Abstract
Bacterial transpeptidase and transglycosylase on the surface are essential for cell wall synthesis, and many antibiotics have been developed to target the transpeptidase; however, the problem of antibiotic resistance has arisen and caused a major threat in bacterial infection. The transglycosylase has been considered to be another excellent target, but no antibiotics have been developed to target this enzyme. Here, we determined the crystal structure of the Staphylococcus aureus membrane-bound transglycosylase, monofunctional glycosyltransferase, in complex with a lipid II analog to 2.3 Å resolution. Our results showed that the lipid II-contacting residues are not only conserved in WT and drug-resistant bacteria but also significant in enzymatic activity. Mechanistically, we proposed that K140 and R148 in the donor site, instead of the previously proposed E156, are used to stabilize the pyrophosphate-leaving group of lipid II, and E100 in the acceptor site acts as general base for the 4-OH of GlcNAc to facilitate the transglycosylation reaction. This mechanism, further supported by mutagenesis study and the structure of monofunctional glycosyltransferase in complex with moenomycin in the donor site, provides a direction for antibacterial drugs design.
Keywords: membrane protein structure, antibiotic discovery, bacterial cell wall synthesis
Transglycosylase (TG; also known as peptidoglycan glycosyltransferase or murein synthase) and transpeptidase (TP) are the two crucial membrane-bound enzymes for the synthesis of bacterial cell wall, which is an essential structure in scaffolding the cytoplasmic membrane and maintaining structural integrity of the bacteria (1). Most antibiotics such as β-lactams (e.g., penicillin and methicillin), glycopeptides (e.g., vancomycin and teicoplanin), and glycolipopeptides inhibit the activity of TP (2). However, because of the mutation on TP and other changes under the pressure of such antibiotics, new bacteria, such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus, emerge and spread (3). In contrast, none of current antibiotics is available to target the other essential and extracellular enzyme TG, perhaps because of the lack of detailed understanding of the transglycosylation process (4–6). Multiple sequence alignment analyses reveal that five motifs of TG in various methicillin-resistant S. aureus and drug-resistant Gram-negative bacteria (e.g., Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa) and Mycobacterium tuberculosis are highly conserved (SI Appendix, Fig. S1), and the polysaccharide backbone of the peptidoglycan always remains unchanged in WT and resistant strains (7). Thus, new antibiotics that target the transglycosylation step may be less prone to resistance development.
In the synthesis of bacterial cell wall, the peptidoglycan transglycosylation by the enzyme TG takes place through polymerization of lipid II substrates (8). The substrate-binding site of TG has been proposed to consist of a glycosyl acceptor site, where the disaccharide monomer lipid II binds, and a glycosyl donor site, where another lipid II binds and accommodates the growing sugar chain (9, 10). The only known natural product that directly inhibits the function of TGs is moenomycin (11), which binds to the glycosyl donor site (12–15). Unfortunately, moenomycin cannot be given to humans because of its poor pharmacokinetic properties (16). The molecular interaction between lipid II and the glycosyl acceptor site of TG is needed to elucidate the mechanism of lipid II polymerization and also, serve as a new basis for the development of inhibitors of TG. In this study, we designed lipid II analogs and used them to cocrystallize with S. aureus membrane-bound monofunctional glycosyltransferase (MGT) by bicelle method (17) to unveil the structure of MGT and together with mutagenesis analysis, elucidate the mechanism of transglycosylation reaction.
Results
Importance of Transmembrane Helix and Strategy for Crystallization.
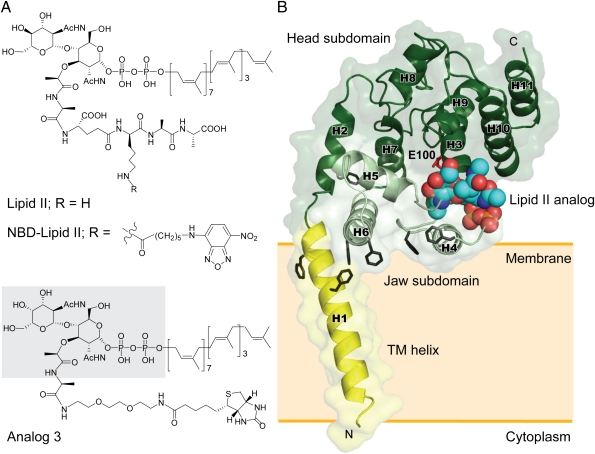
To maintain the structural integral of the interaction between S. aureus MGT (referred to as SaMGT) and lipid II analogs, we used the protein construct of SaMGT containing both the TG domain and the transmembrane (TM) helix for structural and functional analysis. The catalytic activity of SaMGT using lipid II as substrate was measured to be kcat (s−1) = 0.40 ± 0.063, Km (μM) = 9.6 ± 0.76, and kcat/Km (M−1s−1) = (4.15 ± 0.46) × 104 (SI Appendix, Fig. S2). When the TM helix was removed from the SaMGT (referred to as SaMGTΔTM), it was found that the enzymatic activity, measured with the initial velocity, dropped 10 times lower (SI Appendix, Fig. S3). For comparison, we also looked into the penicillin-binding protein 1b from E. coli (referred to as E. coli PBP1b) and used isothermal titration calorimetry to determine the energetic contribution of the TM helix to the binding of moenomycin (SI Appendix, Fig. S4). With the TM helix, SaMGT and E. coli PBP1b exhibited higher entropy of interaction with moenomycin than without the TM helix in SaMGTΔTM and E. coli PBP1bΔTM, suggesting that the TM helix contributes to the hydrophobic interaction. We postulate that the SaMGT, in which the TM helix maintains the proper membrane orientation for enzymatic activity, may facilitate the cocrystallization with the substrate 6-[N-(7-nitrobenzyl-2-oxa-1,3-diazol-4-yl) amino] hexanoyl (NBD)-lipid II (referred to as NBD-lipid II) and lipid II analogs (Fig. 1A and SI Appendix, Fig. S5) (13). Similar to the previously solved E. coli PBP1b structure, we observed that the TM domain maintains a proper membrane orientation in the structures solved in this study: SaMGT-analog (SaMGT in complex with a lipid II analog 3), SaMGT-substrate (SaMGT in complex with NBD-lipid II), SaMGT-moenomycin (SaMGT in complex with moenomycin), and SaMGT-apo (SI Appendix, Fig. S6).
Fig. 1.
Structures of lipid II, lipid II analogs, and SaMGT-analog complex. (A) Chemical structures of lipid II and lipid II analogs. The portion of analog 3 that can be observed on the electron density map is shaded in gray. (B) An overall structure of SaMGT-analog. The TM helix, jaw subdomain, and head subdomain are color-coded in yellow, light green, and dark green, respectively. The lipid II analog 3 is shown as van der Waals spheres. Putative active site, E100 of SaMGT, and aromatic residues are located near the water–membrane interfaces, which are shown as sticks in red and black, respectively. The numbering of helices of SaMGT is indicated as H1–H11. The proposed membrane location is indicated by an orange shaded rectangle. The dissociation constant of analog 3 is ∼12.9 μM.
Overall Structure of SaMGT in Complex with Lipid II Analog.
To understand how NBD-lipid II and other lipid II analogs bind to TG at glycosyl acceptor site, the X-ray diffraction data of SaMGT-substrate and SaMGT-analog were collected to 3.2 and 2.3 Å resolutions, respectively (SI Appendix, Table S1). The electron density of NBD-lipid II in SaMGT is superimposable with analog 3 in SaMGT (SI Appendix, Fig. S7), which suggests that analog 3 is located at the substrate-binding site. The TG domain (residues D71 to R269) of SaMGT-analog exhibits similar 3D fold with other TGs in E. coli PBP1b (13), A. aeolicus peptidoglycan glycosyltransferase (PGT) (14), S. aureus PBP2 (12), and S. aureus MGTE100Q (15) (SI Appendix, Fig. S8). The lipid II analog 3 binds to a position near residues E100 surrounded by helices H3–H5, which has been proposed as the glycosyl acceptor site of TG where lipid II substrate binds (12, 18) (Fig. 1B).
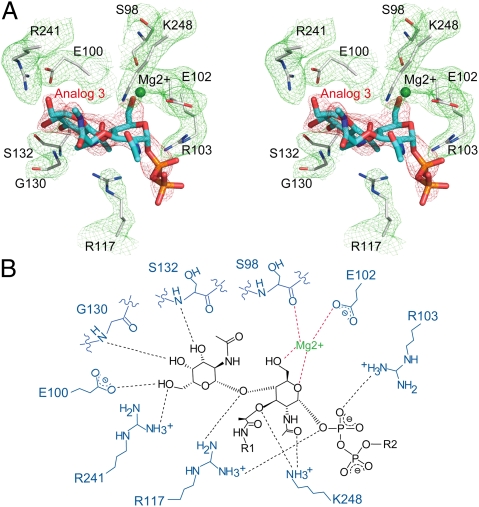
The nine interacting residues in SaMGT surrounding analog 3 within the hydrogen-bonding distance of 3.5 Å can be clearly observed on the 2Fo-Fc electron density map, and the structure of analog 3 can be clearly defined by an omit map generated from analog 3-deleted SaMGT model (Fig. 2A). In the lipid II-analog binding pocket of SaMGT, E100, G130, S132, and R241 bind to the GalNAc residue, in which G130 and S132 bind to the GalNAc through backbone amide group (Fig. 2B). The R241 also forms the salt bridge with E100 as observed in the structure of S. aureus PBP2 (12). R103 and R117 both interact with the pyrophosphate of lipid II analog 3, and R117 also interacts with the β1–4 glycosidic oxygen of lipid II analog 3. Although K113 is near the pyrophosphate, it is not clearly shown in the electron density map (SI Appendix, Fig. S9). Surprisingly, the d-lactyl ether moiety of MurNAc, which is part of the pentapeptide and supposed to participate in the transpeptidation reaction (1), interacts with K248 in the TG domain of SaMGT; moreover, K248 also binds to the N-acetyl group of MurNAc. Two possible Mg2+ cations can be identified in the electron density difference map (σ-cutoff ∼ 4.8σ) with an octahedral coordination manner (O···Mg···O angles near 90° and Mg···O distances near 2.4 Å in SaMGT-analog). One binds E102 and the backbone carbonyl group of S98 with the 5-CH2OH groups and the ester oxygen O1 of MurNAc (Fig. 2B and SI Appendix, Fig. S10). The other Mg2+, which is close to the analog 3 binding pocket of SaMGT, does not directly bind to analog 3; instead, it interacts with T115, G131, and Q136 near H4 (SI Appendix, Fig. S10), in which Q136 has been shown to be important for the activity of SaMGT (15).
Fig. 2.
Stereo and schematic view of interaction between SaMGT and analog 3. (A) An electron density map of binding interaction between analog 3 and SaMGT. The analog 3 and the nine SaMGT residues around the analog 3 are shown as sticks in blue and gray, respectively. The Mg2+ is shown as a ball in green. The 2Fo-Fc map for lipid II analog-contacting residues (σ-cutoff = 1σ) is shown in green mesh. The omit map for analog 3 (σ-cutoff = 3σ) is shown in red mesh. (B) Hydrogen-bonding networks between SaMGT and lipid II analog. R1 and R2 represent portions of analog 3 that cannot be observed in the electron density map. Hydrogen bonds (distance ≤ 3.5 Å) and interactions around Mg2+ are shown as dashed lines in black and red, respectively.
Sequence Alignment, Structural Alignment, and TG Activity of Lipid II Analog-Contacting Residues of SaMGT.
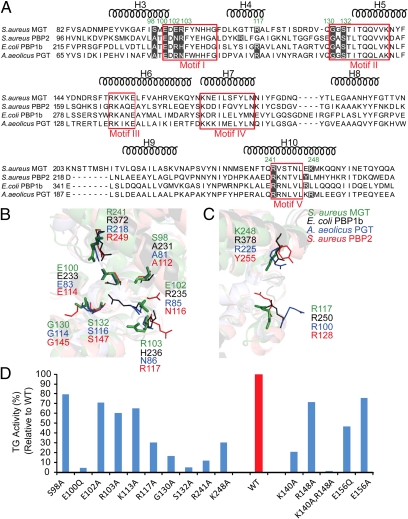
To understand the significance of the lipid II analog-contacting residues, we compared those residues in five conserved motifs of TG (1, 19, 20) and created the mutant within the lipid II analog-contacting residues to determine the TG activity (Fig. 3). The sequence alignment showed that seven residues (S98, E100, E102, R103, G130, S132, and R241) of SaMGT have the corresponding residues in E. coli PBP1b, A. aeilocus PGT, and S. aureus PBP2 (Fig. 3A) that are aligned in the structures (Fig. 3B). However, R117 and K248 do not exactly align in the TG sequences (Fig. 3A); the nearby residues can substitute their functions and also superimpose in the structural alignment (Fig. 3C). We created mutations at the lipid II analog-contacting residues of SaMGT and measured the TG activity of each mutant protein. All mutants at the lipid II analog-contacting residues of SaMGT reduced the TG activity in vitro (Fig. 3D). We also created the mutants of each corresponding lipid II analog-contacting residues in E. coli PBP1b and measured the complementary activity in E. coli JE5702, which is a PBP1b-defective/PBP1a temperature-sensitive strain (SI Appendix, Fig. S11A). Almost all mutants, E233Q, R235A, H236A, R250A, G264A, S266A, R372A, and R378A have reduced activity in E. coli JE5702 compared with the WT E. coli PBP1b. The essential role of the lipid II analog-contacting residues in sequence alignment and structural superimposition of TGs are consistent with the activity assay of each mutant protein in SaMGT and E. coli PBP1b, suggesting that these residues play an critical role for maintaining the activity of TG to catalyze the lipid II polymerization.
Fig. 3.
Sequence alignment, structural alignment, and TG activity of lipid II analog-contacting residues of SaMGT. (A) Sequence alignment of SaMGT-analog with TGs from E. coli PBP1b, A. aeilocus PGT, and S. aureus PBP2. The typical five conserved motifs of TG are enclosed in red squares. Residues that have the potential to interact with analog 3 are shaded in black, and the residues of SaMGT are indicated in green. The helices H1–H11 of SaMGT are indicated above the sequence alignment. (B and C) Structural alignment of SaMGT-analog with TG from E. coli PBP1b (PDB ID code 3FWM, rmsd = 1.694 Å), A. aeilocus PGT (PDB ID code 3D3H, rmsd = 1.020 Å), and S. aureus PBP2 (PDB ID code 2OLV, rmsd = 1.635 Å). Residues from E. coli PBP1b, A. aeilocus PGT, S. aureus PBP2, and SaMGT-analog are color-coded in black, blue, red, and green, respectively. All of the corresponding residues from four TGs are indicated, except the missing G264 and S266 of E. coli PBP1b. (D) Activity of the mutant SaMGT proteins compared with WT SaMGT. The activity of WT SaMGT (WT) is normalized to 100%.
Discussion
Comparison of Binding Modes Between TG-Moenomcin and TG-Lipid II.
To understand the binding modes between TG-moenomycin and TG-lipid II analog, we compared the structures of TG-moenomycin with SaMGT-analog. Both the lipid II analog and moenomycin binding pockets of TGs are conserved (SI Appendix, Fig. S12). Comparing the binding modes of TGs-moenomycin and SaMGT-analog, the binding interactions are similar in the phosphate moiety of moenomycin and lipid II analog 3. The electrostatic surface of SaMGT clearly shows two positively charged patches (SI Appendix, Fig. S13A), one surrounding the phosphate of the moenomycin (where K140 and R148 are located) and the other surrounding the pyrophosphate of the lipid II analog 3 (where R103, K113, and R117 are located) (SI Appendix, Fig. S13B). In addition, the binding interaction in the sugar moieties of moenomycin, which are thought to mimic the sugars of lipid IV, has more hydrogen-bonding interactions with the TGs than lipid II analog 3 (SI Appendix, Fig. S12), suggesting that the stronger binding force in the glycosyl donor site may help stabilize the growing sugar chain and facilitate the growing sugar to shuffle to the glycosyl donor site from the acceptor site after transglycosylation reaction.
K140 and/or R148 as the General Acid in the Bacterial Transglycosylation Reaction.
To clearly define the mechanism of transglycoyslation reaction, we reexamined the residues of general acid from the previously proposed TG-moenomycin structures. The E156 has been thought to be the general acid to protonate the pyrophosphate or through a divalent metal cation, coordinate with the pyrophosphate moiety at the glycosyl donor site in the transglycosylation reaction (12, 19). However, from the available crystal structures (12–15), E156 of SaMGT and the corresponding residues in other TGs are too far away to interact with the phosphate moiety of moenomycin, which is thought to mimic the pyrophosphate of lipid II or lipid IV (distance is 6.6 Å in SaMGT, 7.2 Å in E. coli PBP1b, 8.1 Å in S. aureus PBP2, and 6.6 Å in A. aeolicus PGT), and enzymatic assay indicated that E156Q did not abolish the TG activity (Fig. 3D). From the TG-moenomycin structures, the K140 in SaMGT and the corresponding residues in other TGs are closer to the phosphate moiety of moenomycin (distance is 3 Å in SaMGT, 3.4 Å in E. coli PBP1b, 3.3 Å in S. aureus PBP2, and 3.3 Å in A. aeolicus PGT), and the R148 has been thought to stabilize by E156 to interact with the pyrophosphate (15). Therefore, we measured the TG activity of K140A, R148A, E156A, and E156Q in SaMGT (Fig. 3D). The K140A resulted in lower activity than E156A, E156Q, and R148A; however, the K140A still has 20% TG activity. We thus proposed that K140 and R148 both are used to stabilize the pyrophosphate-leaving group in the transglycosylation reaction, and the double mutant, K140A and R148A, was created to support their role in the transglycosylation reaction. Indeed, the double mutant, K140A and R148A, showed undetectable TG activity (Fig. 3D and SI Appendix, Fig. S11B). Structurally, the hydrophobic residues, V138 and V139, and the aromatic residues, Y142, F143, and Y144, surrounding the K140 would make it easier to protonate the pyrophosphate leaving group.
Mechanism of Lipid II Polymerization.
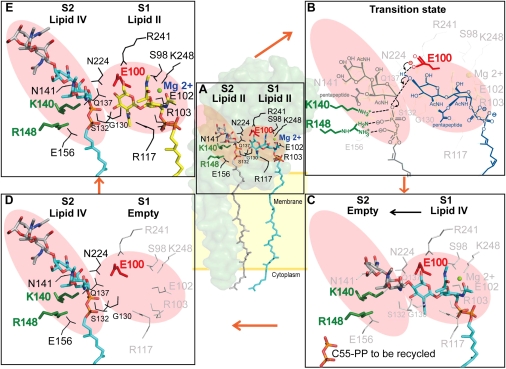
With the comprehensive crystal structures of SaMGT-analog and SaMGT-moenomycin available, we propose a schematic model for the transglycosylation (Fig. 4). The lipid II substrate or the growing glycan chain at the glycosyl donor site (12, 18) is represented by the moenomycin, where GlcNAc and MurNAc correspond to the rings E and F of moenomycin, respectively. The pyrophosphate and undecaprenyl moiety of lipid II or the growing glycan chain corresponds to the phosphate and moenocinol groups. Moreover, lipid II at the glycosyl acceptor site is represented by analog 3, where the GlcNAc corresponds to GalNAc. The substrate-binding site of TG can be divided into the glycosyl acceptor site S1 and glycosyl donor site S2 (Fig. 4A). The catalytic residue E100 is located in S1, and K140 and R148 are located in S2. In the first step of transglycosylation, S98, E102, R103, R117, S132, R241, and K248 of SaMGT interact with lipid II at the glycosyl acceptor site (S1), stabilizing through Mg2+ cations (Fig. 4A). The residues (G130, Q137, K140, N141, R148, and N224) around the conserved motifs II, III, and IV are important for binding lipid II or the growing glycan chain to the glycosyl donor site S2 (12–15, 19–21). The loop from G130 to S132 is used to stabilize the substrate at both S1 and S2. The 4-OH of GlcNAc in lipid II in the S1 site is deprotonated by E100, which is then stabilized by R241 followed by a simultaneous reaction with the C1 of lipid II (or growing glycan chain) in S2 (9) (Fig. 4B), and the K140 and R148 both facilitate the departure of the pyrophosphate leaving group (Fig. 4B). After lipid II (or growing glycan chain) at the glycosyl donor site (S2) is transferred to the lipid II at the glycosyl acceptor site (S1) by forming a β1–4-linked glycan chain (Fig. 4C), the product lipid IV (or growing glycan chain) is shuffled to the glycosyl donor site (S2) (Fig. 4D), and a new lipid II is bound to the glycosyl acceptor site (S1) again (Fig. 4E) for another transglycosylation reaction.
Fig. 4.
Proposed mechanism for lipid II polymerization by TG. (A) Two substrate binding sites: glycosyl acceptor (S1; shaded in red) and donor site (S2; shaded in red). The lipid II polymerization is initiated by accepting two lipid II substrates. (B) The 4-OH of GlcNAc of the lipid II (S1) is deprotonated by E100 (red stick) followed by a simultaneous reaction with the C1 of lipid II (or growing glycan chain) in S2, and the K140 and R148 (green stick) both facilitate the departure of the pyrophosphate-leaving group. (C) Lipid II (or growing glycan chain) at the glycosyl donor site (S2) reacts with the acceptor site of lipid II to form a β1–4-linked glycan chain. (D) The newly formed lipid IV is shuffled to the glycosyl donor site. (E) A new lipid II is docked at the glycosyl acceptor site (S1) again.
Mechanism of Inhibition by Lipid II Analog 3.
From the crystal structure of SaMGT in complex with a lipid II analog, we proposed a design for the lipid II substrate-based antibiotics. By inverting the position 4-OH in the GlcNAc group of lipid II to form GalNAc of analog 3, the binding distance between 4-OH and G130 is reduced (SI Appendix, Fig. S14); therefore, analog 3 can be bound more tightly in the lipid II binding pocket but cannot serve as substrate catalyzed by the E100. The SaMGT-analog structure not only confirms the central importance of the E100 for the deprotonation of 4-OH of GlcNAc in lipid II but also provides insights into lipid II-based antibiotics design.
In summary, although inhibitors are available to interfere with lipid II binding (e.g., Vancomycin, Lantibiotics, Ramoplanin, and Mannopeptimycins), no antibiotics have been developed as TG inhibitors except moenomycin, which is used in animals (4). Recent efforts directed to the synthesis of moenomycin and derivatives (22), elucidation of the moenomycin biosynthetic pathway (23, 24), and crystal structure as well as mechanistic studies on the TG-moenomycin complex (12–15) have opened the door to the design of TG inhibitors (25). In this study, we further design a lipid II analog for the cocrystallization and structural determination of the SaMGT membrane enzyme to reveal the glycosyl acceptor site and elucidate (supported by mutagenesis study) the mechanism of TG reaction. With a better understanding of how lipid II substrates interact with the acceptor and donor sites in the enzyme and the mechanism of the transglycosylation reaction, new structure-based antibiotics targeting this enzyme may be developed.
Materials and Methods
Cloning, Expression, and Purification.
Full-length SaMGT was easily degraded into a slightly smaller protein. After N-terminal sequencing and molecular mass determination by MALDI-TOF MS, we identified the more stable region of SaMGT, which contained amino acids Q28–R269. The DNA of the stable region (residues Q28–R269) of SaMGT was amplified from S. aureus Mu50 genomic DNA and cloned into the pET15b (EMD Biosciences) at the NdeI and BamHI restriction enzyme sites. BL21-CodonPlus (DE3)-RIPL E. coli host cells transformed with expression vectors were grown at 37 °C until O.D.600 (optimal density at 600 nm) reached 0.6; then, protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Anatrace) for 3 h. Cell pellets were resuspended in 20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 0.2 mM lysozyme, 2 mM PMSF, and 20 mM N-decyl-β-d-maltopyranoside (DM) (Anatrace). Cell lysate was centrifuged, and supernatant was loaded onto a Ni-NTA (nickel-nitrilotriacetic acid) affinity column, which was washed with an imidazole gradient from 50 to 150 mM using 20 mM Tris⋅HCl, pH 8.0, and 200 mM NaCl (Buffer A) in the presence of 2 mM DM. Then, the protein was eluted with 300–500 mM imidazole gradient in the Buffer A in the presence of 2 mM DM. Purified protein was desalted in Buffer A in the presence of 2 mM DM and concentrated by Amicon Ultra Centrifugal Filter Devices (MW100K cutoff; Millipore). The N-terminal (His)6 tag was removed by thrombin cleavage (Sigma–Aldrich) at 25 °C overnight. Finally, the size-elusion column (Superdex 200 10/300 GL; GE Healthcare) was used to remove aggregation of the protein for additional assay or crystallization.
Crystallization and Cryoproection.
Crystals were grown by using the handing-drop vapor diffusion method. Tag-free SaMGT was mixed with lipid II analog in a ratio of one to five and mixed with a 20% (wt/vol) (3:1) DMPC (dimyristoylphosphatidylcholine)/Chaps bicellar solution, making a 15.0 mg/mL complex protein/3% (wt/vol) bicelles mixture (17). The SaMGT and lipid II analog mixture were mixed with the equal volume of reservoir solution [100 mM MgCl2, 100 mM Hepes, pH 8.0, 25% (wt/vol) PEG400]. Then, the disk-like crystal formed after 14 d at 4 °C. After the crystal had formed, the reservoir solution was added with PEG400 (Sigma) at a final concentration of 30% (wt/vol), and crystal drop was diffused for 3–7 d before X-ray diffraction. The crystals were immersed in cryoprotectant solution [36% (wt/vol) PEG400] and followed by flash cooling in liquid nitrogen.
Data Collection and Structure Determination.
SaMGT-analog and SaMGT-apo datasets were collected by beamline BL44XU at Japan Synchrotron Radiation Research Institute, SPring-8 (Hyogo, Japan). SaMGT-substrate and SaMGT-moenomycin datasets were collected at beamline BL13B1 and BL13C1, respectively, at National Synchrotron Radiation Research Center (Hsinchu, Taiwan). The datasets were indexed, integrated, and scaled by HKL2000 (26). Molecular replacement was used to search the solution by program PHASER (27). TG domain of 3HZS (15) and TM helix of 3FWM (13) were used as the model templates. The model was manually built using COOT (28). Refinement was performed by Phenix (29) and Refmac (30), with the individual atomic displacement parameters, TLS (Translation, Libration, Screw), and target weight options turned on. The structural figures were generated with the programs PyMOL (www.pymol.org) and ChemDraw (www.cambridgesoft.com).
TG Activity Assay for SaMGT.
For SaMGT activity assay, purified protein was concentrated in Buffer A in the presence of 2 mM DM. NBD-lipid II was used as a substrate to measure TG enzymatic activity with a similar approach as described in our previous studies (13, 31). Assays were performed by incubating NBD-lipid II (1–100 μM), 60 nM S. aureus MGT, 50 mM Tris⋅HCl, pH 8.0, 10 mM MnCl2, 0.085% Decyl PEG (Anatrace), and 15% (vol/vol) MeOH for 0–30 min at 37 °C. The reactions were stopped by adding 200 μM moenomycin, and the polymerized products were digested by adding 13 μM muramidase (Sigma-Aldrich) into the reaction buffer. The NBD-labeled substrates and products were detected and analyzed by anion-exchange column SAX1 (Supelco) on HPLC (Hitachi), and these reactions were monitored at λexcitation = 466 nm and λemission = 535 nm. The elution procedure was a linear gradient of ammonium acetate (20 mM to 1 M) in MeOH. The kinetic parameter was estimated by following the Michaelis–Menten equation.
Assay for Comparing the Activity of SaMGT With or Without TM and Mutant SaMGT Proteins.
For the effect of TM helix of TG in the TG reaction assay, the SaMGT and SaMGTΔTM (residues M1–L64 were removed) were extracted by Buffer A in the presence of 13 mM FOSCHOLINE-14 (Anatrace) and then purified by the same procedures as previously described. The purified SaMGT and SaMGTΔTM were concentrated in Buffer A in the presence of 2 mM DM. The mutant SaMGT proteins of lipid II analog-contacting residues were extracted and purified by Buffer A in the presence of DM (Anatrace) as previously described. The activity assay was carried out by using the following condition: 12 μM NBD-lipid II, 60 nM protein, 50 mM Tris⋅HCl, pH 8.0, 10 mM MnCl2, 0.085% decyl PEG, and 15% (vol/vol) MeOH at 37 °C for time = 0–30 min. The initial velocity was used to compare the activity of SaMGT with SaMGTΔTM.
Isothermal Titration Calorimetry.
The SaMGT and SaMGTΔTM were extracted by Buffer A in the presence of 13 mM FOSCHOLINE-14 (Anatrace) and then purified by the same procedures as previously described. The purified SaMGT and SaMGTΔTM were concentrated in Buffer A in the presence of 2 mM DM. The E. coli PBP1b and PBP1bΔTM (residues M1–L87 were removed) (13) were extracted by 20 mM Tris⋅HCl (pH 8.0) and 300 mM NaCl in the presence of 13 mM FOSCHOLINE-14 (Anatrace) and concentrated in 20 mM Tris⋅HCl, pH 8.0, 300 mM NaCl, and 1 mM N-dodecyl-β-d-maltopyranoside (DDM) (Anatrace). The moenomycin was diluted in the same buffer with protein. The moenomycin (700 μM for SaMGT and 500 μM for E. coli PBP1b) was titrated to the protein (50 μM for SaMGT and 30 μM for E. coli PBP1b) at 25 °C for 2 μL for 19 cycles by isothermal titration calorimeter iTC200 (MicroCal). In addition, the moenomycin was diluted in the buffer as the reference to substrate of the experiment data. The results were analyzed by the MicroCal Origin version 5.0 software accompanied with the instrument.
Site-Directed Mutagenesis.
Mutations of lipid II analog-contacting residues were generated in E. coli PBP1b and S. aureus MGT by site-directed mutagenesis. Other than E100Q and E156Q of S. aureus MGT and E233Q and A231L of E. coli PBP1b, others lipid II analog-contacting residues were changed to Alanine.
Complementary Activity Assay.
The plasmids of encoded PBP1b mutant protein were created by site-directed mutagenesis. Each mutant plasmid was transformed into JE5702 (ponB353, ponAts, and Str), which is a PBP1b-defective/PBP1a temperature-sensitive E. coli host strain [National BioResource Project (NIG, Japan): E. coli]. The 30 °C overnight-cultured transformants were diluted 1:100 with the fresh LB liquid media and then cultured with 0.5 mM isopropyl-β-d-thiogalac-topyranoside at 30 °C and 42 °C. Bacterial growth was examined by O.D.600 for 4 h. The expression level of various mutated PBP1b was detected by the anti-E. coli PBP1b monoclonal antibody.
Supplementary Material
Acknowledgments
We thank C. Lim, M.-F. Hsu, and C.-H. Huang for insightful discussions. X-ray diffraction data were collected at BL13B1 and BL13C1 of the National Synchrotron Radiation Research Center, Taiwan, and BL44XU of the SPring-8, Japan. This work was supported by Academia Sinica and National Science Council, Taiwan Grant 99-2113-M-001-020-MY3 (to C.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in Protein Data Bank, www.pdb.org [PDB ID codes 3VMQ (SaMGT-apo), 3VMR (SaMGT-moenomycin), 3VMS (SaMGT-substrate), and 3VMT (SaMGT-analog)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203900109/-/DCSupplemental.
References
- 1.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: From targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 4.Halliday J, McKeveney D, Muldoon C, Rajaratnam P, Meutermans W. Targeting the forgotten transglycosylases. Biochem Pharmacol. 2006;71:957–967. doi: 10.1016/j.bcp.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 5.Wright GD. Biochemistry. A new target for antibiotic development. Science. 2007;315:1373–1374. doi: 10.1126/science.1140374. [DOI] [PubMed] [Google Scholar]
- 6.Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Ritter TK, Wong CH. Carbohydrate-based antibiotics: A new approach to tackling the problem of resistance. Angew Chem Int Ed Engl. 2001;40:3508–3533. doi: 10.1002/1521-3773(20011001)40:19<3508::AID-ANIE3508>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 8.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welzel P. Syntheses around the transglycosylation step in peptidoglycan biosynthesis. Chem Rev. 2005;105:4610–4660. doi: 10.1021/cr040634e. [DOI] [PubMed] [Google Scholar]
- 10.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J Am Chem Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallhausser KH, Nesemann G, Prave P, Steigler A. Moenomycin, a new antibiotic. I. Fermentation and isolation. Antimicrob Agents Chemother (Bethesda) 1965;5:734–736. [PubMed] [Google Scholar]
- 12.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 13.Sung MT, et al. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci USA. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Y, et al. Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem Biol. 2008;3:429–436. doi: 10.1021/cb800078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heaslet H, Shaw B, Mistry A, Miller AA. Characterization of the active site of S. aureus monofunctional glycosyltransferase (Mtg) by site-directed mutation and structural analysis of the protein complexed with moenomycin. J Struct Biol. 2009;167:129–135. doi: 10.1016/j.jsb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Goldman RC, Gange D. Inhibition of transglycosylation involved in bacterial peptidoglycan synthesis. Curr Med Chem. 2000;7:801–820. doi: 10.2174/0929867003374651. [DOI] [PubMed] [Google Scholar]
- 17.Faham S, Bowie JU. Bicelle crystallization: A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 18.Lovering AL, De Castro L, Strynadka NC. Identification of dynamic structural motifs involved in peptidoglycan glycosyltransfer. J Mol Biol. 2008;383:167–177. doi: 10.1016/j.jmb.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Terrak M, et al. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 20.Terrak M, et al. Importance of the conserved residues in the peptidoglycan glycosyltransferase module of the class A penicillin-binding protein 1b of Escherichia coli. J Biol Chem. 2008;283:28464–28470. doi: 10.1074/jbc.M803223200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuse S, et al. Functional and structural analysis of a key region of the cell wall inhibitor moenomycin. ACS Chem Biol. 2010;5:701–711. doi: 10.1021/cb100048q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor JG, Li X, Oberthür M, Zhu W, Kahne DE. The total synthesis of moenomycin A. J Am Chem Soc. 2006;128:15084–15085. doi: 10.1021/ja065907x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostash B, Saghatelian A, Walker S. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostash B, et al. Complete characterization of the seventeen step moenomycin biosynthetic pathway. Biochemistry. 2009;48:8830–8841. doi: 10.1021/bi901018q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih HW, et al. Combinatorial approach toward synthesis of small molecule libraries as bacterial transglycosylase inhibitors. Org Biomol Chem. 2010;8:2586–2593. doi: 10.1039/c000622j. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz B, Markwalder JA, Seitz SP, Wang Y, Stein RL. A kinetic characterization of the glycosyltransferase activity of Escherichia coli PBP1b and development of a continuous fluorescence assay. Biochemistry. 2002;41:12552–12561. doi: 10.1021/bi026205x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.