Abstract
Legumes and soil bacteria called rhizobia have coevolved a facultative nitrogen-fixing symbiosis. Establishment of the symbiosis requires bacterial entry via root hair infection threads and, in parallel, organogenesis of nodules that subsequently are invaded by bacteria. Tight control of nodulation and infection is required to maintain the mutualistic character of the interaction. Available evidence supports a passive bacterial role in nodulation and infection after the microsymbiont has triggered the symbiotic plant developmental program. Here we identify in Sinorhizobium meliloti, the Medicago symbiont, a cAMP-signaling regulatory cascade consisting of three receptor-like adenylate cyclases, a Crp-like regulator, and a target gene of unknown function. The cascade is activated specifically by a plant signal during nodule organogenesis. Cascade inactivation results in a hyperinfection phenotype consisting of abortive epidermal infection events uncoupled from nodulation. These findings show that, in response to a plant signal, rhizobia play an active role in the control of infection. We suggest that rhizobia may modulate the plant’s susceptibility to infection. This regulatory loop likely aims at optimizing legume infection.
Rhizobia are phylogenetically diverse bacteria that have achieved the ability to enter a nitrogen-fixing symbiosis with legumes (1). Rhizobia elicit the formation on roots of legume host plants of new organs called “nodules,” which rhizobia colonize intracellularly and in which they fix atmospheric nitrogen, to the benefit of the plant. The best-described mode of rhizobia entry into plant roots involves the formation of specific tubular structures called “infection threads” (ITs) that rhizobia initiate at the tip of susceptible root hairs. Extension and branching of ITs in direction of the root cortex ensures bacterial colonization of the forming nodule primordium. Ultimately, rhizobia leave ITs to form intracellular structures called “symbiosomes” that acquire competence in biological nitrogen fixation.
Nodule morphogenesis and rhizobial infection are tightly coordinated, although genetically dissectible, processes (2, 3). Flavonoid compounds present in legume root exudates combined with the rhizobial NodD transcriptional regulator(s) induce the synthesis of substituted lipochitooligosaccharides called “Nod factors,” which are required for nodule organogenesis and IT formation (3, 4). Additional bacterial molecules, such as low-molecular-weight exopolysaccharides, are required for IT elongation. However, their mode of action has not been elucidated thus far (5).
Available evidence indicates that the microsymbiont, after it has initiated the plant developmental program, has a passive role in the interaction that is dominated by the plant (3). Nodule number is regulated negatively by a variety of mechanisms including the systemic autoregulation of nodulation that involves signaling by CLE peptides (6–8). Bacterial infection of root tissues is controlled by the plant at the epidermal cell layer in close coordination with nodule development (2, 9). Hyperinfection, usually associated with abnormal nodule development, has been described in a few legume mutants (9–11). Finally, cysteine-rich plant peptides strictly control bacteroid proliferation and differentiation in galegoid legumes (12).
Here we report a slightly contrasting pattern in the Sinorhizobium meliloti–Medicago symbiosis, showing that the control of epidermal infection actually involves a sustained molecular dialogue between the two partners. Specifically, we have found that a plant signal activates a cAMP regulatory cascade in S. meliloti that, in turn, modulates the extent of epidermal infection, possibly by modifying the plant susceptibility to infection.
Results
Characterization of a cAMP Signal Transduction Cascade in S. meliloti.
S. meliloti sequencing revealed the occurrence of 26 type III purine nucleotidyl (adenylate and guanylate) cyclases in the genome (13). CyaD1 (SMc02176), CyaD2 (SMc04307), and CyaK (SMb20776) share the following structural organization: an amino-terminal signal peptide, a CHASE2 extracellular domain (IPR007890) (14), a set of three membrane-spanning domains, and a cytoplasmic catalytic domain (IPR001054) characteristic of type III adenylate cyclase/guanylate cyclase (AC/GC) enzymes. To characterize the substrate specificity of these putative cyclases, we measured cAMP and cGMP levels in vivo in the wild-type S. meliloti strain and in a cyaD1D2K triple mutant. In the wild-type strain, cAMP levels were ca. 20-fold higher than cGMP levels in vivo. Neither was affected by the simultaneous inactivation of all three cyclases, possibly because of functional redundancy. Instead, expression of a truncated form of CyaD1 in which the periplasmic CHASE2 domain was deleted in frame (pGMI50127; see Table S2) provoked a significant increase in cAMP content in S. meliloti cell extracts (Fig. 1). This increase was consistent with CyaD1 acting in vivo as an AC, possibly with a GC side activity, like most type III enzymes described so far (15). Sequence alignment of CyaD1, CyaD2, and CyaK with well-characterized ACs and GCs showed that a lysine residue (K56 in CyaD1) that is specifically conserved in ACs (16) is conserved in all three cyclases (Fig. S1), thus suggesting that CyaD2 and CyaK also are ACs.
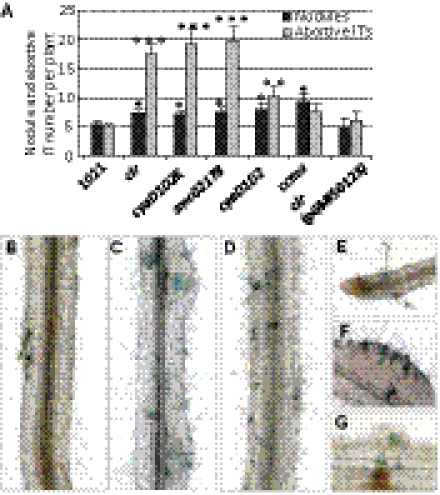
Fig. 1.
Functional characterization of the CyaD1 protein. ELISA measurements of intracellular cAMP and cGMP levels in S. meliloti strains 1021, cyaD1D2K, and clr and in the cyaD1ΔCHASE2-expressing derivative 1021 (pGMI50127). ***P < 0.001.
Next to cyaD1 on the S. meliloti chromosome, we identified a Crp-like transcriptional regulator (Clr; Smc02175) as well as a gene of unknown function, smc02178, that we have found to be a target gene for clr. Clr belongs to a family of 13 Crp-like proteins in S. meliloti that includes the FixK regulators of microoxic respiration and nitrogen-fixation genes whose activity is cAMP independent (17). Instead, the conservation in Clr of residues that bind cAMP in Escherichia coli CRP and other characterized cAMP-binding proteins (Fig. S2) predicted that Clr would be a cAMP-binding protein. We have found that clr is not involved in catabolite repression by succinate in S. meliloti.
A smc02178-lacZ reporter gene fusion was expressed at a very low level in free-living conditions in either synthetic Vincent medium (Fig. 2) or complex [tryptone-yeast extract (TY) or LB medium]. High expression could be driven by exogenously provided cAMP, in a clr-dependent manner, independently of cyaD1cyaD2cyaK (Fig. 2A). Similarly, endogenous production of cAMP by the plasmid construct pGMI50127 (cyaD1ΔCHASE2) led to high constitutive expression of the smc02178 target gene (Fig. 2B). Taken together, these data identify a regulatory cascade in which activated CyaD1, CyaD2, and CyaK cyclases produce cAMP that, together with the Clr transcriptional regulator, drives smc02178 gene expression.
Fig. 2.
cAMP drives clr-dependent activation of the smc02178-lacZ reporter gene fusion ex planta. cAMP was provided either exogenously (A) or endogenously by the constitutive cyaD1ΔCHASE2-expressing plasmid pGMI50127 (B). smc02178-lacZ activity in S. meliloti 1021, clr, and cyaD1D2K strains and in the cyaD1ΔCHASE2-expressing strain (1021 pGMI50127) is expressed in Miller units. Error bars indicate SE; n = 3. The mean value difference between strain 1021 and the cyaD1D2K mutant in A was not statistically significant (t = 0.07).
Plant Signal Triggers Symbiotic Activation of the Cascade.
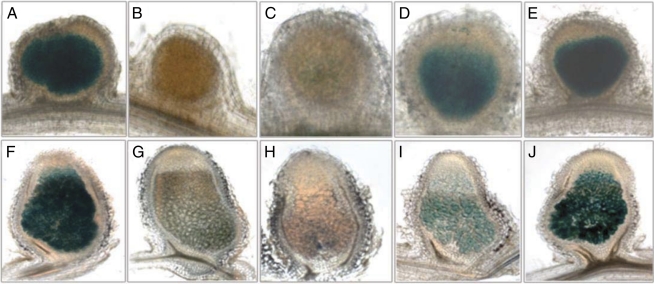
Expression of the smc02178-lacZ reporter gene fusion was observed in a S. meliloti 1021 wild-type background in all infected parts of young [7 d postinfection (dpi)] and mature (14 dpi) nodules of Medicago sativa (Fig. 3 A and F). By contrast, smc02178-lacZ expression was rarely observed in epidermal ITs. Inactivation of the clr gene or triple cyaD1D2K inactivation abolished expression of the smc02178-lacZ fusion in nodules (Fig. 3 B, C, G, and H). Individual inactivation of cyaD1, cyaD2, and cyaK or simultaneous inactivation of cyaD1 and cyaD2 had a partial effect on smc02178-lacZ expression in nodules (Fig. 3 D, E, I, and J), thus indicating that all three cyclases contribute to symbiotic activation.
Fig. 3.
Expression of the smc02178-lacZ reporter gene fusion in M. sativa nodules. Nodules were observed at 7 dpi (A–E) and 14 dpi (F–J) after inoculation with S. meliloti 1021 (A and F), clr (B and G), cyaD1D2K (C and H), cyaK (D and I), and cyaD1D2 strains (E and J) carrying pGD2178.
Nodule crude extracts prepared from 14-dpi-old nodules formed by a wild-type S. meliloti strain induced high expression of the smc02178-lacZ reporter gene fusion ex planta in a S. meliloti 1021 wild-type genetic background (Fig. 4A). Activation did not occur in either a clr mutant background or in a cyaD1D2K triple mutant (Fig. 4A) that excluded activation by the cAMP potentially present in nodules (18). This result suggested that a bona fide inducing signal was present in nodule extracts. Medicago root exudates, the nodulation gene inducer luteolin (4), cytokinin, or proline had no detectable effect on smc02178-lacZ expression. Similarly, low oxygen (19), high sucrose, or NaCl concentrations had no effect. No signaling activity could be detected in nonnodulated portions of inoculated roots or in noninoculated roots of M. sativa (Fig. 4A). By contrast, signaling activity was detected in shoots of both inoculated and noninoculated plants but at a lower specific activity than in nodules (Fig. 4A).
Fig. 4.
A plant signal triggers activation of the cAMP cascade in nodules. smc02178-lacZ expression was monitored ex planta in 1021, clr, and cyaD1D2K background strains after addition of M. sativa tissue extracts. (A) Extracts of nodules or nonnodulated portions of roots and shoots inoculated with S. meliloti 1021 or shoot and root extracts of noninoculated (NI) plants. (B) Bacteria-free nodule extracts of the spontaneously nodulating M. sativa Nar variant trigger cascade activation ex planta.
High inducing activity was present in sterile, spontaneous nodules formed by the nodulation in the absence of rhizobia (Nar) variant of M. sativa (Fig. 4B) (20), thus showing that the signal was of plant origin. Signal activity also was detected in noninvaded nodules induced by the exopolysaccharide-defective exoY mutant of S. meliloti (21) (Fig. S3). Hence signal presence in wild-type Medicago nodules did not require infection by bacteria.
High signaling activity also was detected in nodule and shoot extracts from pea and lotus as well as in the shoots of nonlegumes such as rice (Fig. S4).
Cascade Mutants Are Hyperinfective in M. sativa.
The cyaD1D2K, clr, and smc02178-null mutants induced normal, elongated, pink nodules with the same early kinetics as wild-type 1021. No significant increase in nodule number was observed over a large set of independent experiments (Figs. 5A and 6A; also see Fig. S6). The cyaD1D2K mutant’s nitrogen-fixation ability (as measured by acetylene-reduction assay) and whole-plant growth yield of plants (dry weight) were not statistically different from wild type (Fig. S5). Instead, a distinctive phenotype of the cyaD1D2K, clr, and smc02178 mutants was a three- to fourfold increase in the total number of epidermal ITs formed. This increase was statistically highly significant (P < 0.001) at 14 dpi (Fig. 5A) but could be observed as early as 7 dpi, although with more variation (Fig. S6). As a control, we monitored infection by the S. meliloti ccmA mutant that is completely defective in nitrogen fixation (22). The ccmA mutant formed significantly more nodules than wild type, as expected for a Fix− mutant, but did not display a hyperinfection phenotype (Fig. 5A), thus ruling out the possibility that hyperinfection could result from a slight decrease in nitrogen-fixation efficiency. Complementation assays confirmed that the hyperinfective phenotype was linked genetically to the clr mutation (Fig. 5A). Simultaneous inactivation of cyaD1 and cyaD2 led to an intermediate hyperinfection phenotype (Fig. 5A; P < 0.01), whereas inactivation of cyaK alone had no detectable effect, thus suggesting that the three cyclases contribute to the control of infection.
Fig. 5.
Symbiotic phenotypes of cAMP-cascade mutants on M. sativa. (A) Total number of nodules and total number of ITs formed outside nodules (abortive ITs) at 14 dpi on M. sativa inoculated with 1021 and various mutants, including a ccmA Fix− mutant and the complemented clr(pGMI50128) mutant strain, all carrying a constitutive hemA-lacZ reporter gene fusion (pXLGD4) for visualization. Data were collected from three independent experiments with 10 plants inoculated per bacterial strain. P < 0.05; **P < 0.01; ***P < 0.001. (B–G) M. sativa seedlings were inoculated with 1021 (B), cyaD1D2K (C), clr (D and F), or smc02178 (G) mutants. Multiple infection loci (colored blue) were initiated by mutants along the main root at 7 dpi (B–D) and in a 7-dpi nodule (F). Note the absence of nodule primordium in D. IT formation near the tip of a lateral root is shown in E. Abortive ITs with associated defense reactions (brown accumulation of polyphenolics) were visible at 14 dpi in G. (Magnification: B–D, 10×; E and F, 20×; G, 30×.)
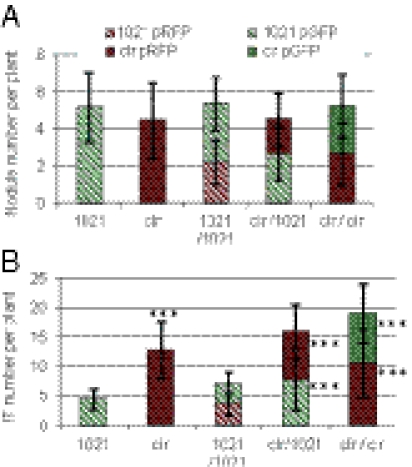
Fig. 6.
Coinoculation experiments of wild-type and clr mutant strains. Nodule (A) and abortive IT (B) numbers in coinoculation experiments (1/1 ratio) of the wild-type and clr mutant strains at 14 dpi. M. sativa seedlings were inoculated with bacteria (4.102) of either strain labeled with a GFP or RFP tag (Table S2). Data were collected from three independent experiments with 10 plants inoculated per bacterial strain. Statistical significance (***P < 0.001) is shown with respect to strain 1021 alone.
The superfluous ITs typically elongated over the entire root hair length but aborted at the interface between the epidermis and the cortex with associated defense reactions (Fig. 5G and Fig. S7). They occurred on roots irrespective of the presence of nodule primordium (Fig. 5 B–D). Nodules with multiple infection foci, a feature that we never observed in the wild-type strain, sometimes were observed (Fig. 5F). Taken together, these findings indicated that hyperinfection is uncoupled from nodule formation. Finally, compared with wild type, the mutants initiated more abortive ITs outside the portion of the root that normally is susceptible to primary infection (23), including on lateral roots (Fig. 5E and Fig. S7).
Bacterial cAMP Cascade May Modulate Plant Susceptibility to Infection.
To compare the symbiotic performances of the wild type and clr mutant, we coinoculated them (1/1 ratio) on M. sativa seedlings after labeling the strains with RFP or GFP fluorescent tags (see Table S2). The numbers of nodules and abortive ITs initiated by each strain were recorded at 14 dpi (Fig. 6). We verified by swapping experiments that the tags did not influence results.
Nodule number was the same for the wild-type and clr strains and did not change upon coinoculation (Fig. 6A), thus confirming that inactivation of the cAMP-signaling cascade had no effect on nodulation. This result also indicated that the two strains had the same competitiveness for nodulation and did not differ in their ability to initiate primary infection events, i.e., the formation of productive ITs leading to nodule formation. Upon coinoculation, the number of abortive ITs initiated by the clr strain was not higher than that initiated by the wild-type strain, as would be expected if the clr strain had a better infectiveness (Fig. 6B). Instead, the number of ITs initiated by the wild-type strain increased significantly upon coinoculation with the clr mutant, so as to match the inoculation ratio (1/1). Conversely, the number of ITs initiated by the clr mutant decreased in the presence of the wild-type strain. Taken together, these findings indicate that the plant did not discriminate between the wild-type and the clr mutant strains for infection. Because of the very low inoculum load (4.102 bacteria of each strain per plantlet), it is highly unlikely that the two strains could complement each other extracellularly, and, indeed, colocalized bacteria were not observed on roots. Instead, these data suggest that the bacterial cAMP cascade may modulate the plant’s sensitivity to infection. Interestingly, the repartition of abortive ITs initiated by the wild-type strain along the root now matched that of the clr mutant, a finding that also suggests an altered sensitivity of the plant root.
Discussion
We have shown here activation by a plant signal of a cAMP cascade in bacteria that are colonizing the forming nodule. Genetic inactivation of the cAMP cascade led to hyperinfection of plant roots, thus suggesting that the biological function of the cAMP cascade is to keep root-hair infection (IT formation) under control. To summarize our findings, we propose a working model in which the cAMP regulatory cascade is activated in bacteria that have penetrated nodule tissues successfully. Activation of this cAMP cascade, in turn, would restrict further infection by external, rhizospheric bacteria by a mechanism that remains to be elucidated (Fig. 7). Inactivation of the cAMP cascade may increase intrinsic bacterial infectiveness or, alternatively, may enhance the root’s susceptibility to infection. Although the two possibilities remain open, we presently favor the second hypothesis, because it accounts more readily for all our observations: (i) it explains how activation of the cAMP cascade in bacteria that already have penetrated nodule tissues may influence infection by external, rhizospheric rhizobia; (ii) it is consistent with the observation that the plant did not discriminate between wild-type bacteria and the clr mutant in coinoculation experiments under conditions where extracellular complementation was unlikely; (iii) it is consistent with the increased occurrence of infection events at atypical locations of the root upon inoculation with cAMP-cascade mutants or upon coinoculation with the wild-type strain and the clr mutant (although this increase could not be quantified precisely). A longer-lasting susceptibility of the plant to infection after inoculation with cAMP-cascade mutants and/or a spatial modulation of the root susceptibility to infection may account for our observations. However, further experiments, including the identification and precise localization of the plant signal in nodules/infected roots as well as the elucidation of the mode of action of the SMc02178 protein, are needed to discriminate definitely between an enhanced infectiveness of cAMP-cascade mutants and/or an enhanced sensitivity of the plant. Whatever the mechanism at work, we speculate that this regulatory loop may have evolved as a sophistication of the symbiotic interaction toward mutualism by optimizing the extent of IT formation and preventing spurious infection events once a sufficient number of neo-formed nodules has been infected successfully.
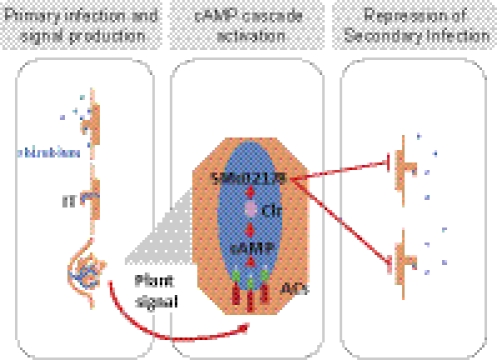
Fig. 7.
Model for the bacterial control of IT formation. Rhizobia (blue) trigger initial IT formation and nodule organogenesis on susceptible host plants. A plant signal synthesized during nodule organogenesis promotes activation of the cAMP cascade in endosymbiotic rhizobia. Activation of the bacterial cAMP cascade in turn inhibits secondary IT formation by rhizospheric bacteria. Genetic disruption of the cascade results in abortive epidermal hyperinfection uncoupled from nodulation.
Signal exchange is central to the establishment and maintenance of the mutualistic interaction between the two partners. Legume signals described so far include flavonoids as elicitors of bacterial nod gene expression (4) and peptides as regulators of nodulation and bacteroid differentiation (24). The results reported here provide evidence of another instance of signaling in this symbiosis. The plant signal was detected in nodules and shoots but could not be detected in roots, whether inoculated or not. This observation argues against signal translocation from shoots to nodules and, instead, tends to support signal synthesis during nodule organogenesis. Biochemical identification of the signal molecule and analysis of the regulation of the gene(s) controlling its synthesis should shed light on the link between signal synthesis, nodule organogenesis, and, possibly, Nod factor signaling. How endosymbiotic bacteria sense the plant signal also remains to be elucidated. Our data suggest that the periplasmic regulatory CHASE2 domain, whose truncation resulted in constitutive activation of the CyaD1 cyclase, contributes to signal sensing, either upon direct interaction with the plant signal molecule itself or indirectly via additional receptor proteins. The specificity of cAMP signaling in S. meliloti also is an intriguing issue, given the high number of ACs in this bacterium. Cellular compartmentalization of cAMP in microdomains may contribute to specificity, as suggested in other systems (25).
Little is known so far about cAMP signaling in the context of plant–microbe interactions. However, cAMP signaling is known to play a major role in the interaction between animal pathogens and their eukaryotic hosts. In those cases, cAMP signaling serves two complementary functions (reviewed in ref. 26). First, it coordinates virulence gene expression with host environment cues. Second, it suppresses host immune responses by manipulating host cAMP-signaling pathways, either by injection of AC toxins and/or effector molecules or by direct secretion of cAMP into macrophages (26). Functional studies of the SMc02178 protein should shed light to the mechanism(s) at work in S. meliloti. The contribution of cAMP signaling to plant–microbe interactions thus may attract further interest in the near future.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in Tables S1 and S2, respectively. S. meliloti strains were grown at 28 °C in rich LB medium supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4, or in modified Vincent synthetic medium with glutamate (0.1%) and mannitol (1%) as nitrogen and carbon sources, respectively (VGM) (27), or in M9 minimal medium with succinate (0.2%) as the carbon source (28). The concentrations of antibiotics used for S. meliloti cultures were 200 μg/mL for streptomycin, 100 μg/mL for neomycin, 10 μg/mL for tetracycline, and 30 μg/mL for gentamicin.
Construction of Plasmids and Mutant Strains.
Primers used for DNA amplification are listed in Table S3. S. meliloti 1021 was used as template for DNA amplification. The cyaD1, clr, and smc02178 single mutants (GMI11561 GMI11567, and GMI11566, respectively) were constructed by site-specific insertional inactivation using the plasmid pVO155 that does not replicate in S. meliloti (29). cyaD1, clr, and smc02178 internal PCR fragments were amplified using D1L-D1R, X2175-B2175, and B2178H-X2178B primers, respectively, cloned into pCR2-1-TOPO and then digested with BamHI and XbaI for cloning into pVO155. The resulting pVO155 derivatives were introduced into E. coli by transformation and then were conjugated in S. meliloti. pVO155 integration at the homologous site in S. meliloti was verified by Southern blot analysis.
For deletion of the cyaD2 and cyaK genes, we used the cre-lox system (30). PCR fragments encompassing the upstream/amino-terminal coding region and the downstream/carboxyl-terminal coding region of cyaD2 were amplified using D2upL-D2upR and D2downL-D2downR as primers (Table S3), digested by EcoRI-KpnI and ApaI-SacI, and cloned into the EcoRI-KpnI and ApaI-SacI restriction sites of pCM351, respectively. The resulting plasmid was introduced into the S. meliloti 1021 cyaD1 mutant (GMI11561) by conjugation. Transconjugants sensitive to tetracycline and resistant to gentamicin were screened. A cyaD1 cyaD2 double-mutant GMI11557 was selected, into which the plasmid pCM157 expressing Cre recombinase was introduced by conjugation. Transconjugants sensitive to gentamicin were screened. Subsequently, pCM157 was cured by screening for tetracycline-sensitive strains. Similarly, the upstream 5′-coding end and the downstream 3′-coding end regions of cyaK were amplified using KupL-KupR and KdownL-KdownR as primers, were digested by PvuII-KpnI and ApaI-SacI, and were cloned sequentially into the PvuII-KpnI and ApaI-SacI restriction sites of pCM351, respectively. The resulting plasmid then was introduced into the S. meliloti cyaD1cyaD2 double mutant (GMI11557) or into the 1021 wild-type strain by conjugation. Transconjugants resistant to gentamicin and sensitive to tetracycline were screened. A cyaD1cyaD2cyaK triple mutant was selected as GMI11558, and a cyaK mutant was selected as GMI11556 (Table S1).
For the construction of the cyaD1ΔCHASE2-expressing plasmid pGIMI50127 (Table S2), the upstream/amino-terminal and the downstream/carboxyl-terminal coding regions of cyaD1 first were amplified using cyaD1Lb-chase2upRb and chase2downLb-cyaD1R as primers (Table S3) and then were digested with BamHI-NotI and NotI, respectively, and were ligated together. The resulting CHASE2-lacking DNA fragment was introduced between blunt-ended HindIII and BamHI sites of pDK5, generating the pDK5::cyaD1ΔCHASE2 intermediate construct. The 1,130-bp KpnI-EcoRI fragment from pDK5::cyaD1ΔCHASE2 was cloned into digested KpnI- and EcoRI-digested pBBR1MCS5, which replicates in both E. coli and S. meliloti, to yield pGIMI50127.
Similarly, the clr-expressing construct pGMI50128 was obtained after PCR amplification of the clr gene-coding region using S. meliloti 1021 genomic DNA as template and REco2175 and LBamH2175 as primers. The PCR fragment was digested with BamHI and EcoRI, ligated into a BamHI-EcoRI–digested pFAJ1708 plasmid, and introduced into E. coli DH5α by transformation. The verified plasmid was digested by XbaI and SstI and cloned into the XbaI-SacI–digested pBBRMcs5 plasmid to yield pGMI50128.
To construct pGD2178, a 436-bp PCR fragment encompassing the smc02178 promoter region was amplified using 2178B and 2178H primers, digested with BamH1 and HindIII, and cloned in the in-frame orientation at the same sites of the lacZ translational fusion plasmid pGD926.
All constructs were verified by PCR and Sanger sequencing in E. coli and by PCR in S. meliloti. Plasmids were transferred from E. coli to S. meliloti by triparental mating using pRK600 as the helper plasmid.
Plant Assays and Plant Extracts and Preparations.
Seeds of M. sativa cv. Europe were surface sterilized, germinated, and allowed to grow in 12-cm2 plates containing slanting nitrogen-free Fahraeus agar medium for 3 d at 22 °C with day/night cycles of 16/8 h. The plants then were inoculated with 2 × 103 bacteria per plant. M. sativa cv Gemini Nar variants were grown under aeroponic sterile conditions as described previously (31). Acetylene-reduction assays were performed as previously described 42 d after rhizobial inoculation (32).
For plant extract preparation, M. sativa seedlings growing on Fahraeus medium in squared plates were inoculated with either S. meliloti 1021 or exoY mutant or were not inoculated. At 14 dpi, nodules, shoots, and nonnodulated portions of the roots of inoculated plants as well as roots and shoots of noninoculated plants were collected and frozen immediately in liquid nitrogen. Frozen nodules, roots, and shoots were crushed with a pestle in Eppendorf tubes, and the resulting material was resuspended in distilled water and centrifuged at 3,000 × g for 8 min. The cleared supernatant was filtered through a 0.22-μm filter (Millipore) and stored at −80 °C, if needed, before assay. Nodule and/or shoot extracts of pea (Pisum sativum Cameor), lotus (Lotus japonicus GIFU), and rice (Oryza sativa Nippon Bare) were prepared similarly. M. sativa root exudates were prepared as previously described (33).
β-Galactosidase Assays.
S. meliloti strains carrying the pGD2178 plasmid were grown at 28 °C in VGM. Overnight cultures were diluted to an OD600 of 0.1 in VGM and grown for an additional 2 h. Cultures (10 mL) then were incubated in microoxic conditions as described (32) or were supplemented with 0.5 mL plant extracts, 1 mL of a 100-fold dilution of Medicago root exudates, 10 mM (final concentrations) of cAMP (A6885; Sigma), 10 μM luteolin (L9283; Sigma), 0.2% proline, 0.15 M sucrose, and 0.3 M NaCl and were grown for an additional 3 h or 24 h. β-Galactosidase activities were measured (in Miller units) using 1 mL of culture, as previously described (34). When plant tissue extracts were used as inducers, the measured activity was corrected for the background activity (i.e., activity of the fusion in absence of plant extract) and divided by the fresh weight of plant material used for the extract.
Cytological Techniques.
Plants were inoculated with S. meliloti strains (wild-type and mutants) carrying the pXLGD4 replicative plasmid (Table S2) that expresses the hemA-lacZ reporter gene fusion constitutively. Entire roots were collected 7 dpi or 14 dpi, fixed with 2% (vol/vol) glutaraldehyde solution for 1.5 h under vacuum, rinsed three times in Z′ buffer [0.1 M potassium phosphate buffer (pH 7.4), 1 mM MgSO4, and 10 mM KCl], and stained overnight at 28 °C under vacuum in Z′ buffer containing 0.08% 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal), 5 mM K3Fe(CN)6, and 5 mM K4Fe(CN)6. Nodules were harvested at 14 dpi, fixed with 2% (vol/vol) glutaraldehyde in Z′ buffer, and then sliced into 80-μm-thick longitudinal sections using a vibrating-blade microtome (VT1000S; Leica) before staining without vacuum overnight at 28 °C. Entire roots or nodule sections were observed under a light microscope.
cAMP and cGMP Measurements.
The bacterial pellet obtained after centrifugation of a 20-mL early exponential phase culture of S. meliloti in M9 medium was washed with 0.9% NaCl and resuspended in 2 mL distilled water. Then 1 mL was used for protein quantification using the Bradford method (Bio-Rad), and 1 mL, supplemented with EDTA (4 mM final concentration), was boiled for 10 min and centrifuged. The resulting supernatant was lyophilized and stored at −20 °C until the test. The lyophilized samples were used to determine intracellular cAMP and cGMP levels with the enzyme immunoassay method according to the protocol given in Amersham kits RPN2251 for cAMP and RPN226 for cGMP.
Statistical Analysis.
Data are presented as means ± SEM unless otherwise indicated. An unpaired, two-sided Student’s t test was used when indicated.
Supplementary Material
Acknowledgments
We thank Drs. J. V. Cullimore, D. Capela, and J. Denarié [Laboratoire des Interactions Plantes-Microorganismes (LIPM)] and our anonymous reviewers for helpful comments on the manuscript; Dr. T. Huguet (LIPM and L'École Nationale Supérieure Agronomique de Toulouse) for the generous gift of M. sativa Nar seeds; and F. Maillet (LIPM) for providing Medicago root exudates. C.F.T. was sponsored by the China Scholarship Council, China Postdoctoral Science Foundation (20100480377), and by the National Basic Research Program of China (“973”, Grant 2010CB126500). C.M.D. holds a doctoral fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche. The authors acknowledge the support of the French Agence Nationale de la Recherche (ANR), under Grant RhizocAMP, ANR-2010-BLAN-1719-01. This work was supported by funds from the Laboratoire d'Excellence entitled Towards a Unified Theory of Biotic Interactions: Role of Environmental Perturbations (TULIP) (ANR-10-LABX-41).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120260109/-/DCSupplemental.
References
- 1.Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Madsen LH, et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nature Communications. 2010;1 doi: 10.1038/ncomms1009. 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oldroyd GED, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 4.Long SR. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001;125:69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li DX, Kinkema M, Gresshoff PM. Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. J Plant Physiol. 2009;166:955–967. doi: 10.1016/j.jplph.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Magori S, Kawaguchi M. Long-distance control of nodulation: Molecules and models. Mol Cell. 2009;27:129–134. doi: 10.1007/s10059-009-0016-0. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto S, et al. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- 9.Murray JD, et al. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 10.Penmetsa RV, Cook DR. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- 11.Vernie T, et al. EFD Is an erf transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. The Plant Cell. 2008;20:2696–2713. doi: 10.1105/tpc.108.059857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 13.Galibert F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 14.Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in Bacteria and Archaea. J Bacteriol. 2003;185:285–294. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder JU. Class III adenylyl cyclases: Molecular mechanisms of catalysis and regulation. Cell Mol Life Sci. 2006;63:1736–1751. doi: 10.1007/s00018-006-6072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marden JN, Dong QA, Roychowdhury S, Berleman JE, Bauer CE. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol Microbiol. 2011;79:600–615. doi: 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batut J, Daveran-Mingot ML, Jacobs MDJ, Garnerone AM, Kahn D. FixK, a gene homologous with FNR and CRP from Escherichia coli, regulates nitrogen-fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989;8:1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terakado J, Fujihara S, Yoneyama T. Changes in cyclic nucleotides during nodule formation. Soil Sci Plant Nutr. 2003;49:459–462. [Google Scholar]
- 19.Soupene E, Foussard M, Boistard P, Truchet G, Batut J. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc Natl Acad Sci USA. 1995;92:3759–3763. doi: 10.1073/pnas.92.9.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truchet G, et al. Alfalfa nodulation in the absence of Rhizobium. Mol Gen Genet. 1989;219:65–68. [Google Scholar]
- 21.Cheng HP, Walker GC. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capela D, Filipe C, Bobilk C, Batut J, Bruand C. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: A transcriptomic dissection. Mol Plant Microbe Interact. 2006;19:363–372. doi: 10.1094/MPMI-19-0363. [DOI] [PubMed] [Google Scholar]
- 23.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batut J, Mergaert P, Masson-Boivin C. Peptide signalling in the rhizobium-legume symbiosis. Curr Opin Microbiol. 2011;14:181–187. doi: 10.1016/j.mib.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Jarnaess E, Tasken K. Spatiotemporal control of cAMP signalling processes by anchored signalling complexes. Biochem Soc Trans. 2007;35:931–937. doi: 10.1042/BST0350931. [DOI] [PubMed] [Google Scholar]
- 26.McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat Rev Microbiol. 2011;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A, et al. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact. 2004;17:292–303. doi: 10.1094/MPMI.2004.17.3.292. [DOI] [PubMed] [Google Scholar]
- 28.Dephilip P, Batut J, Boistard P. Rhizobium meliloti FixL is an oxygen sensor and regulates Rhizobium meliloti nifA and fixK genes differently in Escherichia coli. J Bacteriol. 1990;172:4255–4262. doi: 10.1128/jb.172.8.4255-4262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oke V, Long SR. Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol. 1999;2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 30.Marx CJ, Lidstrom ME. Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 31.Ampe F, Kiss E, Sabourdy F, Batut J. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 2003;4:R15. doi: 10.1186/gb-2003-4-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bobik C, Meilhoc E, Batut J. FixJ: A major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J Bacteriol. 2006;188:4890–4902. doi: 10.1128/JB.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulligan JT, Long SR. Induction of Rhizobium meliloti nodC expression by plant exudate requires NodD. Proc Natl Acad Sci USA. 1985;82:6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller G. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.