Abstract
Copy number variants (CNVs) are a recently recognized class of human germ line polymorphisms and are associated with a variety of human diseases, including cancer. Because of the strong genetic influence on prostate cancer, we sought to identify functionally active CNVs associated with susceptibility of this cancer type. We queried low-frequency biallelic CNVs from 1,903 men of Caucasian origin enrolled in the Tyrol Prostate Specific Antigen Screening Cohort and discovered two CNVs strongly associated with prostate cancer risk. The first risk locus (P = 7.7 × 10−4, odds ratio = 2.78) maps to 15q21.3 and overlaps a noncoding enhancer element that contains multiple activator protein 1 (AP-1) transcription factor binding sites. Chromosome conformation capture (Hi-C) data suggested direct cis-interactions with distant genes. The second risk locus (P = 2.6 × 10−3, odds ratio = 4.8) maps to the α-1,3-mannosyl-glycoprotein 4-β-N-acetylglucosaminyltransferase C (MGAT4C) gene on 12q21.31. In vitro cell-line assays found this gene to significantly modulate cell proliferation and migration in both benign and cancer prostate cells. Furthermore, MGAT4C was significantly overexpressed in metastatic versus localized prostate cancer. These two risk associations were replicated in an independent PSA-screened cohort of 800 men (15q21.3, combined P = 0.006; 12q21.31, combined P = 0.026). These findings establish noncoding and coding germ line CNVs as significant risk factors for prostate cancer susceptibility and implicate their role in disease development and progression.
Keywords: cancer genetics, functionally active DNA loci, cancer predisposition, metabolism, chromosome looping
Prostate cancer is a leading cause of cancer death in males throughout the world (1) and demonstrates the largest estimated effect of heritability among the most common tumor types, as determined by a Scandinavian twin-based registration study (2). Emerging insights into the genetics of constitutional disease etiology demonstrate that germ line polymorphisms in the form of copy number variants (CNVs) (3, 4), both de novo and inherited, are associated with diseases, including Alzheimer's, Parkinson, mental retardation, autism, and schizophrenia (5). CNVs also confer risk to developing cancers, such as neuroblastoma, and are enriched in Li-Fraumeni syndrome (6, 7). Therefore, given the strong heritable nature of prostate cancer, we sought to detect clinically-informative prostate cancer risk CNVs in the setting of widespread prostate-specific antigen (PSA) screening, as practiced in the United States (8) and Western Europe (9). To this end, we examined peripheral blood samples from the Tyrol Early Prostate Cancer Detection Program (10, 11). This cohort is a population-based prostate cancer screening program started in 1993 that intended to evaluate the utility of intensive PSA screening in reducing prostate cancer-specific death. We here report the results of a comprehensive large scale, unbiased study using this patient population to study the contribution of germ line CNVs toward prostate cancer risk.
Results
All men enrolled in the program came from the same “at risk” population, characterized by elevated age-adjusted PSA levels (10, 11). Cases were defined as men with biopsy-confirmed prostate cancer, and controls were defined as men with benign prostate biopsy results and no cancer diagnosis on available follow-up data. Demographics are presented in Dataset S1, Table S1. Cases and controls were profiled using the Affymetrix 6.0 Whole Genome SNP Array platform. After applying strict data-quality filters (12), the final study cohort consisted of 1,903 unrelated individuals of Caucasian origin (867 cases and 1,036 controls).
SNPs Associated with Prostate Cancer Risk.
Because several past genetic studies have reported on SNPs associated with risk for prostate cancer (13), we first queried 53 such reported risk SNPs in our study cohort (SNP selection criteria in Methods) and replicated associations for 22 of them (Dataset S1, Table S2), including SNPs at 3q21.3, 4q24, 6q25.3, 7p15.2, 7q21.3, 8q24.21, 11p15.5, 11q13.2, 11q13.3, 12q13.13, and 17q24.3. The strongest signal was detected for prostate cancer and for aggressive (14) prostate cancer at 11q13.3 [rs7130881; odds ratio (OR) = 1.52, P = 8 × 10−5 and OR = 1.624, P = 6 × 10−5, respectively]. The risk SNP replication rate is comparable to other recent studies (15, 16) and could be attributed to cohort and disease heterogeneity (17, 18).
CNVs.
We systematically identified CNVs along the genome of each individual from the Tyrol cohort (see Methods). This process resulted in a map of 2,611 CNVs, where 11.5% of CNVs were smaller than 20 kb in size and 41.7% of CNVs mapped to gene-coding areas. The vast majority of CNVs (94.6%) were present in the population with two or three copy number states and overall 34.1% of the CNVs exhibited minor allele frequencies ≤ 5%. Dataset S1, Table S3 summarizes the CNV characteristics (i.e., size, frequency).
To prioritize association studies of the identified CNVs, we reasoned that new functionally important risk markers for prostate cancer would be deletions with moderate or low frequency. Deletions are often associated with diseases (19), and less-common variants generally have a higher impact in driving human diseases (20). In addition, low-frequency CNVs are not well-tagged by SNPs (21) and, therefore, would carry distinct information from previously studied common SNPs that have been evaluated for their potential association with prostate cancer (22). We anticipated that functionally active low-frequency deletions might unravel an as yet unexplored portion of the genetic background.
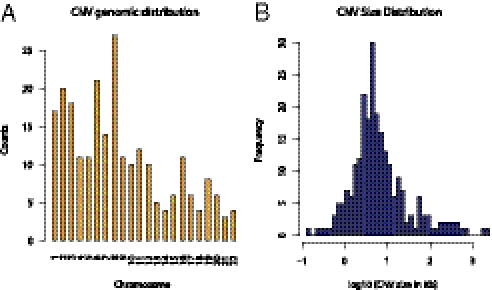
Therefore, priority was given to CNVs that had functional constraints, including potentially altering gene function (e.g., gene-coding variants and variants overlapping regulatory elements). Regulatory elements are characterized by the presence of consistent H3K4Me1 histone marks in the ENCODE ChIP DNA sequencing (ChIP-seq) data (23). Overall, 238 deletion CNVs fulfilled the selection criteria, including functional constraints with minor allele frequencies below or equal to 10%, and were deemed candidates for the association analysis with the disease state. The genomic characteristics of the 238 CNVs are summarized in Fig. 1 and Fig. S1. The variants are distributed along the autosomal chromosomes, with median size range between 10 and 100 Kb, with about 75% mapping to coding regions and 25% overlapping enhancer sites. To help prioritize the risk variants for subsequent in vitro functional assays, each gene-coding variant from the 238 set was also characterized for transcript abundance differentiation among the copy number states using human benign and cancer prostate tissues. About 10% of the gene-coding variants showed significant association with gene transcript levels.
Fig. 1.
Genomic characteristics of the transcriptionally active, low-frequency, deletion CNVs selected for prostate cancer risk association analysis. (A) Genomic location along the 22 autosomal chromosomes. (B) CNV size distribution.
Table 1 shows the characteristics of the top CNVs identified as susceptibility markers for prostate cancer in the Tyrol cohort upon age and preoperative PSA adjustment (P ≤ 0.01 and false-discovery rate < 0.2). Dataset S1, Table S4 includes extended association results for all of the 238 variants included in the analysis.
Table 1.
Low-frequency deletion CNVs associated with prostate cancer risk in the Tyrol cohort (P ≤ 0.01, false-discovery rate < 0.2)
| Allele test (age- and PSA-adjusted) |
||||||
| CNV genomic region (GRCh37/hg19) | Gene symbol/Entrez gene ID | Minor allele frequency | Risk | P | OR (CI 97.5%) | False-discovery rate |
| chr15:54197663–54203357 | — | 0.030 | Deletion | 0.0008 | 2.78 (1.55–5.14) | 0.087 |
| chr19:36828985–36847652 | ZFP14/57677 | 0.050 | Deletion | 0.0018 | 0.49 (0.31–0.76) | 0.095 |
| chr12:86428508–86435479 | MGAT4C/25834 | 0.014 | Deletion | 0.0026 | 4.80 (1.86–14.89) | 0.097 |
| chr8:3718722–3720335 | CSMD1/64478 | 0.038 | Deletion | 0.0041 | 0.47 (0.27–0.78) | 0.108 |
| chr8:145685468–145691484 | CYHR1/50626 | 0.012 | Deletion | 0.0049 | 6.24 (1.97–27.63) | 0.111 |
| chr7:4089162–4092270 | SDK1/730351 | 0.009 | Deletion | 0.0106 | 3.97 (1.39–11.75) | 0.175 |
The CNV that showed the strongest association with prostate cancer risk maps to a noncoding area on 15q21.3 (P = 7.7 × 10−4, OR = 2.78); that is, the risk allele (deletion allele) poses 2.8-times higher odds of having prostate cancer compared with the normal allele. Interestingly, this variant also scores as significant for aggressive prostate cancer association (P = 0.009, OR = 2.36). Among the top-ranked gene coding significant risk variants (Table 1), α-1,3-mannosyl-glycoprotein 4-β-N-acetylglucosaminyltransferase C (MGAT4C) CNV (P = 2.6 × 10−3, OR = 4.8), located on 12q21.31, also exhibited significant deregulation of the gene transcript expression with respect to copy number state (where ZFP14 did not), and was therefore prioritized for further investigation together with the 15q21.3 locus.
These two genomic regions, 15q21.3 and 12q21.31, have both been previously recognized as copy number variable by Conrad et al. (24) and by other high-resolution/sequencing studies, including Pang et al. (21) and Ju et al. (25). The estimated minor allele frequencies for the HapMap samples of Utah residents with ancestry from northern and western Europe (CEU) origin were equal to 0.017 [95% confidence interval (CI) 0.004, 0.054] and 0.018 (95% CI: 0.005, 0.056) for the 15q21.3 and 12q21.31 CNV regions, respectively. These data were consistent with the frequency of the exposured allele in the control set of our study, 0.0289 (95% CI: 0.0220, 0.0377) and 0.0131 (95% CI: 0.0087, 0.0196), respectively. As expected (21), the two low-frequency variants are not in linkage disequilibrium with known SNPs and thus have not been previously explored as risk polymorphisms for prostate cancer. Fig. S2 shows the University of California at Santa Cruz (UCSC) browser view of the haplo-block information for these two regions.
Replication of Prostate Cancer Risk Variants in Independent PSA Screening Cohort.
The association of the two risk variants with prostate cancer was then queried in an independent United States PSA-screening cohort from the Early Detection Research Network (26) (EDRN) consisting of 346 cases and 454 controls (Dataset S1, Table S5) from three recruitment sites. The 15q21.3 variant showed significant association with the aggressive phenotype (P = 0.05) consistent with the Tyrol cohort data and with prostate cancer risk in the Cornell subcohort (P = 0.005). The gene-coding variant mapping to MGAT4C showed consistent association with the aggressive phenotype (P = 0.067), with a stronger signal observed when the control set was confined to men of greater age (P = 0.037). The analysis for prostate cancer risk and aggressive phenotype in the combined Tyrol and EDRN cohorts showed significant association for both the 15q21.3 variant (P = 0.006 and P = 0.052, respectively) and the 12q21.31 variant (P = 0.026 and P = 0.059, respectively). Overall, the 15q21.3 and 12q21.31 associations detected in the Tyrol cohort were recapitulated in the EDRN series.
Role of the Noncoding Risk CNV.
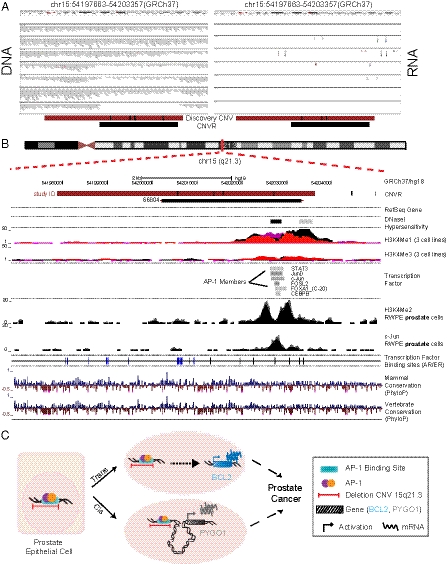
To investigate the potential functional role of the noncoding CNV, 15q21.3, in conferring prostate cancer risk, we analyzed the recently published next-generation sequencing data from seven human prostate cancers (27) to identify any potential nonannotated transcribed gene at chr15q21.3. This analysis did not show any evidence of transcript expression (Fig. 2A). Hence, we surmised this region was likely a distal regulatory element. Consistent with the presence of a regulatory element (H3K4me1), ENCODE ChIP-seq data also showed that the locus was bound by three members of the activator protein 1 (AP-1) complex of transcription factors (c-Jun, JunD, and FOSL2) in various cell lines. We therefore interrogated this locus in RWPE1 benign prostate cells by ChIP-seq (see SI Methods) where we detected a H3K4me2 double peak and confirmed c-Jun binding signal (Fig. 2B). In addition, this locus harbors several putative androgen and estrogen receptor binding sites (28, 29), suggesting that it may be under hormonal regulation.
Fig. 2.
Genomic region for the noncoding CNV on chromosome region 15q21.3 associated with aggressive prostate cancer risk. (A) DNA and RNA views of seven individuals at chr15q21.3locus. Paired DNA (Left) and RNA (Right) data are visualized for seven individuals with different numbers of DNA copies (two copies, one-copy loss, two-copy loss) at the locus of interest (27). No transcripts are present. (B) The tracks include the following information: Prostate cancer-associated CNV on 15q21.3 (dark red; coordinates include midpoint distance to neighboring marker on the genotyping platform; black vertical bars on platform indicate probe locations); the locus corresponding to Variation_66804 in Conrad et al. (24) (black); RefSeq track for validated transcripts; ENCODE DNasel hypersensitivity; ENCODE mono and trimethylation tracks, H3K4Me1 and H3K4Me3, consistent with enhancer activity (48); ENCODE transcription factor-binding signal (intensities are proportional to binding support); H3K4Me2 and c-Jun ChIP-seq tracks in RWPE cells; predicted androgen receptor (AR) (black) and estrogen receptor (ER) (blue) binding sites (28, 29); and sequence conservation tracks. The positions are to scale and adapted from UCSC genome browser, Feb2009 (GRCh37/hg19) assembly (http://genome.ucsc.edu/). (C) Summary of functional effects of the intergenic 15q21.3 prostate cancer risk locus. This CNV overlaps a potential regulatory element with AP-1 transcription factor binding sites. Our data support a trans- and cis-regulatory role for this region. Presence of deletion in this CNV region is associated with expression of target genes both in cis and trans, potentially leading to prostate cancer predisposition.
To understand the function of this enhancer region, we carried out an association analysis between the CNV deletion allele (risk allele) and transcript expression levels of the AP-1 trans-target genes (30) by querying data generated from human benign and cancer prostate tissues (Dataset S1, Table S6). This analysis demonstrated that the presence of the risk allele in both the prostate tissue datasets was significantly correlated with transcript expression of the proto-oncogene B-cell lymphoma 2 (BCL2), an AP-1 target gene located on 18q21.33 (Fig. S3). Individuals carrying the risk allele showed higher transcript expression.
Next, we used a nonbiased approach of chromosome conformation capture, referred to as Hi-C (31) (see SI Methods) to identify potential long-range cis-gene interactions. We used the benign prostate epithelial cell line, RWPE1, for performing the chromatin-interaction experiment. This study revealed significant interactions between the locus on 15q21.3 and multiple gene promoters and microRNA promoter sites within a 50-Mb window (Dataset S1, Table S7). Gene-set enrichment analysis showed transcriptional regulation activity as being significantly overrepresented in our gene set (P = 0.017). Association analysis was performed between the CNV copy number states and transcript expression of the cis-interacting genes. Among the genes with consistent transcript level association, pygopus homolog 1 (PYGO1) gene showed the strongest signal (P = 0.0002) (Dataset S1, Table S7).
Taken together, these data suggest that the noncoding locus identified as protective against prostate cancer development is a regulatory region, potentially involved in the active regulation of both cis- and trans-gene expression (Fig. 2C).
Functional Characterization of the Coding Risk CNV.
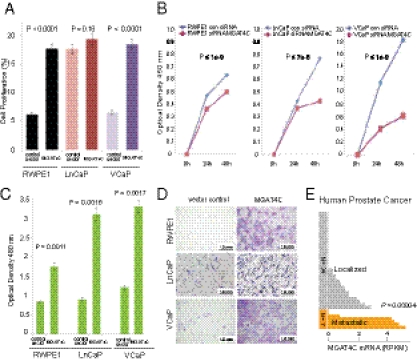
Next, we carried out functional characterization of the gene coding CNV associated with prostate cancer risk involving the MGAT4C gene. Association analysis demonstrated an intronic deletion in MGAT4C to be the prostate cancer risk allele. MGAT4C transcript abundance analysis suggested an inverse association between copy number states and transcript levels in human prostate tissues (both benign and tumor) and in lymphoblastoid cells (P = 0.049, P = 0.041, and P = 0.032, respectively) (Fig. S4). This risk CNV in the intronic region of MGAT4C likely overlaps a repressor/insulator element, such that a deletion in this region leads to transcript overexpression, a scenario seen in several other instances (32–34). Two recent studies (34, 35) identified several deletion CNVs that were associated with increased transcript expression, similar to our observations with MGAT4C. To evaluate the effect of changes in transcript expression, we carried out in vitro experiments using prostate cell lines (SI Methods). Overexpression of MGAT4C in benign (RWPE1) and cancer (VCaP) prostate cell lines (Fig. S5), resulted in significant increase in cell proliferation (P < 0.0001) (Fig. 3A) and migration (P = 0.0011 and P = 0.0017, respectively) (Fig. 3 C and D). Knock down of MGAT4C expression with siRNA in one benign (RWPE1) and two prostate cancer cell lines (LNCaP and VCaP) resulted in significant decrease in proliferation (P ≤ 1 × 10−9, P < 1 × 10−7, and P < 1 × 10−9, respectively) (Fig. 3B). We next asked if MGAT4C levels vary with human prostate cancer progression and found that transcript levels were significantly higher in metastatic versus localized prostate cancers (P = 0.00004) (36) (Fig. 3E). Taken together, these data show that transcript overexpression associated with the deletion CNV results in changes associated with tumor progression.
Fig. 3.
Tumorigenic effects of MGAT4C on prostate cell lines. (A) Proliferation rate at 24 h in RWPE1, LNCaP, and VCaP cells transfected with MGAT4C. (B) Proliferation rate at 24 and 48 h in RWPE1, LNCaP, and VCaP cells following knockdown of MGAT4C expression. (C) Migration of MGAT4C overexpressing RWPE1, LnCaP, and VCaP cells compared with vector control cells (Boyden Chamber Assay). (D) Representative images of migrated cells for each cell line. (E) MGAT4C expression levels in human localized and lethal prostate cancer samples.
Discussion
In this unique large-scale study to investigate the association of functional CNVs and prostate cancer, we identified and characterized two low-frequency risk CNVs located on 15q21.3 and 12q21.31. The first risk CNV overlaps a noncoding gene desert area. Such noncoding CNVs might indirectly modulate gene transcription through distal regulatory elements and long-range interactions. Targeted disruption of enhancers has been shown to exert tumorigenic effects through remote transcriptional dysregulation in acute myeloid leukemia (37). We demonstrated that this risk CNV might be a regulatory site, as evidenced by the presence of experimentally determined AP-1 binding sites. AP-1 is a dimeric transcription factor that includes the JUN, FOS, activation transcription factor, and musculo-aponeurotic-fibrosarcoma protein families, with FOS and JUN being the most prevalent proteins in mammalian cells. AP-1 exerts both oncogenic and tumor-suppressive roles and does so by regulating genes involved in cell proliferation, differentiation, apoptosis, angiogenesis, and tumor invasion (30). FOS and JUN expression has been linked to prostate cancer progression, with JUN also being associated with relapse-free survival (38).
There are several mechanisms by which this noncoding risk CNV may lead to prostate carcinogenesis. The first evidence is based on the modulation of gene transcription in the presence of the noncoding risk CNV. To test the hypothesis that the risk locus bound by AP-1 family members exerts an effect in a tissue-specific manner in prostate (39), we queried mRNA expression of AP-1 target genes with respect to the risk-allele status in tumor and benign prostate tissues. Strikingly, we observed a statistically significant association between the expression of the BCL2 oncogene, an AP-1 target gene (30), and the noncoding CNV status in both the prostate tissue sets. Although not definitive, this result indicates that the noncoding CNV may play a role in tumor initiation and maintenance through control of BCL2.
The second evidence that suggests a potential functional role of this CNV comes from analysis of chromatin cis-interactions deduced from Hi-C data. We found cis-genes, the mRNA expression of which correlates with CNV status, and that are predicted to physically interact with the noncoding CNV according to Hi-C. These genes include PYGO1, one of the Pygopus proteins that are involved in Wnt signaling, where they act as coactivators by binding to the β-catenin complex. PYGO2, a member of the Pygopus family, has been shown to be overexpressed in breast cancer (40). Interestingly, an additional gene that was shown to interact with the risk CNV by Hi-C, Ras-related protein Rab-27A (RAB27A), a small GTPase, was seen to be differentially expressed in aggressive prostate cancer (41) and also, more recently, identified as a “driver” oncogene in aggressive melanoma (42). Overall, our finding suggests that the risk locus, which has all of the hallmarks of an AP-1–bound enhancer, often localizes with a transcription factory that also contains multiple genes involved in transcriptional regulation activity. Under some conditions, such as when AP-1 and its factors are active, this functionally coherent gene set may then be activated or repressed by our risk locus.
The other prostate cancer risk CNV we identified and followed-up on maps to an intron of the gene MGAT4C. This gene belongs to the family of glycosyltransferases, two of whose members, MGAT4A and MGAT4B, have been shown to be differentially regulated in pancreatic cancer (43). Glycosyltransferases are involved in the transfer of N-acetylglucosamine to the core mannose residues of N-linked glycans. By regulating N-glycan branching of cell adhesion molecules, such as E-cadherin, these enzymes play an important role in maintaining cell-cell adhesion in normal epithelia where the extent of branching is inversely correlated with epithelial cell adhesion (44). MGAT4C promotes branching, which results in reduced cell-cell adhesion (44). From our results, it appears that the presence of a germ line deletion CNV at this locus results in overexpression of the gene and, hence, demonstrates a mechanistic basis for the observed increase in invasion of prostate cells in the in vitro cell line assays. Hence, our results strongly support a role whereby MGAT4C could promote disease initiation and progression by reducing cell-cell adhesion and resulting in increased cell motility and migration.
Taken together, our findings demonstrate association of two low-frequency functionally active CNVs with prostate cancer risk. Functional characterization of these risk CNVs show that gene coding and noncoding “gene desert” germ line CNVs may directly or indirectly modulate the transcriptome machinery of known oncogenic pathways in prostate cancer enabling carcinogenesis. The genes and loci identified in this study are candidates for further functional investigation and for replication in independent cohorts and provide alternative information in the assessment of prostate cancer risk, so far limited to SNP variants. In vivo studies in transgenic mice or zebrafish model systems will potentially help elucidate the role of CNVs in disease development, as recently shown for type 2 diabetes and obesity (45).
Methods
The Tyrol PSA Screening Cohort represents a population-based PSA testing cohort in asymptomatic men initiated in 1993 and intended to evaluate the utility of intensive PSA screening in the reduction of prostate cancer-specific death. Cases were defined as men with biopsy-confirmed prostate cancer. Controls were defined as men with a benign prostate biopsy result and no cancer diagnosis in available follow-up data. The demographics and full description of this cohort and of sample collection and preparation for this study are available in Dataset S1, Table S1 and the SI Methods, respectively.
EDRN PSA-Screening Cohorts consists of three Prostate Cancer Clinical Validation Center institutions of Beth Israel Deaconess Medical Center (“Harvard”, Harvard University, Cambridge, MA), the University of Michigan (“Michigan,” Ann Arbor, MI), and Weill Cornell Medical College (“Cornell,” New York, NY). Using a common research protocol at the three institutions, men are prospectively enrolled who are at risk for prostate cancer in three catchment areas in the United States: Boston, MA, Southeast Michigan, and New York, NY, respectively. For this cohort, cases are defined as men diagnosed with prostate cancer and controls are men who have undergone prostate needle biopsy without any detectable prostate cancer and no prior history of prostate cancer. The demographics, cohort collection protocol of men enrolled in this trial and sample preparation are presented in Dataset S1, Table S5 and SI Methods, respectively. A total of 800 DNA samples from Caucasian individuals (based on self-declaration) (346 cases and 454 controls) that passed all quality controls were included in the validation study.
CNV Detection and Selection of CNVs for Prostate Cancer Risk Association Analysis.
For CNV characterization, the across-sample detection approach from Banerjee et al. (35) was applied. This approach takes advantage of the polymorphic signal across the entire sample set and helps improve on CNV detection. To benchmark the CNV detection algorithm and genotyping approach in the current study, we quantitatively compared its performance with the data from Conrad et al. (24), verified copy number states using high-coverage DNA sequencing data (27, 46), and performed qPCR on a selected set of CNVs (SI Methods, and Figs. S6 and S7). With the intent to ultimately query a comprehensive and well-characterized set of CNVs, we combined the variants detected by the across-sample approach (35) with variants from Conrad et al. (24), for a total number of 2,611 CNVs. All CNVs were genotyped as in Banerjee et al. (35).
The inclusion criteria for biallelic deletion, low-frequency (≤ 10%), gene coding, or functionally active (histone mark analysis) CNVs led to the selection of 238 variants (Dataset S1, Table S3B).
Quantitative PCR Validation for CNVs.
As additional validation step, a subset of CNVs was selected for genotype verification using TaqMan copy number assays (Applied Biosystems) as in Setlur et al. (47) (Fig. S6). The quantitative PCR assays developed for the prostate cancer risk CNVs at 15q21.3 and 12q21.31 were used on the EDRN cohort. Details are included in SI Methods.
Risk SNP Selection and Association Analysis.
A total of 56 published prostate cancer risk SNPs was queried in the Tyrol cohort, including variants from 10 studies. Dataset S1, Table S2 includes details on the original studies and associations for each SNP. Details on SNP selection, quality controls and association tests are reported in SI Methods. We considered an SNP to be concordantly associated so long as one of the test P value was ≤ 0.05 and the risk allele identified within the Tyrol cohort consistent with the reported risk allele.
Cell Lines for Functional Assays.
Human Prostate cell lines RWPE1, VCaP, LnCaP (Clone FGC) were purchased from American Type Culture Collection (ATCC); RWPE1: CRL-11609; LnCaP: CRL-1740; VCaP: CRL-2876. Details on functional assays are reported in SI Methods.
Description of Clinical Human Prostate Cancer Cohort and Transcript Data.
All prostate tissue samples were collected as part of Institutional Review Board-approved protocols at Weill Cornell Medical College. Details on human samples are reported in SI Methods.
Supplementary Material
Acknowledgments
We thank R. Kim for the Weill Cornell Institutional Biobank; W. M. Hussain, A. Romanel, and R. R. Hossain for technical support; H. Hauffe, C. Seifarth, and E. Steiner for organizing the clinical data; D. Govindaraju for his insightful comments on epistatic interactions; and the 1000 Genome Project, the International HapMap Consortium, and the Encyclopedia of DNA Elements (ENCODE) Project (locus-specific information). This work was supported in part by Early Detection Research Network National Cancer Institute Grants UO1CA113913 (to M.A.S., J.T.W., D.S.S., and M.A.R.) and U01CA11275 (to F.D., A.M.C., and M.A.R.); the Starr Cancer Consortium (F.D., S.R.S., S.B., C.L., and M.A.R.); Grant NCI-CA-R01-116337 (to F.D., S.B., and M.A.R.); the Clinical and Translational Science Center (F.D.); Department of Defense Grant PC094516 (to F.D.); and National Human Genome Research Institute Grants P41HG004221 and UO1HG005209 (to C.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Genotype and phenotype data reported in this paper have been deposited in the database of Genotypes and Phenotypes, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000487.v1.p1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117405109/-/DCSupplemental.
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer—Analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlien A, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2008;105:11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schröder FH, et al. ERSPC Investigators Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 9.Andriole GL, et al. PLCO Project Team Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartsch G, et al. Tyrol Prostate Cancer Screening Group Tyrol Prostate Cancer Demonstration Project: Early detection, treatment, outcome, incidence and mortality. BJU Int. 2008;101:809–816. doi: 10.1111/j.1464-410X.2008.07502.x. [DOI] [PubMed] [Google Scholar]
- 11.Oberaigner W, et al. Reduction of prostate cancer mortality in Tyrol, Austria, after introduction of prostate-specific antigen testing. Am J Epidemiol. 2006;164:376–384. doi: 10.1093/aje/kwj213. [DOI] [PubMed] [Google Scholar]
- 12.Oldridge DA, Banerjee S, Setlur SR, Sboner A, Demichelis F. Optimizing copy number variation analysis using genome-wide short sequence oligonucleotide arrays. Nucleic Acids Res. 2010;38:3275–3286. doi: 10.1093/nar/gkq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 2011;20:1599–1610. doi: 10.1158/1055-9965.EPI-11-0312. [DOI] [PubMed] [Google Scholar]
- 14.D'Amico AV, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 15.Jin G, et al. International Consortium for Prostate Cancer Genetics Validation of prostate cancer risk-related loci identified from genome-wide association studies using family-based association analysis: evidence from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum Genet. 2011 doi: 10.1007/s00439-011-1136-0. PMID: 22198737 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher FR, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niculescu AB, Le-Niculescu H. The P-value illusion: How to improve (psychiatric) genetic studies. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:847–849. doi: 10.1002/ajmg.b.31076. [DOI] [PubMed] [Google Scholar]
- 18.Lapointe J, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 19.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 20.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 21.Pang AW, et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11:R52. doi: 10.1186/gb-2010-11-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craddock N, et al. Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birney E, et al. ENCODE Project Consortium NISC Comparative Sequencing Program Baylor College of Medicine Human Genome Sequencing Center Washington University Genome Sequencing Center Broad Institute Children's Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad DF, et al. Wellcome Trust Case Control Consortium Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju YS, et al. Reference-unbiased copy number variant analysis using CGH microarrays. Nucleic Acids Res. 2010;38:e190. doi: 10.1093/nar/gkq730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre SJ, et al. Validation in a multiple urology practice cohort of the Prostate Cancer Prevention Trial calculator for predicting prostate cancer detection. J Urol. 2009;182:2653–2658. doi: 10.1016/j.juro.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farré D, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messeguer X, et al. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 30.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaloy C, et al. Deletion of WNK1 first intron results in misregulation of both isoforms in renal and extrarenal tissues. Hypertension. 2008;52:1149–1154. doi: 10.1161/HYPERTENSIONAHA.108.120899. [DOI] [PubMed] [Google Scholar]
- 33.Jin W, Bi W, Huang ES, Cote GJ. Glioblastoma cell-specific expression of fibroblast growth factor receptor-1beta requires an intronic repressor of RNA splicing. Cancer Res. 1999;59:316–319. [PubMed] [Google Scholar]
- 34.Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21:2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee S, et al. A computational framework discovers new copy number variants with functional importance. PLoS ONE. 2011;6:e17539. doi: 10.1371/journal.pone.0017539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steidl U, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang X, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2132–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 39.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 40.Andrews PG, Lake BB, Popadiuk C, Kao KR. Requirement of Pygopus 2 in breast cancer. Int J Oncol. 2007;30:357–363. [PubMed] [Google Scholar]
- 41.Setlur SR, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ide Y, et al. Aberrant expression of N-acetylglucosaminyltransferase-IVa and IVb (GnT-IVa and b) in pancreatic cancer. Biochem Biophys Res Commun. 2006;341:478–482. doi: 10.1016/j.bbrc.2005.12.208. [DOI] [PubMed] [Google Scholar]
- 44.Vagin O, Tokhtaeva E, Yakubov I, Shevchenko E, Sachs G. Inverse correlation between the extent of N-glycan branching and intercellular adhesion in epithelia. Contribution of the Na,K-ATPase beta1 subunit. J Biol Chem. 2008;283:2192–2202. doi: 10.1074/jbc.M704713200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragvin A, et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci USA. 2010;107:775–780. doi: 10.1073/pnas.0911591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durbin RM, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setlur SR, et al. Genetic variation of genes involved in dihydrotestosterone metabolism and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:229–239. doi: 10.1158/1055-9965.EPI-09-1018. [DOI] [PubMed] [Google Scholar]
- 48.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.