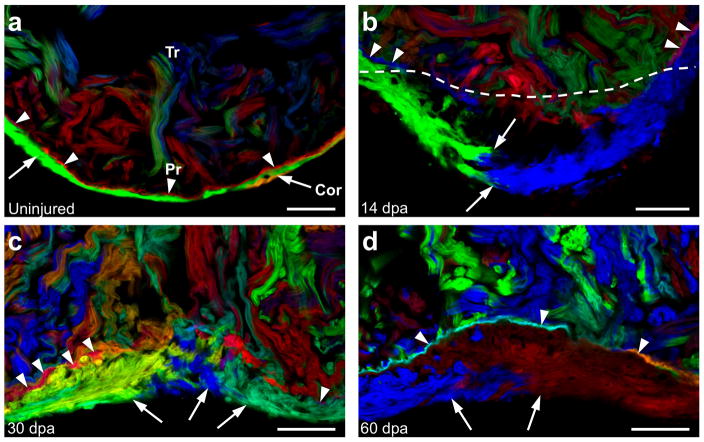
Zebrafish possess a robust capacity for heart regeneration throughout life18, based on the ability to activate the proliferation of spared cardiomyocytes after injury11,19,20. We examined the regenerative potentials of primordial and cortical muscle by amputating ventricular apices from 90 dpf cmlc2:CreER; priZm animals that had been labeled at 2 dpf. At 14 days post-amputation (dpa), we detected growth of adjacent cortical muscle clones in lateral and radial directions into the wound area (Fig. 4a, b), but the primordial layer lagged behind the amputation site. By 30 dpa, as the wall was reconstructed with clonal patches of cortical cardiomyocytes, clones of single-cardiomyocyte thickness contiguous with the primordial layer first became detectable in the regenerate (Fig. 4c). By 60 dpa, the primordial layer was largely restored as a complete structure positioned between cortical and trabecular muscle (Fig. 4d). Thus, regenerating primordial muscle undergoes restricted lateral expansion as during morphogenesis, while cortical muscle regeneration is less constrained and assumes the primary component of the new wall. Interestingly, these events occur in a temporally reversed manner compared to initial morphogenesis, with the underlying primordial layer regenerating subsequent to cortical muscle.
Figure 4. Regeneration of cortical and primordial muscle after injury.
a, Section of an uninjured ventricular apex, indicating the primordial (arrowheads) and cortical (arrows) layers. b, Regenerating ventricular apex at 14 days after resection (dpa). Cortical muscle clones converge within the injury site, while the primordial layer lags behind. Dashed line indicates amputation plane. c, 30 dpa ventricular apex, indicating multiple cortical clones and an incomplete primordial layer. d, Regenerated ventricular apex at 60 dpa, containing cortical muscle overlying a mostly contiguous layer of primordial muscle. n=6 animals for each timepoint. Scale bars, 50 μm
Origins of dominant cardiomyocyte clones
To determine when cortical cardiomyocytes originate during heart morphogenesis, we initiated color labeling at 30 dpf, after formation of juvenile structure but 1–2 weeks prior to emergence of cortical muscle. Strikingly, 90 dpf ventricles that had been incubated briefly with 4-HT at 30 dpf displayed surface representation by large clonal patches (Fig. 5a and Supplementary Fig. 9), with largest clones present at the chamber base. We quantified the size and number of clones in ventricles with basal clones of recombined colors (11 of 20 animals). Our data indicated that adult cortical myocardium arises in patches of diverse sizes from an average of 8.6 ± 0.7 labeled 30 dpf cardiomyocytes (Fig. 5b, c), a number similar to that observed after labeling at 2 dpf.
Figure 5. Origins of clonally dominant cardiomyocytes.
a, 90 dpf ventricular surface, after 30 dpf 4-HT labeling. b, %Surface area occupied by 90 dpf clones (58 clones, 11 ventricles). c, Surface clones per ventricle (n=11). d–g, 8 wpf ventricular surface (d, f) and confocal slice (e, g), after 30 dpf 4-HT labeling. Cyan cortical muscle (arrows, f, g) overlies primordial muscle (arrowheads) that is largely red/unlabeled, and cyan trabeculae (arrow, g). h–k, 8 wpf ventricular surface (h) and confocal slices (i–k) after limited 4-HT labeling, showing no obvious primordial muscle labeling (arrowheads, i–k). Arrows indicate green (h, i (left); j) and hazel (h, i (right); k) cortical/trabecular clones. All cortical muscle in h is outlined in white. Scale bars, 50 μm
In some ventricles assessed 4 weeks after labeling (8 wpf), we observed trabecular muscle of the same color near the emergent cortical clone (Fig. 5e, g). To confirm this association, we examined sections of 6–7 wpf ventricles (labeled at 2 dpf), and consistently found nearby trabecular muscle of the same color as the small, basal cortical clone (13 of 13 ventricles; see cyan clone in Fig. 3c). We also could identify cases from these examples in which trabecular and cortical muscle of one color connected through an apparent breach in the primordial layer (Supplementary Fig. 10). These observations suggested a clonal relationship between trabecular and cortical cardiomyocytes in the maturing zebrafish ventricle.
We noticed lower color recombination in the primordial layer (~15%) than in the trabecular and cortical muscle lineages (~61%) at 8 wpf (Fig. 5d–g). Taking advantage of this differential labeling efficiency, we titrated 4-HT and identified a low dose that induced sparse recombination at 30 dpf in trabeculae and no obvious recombination in primordial muscle. Importantly, even with very limited labeling, cortical clones of a recombined color were still discernable in 3 of 24 ventricles at 8 wpf (Fig. 5h, i), a finding that reaffirmed a trabecular source for the cortical lineage. As with other samples mentioned above, images of confocal slices from these ventricles could identify single-color clones containing both cortical and trabecular muscle, connecting through an apparent breach in primordial muscle of unrecombined color (Fig. 5i–k).
In total, our findings indicate a dynamic mechanism that generates a final ventricular muscle lineage and completes adult cardiac structure. Trabecular cardiomyocytes penetrate the primordial layer in rare, spatially segregated events at the juvenile stage, seed the ventricular surface, and undergo expansion to create the cortical myocardium.
Discussion
Here, we applied a multicolor clonal analysis and discovered unsuspected cellular mechanisms guiding heart morphogenesis in zebrafish. We found that three ventricular myocardial lineages are present in the adult form: primordial, trabecular, and cortical muscle, created in order from each other. Our data indicate that the innermost trabecular lineage is initiated predominantly by delamination from the embryonic primordial layer, as proposed in an earlier study17. Next, the outermost cortical layer is created as juveniles mature to adults, emerging wholly or in part via an ‘inside-out’ mechanism from trabeculae. Here, one or more particularly expansive juvenile cardiomyocytes access the ventricular surface at each of only ~8 sites per animal.
Zebrafish share several aspects of cardiac development with higher vertebrates, and the multicolor results we report here indicate some basic commonalities with results of single-marker clonal analysis performed previously in early mouse and chick embryos. For instance, cardiomyocytes were also observed in coherent clonal populations during cardiac growth in these systems, after an early phase of dispersion9,10. Most unpredicted is that the zebrafish ventricle maintains a primordial layer of single cardiomyocyte thickness without noticeable cell division in the z-plane, and that wall thickening instead occurs after seeding of a separate cortical lineage by dominant clones. It will be interesting to determine the evolutionary distribution of aspects of this uncovered morphogenetic mechanism within other bony fish and among other classes of vertebrates like mammals.
The origins and diversity of clonal patterns of cortical muscle we report here are more consistent with a stochastic model of source cell selection than a hierarchical or predetermined model. Additionally, the observation of clonal dominance in cardiomyocytes is reminiscent of stem cell compartments that drift toward clonality during homeostatic tissue maintenance, events that have been explained by stochastic models21–24. We suspect that molecular and/or physiologic cues enable the emergence and proliferative capacity of cortical cardiomyocytes, ostensibly with preferential influence at the ventricular base. It is likely that clonal dominance behavior is a recurring mechanistic strategy to help shape vertebrate organs.
METHODS SUMMARY
Wild-type or transgenic zebrafish of the EK/AB background were used for all experiments and maintained at 3 fish/liter. Ventricular resection surgeries were performed as described previously18. All published transgenic strains used here are listed in the separate Methods section and were analyzed as hemizygotes. Tg(β-act2:Brainbow1.0L)pd49 was generated and 4-HT labeling experiments were performed as described in Methods. Hearts were extracted at the indicated timepoints and fixed in 4% paraformaldehyde. After rinsing, the atrium was removed and the ventricle was placed on a cover slip in Fluoromount G. Another cover slip was used to gently compress the ventricle, allowing imaging of both ventricular surfaces. For surface images of whole-mounted ventricles, the z-position was adjusted until only surface muscle was visible. For confocal slices through whole-mounted hearts, the z-position was adjusted through the ventricle until trabecular muscle could be visualized. For histological analysis, 50 μm cryosections were mounted with Fluoromount G. Images from all multicolor samples were acquired using a Leica SP5 AOBS microscope equipped with X20 (0.7 NA) and X40 (1.25 NA) objectives. Antibodies were not used to enhance fluorescence. 458 nm, 515 nm, and 561 nm lasers were used to excite CFP, YFP, and RFP, respectively. Each channel was acquired sequentially and imported into ImageJ, where channels were overlaid. Uniform contrast and brightness adjustments were made using Adobe Photoshop. To visualize the entire outer surface area, images were joined using Photoshop. To quantify clone area, the size of a given clone in μm2 was traced using ImageJ software. The percentage area occupied by a clone was calculated by dividing its measured area by the total surface area of both sides of the ventricle.
METHODS
Transgenic lines
Published transgenic strains used in this study were (Tg(cmlc2:CreER)pd10)11, (Tg(gata5:loxP-mCherry-STOP-loxP-nucEGFP)pd40)11, (Tg(cmlc2:EGFP)f1)25, (Tg(cmlc2:nucDsRed2)f2)25, and (Tg(β-actin:HRAS-EGFP)vu119)26. Experiments with zebrafish were approved by Institutional Animal Care and Use Committee at Duke University.
Construction of priZm
The Brainbow 1.0L plasmid was digested with NheI and partially digested with NotI to obtain the entire cassette, and subcloned downstream of the 9.8 kb zebrafish β-actin2 promoter from the β-act2:RSG construct11. The resulting plasmid was linearized by I-SceI digestion and injected into one-cell zebrafish embryos. Red fluorescent protein (RFP) is the initial reporter cassette and default expression state from this construct. Two paired sites, lox2272 and loxp, exist in the construct that enable Cre recombinase-mediated switching to expression of either Cyan (CFP) or Yellow fluorescent proteins (YFP).
Thirty founder lines were isolated, and Tg(β-act2:Brainbow1.0L)pd49 was used for this study. The zebrafish β-actin2 promoter does not drive expression in epicardial or endocardial cells11, facilitating clear visualization of priZm cardiomyocytes.
4-HT labeling
For 4-HT labeling of cmlc2:CreER; priZm embryos, 2 dpf embryos were placed in egg water with 4-HT added to a final concentration of 4 μM, from a 1 mM stock made in 100% ethanol. Embryos were treated for 6 hours, rinsed once, and placed in fresh egg water. This labeling protocol was highly reproducible and induced recombination and appearance of diverse, non-red colors on half of the ventricular surface, calculated by digital quantification of red and non-red surface areas from whole-mount images of 30 dpf ventricles (50.0%; n = 10). By contrast, increased or reduced presence of 4-HT led to higher and lower recombination frequencies, respectively, that each reduced color diversity.
For labeling of cmlc2:CreER; priZm juveniles at 30 dpf, animals were incubated in 1μM 4-HT in aquarium water for 3 hours. To specifically label trabecular muscle at 30 dpf, animals were incubated in 0.25 μM 4-HT for 1 hour.
Embryonic ventricular cardiomyocyte quantification and proliferation indices
cmlc2:nucDsRed2; cmlc2:EGFP double transgenic animals were raised to 3 dpf, fixed, sectioned at 30 μm, and stained with an antibody to DsRed2. Confocal stacks of the entire heart from each embryo were taken, and the number of myocytes within the ventricular wall was determined from 3D projections generated using Imaris. To determine effects of 4-HT on cardiomyocyte proliferation, we treated 2 dpf embryos with 4 μM 4-HT for 6 hrs and fixed at 3 dpf. Ten μm sections were stained with Mef2 and PCNA as described27, and cardiomyocyte proliferation indices for each group were calculated from 3 ventricular sections. The numbers of Mef2+ and Mef2+/PCNA+ cells were manually counted with the aid of ImageJ software.
Examination of effects of color combinations on cardiomyocyte clone size
To examine the possibility that recombined color combinations had differential effects on cardiomyocyte proliferation, we examined color diversity in the largest and smallest cortical clones of 90 dpf ventricles that were labeled at 2 or 30 dpf. From 21 ventricles, 16 unique colors were represented in the 21 large basal clones. Seventeen unique colors were represented in 21 of the smallest clones. Individual colors were observed in both small and large clone groups, including two shades of gray (in which the clone is expressing all three fluorescent proteins). This distribution was consistent with the idea that color combinations do not have significant effects on proliferative ability.
Supplementary Material
Acknowledgments
We thank K. Kikuchi for generating cmlc2:CreER animals and for advice; J. Burris, A. Eastes, P. Williams, and N. Blake for zebrafish care; A. Dickson for artwork; B. Hogan and Poss lab members for comments on the manuscript; and S. Johnson and Y. Gao for imaging advice. V.G. was supported by a National Heart, Lung, and Blood Institute (NHLBI) Medical Scientist Training Program supplement. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute. This work was supported by grants from NHLBI (HL081674) and American Heart Association to K.D.P.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. V.G. and K.D.P. designed experimental strategy, analyzed data, and prepared the manuscript. V.G. performed all of the experiments.
Author Information. The authors declare no competing financial interests.
References
- 1.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y, et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 7.Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- 8.Stainier DY, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I Myocardial fate map and heart tube formation. Development. 1993;119:31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- 9.Mikawa T, Borisov A, Brown AM, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev Dyn. 1992;193:11–23. doi: 10.1002/aja.1001930104. [DOI] [PubMed] [Google Scholar]
- 10.Meilhac SM, et al. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130:3877–3889. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auman HJ, et al. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011 doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazic S, Scott IC. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev Biol. 2011;354:123–133. doi: 10.1016/j.ydbio.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Hu N, Yost HJ, Clark EB. Cardiac morphology and blood pressure in the adult zebrafish. Anat Rec. 2001;264:1–12. doi: 10.1002/ar.1111. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137:3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell. 2010;7:214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 24.Doupe DP, Klein AM, Simons BD, Jones PH. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Burns CG, et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- 26.Cooper MS, et al. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Dev Dyn. 2005;232:359–368. doi: 10.1002/dvdy.20252. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.