With great interest, we read the work by Liu et al. (1), which described the development of an alkyne analog of puromycin for labeling and imaging of newly synthesized peptides. Available techniques for this type of measurement are very limited, and the work by Liu et al. (1) claimed that an alternative method, which is based on antipuromycin immunohistochemistry (SUnSET) (2), results in a subcellular localization of nascent peptides that is not consistent with the expected location of newly synthesized proteins [e.g., “antipuromycin staining shows the strongest signal at the plasma membrane” (1)]. For the following reasons, we have serious concerns with this potentially misleading claim. First, the claim is largely based on studies in which azidohomoalanine (Aha)/homopropargylglycine (Hpg) analogs of methionine were used to identify the localization of newly synthesized proteins. However, there is currently no evidence that the Aha/Hpg–methionine techniques reveal the true localization pattern in cells. On this point, it is important to consider that the Aha/Hpg–methionine technique uses an initial preincubation period to deplete the intracellular stores of methionine, and this stress might alter the subcellular localization of newly synthesized proteins. Furthermore, a direct comparison of the localization patterns with the alkyne analog of puromycin, SUnSET, and Aha/Hpg–methionine techniques has not been performed. Therefore, any presumed differences in the localization pattern observed with these techniques might simply be because of differences in the experimental conditions (e.g., differences in cell type, culture conditions, fixation conditions, etc.). Second, the study by Schmidt et al. (2) used the SUnSET technique to reveal that the majority of nascent peptides are located in the cytoplasm of cultured cells and not the plasma membrane as claimed in the work by Liu et al. (1) (Fig. 1). Third, the work by David et al. (3) recently used the SUnSET technique to show that nascent peptides are colocalized with the ribosomal P protein and the multiaminoacyl–tRNA synthetase complex at the endoplasmic reticulum level of HeLa cells and in vaccinia virus factories. Finally, the work by Goodman et al. (4) used the SUnSET technique to examine protein synthesis in mature skeletal muscle fibers in vivo and found a prominent signal beneath the plasma membrane along with a more diffuse signal in the intermyofibrillar region. This finding was not surprising, because previous studies have shown that the majority of 28S rRNA and various ribosomal proteins are located along the periphery of skeletal muscle fibers, with smaller amounts located among the myofibrils (4, 5) (Fig. 1). In summary, multiple experiments have shown that the location of newly synthesized peptides identified with the SUnSET technique is highly consistent with the expected location of their synthesis. Therefore, we believe that a direct comparison of the different techniques under identical conditions should be performed before any claims about problems/differences in the localization patterns are considered valid. If such differences do exist, then an important challenge for future studies will be to determine which technique accurately reveals the true localization of newly synthesized proteins.
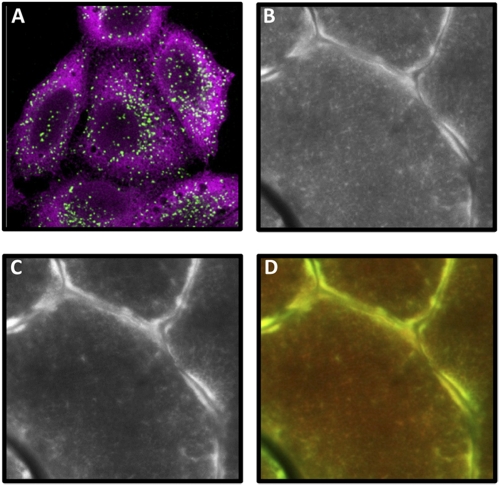
Fig. 1.
(A) Confocal immunofluorescence imaging of HeLa cells labeled with puromycin for 10 min. Puromycin-labeled proteins detected with antipuromycin 12D10 antibody (magenta) are mostly localized on the endoplasmic reticulum (ER) and in the cytosol as previously described (2, 3). Late endosomes and lysosomes [lysosomal-associated membrane protein 1 (LAMP1); green] are shown to delimit the cells cytoplasm. (B–D) A cross-section of skeletal muscle (tibialis anterior) from a puromycin-treated mouse was subjected to immunohistochemistry for puromycin-labeled proteins (B) and the ribosomal S6 protein (C) as previously described (4). (D) A merge of the images from B (red) and C (green) reveals extensive colocalization between the signals for puromycin and the ribosomal S6 protein. Furthermore, the most prominent signal was consistently found near the periphery of the individual fibers, with a more diffuse reticular signal found in the intermyofibrillar regions.
Acknowledgments
This work was supported by La Ligue Nationale Contre le Cancer Grant ANR 07-MIME-005 “DC-TRANS” (to P.P.) and National Institutes of Health Grant AR057347 (to T.A.H.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 3.David A, et al. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J Biol Chem. 2011;286:20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman CA, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habets PE, et al. RNA content differs in slow and fast muscle fibers: Implications for interpretation of changes in muscle gene expression. J Histochem Cytochem. 1999;47:995–1004. doi: 10.1177/002215549904700803. [DOI] [PubMed] [Google Scholar]