Abstract
Cell-based studies support the existence of two promoters on the heavy strand of mtDNA: heavy-strand promoter 1 (HSP1) and HSP2. However, transcription from HSP2 has been reported only once in a cell-free system, and never when recombinant proteins have been used. Here, we document transcription from HSP2 using an in vitro system of defined composition. An oligonucleotide template representing positions 596–685 of mtDNA was sufficient to observe transcription by the human mtRNA polymerase (POLRMT) that was absolutely dependent on mitochondrial transcription factor B2 (TFB2M). POLRMT/TFB2M-dependent transcription was inhibited by concentrations of mitochondrial transcription factor A (TFAM) stoichiometric with the transcription template, a condition that activates transcription from the light-strand promoter (LSP) in vitro. Domains of TFAM required for LSP activation were also required for HSP2 repression, whereas other mtDNA binding proteins failed to alter transcriptional output. Binding sites for TFAM were located on both sides of the start site of transcription from HSP2, suggesting that TFAM binding interferes with POLRMT and/or TFB2M binding. Consistent with a competitive binding model for TFAM repression of HSP2, the impact of TFAM concentration on HSP2 transcription was diminished by elevating the POLRMT and TFB2M concentrations. In the context of our previous studies of LSP and HSP1, it is now clear that three promoters exist in human mtDNA. Each promoter has a unique requirement for and/or response to the level of TFAM present, thus implying far greater complexity in the regulation of mammalian mitochondrial transcription than recognized to date.
Keywords: mitochondria, gene expression, initiation, gel-shift assay, footprinting
Human mitochondrial DNA (mtDNA) encodes 13 proteins essential to formation of a functional electron transport chain and ATP synthase. In addition, mtDNA encodes rRNAs and tRNAs required for translation of the 13 mRNAs for the mtDNA-encoded proteins. One strand of mtDNA is referred to as the heavy strand, and the other strand is referred to as the light strand. Transcription of the heavy strand produces 2 rRNAs, 12 mRNAs, and 14 tRNAs, whereas transcription of the light strand produces only 1 mRNA and 8 tRNAs. Given this genome organization, at least two promoters are needed: a light-strand promoter (LSP) and a heavy-strand promoter (HSP). The works by Montoya et al. (1, 2) identified two transcripts from the heavy strand. They differed both in the site of initiation and the site of termination (2). One transcript initiated 25 bp upstream of the phenylalanyl tRNA gene and frequently ended immediately after the 16S rRNA gene. The other transcript initiated immediately upstream of the 12S rRNA gene but terminated at an undefined position after the threonyl tRNA gene, thus producing a near genome-length polycistronic RNA. The former promoter has been termed HSP1; the latter promoter has been termed HSP2 (Fig. 1A).
Fig. 1.
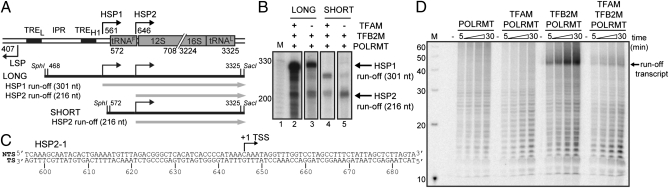
HSP2 transcription in vitro. (A) Schematic of mtDNA promoters: LSP, HSP1 and HSP2, interpromoter region, TRE for LSP and HSP1, and nearby genes. This region of mtDNA was used to produce LONG and SHORT templates capable of producing the indicated runoff transcripts. (B) Runoff transcription from the LONG or SHORT templates and indicated factors. All experimental details for this experiment and subsequent experiments are provided in SI Materials and Methods. M designates size markers. (C) Sequence of DNA oligonucleotide representing HSP2; the proposed TSS is indicated by +1. (D) Runoff transcription from the HSP2-1 template and indicated factors.
Basal transcription initiation on mtDNA was thought to use three factors regardless of the promoter: mtRNA polymerase (POLRMT), transcription factor A (TFAM), and transcription factor B2 (TFB2M). The long-standing paradigm was that TFAM binds to a site in the promoter upstream of the transcription start site and recruits a complex POLRMT/TFB2M by an interaction of the carboxyl-terminal tail of TFAM with TFB2M (3). However, we recently showed that basal mitochondrial transcription is not absolutely dependent on TFAM (4). This observation suggested that the two-component transcription system found in lower eukaryotes had acquired an additional layer of regulation in mammals that is mediated by TFAM (4). TFAM is a member of the high mobility group (HMG) box family of DNA binding, bending and wrapping proteins (3). For LSP and HSP1, TFAM-mediated activation has been linked directly to the ability of TFAM to impose a U-turn in the TFAM-responsive element (TRE) to which it binds (5–7).
The robustness and specificity of transcription from LSP in vitro encouraged the use of this promoter to study mitochondrial transcription in vitro both in cell-free (8) and recombinant (9–11) systems. Most of these studies showed that the strength of LSP is substantially greater than HSP1 in vitro (9), although HSP1 seems to be stronger than LSP in cells (2). We recently resolved this apparent contradiction by using an oligonucleotide template that included LSP, HSP1, and the intervening interpromoter region (IPR), a better mimic of the mtDNA control region (4). In the absence of TFAM, significantly more transcription was observed from HSP1 than from LSP (4). However, concentration of TFAM equivalent to template caused full activation of transcription from LSP without changing the level of transcription from HSP1. When the concentration of TFAM exceeded the concentration of the template by 5- to 10-fold, transcription from LSP was inhibited substantially, but transcription from HSP1 was still activated substantially. These observations further strengthened the hypothesis that a TFAM-regulated, two-component transcription system is operative in human mitochondria.
The least understood promoter is HSP2. The first reports of its existence were published in the early 1980s (1, 2). However, evidence of its activity using a cell-free system took nearly 25 years to acquire (12). Recently, however, the work by Litonin et al. (13) called into question the existence of HSP2 based on the inability to observe transcription from HSP2 using recombinant proteins under conditions in which transcription from LSP is robust. We confirm herein the existence of HSP2 using a highly purified transcription system in vitro (14). We find that POLRMT and TFB2M together are necessary and sufficient for transcription from HSP2 and that TFAM represses transcription from HSP2 by binding to TREs flanking the transcription start site, thus preventing productive binding of POLRMT and TFB2M. Importantly, the domains of TFAM required for transcriptional activation of LSP are the same as those required for transcriptional repression of HSP2. Our inability to identify a single condition that supports maximal transcription from all three promoters in vitro may underpin an unappreciated mechanism for differential expression of the human mitochondrial genome in vivo. We discuss the possibility that such a TFAM-regulated program may permit the energy-producing demands of the mitochondria to be maintained without a commitment to biogenesis.
Results
Transcription from HSP2 in Vitro.
Whether expression of the human mitochondrial genome requires two or three promoters is fundamental to our understanding of mitochondrial biology. We have shown that POLRMT, TFAM, and TFB2M produced in bacteria are capable of bona fide initiation of transcription from LSP and HSP1 using either linearized plasmid or synthetic oligonucleotide templates in vitro (14). We used that system here to determine if transcription from HSP2 could be observed in vitro.
Two plasmid-derived fragments were evaluated. The first fragment spanned from position 468 to position 3,325 of mtDNA, which is referred to as LONG in Fig. 1A. The other spanned from position 572 to position 3,325 of mtDNA, which is referred to as SHORT in Fig. 1A. Both constructs omitted mtDNA sequences from 709 to 3,223 (Fig. 1A). The LONG template contains both heavy-strand promoters; the SHORT template only contains HSP2. Assuming the transcription start sites (TSSs) for HSP1 (561) and HSP2 (646) previously reported (12), the HSP1 runoff transcript should be 301 nt in length, and the runoff transcript from HSP2 should be 216 nt in length (Fig. 1A). In the presence of all three factors, a ladder of transcription products was evident that complicated interpretation of the experiment when the LONG template was used (Fig. 1B, lane 2). However, omission of TFAM revealed two bands of the expected size (Fig. 1B, lane 3). We have shown previously that transcription from HSP1 can be TFAM-independent. This experiment, therefore, suggested the possibility that transcription from HSP2 may also be TFAM-independent. To determine if the band migrating around 216 nt was indeed a product of transcription from HSP2, we used the SHORT template. In the presence of all three components, the 216-nt product was formed; however, a product consistent with the entire length of the template was also visible (Fig. 1B, lane 4). When TFAM was omitted, the primary transcription product was consistent with the product expected from HSP2 (Fig. 1B, lane 5).
Next, we turned to an oligonucleotide template. The core of LSP and HSP1 is contained from 50 bp upstream to 40 bp downstream of the TSS (14). We used a fragment spanning from position 596 to position 685 (Fig. 1C) (Table S1). Neither POLRMT alone nor combined with TFAM was sufficient for anything more than nonspecific transcription (Fig. 1D). Use of POLRMT and TFB2M was quite robust in production of a runoff transcript that migrated between the 40- and 50-nt markers (Fig. 1D). Unexpectedly, addition of TFAM at a concentration equivalent to POLRMT and TFB2M substantially reduced the amount of runoff product observed (Fig. 1D). This same condition leads to activation of transcription from LSP when an oligonucleotide template is used (14). These data confirm the existence of HSP2 and suggest that TFAM is a potent repressor of transcription from this promoter.
Position 644 as a Start Site for Transcription from HSP2 in Vitro.
The runoff product of transcription from HSP2 migrates slower than expected for a 40-nt RNA (Fig. 1D). To show that the runoff product originated from a site in the vicinity of the expected start site, we evaluated three additional constructs. We removed 10 bp upstream of the TSS (Fig. 2A, HSP2-2), and we added 10 bp upstream of the TSS (Fig. 2A, HSP2-3). We also extended the templating region by 10 bp (Fig. 2A, HSP2-4) (Table S1). In all cases, runoff products in the appropriate size range were observed (Fig. 2B, lane −TFAM), and transcription was still inhibited by the presence of TFAM (Fig. 2B, lane +TFAM). When the central 24 bp of HSP2 were randomized (Fig. S1A), this template failed to produce a 40-nt runoff product (Fig. S1B, HSP2-Random). Fortuitously, a sequence resembling the proposed TSS was present 14 bp upstream that supported some transcription (Fig. S1B, HSP2-Random). Changing the nucleotide sequence around the TSS of the plasmid-derived template also abrogated transcription (Fig. S1C). We conclude that the start site is located in the expected region.
Fig. 2.
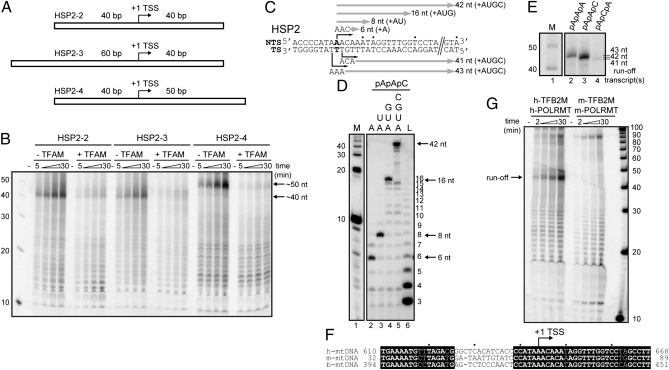
Specificity of HSP2 transcription in vitro. (A) HSP2 transcription templates designed to assess the specificity of initiation. (B) Runoff transcription from the templates shown in A by POLRMT-TFB2M in the absence or presence of TFAM. (C) Sequence surrounding HSP2; shown are the expected start sites and sizes of RNA transcripts when using pApApC, pApApA, or pApCpA trinucleotide primer for initiation with the indicated nucleotide(s). (D) Transcription by POLRMT-TFB2M from the HSP2-1 template in the presence of pApApC and the indicated nucleotide(s). (E) Runoff transcription from the HSP2-1 template in the presence of pApApA, pApApC, or pApCpA trinucleotide primer and all four nucleotides. (F) Alignment of human HSP2 with corresponding mouse and bovine sequences reveals a region of high interspecies differences from −18 to −6. (G) Mouse POLRMT-TFB2M fails to produce a promoter-specific runoff transcript from human HSP2 indicating specificity of human POLRMT-TFB2M.
Attempts to directly map the 5′ end of the product RNA using 5′ RACE failed. We were, therefore, forced to use indirect methods to identify the TSS. Di- and trinucleotide primers capable of hybridizing to the templating bases can be used as primers for RNA synthesis by POLRMT (11, 14). The TSS proposed in the work by Martin et al. (12) is 3′-GTT-5′, which corresponds to an RNA 5′ end of 5′-CAA-3′. Therefore, the use of a CA dinucleotide primer should lead to production of a 40-nt product using the HSP2 oligonucleotide template. To permit this experiment to be interpreted, we labeled the primer with 32P. The labeled CA primer failed to produce a runoff transcript (Fig. S2A, pCpA). Both an AA dinucleotide primer and an AAA trinucleotide primer produced runoff products (Fig. S2A, pApA and pApApA). The inability of the CA dinucleotide to prime runoff transcription and the apparent mobility of the runoff exceeding 40 nt led us to conclude that the TSS was at position 644 and/or 643, upstream of the position observed in the work by Martin et al. (12).
RNA may not migrate at the expected size relative to a marker because of sequence differences that, for example, prevent complete denaturation. To produce RNA of the size and sequence expected for initiation from position 644, we fused this sequence to LSP (Fig. S2B, LSP-HSP2 template). The LSP template produced a 40-nt product (Fig. S2C, lane 2). The LSP-HSP2 template produced a 42-nt product (Fig. S2C, lane 3), and this product migrated at the same position as one of the products observed using the HSP2 template (Fig. S2C, lane 4). We conclude that more than one TSS exists for HSP2 in vitro; one TSS is at position 644. The work by Zollo et al. (15) reached the same conclusion.
We used a pApApC trinucleotide primer to further document the specificity of initiation from position 644. As shown in Fig. 2C, transcription products of specific lengths are predicted when pApApC is used with specific combinations of nucleotides. Consistent with position 644 as a start site, products of the expected length were observed (Fig. 2D). When a pApApA primer was used, a 43-nt product was formed (Fig. 2E, lane 2). A pApCpA primer yielded a 41-nt product (Fig. 2E, lane 4). Again, the pApApC primer yielded the most product (Fig. 2E, compare lane 3 with lanes 2 and 4). Collectively, these data confirm specificity of initiation from HSP2, with position 644 serving as the primary TSS in vitro.
The adenylate residue at position 644 of human mtDNA and adjacent nucleotides are conserved in mouse and bovine species (Fig. 2F). Interestingly, the POLRMT and TFB2M pair from mouse failed to produce the 42-nt product using the human HSP2 template (Fig. 2G). A nonspecific product of template length (90 nt) was observed (Fig. 2G). Importantly, the combination of POLRMT, TFB2M, and TFAM from mouse was active for promoter-specific transcription using a mouse LSP template (Fig. S3). We conclude that the initiation from HSP2 observed here is not just a reflection of the presence of a cryptic TSS capable of being recognized by any POLRMT in vitro.
Repression of HSP2 Transcription by TFAM.
TFAM has two well-documented activities. It serves as a transcriptional activator for LSP and HSP1 (16, 17) and a packaging protein of the mtDNA nucleoid (18). TFAM has three functional domains: two HMG boxes and a carboxyl-terminal domain (CTD) (Fig. 3A). The CTD seems to be uniquely important for the transcriptional functions of TFAM (8). Portions of the CTD seem to govern DNA binding specificity/affinity (5, 19, 20), and portions seem to govern interactions with proteins (for example, TFB2M) (3). Therefore, we asked if the CTD was important for transcriptional repression of HSP2 by TFAM. Transcription from HSP2 was evident in the absence of TFAM (Fig. 3B, −TFAM). Again, transcription from HSP2 was inhibited by the presence of TFAM (Fig. 3B, +TFAM) but not by the presence of TFAM lacking the terminal 26-aa residues (Fig. 3B, +TFAM-ΔCT26). Interestingly, the terminal 10-aa residues were not required for repression (Fig. S4A, +TFAM-ΔCT10).
Fig. 3.
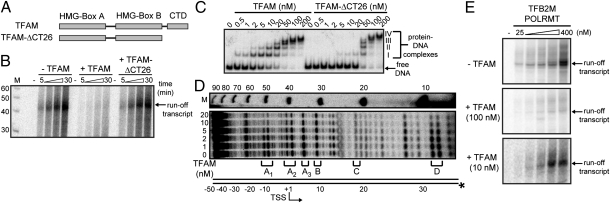
Repression of HSP2 transcription by TFAM in vitro. (A) TFAM uses two HMG boxes (Boxes A and B) for DNA binding and CTD for transcriptional activation. The activation portion of the CTD is deleted in TFAM-ΔCT26. (B) TFAM-ΔCT26 does not inhibit runoff transcription from the HSP2-1 template by POLRMT-TFB2M. (C) EMSA of HSP2-1 template over a range of TFAM concentrations shows formation of four complexes (I–IV), with complex I present from 0.5 nM. Formation of complex I is impaired for TFAM-ΔCT26. (D) DNase I footprinting of TFAM-HSP2-1 complexes reveals protection in the regions designated as A1, A2, A3, and D without protection in regions B and C. A schematic of HSP2-1 is shown on the left, and size markers (M) are on the right. (E) Runoff transcription from the HSP2-1 template (100 nM) in the absence or presence of 100 or 10 nM TFAM, which are values that are 10-fold higher than or equivalent to the equilibrium dissociation constant, respectively. In this experiment, product formed at 30 min over the indicated range of POLRMT-TFB2M concentrations.
The work by Wong et al. (20) details an extensive biophysical characterization of TFAM-ΔCT26. The only effects of the deletion noted in the work by Wong et al. (20) were related to the loss of the monomer–dimer equilibrium observed for this protein and a twofold reduction in its ssDNA binding activity (without a significant change of its dsDNA binding activity) (20). We used an EMSA to investigate the DNA binding properties of TFAM and TFAM-ΔCT26 (Fig. 3C). Four protein–DNA complexes (I–IV) were formed with both proteins using the HSP2 template as a probe (Fig. 3C). However, formation and/or stability of complex I was reduced substantially by deleting the carboxyl-terminal 26-aa residues of TFAM (Fig. 3C). When a probe containing the randomized version of the sequence for the LSP template was used, formation and/or stability of all four complexes were diminished for TFAM (Fig. S4B). We conclude that formation and/or stability of complex I require sequences in HSP2 and the TFAM CTD. Therefore, it is possible that the reduced level of complex I formed on HSP2 by TFAM-ΔCT26 leads to the inability of this derivative to repress transcription from HSP2. Consistent with this interpretation, formation of complex I on the LSP template (Fig. S5A) correlates with the ability of TFAM to activate transcription from LSP (Fig. S5 B and C). Other mtDNA binding proteins, particularly the ssDNA binding protein and termination factor, produced no effect on HSP2 transcription and failed to bind to the HSP2 template when the proteins were present in the 1–10 nM range (Fig. S6).
Complex I of LSP corresponds to the well-documented TRE observed by DNase I footprinting (Fig. S5D) (14). Therefore, we asked if we could identify a TRE in HSP2 by DNase I footprinting. We observed two sites of protection. The site most clearly protected by TFAM was located at a position between −15 and −10 relative to the TSS (Fig. 3D, region A1). Protection at sites around +1 and +5 was also evident (Fig. 3D, regions A2 and A3, respectively). Protection was also observed downstream of the TSS (for example, at +30) (Fig. 3D, region D). Under these same conditions (2–20 nM TFAM), complexes I and II were the most abundant complexes formed (Fig. 3C). All of the protection observed on HSP2 was less pronounced than the protection observed on LSP (Fig. S5D). Worth noting, the DNase I accessibility of several regions did not change in this experiment (Fig. 3D, regions B and C). We conclude that TREs exist in HSP2 that are located in positions that should be capable of inhibiting binding by POLRMT-TFB2M.
We reasoned that if the mechanism of repression by TFAM relates to competition with POLRMT-TFB2M for binding to the promoter, then TFAM repression should be reduced by increasing the concentration of POLRMT-TFB2M present in the reaction. POLRMT-TFB2M at a concentration of 400 nM fully overcame repression by 10 nM TFAM [Fig. 3E, +TFAM (10 nM)]. However, this same concentration of POLRMT-TFB2M was insufficient to overcome 100 nM TFAM [Fig. 3E, +TFAM (100 nM)]. Also consistent with a competitive mechanism was the finding that the inhibitory potential of TFAM on HSP2 transcription was antagonized by the presence of a 28-bp DNA oligonucleotide containing the LSP TRE (Fig. S7, TFAM-BS). A 28-bp DNA oligo of random sequence was not as potent of an antagonist, and in fact, it was inhibitory under conditions in which the TFAM-BS was stimulatory (Fig. S7, compare TFAM-BS with random). We conclude that TFAM represses transcription from HSP2 by using a classical paradigm for transcriptional repression in which the repressor binding site overlaps the polymerase binding site in the promoter (21).
Functional Conservation of Mammalian TFAM.
The work by Gaspari et al. (10) has shown that an interspecies exchange of POLRMT does not support transcription from LSP, but interspecies exchange of TFAM does support transcription from LSP. Our finding that mouse POLRMT-TFB2M does not support transcription from HSP2 is consistent with the data for LSP (Fig. 2F) (10). Evaluating the exchangeability of TFAM was of interest to us, because the most divergent portion of the TFAM CTD is in the last 10-aa residues (Fig. 4A), which are not required for transcriptional repression (Fig. S4A) or transcriptional activation (Fig. S5E). Interestingly, TFAMs from mouse and bovine species were able to inhibit transcription from HSP2 (Fig. 4B), and these noncognate TFAMs were able to form complexes I and II on human HSP2 as efficiently as human TFAM (Fig. 4C). Similar observations were made with mouse TFAMs on human LSP (Fig. S5). These data illuminate a role for residues 220–236 of TFAM in the formation of complexes with TREs of mtDNA, whether these interactions are activating or repressing. Functionally, these interactions are likely conserved and exchangeable between mammalian species.
Fig. 4.
Repression of HSP2 transcription by TFAM orthologs in vitro. (A) Alignment of carboxyl-terminal sequences of TFAMs from human, mouse, and bovine species. (B) TFAMs from mouse and bovine inhibit runoff transcription from the HSP2-1 template by POLRMT-TFB2M. (C) EMSA of HSP2-1 template over a range of mouse and bovine TFAM concentrations shows formation of four complexes (I–IV), with complex I present from 0.5 nM as observed for human TFAM in Fig. 3C.
Discussion
Implicit in the elaboration of a genome to include multiple promoters is the existence of regulated gene expression. The multicellular eukaryote, Caenorhabditis elegans, has one of the smallest mitochondrial genomes known and is thought to use a single promoter for expression of a single polycistronic RNA. Therefore, there is likely a biological reason that mammals have expanded their mitochondrial genomes to include a control region that is home to at least two promoters: LSP and HSP1 (3). The initial objective of this study was to determine if we could confirm the existence of HSP2, a promoter predicted by cell-based studies (1, 2, 12) but disputed by some biochemical studies (13).
All of the experiments reported here are consistent with the existence of a second heavy-strand promoter (Figs. 1 and 2). Transcription from HSP2 is strictly dependent on POLRMT and TFB2M (Fig. 1). Our experiments suggested TSSs at positions 643 and/or 644, with position 644 preferred in our in vitro system (Fig. 2D and Fig. S2). Our observations are consistent with the S1 mapping and 5′ RACE experiments of HSP2 in vitro transcripts in the work by Zollo et al. (15). However, these start sites do not agree with the TSS at position 646 inferred from S1 mapping of HSP2 transcripts isolated from cells (12). The reason for this difference is unclear. One interesting possibility is that start site selection at HSP2 may be a regulated event (22). For example, mtDNA topology, additional transacting factors, and/or nucleotide pools could alter the TSS (23–25). If initiation from this promoter is as variable in vivo as it is in vitro, then it is also possible that some technical procedural nuance led to loss of 5′-A terminated transcripts or enrichment of the 5′-C terminated transcripts. The use of multiple start sites from position −3 to +3 relative to a primary start site is well-documented in the nuclear genomes of mammals (26).
The orthologous mouse POLRMT-TFB2M pair was not able to initiate transcription from human HSP2 (Fig. 2F). Although this observation suggests that the ability of human POLRMT-TFB2M to use this promoter is not fortuitous, the factor–promoter incompatibility is surprising. HSP2 is located within the genes for tRNAF and 12S rRNA, and these genes are well-conserved across mammalian species. Interestingly, the region from −18 to −6 of HSP2 exhibits substantial interspecies variability (Fig. 2E). The variable region of the sequence encodes the TψC stem loop of tRNAF; equivalent structures are predicted for all of the sequences. We propose that this sequence is responsible for the observed species specificity. We do not know the selective pressure that has led to a need to maintain stringent factor–promoter specificity, but this factor–promoter specificity extends to LSP and HSP1 (10). The POLRMT-related bacteriophage T7 RNA polymerase uses an aminoterminal domain to recognize sequences from −17 to −12 of its promoter (27). It is tempting to speculate that within −18 to −8 of HSP2 is a region recognized by POLRMT and/or TFB2M.
The most unexpected and exciting finding of this study is the observation that TFAM is not only a transcriptional activator but also a transcriptional repressor at concentrations that activate other promoters. This inhibitory activity is manifested on HSP2 (Figs. 1–4). The residues of TFAM known to be uniquely required for transcriptional activation, particularly the CTD, are all required for repression (Fig. 3B and Fig. S4A). We interpret this observation to mean that the two activities are not separable. You cannot have activation of LSP and/or HSP1 without repression of HSP2, at least in vitro.
Footprinting experiments performed in the past by us and others on LSP have shown that POLRMT-TFB2M protects from −18 to +10 of this promoter (10, 14). There is no reason to believe that the conformation used by POLRMT-TFB2M for initiation from HSP2 would be different. Therefore, the binding sites for TFAM at −10, +1, and perhaps even +5 of HSP2 suggested by our footprinting experiments should interfere with POLRMT-TFB2M binding (Fig. 3D). Consistent with this model, a 40-fold excess of POLRMT-TFB2M relative to TFAM precludes inhibition, but a fourfold excess does not (Fig. 3E).
Recently, several laboratories have joined forces to study the human mitochondrial transcriptome in a variety of cell lines (28). As a part of that study, an in vivo DNase I footprinting experiment was performed with essentially single-nucleotide resolution (28). That study revealed strong sites of protection in HSP2 and adjoining sequences (Fig. S8) (28). All of those sites observed by us in vitro (Fig. 3D) were observed in vivo (Fig. S8). Whether or not TFAM binding protects these sites in vivo is not known but could be determined using ChIP-seq methodologies. In addition, the relevance of the protected sites to gene expression is not known but will not be straightforward to address experimentally given the absence of a reverse genetics system for mtDNA.
This study reinforces and clarifies the work of others that has pointed to the CTD of TFAM as a critical domain for transcriptional functions (5, 8, 20). Deletion of the terminal 10-aa residues from TFAM supports transcriptional repression (Fig. S4) and activation (Fig. S5). However, deleting an additional 16-aa residues eliminates both transcriptional repression (Fig. 3B) and activation (Fig. S5 B and C). These last 10-aa residues of TFAM vary from one species to another, but the preceding 16-aa residues do not (Fig. 4A). Because mouse and bovine TFAM repress transcription from HSP2 (Fig. 4B) and activate transcription from LSP (Fig. S5 B and C), we conclude that residues 220–236 of TFAM are those residues most essential to CTD function.
Two groups independently solved the structures for TFAM complexed with the TRE from LSP (6, 7). Residues 232–236 interact with DNA (Fig. 5A). The conformation of this portion of the CTD is likely stabilized by the salt bridges between Arg-227, Glu-148, and Asp-229 (Fig. 5B). In addition, residues 225–229 of one molecule may pack against the same residues on a second molecule in a TFAM dimer (Fig. 5B) (7). Residues of the helix between the two HMG boxes also contribute to interaction between the two TFAM-DNA complexes. All of these residues are conserved (Fig. 4A). Therefore, these interactions lend a nice structural explanation for the requirement of the CTD of TFAM for its transcriptional functions. Analytical ultracentrifugation experiments detailed in the work by Wong et al. (20) revealed a monomer–dimer equilibrium for TFAM that was lost when the CTD was deleted. The interface suggested here may be responsible for TFAM dimerization. Interactions between TFAM monomers bound at different positions of HSP2 could contribute further to the efficiency with which TFAM represses this promoter.
Fig. 5.

Interactions of the TFAM CTD. Structural models were produced using Protein Data Bank ID code 3TQ6 (7). (A) TFAM residues 232–236 (green) interact with the phosphodiester backbone of bound DNA (red). (B) Two TFAM-DNA complexes are present in the asymmetric unit and designated here as chains A (dark green) and B (light green). Structural integrity of the CTD of each monomer benefits from interaction of Arg-227 in each monomer with both Asp-229 and Glu-148 of the same monomer. The CTD of one monomer packs against the CTD of a second monomer, perhaps creating a mechanism for association between TFAM-DNA complexes.
Our data would suggest that the presence of any TFAM should lead to repression of HSP2. If this repression were the case, then mRNAs would never be produced. Clearly, all mitochondria contain TFAM, although nucleoids vary in their TFAM levels in vivo (29). TFAM is a substrate for degradation by the mitochondrial Lon protease and perhaps contributes to TFAM depletion (30). For this mechanism to work in vivo, a mechanism may be needed to specifically control the functions of TFAM in transcription without altering protein levels globally. Posttranslational modification of the CTD could serve such a function. One report of TFAM acetylation exists, although this study did not map the site of this modification (31). It is also now clear the cytosines of human mtDNA are both methylated and hydroxymethylated (32). Epigenetic marks such as these marks could also influence the sites to which TFAM binds and the ensuing transcriptional response.
In going from a disabled CTD or low TFAM level to a fully enabled CTD or a high TFAM level, the transcriptional program would go from HSP2 > HSP1 >>> LSP to LSP >> HSP1 >> HSP2 followed by HSP1 >>> LSP1 >>> HSP2. HSP2 is required for maintenance of the electron transport chain. LSP and HSP1 are required for mitochondrial biogenesis. LSP produces the primer for replication of the heavy strand (3). Full activation of HSP1 will produce an abundance of 12S and 16S rRNAs, likely to double the mass of ribosomes before fission. Such a dichotomy makes biological sense, because it would uncouple a transcriptional program supporting mitochondrial homeostasis from one supporting mitochondrial biogenesis. One caveat to this hypothesis is that LSP-driven expression of ND6 protein and tRNAs would need to be stable enough to not require transcription of these genes when HSP2 is the primary promoter operating. This hypothesis also assumes termination of transcription after completion of 16S rRNA when initiated from HSP1 (12). This mechanism would predict that each promoter would be affected differently in experiments in which TFAM levels are increased or decreased. Unfortunately, none of the studies performed to date that modulate the intracellular TFAM level have evaluated transcripts from all three promoters (33, 34).
The past few years have witnessed a revival in molecular studies of human mitochondrial transcription because of the creation of more robust in vitro systems (11, 14). The TFAM-regulated, two-component system documented recently may be even more complex than previously imagined (4). The response of each promoter to TFAM is unique. Here, we have shown that TFAM is no longer only a transcriptional activator but also a transcriptional repressor. It will be important to determine if the promoter-specific, TFAM-dependent regulation observed in vitro also occurs in cells.
Materials and Methods
Materials.
Recombinant proteins and substrates were produced as described previously (14). All other regents were of the highest grade available. A complete description of materials and corresponding sources is given in SI Materials and Methods.
Assays.
In vitro transcription assays (14), DNase I footprinting (14), and EMSA (19) were performed as described previously. Detailed protocols are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Neal Sondheimer for sharing data before publication and comments on the manuscript and Eric Smidansky for comments on the manuscript. This work was supported in part by the Paul Berg Endowment of the Eberly College of Science, National Institutes of Health (NIH) Postdoctoral Fellowship DK-091042 (to M.B.), and NIH Grant HL-059655 (to G.S.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118710109/-/DCSupplemental.
References
- 1.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 3.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: Multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 4.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malarkey CS, Bestwick M, Kuhlwilm JE, Shadel GS, Churchill ME. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubio-Cosials A, et al. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 8.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 9.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 10.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 13.Litonin D, et al. Human mitochondrial transcription revisited: Only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodeiro MF, et al. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter (HSP2) of mtDNA. Proc Natl Acad Sci USA. 2012;109:6508–6512. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 18.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 19.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong TS, et al. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis M. A tale of two repressors. J Mol Biol. 2011;409:14–27. doi: 10.1016/j.jmb.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandelin A, et al. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 27.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer TR, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown TA, et al. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc Natl Acad Sci USA. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinardo MM, et al. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem Biophys Res Commun. 2003;301:187–191. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- 32.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanki T, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.