Abstract
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibirets1, and for the use of cytoplasmic GFP to image the fine processes of neurons.
Elements in the 5′ and 3′ untranslated regions (UTRs) of mRNAs can promote translation and, thereby, increase protein production [reviewed elsewhere (1, 2)]. The nucleotides in the 5′-UTR immediately upstream of the initiating ATG codon have been shown to profoundly affect the level of translation initiation in vertebrates (3–5) and Drosophila (6, 7). In addition, many plant and insect viral mRNAs contain sequences of 20–70 bp in their 5′-UTRs that act in cis to increase translational efficiency (8–10), one of the most studied being omega (Ω) from tobacco mosaic virus (TMV) (11, 12). These translational enhancers can be transferred to other mRNAs and have been widely used to increase protein yields in in vitro extracts derived from cultured insect cells, wheat germ, and rabbit reticulocytes (9, 13, 14).
Sequence elements within the 3′-UTR, such as the polyadenylation signal (AATAAA) and the “GT-rich” element, contribute to the efficient termination of transcription and polyadenylation (1, 15–18). The poly(A) tails themselves are important for mRNA stability (19) and promote translation initiation (20, 21) through cooperative interaction of bound proteins with the 5′ cap (2, 22–24). In baculovirus insect protein expression systems, the 3′-UTR from the Autographa californica nucleopolyhedrovirus (AcNPV) p10 gene (25) increases the efficiency of both polyadenylation and expression of heterologous genes relative to the simian virus 40 (SV40) 3′-UTR (26).
GAL4- or LexA-driven transgenes in Drosophila have generally used the 3′-UTR corresponding to the SV40 early polyadenylation signal (27, 28). This UTR provides sufficient expression of most transgenes. However, in some cases, greater levels of expression are required; this has generally been addressed by including multiple copies of the transgene in the genome. For example, compared with membrane-localized proteins, cytoplasmic fluorescent proteins require higher levels of expression to achieve similar brightness in fine cellular processes because of unfavorable surface area-to-volume ratios. For this reason, it has not been feasible to visualize the finest processes of neurons with single copies of a gene encoding cytoplasmic GFP (29, 30). Likewise, effectors of neuronal cell function, such as temperature-sensitive mutants of dynamin encoded by the Shibire gene, can require high levels of protein expression to silence synaptic transmission; for example, the widely used UAS-Shibirets1 stock (Kitamoto III) (31) contains multiple transgene copies. In addition, although the SV40 UTR provides robust expression in somatic cells, it performs poorly in the female germline (32).
Previous attempts to increase protein expression levels in Drosophila using posttranscriptional methods have met with limited success. Addition of the WPRE element, a posttranscriptional regulatory element derived from a woodchuck hepatitis virus (33), to the 3′-UTR increased expression of cytoplasmic GFP by several fold (Ref. 29 and this report), whereas substituting the UTR from the Drosophila his2Av gene for the SV40 UTR yielded a twofold increase in the expression of a transmembrane protein (34). Inclusion of a small intron in the 5′-UTR also results in a modest increase in expression (29, 34).
In this report, we demonstrate that sequences derived from the 5′-UTR (8–14) and 3′-UTR (25, 26) of viral mRNAs, as well as from the abundantly expressed lobster tropomyosin gene (35), are able to function in Drosophila to enhance protein production. By using 5′- and 3′-UTR elements in combination, increases of >20-fold have been achieved, allowing single transgenes to achieve protein expression levels that previously required multiple transgenes, thereby greatly facilitating genetic strain construction. We also show that the 3′-UTR from the Autographa californica nuclear polyhedrosis virus (AcNPV) p10 gene functions efficiently in the female germline.
Results and Discussion
We first asked whether known enhancers of translation efficiency located in the 5′-UTR would function in Drosophila. We compared six sequences: (i) a 7-bp consensus of sequences upstream of the initiating codon of Drosophila genes, derived by Cavener and Ray (6), that we had used in previous constructs [for example, pJFRC13 (29)]; (ii) a synthetic AT-rich 21-bp sequence (Syn21) made by combining the Cavener consensus sequence with elements from the Malacosoma neustria nucleopolyhedrovirus (MnNPV) polyhedrin gene (9); (iii) the 21 bp immediately preceding the initiation codon of the lobster tropomyosin gene [L21 (35)]; (iv) a 10-bp consensus translation-initiation sequence derived from 23 different AcNPV genes combined with the upstream sequence of AcNPV p10 ATG (10); (v) the 49 bp upstream of initiation codon of the Ecotropis obliqua nucleopolyhedrovirus (EoNPV) polyhedrin gene (9); and (vi) the 68-bp Ω element from TMV (11). These constructs are diagrammed in Fig. 1.
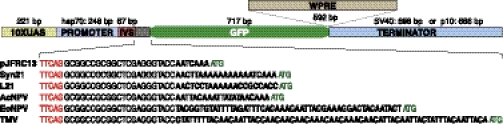
Fig. 1.
Structures of UAS-GFP constructs used in this study. Most components have been held constant and correspond to those in the previously described pJFRC13 vector (29), including the binding sites for the transactivator GAL4 (10XUAS), the promoter (a fragment of the hsp70 gene), a small intron (IVS; derived from the Drosophila myosin heavy chain gene), a GFP coding region optimized for Drosophila codon use, and the transcription terminator sequence derived from SV40. The sizes of these components are indicated. In the constructs used to assay the effects of 5′-UTR elements on GFP levels (see Fig. 2), the nucleotides immediately upstream of the ATG initiating codon of GFP in pJFRC13 have been replaced. The nucleotide sequences of this region, which spans from the end of the IVS (TTCAG, shown in red) to the initiating ATG (shown in green), are shown for each of the constructs. Only the nucleotides shown in bold differ between constructs: pJFRC13 (29); Syn21, pJFRC80 (see Methods); L21, pJFRC83 (35); AcNPV, pJFRC84 (10); EoNPV, pJFRC85 (9); and TMV, pJFRC86 (11). To make the constructs used to test the effects of altering the 3′-UTR, either the WPRE element (33) was placed between the GFP gene and the SV40 terminator (GFP-WPRE) or the SV40 terminator was replaced by the terminator from the AcNPV p10 gene (GFP-p10) (25). The constructs were all inserted in the same genomic location (attP2) to equalize extrinsic effects on expression.
Fig. 2 shows the expression patterns observed when each of these constructs is driven by R66A12-GAL4 (36). When crossed to our standard cytoplasmic GFP reporter (pJFRC13), R66A12-GAL4 drove moderate expression in a pair of neurons in each segment of the larval ventral nervous system (Fig. 2A). Addition of each of the translational enhancers to pJFRC13 increased the level of GFP expression in these cells to a similar extent and also revealed weakly expressing cells that were barely detected with pJFRC13 (Fig. 2 B–F). We chose to use the Syn21 sequence in future constructs because its small size facilitates incorporation into transgenes using oligonucleotide synthesis.
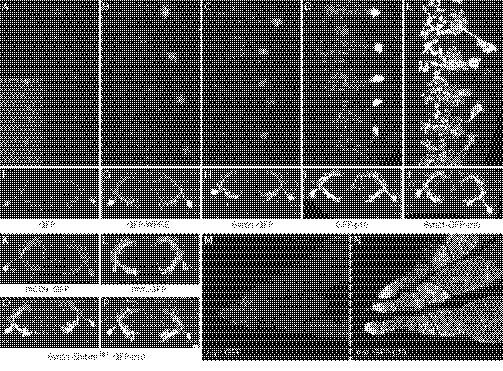
Fig. 2.
Short 5′-UTR sequence elements derived from a variety of viral and cellular mRNAs can increase protein expression in Drosophila. Ventral nerve cords of third-instar larvae that each carry the R66A12-GAL4 driver, paired with a different 10XUAS-GFP construct, are shown; the preparations have been stained with an antibody against GFP. R66A12 drives expression in a sparse set of segmentally repeated neurons, allowing the cell bodies (located laterally) and neurites (located centrally) of individual neurons to be resolved. (A) Expression level obtained with pJFRC13 (29). (B–F) Increased expression of GFP obtained by including in the 5′-UTR the additional sequence: Syn21 (B); L21 (C); AcNPV (D); EoNPV (E); and TMV (F). See Fig. 1 for a more detailed description of the constructs.
We next turned our attention to the 3′-UTR. We determined the effects of adding the WPRE element upstream of the SV40 3′-UTR and of replacing the SV40 UTR with that of the AcNPV p10 gene in the context of pJFRC13 (Fig. 3 A and F; see diagram in Fig. 1 and Table 1 for more details on the structure of these constructs). Confirming our previous results (29), adding the WPRE to the 3′-UTR (Fig. 3 B and G) produced a significant increase in GFP expression; the increase appeared similar to that obtained by including the Syn21 sequence in the 5′-UTR (Fig. 3 C and H). Replacing the SV40 3′-UTR with that of the p10 gene had an even larger positive effect (Fig. 3 D and I). The combination of the Syn21 element in the 5′-UTR with the WPRE element did not give any increase in expression over Syn21 alone, but its combination with the p10 3′-UTR produced the highest levels of expression (Fig. 3 E and J).
Fig. 3.
Enhancement of protein expression levels by 3′-UTR elements. (A–E) Increase in the level of cytoplasmic GFP generated from 10XUAS constructs by the addition of various 5′- or 3′-UTR elements was determined by measuring native GFP fluorescence using the photomultiplier tube (PMT) of a Zeiss 510 confocal microscope. Dissected nervous systems were imaged under identical conditions except for the Inset in A, which was imaged at higher gain. (A) Level of expression obtained with pJFRC13 (Fig. 1) (29), which was used as a baseline to measure the level of enhancement produce by addition of the other sequence elements. (B) Increase of 6.1-fold in cell body GFP fluorescence was observed when the WPRE element was added to the 3′-UTR in pJFRC14 (29). (C) Increase of 7.5-fold was observed by the addition of the Syn21 element to the 5′-UTR (pJFRC80). (D) p10 element produces a 23-fold increase when added to the 3′-UTR (pJFRC28). (E) Further increases in expression are observed when both the Syn21 and p10 elements are present (pJFRC81). (F–J) Pairs of neurons, viewed transversely, from the same series of genotypes, following staining with anti-GFP antibody. (K and L) Specimens imaged as in F–J using vectors that contain the same UTR elements as pJFRC13 but that express membrane-targeted GFP, either mCD8::GFP (K; pJFRC2; 29) or myr::GFP (L; pJFRC12; 29). M (pJFRC12) and N (pJFRC29) show that the p10 element can also increase GFP expression in the female germline. (O and P) Antibody staining for GFP in lines carrying a 20XUAS-Syn21-Shibire[ts1]::GFP-p10 construct (pJFRC101) inserted at the VK00005 (O) or attP2 (P) genomic integration sites. The R66A12-GAL4 driver was used to generate the data shown in A–L and O and P. The R34C10-GAL4 driver was used for M and N.
Table 1.
Description of constructs
| Name | Description |
| pJFRC2 | 10XUAS-IVS-mCD8::GFP |
| pJFRC12 | 10XUAS-IVS-myr::GFP |
| pJFRC13 | 10XUAS-IVS-GFP |
| pJFRC14 | 10XUAS-IVS-GFP-WPRE |
| pJFRC27 | 13XLexAop2-IVS-GCaMP3-p10 |
| pJFRC28 | 10XUAS-IVS-GFP-p10 |
| pJFRC29 | 10XUAS-IVS-myr::GFP-p10 |
| pJFRC57 | 13XLexAop2-IVS-GFP-p10 |
| pJFRC59 | 13XLexAop2-IVS-myr::GFP-p10 |
| pJFRC65 | 13XLexAop2-IVS-GFP-aequorin-p10 |
| pJFRC80 | 10XUAS-IVS-Syn21-GFP |
| pJFRC81 | 10XUAS-IVS-Syn21-GFP-p10 |
| pJFRC83 | 10XUAS-IVS-L21-GFP |
| pJFRC84 | 10XUAS-IVS-AcNPV-GFP |
| pJFRC85 | 10XUAS-IVS-EoNPV-GFP |
| pJFRC86 | 10XUAS-IVS-TMV-GFP |
| pJFRC90 | 20XUAS-IVS-Syn21-mPA-p10 |
| pJFRC91 | 20XUAS-IVS-Syn21-mSPA-GFP-p10 |
| pJFRC92 | 20XUAS-IVS-Syn21-mC3PA-GFP-p10 |
| pJFRC93 | 13XLexAop2-IVS-Syn21-mPA-p10 |
| pJFRC94 | 13XLexAop2-IVS-Syn21-mSPA-GFP-p10 |
| pJFRC95 | 13XLexAop2-IVS-Syn21-mC3PA-GFP-p10 |
| pJFRC96 | 20XUAS-IVS-GFP-aequorin-p10 |
| pJFRC97 | 20XUAS-IVS-GCaMP3-p10 |
| pJFRC98 | 20XUAS-IVS-Shibire[ts1]-p10 |
| pJFRC99 | 20XUAS-IVS-Syn21-Shibire[ts1]-p10 |
| pJFRC100 | 20XUAS-TTS-Shibire[ts1]-p10 |
| pJFRC101 | 20XUAS-IVS-Syn21-Shibire[ts1]-GFP-p10 |
| pJFRC104 | 13XLexAop2-IVS-Syn21-Shibire[ts1]-p10 |
A description of constructs referred to in this report is shown. pJFRC2, pJFRC12, pJFRC13, and pJFRC14 were described previously (29). See Methods and Ref. 29 for additional information. The photoactivatable GFP variants used to construct pJFRC90 to pJFRC95 have been described previously: mPA (53), mSPA (40), and mC3PA (40).
We quantified these expression levels in two ways. Firstly, we used quantitative confocal microscopy. Five nervous systems of each genotype were optically sectioned at constant gain through the abdominal region that contained the cell bodies shown in Fig. 3 A–E to record the level of native GFP fluorescence. Five to 10 cell bodies were chosen from each confocal stack and the average pixel intensity within the brightest optical section for each cell body was determined. The average cell body intensity was then calculated for each nervous system, and a final average then computed for each genotype. Addition of the WPRE element increased expression to 6.1 times that observed for pJFRC13, Syn21 resulted in a 7.5-fold increase, and the p10 3′-UTR yielded a 23-fold increase. Secondly, we measured expression levels by measuring the GFP fluorescence in extracts prepared from dissected nervous systems and subjected to native polyacrylamide gel electrophoresis (see Methods for details). In this assay, we used R57C10-GAL4 (36), a neuronal synaptobrevin promoter-fusion GAL4 line that is expressed very broadly in the nervous system. Syn21 increased expression of GFP by a factor of 7.4 ± 0.3 (n = 3); the p10 3′-UTR increased expression by a factor of 16.9 ± 0.9 (n = 3); and, in combination, Syn21 and the p10 3′-UTR increased expression by a factor of 22.4 ± 4.3 (n = 3). These results are consistent with those obtained by quantitative microscopy and confirm that the p10 3′-UTR can increase protein expression by more than a factor of 10 on its own and by a factor of 20 when in combination with the 5′-UTR element Syn21.
In certain cases, it has not been possible to obtain sufficiently high expression of a protein from a single transgene, requiring the insertion of the transgene at multiple chromosomal locations. However, the requirement for multiple gene copies greatly complicates the generation of complex genotypes, where many different transgenes must be brought together in the same animal. The higher levels of expression that we have demonstrated from single transgenes that incorporate the Syn21 and p10 translational enhancers have allowed us to overcome this limitation, as demonstrated by the constructs described below and listed in Table 1.
Neurophysiological experiments are best performed using cytoplasmic markers and probes of activity, because exogenous highly expressed membrane-associated proteins are considered more likely to interfere with the electrical properties of the cell. However, it has been difficult to achieve expression levels of cytoplasmic markers sufficient to visualize fine neuronal processes, because of their small volume. As a result, experimenters have often resorted to fusions of GFP to transmembrane proteins, such as CD8 (37) and CD4 (34), or to a protein domain that becomes N-myristoylated (29, 38). The increased levels of expression of cytoplasmic markers we can obtain by including the p10 UTR permits the fine processes of the neurons to be clearly visualized (compare Fig. 3 I, K, and L). We also observed that, unlike the SV40 UTR (32), the p10 3′-UTR supports robust expression in the female germline (Fig. 3 M and N).
Likewise, we have constructed vectors for expressing several other proteins that have previously required multiple transgene copies to yield effective concentrations, including photoactivatable GFP (39, 40), GFP-Aequorin (41), and GCaMP3 (42). Single-copy insertions of these constructs achieve sufficient expression levels to perform well, based on initial tests performed in a number of laboratories. We also generated LexA-controlled versions of many of these constructs (see Table 1).
Finally, we made a series of constructs expressing temperature-sensitive variants of the dynamin protein, a GTPase required for synaptic vesicle recycling, encoded by the Shibire gene (43–46). Dynamin is among the most abundant gene products in the brain; its transcript ranks in the top 3% of brain mRNAs (47). The temperature-sensitive dynamin encoded by the transgene acts as a dominant negative at the nonpermissive temperature and must be expressed at high levels to poison the function of endogenous dynamin. Achieving these levels with most GAL4 drivers has required multiple insertions of a UAS-Shibirets1 transgene (31, 48), with the commonly used effective stocks, such as the Kitamoto III stock, containing several copies. The need for multiple insertions greatly complicates stock construction and standardization of genetic background, which can be a critical factor in behavioral experiments.
To express the Shibirets1 protein at high levels, we made constructs in which the p10 3′-UTR and, in some cases, the Syn21 translation enhancer were included (see Table 1). We also made a version in which Shibirets1 was fused at its C terminus to GFP (pJFRC101); when crossed to the R66A12-GAL4 line, expression in the expected pattern and level was observed (Fig. 3 O and P). To allow for even higher expression from a single chromosomal site, we also constructed a tandem dimer of two 20XUAS-Shibire[ts1]-p10 constructs (pJFRC100).
In addition to the widely used Shibirets1 allele, we also made constructs expressing the Shibirets2 (45), ShibireEM33 (46), and Dynamin4ts1 alleles (49). (The Dynamin4 isoform is a splice product of the Shibire gene that results in a C-terminal extension of 48 amino acids.) We found that the ShibireEM33 protein reduced viability when used with the broadly expressed driver R57C10-GAL4, even at the permissive temperature. We also observed pronounced swelling of fine processes when a GFP fusion to ShibireEM33 was expressed using R66A12-GAL4. The Shibirets2 and Dynamin4ts1 constructs did not decrease viability but did not perform as well as those made with Shibirets1; we, therefore, decided to proceed only with Shibirets1 allele.
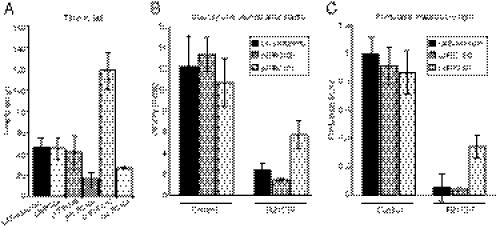
We first tested the Shibirets1 constructs by crossing them to R57C10-GAL4 and measuring the amount of time at restrictive temperature (30.6 °C) required to induce flies to fall off the walls of the vial (Fig. 4A). Flies expressing a single copy of the Shibirets1 transgene with either the p10 UTR (pJFRC98) or with both the Syn21 and p10 sequences (pJFRC99) became incapacitated in a period comparable to the most effective published stock from Kitamoto [UAS-Shibirets1 (Kitamoto III)] (31). Flies expressing the tandem Shibire[ts1]-p10 construct (pJFRC100) were affected more rapidly than the Kitamoto stock (n = 4; P < 0.05). The construct expressing a Shibire[ts1]::GFP fusion gene (pJFRC101) had a reduced effect, suggesting that the GFP fusion had either decreased its expression or impaired its ability to compete with endogenous dynamin. We also generated a LexAop2-Syn21-Shibire[ts1]-p10 construct (pJFRC104), which functioned effectively when crossed to R57C10-LexA, meaning LexA drivers can now be used with the Shibire[ts1] effector. We then assayed a subset of the constructs in assays for startle response (Fig. 4B) and phototaxis (Fig. 4C), which used a GAL4 driver with more limited expression (R21C07-GAL4); pJFRC100 performed similarly to the Kitamoto stock and pJFRC101 was weaker (n = 6).
Fig. 4.
Assays of the effectiveness of Shibirets1 constructs. (A) Ability of various pJFRC constructs (in VK00005) that express Shibirets1 to induce loss of motor control was compared with that of the Kitamoto stock. pJFRC98, pJFRC99, pJFRC100, and pJFRC101 were crossed to a pan-neuronal GAL4 driver, R57C10-GAL4, and pJFRC104 was crossed to R57C10-LexAp65 (in VK00027). The times for flies to fall from the side of the vial at 30.6 °C are shown (mean ± SD). The progeny of control crosses of the same effectors to the BDPGal4U line, which does not express significant amounts of GAL4 in the adult nervous system (36), were all still on the side of the vial after 360 seconds, when the experiment was terminated. The pJFRC effector constructs were inserted into the VK00005 genomic site; see Table 1 for details of the constructs. (B) Effectiveness of the pJFRC100, pJFRC101, and Kitamoto stocks in decreasing locomotor response to startle was compared (all values are mean ± SD). The effectors were each crossed to the control GAL4 driver, BDPGal4U, or to a driver line, R21C07-GAL4, that was expressed in the optic lobes and a small proportion of central brain and ventral nerve cord neurons. Flies normally respond to a startle by an increase in locomotion. The extent to which each of the effector lines was able to suppress this response is shown. (C) Flies show a strong phototaxis response to UV light. The same genotypes as in B were assayed for phototaxis. A phototaxis score was calculated by averaging the median displacement of six tubes of flies toward lights and normalizing by the length of the tubes (mean ± SD). This is a normalized metric for the strength of attraction toward light where a value of one indicates that the population of flies congregates as close to the light as possible and a value of zero indicates no attraction.
The availability of a series of Shibire effector constructs that vary in expression levels allows the use of the weakest construct that yields a behavioral effect with a given GAL4 driver. This can be important because very high levels of Shibire expression have been shown to be toxic even at the permissive temperature (50).
In conclusion, we have shown that several established translational enhancers from other species are effective in Drosophila and can be used to increase protein yields by a factor of more than 20. We have used those elements to generate a series of enhanced expression vectors and have demonstrated their utility in a variety of applications.
Methods
Molecular Biology and Drosophila Genetics.
Standard methods were used to generate the constructs used in this study, as described elsewhere (29, 36). The p10 UTR, C-terminal extension of dynamin4, and three small regions of Shibire, which encode the ts2 mutation, the EM33 mutation, or a small fragment corresponding to but lacking the ts1 mutation, were synthesized by DNA2.0. The 666-bp p10 UTR was cloned 5′-XbaI to 3′-FseI into pJFRC13 and pJFRC12 after removal of the SV40 UTR to generate pJFRC28 and pJFRC29, respectively. Synthetic oligonucleotides (IDT) for Syn21, L21, AcNPV, EoNPV, and TMV were used to amplify Drosophila codon-optimized GFP from pJFRC13 and the products cloned 5′-KpnI and 3′-XbaI into the pJFRC13 backbone in place of the GFP gene. pJFRC98 and pJFRC99 were generated by PCR amplifying the Shibirets1 gene (gift of Jon-Michael Knapp, Janelia Farm Research Campus, Ashburn, VA), alone or with Syn21, and cloning the products as 5′-XhoI to 3′-KpnI fragments into a modified version of pJFRC7 that contained 20× UAS sites and the p10 UTR. pJFRC100 was generated as follows: pJFRC98 was cut with FseI and ligated with the insulated spacer described previously (29) to generate 20XUAS-IVS-Shibire[ts1]-p10-INS. Then, a second aliquot of pJFRC98 was cut with HindIII, made blunt using PfuUltra High-Fidelity enzyme (Agilent Technologies), cut with PmeI, gel-purified, and cloned into PmeI-digested 20XUAS-IVS-Shibire[ts1]-p10-INS. pJFRC101 was generated by triple ligation of the following: the Shibirets1 coding region, PCR amplified to include 3 C-terminal glycine residues and 5′-XhoI and 3′-ScaI flanking restriction sites; Drosophila codon-optimized GFP from pJFRC13, amplified 5′-ScaI to 3′-KpnI; and XhoI- plus KpnI-cut pJFRC98. The resultant Shibire[ts1]::GFP fusion contains a 5-aa linker consisting of GGGST between the Shibire protein and GFP. pJFRC104 was generated by cloning the 5′-XhoI to 3′-FseI fragment of pJFRC99 that contains Shibirets1 and the p10 UTR into pJFRC19, after removal of the myr::GFP gene and the SV40 UTR. Plasmid constructs pJFRC28 and pJFRC81 are available from Addgene. To generate constructs expressing another protein instead of GFP, one would simply amplify the gene encoding that protein with a 5′ primer beginning with a KpnI site and a 3′ primer with an XbaI site and clone into KpnI- plus XbaI-digested pJFRC28. To add the Syn21 sequence, the Syn21 sequence would be included in the 5′ primer.
The DNA fragments contained in R21C07, R66A12, and R57C10 were derived from the nAcRα-7E (PCR primers: gacgactgtcacctgcgagtgtaag and gcaagaagccaagttgtcatgcggt), Atpα (PCR primers: caccccgcgtatcctgtggattgttgcat and cctcgttctaagattccctctcaag), and n-syb (PCR primers: atttcccaccccttggccatcggca and gttctagagggttgcgctctcagtg) genes, respectively. Transgenic lines were generated by Genetic Services. Insertions of the transgenes into the attP2 site were used in all experiments, unless otherwise noted in the text.
Histological Methods.
Dissected tissues from animals grown at 25 °C were fixed for 1 h in 4% (vol/vol) buffered formaldehyde, and then repeatedly rinsed in PBS with 1% Triton-X 100 (PBS-TX). In most instances, GFP was detected immunologically by incubating the tissues overnight in a 1:1,000 dilution of a rabbit anti-GFP antiserum (Invitrogen) in PBS-TX, followed by rinses and overnight incubation with donkey Alexa488–anti-rabbit IgG (Invitrogen) in PBS-TX. Following multiple rinses, tissues were mounted on a poly-l-lysine-coated cover-slip, dehydrated through an ethanol series, cleared in xylene, and mounted in DPX Mountant (Electron Microscope Sciences). For detection of native GFP, fixed tissues were rinsed three to four times in PBS-TX, attached to a poly-l-lysine-coated coverslip, and mounted in VectaShield (Vector Labs).
Confocal Z-stacks were collected at 63× and 0.5-μm intervals using a Zeiss 510 confocal microscope. During the imaging of a series of genotypes, the gain was kept constant, so that images could be compared within each series. The image stacks were processed using Image J (National Institutes of Health) to produce Z-projections and cross-sectional views of each genotype.
For the quantification of native GFP levels, we collected 63× Z-stacks for five nervous systems per genotype. Five to 10 cell bodies were chosen from each confocal stack. We chose the brightest optical section for each cell body, selected the pixels within that section and determined the average pixel intensity using the Measure tool from Image J. The average cell body intensity was then calculated for each nervous system, and a final average was then computed for each genotype.
Native Polyacrylamide Gel Electrophoresis and In-Gel Fluorescence Quantification.
Dissected central nervous systems from 20 larvae of each genotype grown at 25 °C were pooled and placed in 50 μL of lysis buffer (Cell BioLabs; GFP Quantification Kit) containing a 1:100 dilution of a mixture of protease inhibitors (Sigma-Aldrich; catalog no. P 1860). The samples were homogenized using a sterile rod in a 1.5-mL conical tube, followed by two sonication bursts (20 s at ∼24 W). Following centrifugation at room temperature for 30 min at 15,000 × g, a 15-μL aliquot of each supernatant was mixed 1:1 with a buffer containing 100 mM Tris (pH 6.8), 10% (vol/vol) glycerol, 0.05% bromophenol blue, 10 mM 2-mercaptoethanol, and 2% (wt/vol) octyl-β-d-glucopyranoside (Anatrace). The samples were loaded onto a gradient (4–20%) native polyacrylamide gel and subjected to electrophoresis. Once the run was complete, fluorescence was imaged using a FOTO/Analyst workstation equipped with a diode array (Fotodyne). Quantification of the fluorescence intensity was carried out using the TotalLab Quant software. No fluorescence was detected in the pellet following centrifugation or at the stacking gel indicating that the vast majority of the GFP migrated into the separating gel.
Behavioral Assays.
To assay the efficacy of the Shibirets1 constructs, we measured the time after shift to an elevated temperature it took for flies to lose motor control and fall off the side of a vial. Flies were reared at room temperature (∼22 °C). Two- to 5-d-old flies were anesthetized with carbon dioxide; groups of 20 flies, approximately equal numbers of males and females, were put into new vials and allowed 1 d to recover. Fresh vials were preheated for 24 h at 30.6 °C. Flies were quickly transferred from room temperature vials to the preheated vials and observed for 6 min at 30.6 °C. Time to falling was recorded, scored as the amount of time from transfer to time it took for all 20 flies to fall from the top or sides to the bottom.
We also measured the efficacy of the Kitamoto, pJFRC100, and pJFRC101 stocks to effect phototaxis and locomotion using assays similar to those developed by others (51, 52). Five male R21C07-GAL4 flies were crossed to 5 females from each effector line, and the resulting offspring were aged between 3–7 d and sorted into 6 groups of 15 males of each genotype on a cold plate and then assayed in separate tubes. Flies were starved on agar between 1 and 4 h before assaying. The differences for locomotor response at 1 s after a startle stimulus and for phototaxis to UV light were compared with the control cross of the pBDPGal4U line (36), to the same effectors.
Acknowledgments
We thank William Rowell and Wyatt Korff for performing the behavioral assays shown in Fig. 4 B and C and the Janelia Farm Fly Olympiad Project Team for developing these assay methods, Hongjin Zheng and Tamir Gonen for the gel-based quantification of GFP levels, Hui-Min Chen and Tzumin Lee for data shown in Fig. 3 M and N, and Karen Hibbard for stock construction. Robin Harris provided critical feedback and shared preliminary results. The Janelia Farm Molecular Biology Shared Resource performed DNA sequencing. Transgenic lines were generated by GSI, Inc. Crystal Sullivan provided administrative support.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. The scanning model for translation: An update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir MP, Rice MD. TRII: A Probabilistic Scoring of Drosophila melanogaster Translation Initiation Sites. EURASIP J Bioinform Syst Biol. 2010;2010:814127. doi: 10.1155/2010/814127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner R, Foster GD. The potential exploitation of plant viral translational enhancers in biotechnology for increased gene expression. Mol Biotechnol. 1995;3:225–236. doi: 10.1007/BF02789333. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, et al. Performance of expression vector, pTD1, in insect cell-free translation system. J Biosci Bioeng. 2006;102:69–71. doi: 10.1263/jbb.102.69. [DOI] [PubMed] [Google Scholar]
- 10.Ranjan A, Hasnain SE. Influence of codon usage and translational initiation codon context in the AcNPV-based expression system: Computer analysis using homologous and heterologous genes. Virus Genes. 1995;9:149–153. doi: 10.1007/BF01702657. [DOI] [PubMed] [Google Scholar]
- 11.Gallie DR, Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallie DR. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 2002;30:3401–3411. doi: 10.1093/nar/gkf457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawasaki T, Hasegawa Y, Tsuchimochi M, Kasahara Y, Endo Y. Construction of an efficient expression vector for coupled transcription/translation in a wheat germ cell-free system. Nucleic Acids Symp Ser. 2000;2000:9–10. doi: 10.1093/nass/44.1.9. [DOI] [PubMed] [Google Scholar]
- 14.Endo Y, Sawasaki T. High-throughput, genome-scale protein production method based on the wheat germ cell-free expression system. J Struct Funct Genomics. 2004;5:45–57. doi: 10.1023/B:JSFG.0000029208.83739.49. [DOI] [PubMed] [Google Scholar]
- 15.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 16.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 2002;3:REVIEWS0004. 2002 doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 18.Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 19.Bönisch C, Temme C, Moritz B, Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J Biol Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 20.Jackson RJ, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 21.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 23.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 24.Sachs AB, Varani G. Eukaryotic translation initiation: There are (at least) two sides to every story. Nat Struct Biol. 2000;7:356–361. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- 25.Smith GE, Vlak JM, Summers MD. Physical Analysis of Autographa californica Nuclear Polyhedrosis Virus Transcripts for Polyhedrin and 10,000-Molecular-Weight Protein. J Virol. 1983;45:215–225. doi: 10.1128/jvi.45.1.215-225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Oers MM, Vlak JM, Voorma HO, Thomas AA. Role of the 3′ untranslated region of baculovirus p10 mRNA in high-level expression of foreign genes. J Gen Virol. 1999;80:2253–2262. doi: 10.1099/0022-1317-80-8-2253. [DOI] [PubMed] [Google Scholar]
- 27.Carswell S, Alwine JC. Efficiency of utilization of the simian virus 40 late polyadenylation site: Effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito K, Okada R, Tanaka NK, Awasaki T. Cautionary observations on preparing and interpreting brain images using molecular biology-based staining techniques. Microsc Res Tech. 2003;62:170–186. doi: 10.1002/jemt.10369. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 32.Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 33.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han C, Jan LY, Jan Y-N. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci USA. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sano K, Maeda K, Oki M, Maéda Y. Enhancement of protein expression in insect cells by a lobster tropomyosin cDNA leader sequence. FEBS Lett. 2002;532:143–146. doi: 10.1016/s0014-5793(02)03659-1. [DOI] [PubMed] [Google Scholar]
- 36.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 38.Mauss A, Tripodi M, Evers JF, Landgraf M. Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 2009;7:e1000200. doi: 10.1371/journal.pbio.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta SR, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- 40.Ruta V, et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 41.Martin J-R, Rogers KL, Chagneau C, Brûlet P. In vivo bioluminescence imaging of Ca signalling in the brain of Drosophila. PLoS ONE. 2007;2:e275. doi: 10.1371/journal.pone.0000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigliatti TA, Hall L, Rosenbluth R, Suzuki DT. Temperature-sensitive mutations in Drosophila melanogaster. XIV. A selection of immobile adults. Mol Gen Genet. 1973;120:107–114. doi: 10.1007/BF00267238. [DOI] [PubMed] [Google Scholar]
- 44.Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- 45.van der Bliek AM, et al. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant D, Unadkat S, Katzen A, Krishnan KS, Ramaswami M. Probable mechanisms underlying interallelic complementation and temperature-sensitivity of mutations at the shibire locus of Drosophila melanogaster. Genetics. 1998;149:1019–1030. doi: 10.1093/genetics/149.2.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 48.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 49.Chen MS, et al. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Bellido PT, Wardill TJ, Kostyleva R, Meinertzhagen IA, Juusola M. Overexpressing temperature-sensitive dynamin decelerates phototransduction and bundles microtubules in Drosophila photoreceptors. J Neurosci. 2009;29:14199–14210. doi: 10.1523/JNEUROSCI.2873-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu Y, Nern A, Zipursky SL, Frye MA. Peripheral visual circuits functionally segregate motion and phototaxis behaviors in the fly. Curr Biol. 2009;19:613–619. doi: 10.1016/j.cub.2009.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc Natl Acad Sci USA. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]