Abstract
During transcription, RNA polymerase II (RNAPII) must select the correct nucleotide, catalyze its addition to the growing RNA transcript, and move stepwise along the DNA until a gene is fully transcribed. In all kingdoms of life, transcription must be finely tuned to ensure an appropriate balance between fidelity and speed. Here, we used an optical-trapping assay with high spatiotemporal resolution to probe directly the motion of individual RNAPII molecules as they pass through each of the enzymatic steps of transcript elongation. We report direct evidence that the RNAPII trigger loop, an evolutionarily conserved protein subdomain, serves as a master regulator of transcription, affecting each of the three main phases of elongation, namely: substrate selection, translocation, and catalysis. Global fits to the force-velocity relationships of RNAPII and its trigger loop mutants support a Brownian ratchet model for elongation, where the incoming NTP is able to bind in either the pre- or posttranslocated state, and movement between these two states is governed by the trigger loop. Comparison of the kinetics of pausing by WT and mutant RNAPII under conditions that promote base misincorporation indicate that the trigger loop governs fidelity in substrate selection and mismatch recognition, and thereby controls aspects of both transcriptional accuracy and rate.
Keywords: optical trap, optical tweezers, Pol II
The transcription of genetic information stably encoded in DNA into a transient RNA message occurs with remarkable fidelity. RNA polymerase (RNAP) incorporates nucleotides into nascent RNA chains at rates of 10–70 s-1 (1, 2), but only inserts the wrong nucleotide approximately once per 100,000 bases, on average (3). Although the energetics of base-pairing is fundamental to transcription fidelity, the discrimination for correctly matched nucleotide substrates is kinetically controlled, and accomplished by active site conformational changes centered on the trigger loop (TL) (4–7). The TL is an evolutionarily conserved mobile element that stabilizes substrate NTPs in the active site of a variety of multisubunit RNA polymerases, including eukaryotic RNAP I, II, and III, as well as bacterial and archaeal RNAP (8). In the case of RNAPII, alteration of the TL has important consequences for activity and fidelity. TL residue His1085, for example, interacts with the bound NTP substrate, and is fully conserved among multisubunit RNA polymerases. The importance of this residue is underscored by the lethality of the H1085A substitution in yeast (7) and catalytic defects resulting from substitutions at this position for both the yeast and bacterial enzymes (7, 9–11). Additionally, a substitution for residue Glu1103 of the TL has been found not only to promote nucleotide misincorporation (6, 7), but also to increase the elongation rate (6, 7, 12), properties that are jointly consistent with the notion of kinetic proofreading (13). To explore further the interplay between elongation rate and transcriptional fidelity, we determined the effect of E1103G and H1085A substitutions on the nucleotide-addition cycle (NAC) using single-molecule assays.
A key step in the NAC for all polymerases that remains poorly characterized is translocation along the DNA. Traditional ensemble techniques do not observe enzyme motions directly, but instead report the average positions of populations of molecules. This limitation can be problematic, owing to the intrinsic variability in synthesis rates, and for RNAPII, the inherently stochastic nature of transcriptional pausing (1), which can obscure the underlying kinetics of translocation. Here, we describe a single-molecule assay capable of directly observing the motions of individual, elongating RNAPII molecules at subnanometer resolution in real time. Using the assay, we were able to identify and exclude from analysis transcriptional pauses as short as 1 s, and thereby to determine the “pause-free” velocity of an individual molecule, which furnishes an accurate measure of its raw elongation rate. Our results for the pause-free velocity as a function of applied force favor a Brownian ratchet model for transcript elongation, in which the incoming NTP substrate is able to bind in either the pre- or posttranslocated state of RNAPII. By examining the effect of two TL mutant enzymes—containing either E1103G or H1085A/E1103G substitutions—on pause-free velocity, we show that the TL affects NTP binding, translocation and catalysis in the NAC. Furthermore, the effect of the E1103G substitution on transcription provides evidence that the TL promotes fidelity by regulating nucleotide selection and pausing during elongation, which aids in the recognition of misincorporation events.
Results and Discussion
Force-Velocity Relationships Distinguish Between Competing Models for Ordering of Events in the Nucleotide-Addition Cycle.
Because every NAC involves translocation along the DNA template, the elongation rates of RNA polymerase molecules are sensitive to modulation by force, which can either assist or hinder translocation, depending upon the experimental geometry of the assay (Fig. 1A). The magnitude of any such modulation, assessed by determining the force-velocity (F-v) relationship, is affected by the step size of RNAPII and by its equilibrium between pre- and posttranslocated states. The F-v relationship is also sensitive to the temporal ordering of the translocation event with respect to NTP binding during the NAC when measured as a function of NTP concentration (14), and this placement has been the subject of some controversy. Kinetic studies have suggested that NTP binding may precede, and possibly promote, translocation itself (15–17). Because most substrate-bound RNAPII crystal structures are in the posttranslocated state, they do not speak to this issue, but the identification of a nonspecific NTP-binding site in some of these structures suggests that substrate binding might precede translocation (18).
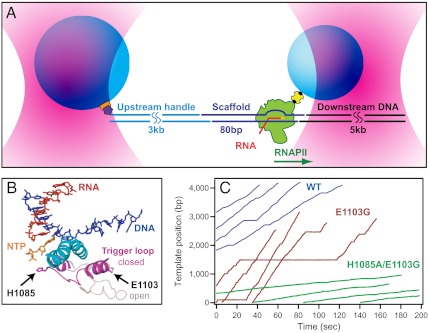
Fig. 1.
Single-molecule assay and transcription records. (A) Experimental geometry for the DNA-pulling optical-trapping assay, based on a “dumbbell” arrangement (not to scale). RNAPII (green) is initiated on a nucleic-acid scaffold (dark blue) that includes template and nontemplate DNA strands plus a short RNA transcript (red). An upstream DNA handle (cyan) and downstream template (black) are ligated to either end of the scaffold. Two polystyrene beads (light blue) are held in separate optical traps (pink). RNAPII is attached to the smaller bead via a biotin:avidin linkage (yellow and black); the DNA handle is attached to the larger bead via a digoxigenin:antibody linkage (purple and orange). Direction of transcription is shown (green arrow); in this orientation, tension assists translocation. (B) Schematic of the RNAPII active site from overlays of two crystal structures, Protein Data Bank (PDB) IDs 2E2H (4) and 1Y1V (41). During transcription, the TL is proposed to fluctuate between a catalytically active “closed” (magenta, 2E2H) and inactive “open” (light pink, 1Y1V) conformation (in this case, restrained by the presence of TFIIS; not displayed). During this transition, the TL comes into proximity with the bridge helix (cyan) and the incoming NTP (orange). The positions of the two TL mutations studied are indicated. (C) Representative records of elongation acquired under 10 pN assisting load for WT RNAPII (blue) and two TL mutant enzymes, E1103G (red) and H1085A/E1103G (green).
We determined the F-v relationships for wild-type (WT) RNAPII and two specific TL mutant enzymes, E1103G and H1085A/E1103G (Fig. 1B) at both saturating and subsaturating NTP levels (1 mM or 0.1 mM of ATP, CTP, GTP, and UTP, respectively). In these experiments, we were able to modulate force on the DNA from 20 pN (assisting load) to -3 pN (hindering load) before either linkage rupture or irreversible backtracking of RNAPII occurred, respectively. We note that the range of hindering forces is limited because yeast RNAPII has a higher propensity to backtrack at relatively small hindering loads compared to bacterial RNAP, consistent with a previous report (19).
Glu1103 lies within a region of the TL that is distal from the enzyme active site (Fig. 1B), and biochemical data suggest that E1103G likely affects catalysis via TL dynamics, by biasing the TL toward a more closed, catalytically active conformation (6, 7). Previous ensemble measurements have shown that E1103G leads to an increase in the overall elongation rate (6, 7, 12), which we confirmed in our single-molecule experiments at saturating NTP in the presence of 10 pN assisting force (Fig. 1C). We also acquired F-v data in the presence of 100 mM ammonium ion, which is known to increase elongation rates in vitro to levels closely resembling transcription rates estimated in vivo (see below) (20–22).
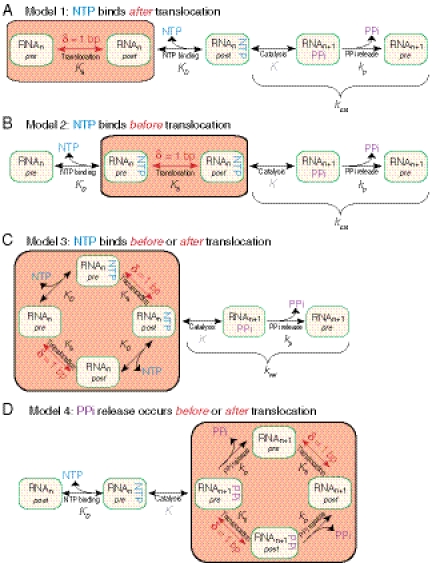
We considered four different pathways for the NAC to distinguish among the competing models for NTP binding and translocation (Fig. 2), each of which has been proposed previously for multisubunit RNA polymerases, based on biochemical or single-molecule kinetic data (14, 16, 23–28). In Model 1 (Fig. 2A), RNAPII oscillates back and forth between the pre- and posttranslocated states with thermal motion. That motion is ultimately rectified in the posttranslocated state by NTP binding (23, 24, 27), after which the NTP is covalently linked into the nascent transcript by a condensation reaction catalyzed by RNAPII, and the NAC is completed with pyrophosphate (PPi) release. In Model 2 (Fig. 2B), substrate binding occurs prior to translocation, suggesting that the incoming NTP drives translocation (16), and the thermal motion between the pre- and posttranslocated states is rectified by the NTP condensation reaction.
Fig. 2.
Reaction pathway models of the nucleotide addition cycle. Four variants of the Brownian ratchet model for elongation are presented. Each model differs in the placement of the force-dependent translocation step in relation to the NTP-binding, catalysis, and phosphate-release steps. Equilibrium constants (for reversible steps) and forward rate constants (for essentially irreversible steps) are shown. Groups of states corresponding to the translocation event(s) are shaded (orange).
Model 3 (Fig. 2C), where the incoming NTP can bind in either the pre- or posttranslocated state, is supported by single-molecule studies of Escherichia coli RNAP (14) and is consistent with studies of RNAPII that posit the existence of a secondary NTP binding site, distinct from the primary nucleotide-addition site (18). (We note that although Model 3 has additional states in the reaction pathway, fits involve the same number of free parameters, m = 3, as Models 1 and 2, due to the assumption that NTP binding to the secondary site is energetically equivalent to active site, which leads to identical equilibrium constants for NTP binding pre- and posttranslocation.) Model 4, in which PPi release can take place from either the pre- or posttranslocated state (Fig. 2D), is inspired by kinetic studies demonstrating that the rate of PPi release is stimulated by the incoming cognate NTP (26). (Similar to the assumption made for Model 3, PPi release is assumed to be equally likely from either the pre- or posttranslocated state, resulting in a F-v relationship with the same number of free parameters as Models 1, 2, and 3.)
Global fits were performed against the models for all the force-velocity data acquired with wild-type and mutant enzymes: this procedure implicitly assumes that enzymes function via the same reaction pathway, regardless of mutation, albeit with different rates, and is justified by the goodness of the fit. The best-fit model (Model 3, Fig. 2C) placed the translocation step at the start of the NAC, prior to catalysis, with the incoming NTP able to bind RNAPII in either its pre- or posttranslocated state (reduced χ2 = 0.52, ν = 33, N = 45, P-value = 0.99; the three other models returned P-values < 0.005; Fig. S1). In particular, the F-v data did not fit an alternative model where NTP binding is required for forward translocation (16) (Fig. 2B), nor did they support a model where NTP binds exclusively to the posttranslocated state (Fig. 2A). The existence of a secondary site for NTP binding, posited previously in the form of a nontemplated “entry,” or “E-site” (18), a templated “preinsertion” site (29), or along exposed bases in the downstream DNA template (15), is consistent with our best-fit model.
Assuming a Boltzmann-type dependence for the load-sensitive translocation step, the F-v relationship for Model 3 can be expressed as (for additional description, see ref. 14, SI Text, and Fig. S2)
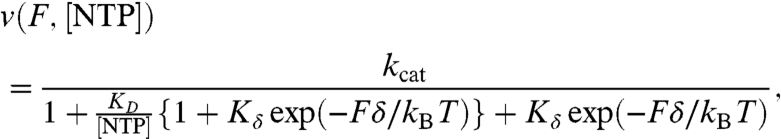 |
[1] |
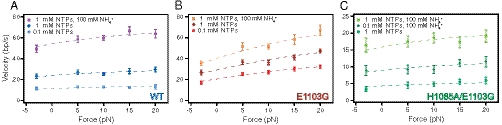
where kcat is the rate of nucleotide condensation followed by PPi release (these steps have been combined into a single rate; see SI Text), KD is the NTP dissociation constant, and Kδ describes the equilibrium between pre- and posttranslocated states (Kδ = [pre]/[post]). The effective step size, δ, is a parameter corresponding to the distance between translocation states; this was fixed to 1 bp (0.34 nm) based on a previous measurement (14). We observed a relatively weak effect of force on the pause-free velocity for the WT RNAPII (Fig. 3A), a dependence that had not been observed previously with RNAPII, presumably due to the limited range of forces that was explored and the larger experimental uncertainties (19). Based on the fit, WT RNAPII is biased toward its posttranslocated state (Table 1), indicating that the translocation step is not rate-limiting for the advance of the WT enzyme. The apparent KD for WT RNAPII is somewhat higher than that suggested by some previous reports (6); however, those determinations were carried out for a single NTP species binding to a given template position, whereas the present value corresponds to an average of all four NTPs measured over thousands of template positions.
Fig. 3.
Force-velocity curves. Average pause-free elongation rates (errors as SEM) measured as functions of force for (A) WT, (B) E1103G, and (C) H1085A/E1103G RNAPII at saturating (1 mM) and subsaturating (0.1 mM) NTPs, as well as in the presence of 100 mM ammonium chloride, which was added to our standard elongation buffer. Global fits to the two-binding site Brownian ratchet model (Model 3, Fig. 2C) are shown (dashed lines).
Table 1.
Trigger loop affects catalysis, NTP binding, and translocation equilibrium. Global fit parameters (errors as SD) obtained from modeling of F-v curves in Fig. 3 to the two-binding site Brownian ratchet model of Fig. 2C.
| WT | E1103G | H1085A/E1103G | |
| kcat | 34 ± 2 s-1 | 56 ± 2 s-1 | 6 ± 1 s-1 |
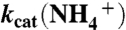 |
77 ± 3 s-1 | 77 ± 3 s-1 | 22 ± 2 s-1 |
| KD | 140 ± 16 μM | 49 ± 6 μM | 94 ± 17 μM |
| Kδ | 0.2 ± 0.1 | 0.8 ± 0.1 | 0.3 ± 0.1 |
| δ | 0.34 nm (fixed) | 0.34 nm (fixed) | 0.34 nm (fixed) |
Role of Ammonium in Transcript Elongation.
A stimulatory effect of ammonium on the overall rate of transcription has been previously reported (20–22), but the mechanism for its action is still unknown. To assess the role of ammonium on individual steps in the NAC, we added 100 mM ammonium chloride to the buffer (in addition to the 130 mM KCl already present) for transcription reactions of WT RNAPII and its TL mutants. This additional set of conditions allowed us to discriminate better among the different models for the NAC, as well as to observe transcription by the H1085A/E1103G mutant enzyme at low NTPs, a condition which would otherwise produce rates too slow to observe practically in single-molecule assays due to residual amounts of baseline drift. Ammonium increased the pause-free velocity by factors of 1.5 to 4 for WT and mutant RNAPII (Fig. 3 A–C). Based on fits to Model 3 of the NAC, ammonium ion increases the pause-free elongation rate by increasing the value of kcat for the both WT and mutant RNAPII, leaving KD, and Kδ substantially unchanged (Table 1). Furthermore, the maximum pause-free velocities in the presence of ammonium ion for the WT and E1103G were statistically indistinguishable, suggesting the existence of a common, rate-limiting step under conditions where kcat is rapid and an assisting load is applied. Taken together, the results indicate that ammonium ion primarily alters the rate of the catalytic step of the nucleotide-addition cycle, and acts independently of trigger loop changes to E1103 or H1085.
Substitutions in the TL Affect NTP Binding, Catalysis, and Translocation.
The force-velocity dependence of the TL mutant enzymes (Fig. 3 B and C) confirms that the TL is involved in catalysis (5, 9, 10), because the E1103G mutation produced a 1.6-fold increase in kcat over the WT (Table 1). We also observed more than a 2.5-fold decrease in KD compared to the WT, indicating that the incoming NTP is more tightly bound to the active site. This reduction may be attributable to a preferential closure of the mutant TL around the bound substrate. The increases observed in both NTP-binding affinity and catalysis were accompanied by an inhibitory effect on translocation, because the E1103G mutant enzyme was found to be significantly more biased toward its pretranslocated state than the WT. A pretranslocational bias is somewhat counterintuitive, in view of the increased elongation rate for the mutant enzyme. However, it can be understood because the TL substitution promotes a net forward translocation of RNAPII. In the case of E1103G, the combination of tighter NTP binding and a faster catalytic rate (Table 1) lead to an increase in the elongation rate despite an inhibition of the translocation step. It was previously observed that E1103G displayed a bias for the pretranslocated state in the absence of the correct NTP when tested at a single template position, and that this bias shifted toward the posttranslocated state in the presence of the correct NTP (6). By contrast, our best-fit model of the NAC, which supplies uncoupled measures of translocation (Kδ) and NTP binding (KD) over many different template positions, instead suggests a pretranslocation bias for E1103G, even at high NTP levels.
If the observed properties of the E1103G substitution were attributable to a stabilization of the closed state of the TL (dependent upon a bound, template-matched NTP), which tends to be more open in the WT, we reasoned that a compensatory destabilization of this closed state might reverse some, or all, of the mutant enzyme properties. Guided by structural data for the TL and RNAPII active site (4, 18), we altered a second residue of the TL, His1085, which, through interaction with NTP substrates, promotes catalysis and likely favors the closed conformation of the TL (4, 5, 7, 9–11). H1085A lethality is suppressed by combination with E1103G, but the transcriptional and phenotypic properties of the double substitution suggest mutual suppression (11). As anticipated, based on compromise of the critical substrate-interacting sidechain, the H1085A/E1103G double-mutant enzyme exhibited a significantly slower elongation rate than either the WT or E1103G single-mutant enzymes, and therefore returned a reduced value for kcat (Table 1). However, in several other key respects, the WT characteristics were restored. Notably, the load dependencies of the F-v curves for the H1085A/E1103G and WT enzymes were similarly flat (Fig. 3C). Moreover, the nucleotide binding affinity (KD) of the double-mutant enzyme was similar to that of WT, and its translocation equilibrium (Kδ) was statistically indistinguishable (Table 1). Taken together, these results suggest that the TL serves to affect not only the rate of catalysis, as previously found, but also the NTP-binding affinity and the translocation equilibrium. An inhibitory effect of TL closure on translocation was recently proposed, based on structural modeling and computational studies (30), and is consistent with the proposed “two-ratchet model” for RNAP translocation (27). In that proposal, the closed state of the TL, which is dependent upon a match between the incoming NTP and the template DNA, precludes translocation. However, as the correct NTP is condensed into the nascent RNA, forward translocation is facilitated by the opening of the TL, induced by thermal motion, likely along with a corresponding alteration in the conformation of the bridge helix (31). The single-molecule data presented here provide experimental evidence consistent with such TL-gated translocation. This gating is analogous to models of translocation in DNA polymerases, where the reversals of domain movements coupled to substrate selection and catalysis have been implicated in the subsequent translocation step, but by and large have been experimentally inaccessible (32).
Pausing Kinetics Imply a Role for the Trigger Loop in Transcriptional Fidelity.
Because pausing has been implicated in RNAP proofreading (33, 34), we examined the characteristics of transcriptional fidelity and pausing behavior in WT RNAPII and its TL mutants. Misincorporation of certain bases (such as the GTP analog, ITP) by RNAP results in backtracking along the DNA template, leading to a relatively long-lived, paused state. The backtracking motion displaces the 3′-end of the nascent RNA from the enzyme active site, permitting transcription factors (such as TFIIS in eukaryotes or Gre factors in bacteria) to activate cleavage of the most recently incorporated bases, thereby removing the error and restoring a new 3′ end of RNA in the active site, allowing transcription to resume (33–35).
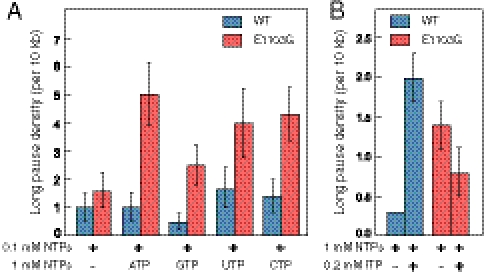
The RNAPII pause lifetime distribution could be subdivided into two categories, the majority of which (∼95%) were “short,” with an average time of ∼1.1 s, and the remainder of which (∼5%) were “long” (> 15 s) (Fig. S3). WT RNAPII entered these long pauses approximately once every 10 kb at subsaturating NTP concentrations, a rate that corresponds closely to the frequency for the incorporation of an incorrect nucleotide substrate, as measured by ensemble assays in vitro (36). It has been shown previously that E1103G promotes NTP misincorporation (6, 7). Consistent with that finding, we observed a slight, but not statistically significant, increase in the long pause density (LPD, number of long pauses/kb) for the E1103G mutant enzyme over the WT enzyme at 0.1 mM NTPs (Fig. 4A).
Fig. 4.
Substrate selection and mismatch recognition. (A) Long pause density (long pauses per 10 kb transcribed; errors as SEM) for the WT (blue) and E1103G mutant enzymes (red) at subsaturating (0.1 mM) NTPS under the conditions indicated. The long pause density increases for the mutant enzyme when a single NTP is present in excess, indicative of a deficiency in substrate selectivity. (B) The inclusion of 200 μM ITP in a saturating NTP buffer (1 mM NTPs) increases the long pause density for the WT (blue), but decreases the density for the E1103G mutant enzyme (red), indicative of a failure in mismatch recognition for ITP incorporation events.
To ascertain whether the increase in long pausing observed for the RNAPII mutant was caused by an increased rate of mismatches arising from a deficiency in substrate selection, we added 1 mM additional nucleotide for each NTP, in turn to subsaturating NTPs (0.1 mM NTPs), which serves to promote mismatching of the excess nucleotide by mass action. The LPD for E1103G increased nearly fivefold upon the addition of excess ATP and about threefold in the presence of excess CTP or UTP (Fig. 4A). Only a modest increase in LPD was observed in the presence of excess GTP. By contrast, the LPD of wild-type RNAPII was comparatively insensitive to excess NTPs (Fig. 4A) and reached a minimum in the presence of additional GTP. These results imply that the E1103G mutant enzyme differentially undertakes long pauses dependent on the specific excess NTP. In particular, the inferred misincorporation events involving GTP are less frequent, or less able to trigger a long pause, or both. This finding is consistent with a mismatch-specific proofreading mechanism proposed previously (33, 36, 37), and may be attributable to specific interactions between the TL and the misincorporated 3′ end of the RNA, which could trigger long pauses.
Inosine triphosphate (ITP), a GTP analog that forms a weak Watson-Crick pair with cytosine and has a similar binding constant and rate of incorporation into RNA, also inhibits next-nucleotide-addition rates in a manner similar to that of certain mismatched base pairs. This property has been exploited previously to investigate postincorporation nucleotide discrimination and removal by RNAPII (38). At saturating concentration of NTPs (1 mM) in the absence of ITP, E1103G showed a clear increase in LPD over WT (Fig. 4B). For the WT RNAPII, the addition of 200 μM ITP to transcription reactions containing 1 mM NTPs significantly increased the LPD (Fig. 4B), as previously described for E. coli RNAP (34), which is indicative of misincorporation-induced pause enhancement. Perhaps surprisingly, the LPD for E1103G decreased in the presence of ITP, despite the fact that the mutant enzyme is able to incorporate ITP into transcripts to the same extent as the WT (Fig. S4). Taken together, these results suggest that the TL promotes transcriptional fidelity in two distinct ways. First, the TL helps to ensure correct NTP selection from among a pool of mixed substrates. Mutation of the TL (E1103G) generally leads to more mismatches being added to the transcript. Second, the TL functions in the efficient recognition of miscorporation events by RNAPII once a selection error has been made, as evidenced by backtracking (38). The E1103G mutant enzyme also appears to be deficient in this regard, because the misincorporation of ITP fails to be recognized, and therefore fails to produce a corresponding increase in the proportion of long pauses.
Conclusion
The observation of elongation by individual RNAPII molecules has provided additional insights into eukaryotic transcription mechanisms, and it offers the ability to separate effects on catalysis from those on translocation, which is critical for the dissection of the RNAPII mechanism. Based on high-resolution measurements of the load dependence of the elongation rate, we find support for a biochemical pathway where incoming NTPs can associate with RNAPII in either its pre- or posttranslocated state. Both single-molecule and bulk biochemical data, comparing wild-type behavior with substitutions in the trigger loop, suggest that this key subdomain of RNAPII functions not only to regulate the basic elongation rate, via changes to the translocation bias and catalytic rate, but also plays a critical role in both NTP substrate selection and mismatch recognition. In particular, substrate selection defects were characterized under conditions of competing levels of matched and mismatched NTPs. The experiments also revealed significant differences in the recognition of ITP misincorporation by the TL mutant enzyme, implicating the TL in the promotion of pausing associated with proofreading repair. We conclude that the sequence of the trigger loop evolved to strike a critical balance between the opposing constraints of catalytic prowess and transcriptional fidelity.
Materials and Methods
A detailed description of materials and methods is given in SI Text.
Transcription Initiation.
Biotinylated, 12-subunit Saccharomyces cerevisiae RNAPII (prepared as described in SI Text) was assembled stepwise onto a DNA:RNA scaffold using a modification of a previous method (39), allowing us to circumvent the need for initiation factors normally required to assemble a transcriptional elongation complex. The 2.7 kb upstream and 4.8 kb downstream DNA “handles” were ligated to the ends of the template DNA by incubating the RNAPII scaffolded complexes (5 nM) with these DNA fragments (20 nM each) in elongation buffer (25 mM Hepes-KOH, pH 8.0, 130 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.15 mM EDTA, 5% glycerol and 25 μg acetylated bovine serum albumin/mL) in the presence of ligase (2 K units) and ATP (1 mM) at 12 °C for 1 h.
DNA-Pulling Optical-Trapping Assay.
Single elongation complexes were tethered between 2 polystyrene beads (0.6 μm and 0.73 μm diameter), each held in a separate optical trap, forming a “dumbbell.” By labeling either the upstream or downstream DNA with digoxigenin, we could apply a controlled load that either assisted or hindered forward translocation of RNAPII (the assisting-load version of the assay is depicted in Fig. 1A). Data were acquired using a dual-beam optical-trapping microscope described previously (14), and an oxygen-scavenger system was used in buffers to reduce photodamage (40).
Data Collection and Analysis.
Records of position data were acquired at 2 kHz using custom software (written in Labview), filtered at 1 kHz by an 8-pole lowpass Bessel filter, and analyzed using custom software (written in Igor Pro). The distance between optically trapped beads was converted into the DNA contour length (14) and pauses were identified and scored as described previously (34). Pause-free elongation velocities of single RNAPII molecules were determined by fitting the velocity distribution to a sum of two Gaussians (one corresponding to pausing, and the other to active elongation) as previously described (1).
Supplementary Material
ACKNOWLEDGMENTS.
We thank D. Koslover and V. Schweikhard for help with data collection, D. Larson for critical reading of the manuscript, and J. Gelles and members of the Block lab for helpful discussions. This work was supported by a National Science Foundation Graduate Research Fellowship (to M.H.L.) and grants from the National Institute of General Medical Sciences (to S.M.B., R.L., and R.D.K.).
Note Added in Proof.
Additional information on materials and methods may be found in ref. 42.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200939109/-/DCSupplemental.
References
- 1.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 2.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ninio J. Connections between translation, transcription and replication error-rates. Biochimie. 1991;73:1517–1523. doi: 10.1016/0300-9084(91)90186-5. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 6.Kireeva ML, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jokerst RS, Weeks JR, Zehring WA, Greenleaf AL. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 9.Yuzenkova Y, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan CD, Jin H, Zhang I, Belyanin A. Dissection of pol II trigger loop function and pol II-activity dependent control of start site selection in vivo. PLoS Genetics. 2012 doi: 10.1371/journal.pgen.1002627. 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malagon F, et al. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Nedialkov YA, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Burton ZF. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J Mol Biol. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 18.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 20.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 21.Sluder AE, Price DH, Greenleaf AL. Elongation by Drosophila RNA polymerase II. Transcription of 3′-extended DNA templates. J Biol Chem. 1988;263:9917–9925. [PubMed] [Google Scholar]
- 22.Gu W, Reines D. Identification of a decay in transcription potential that results in elongation factor dependence of RNA polymerase II. J Biol Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 24.Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 25.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 27.Bar-Nahum G, et al. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 28.Erie DA, Kennedy SR. Forks, pincers, and triggers: The tools for nucleotide incorporation and translocation in multi-subunit RNA polymerases. Curr Opin Struct Biol. 2009;19:708–714. doi: 10.1016/j.sbi.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temiakov D, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Feig M, Burton ZF. RNA polymerase II flexibility during translocation from normal mode analysis. Proteins. 2010;78:434–446. doi: 10.1002/prot.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 32.Golosov AA, Warren JJ, Beese LS, Karplus M. The mechanism of the translocation step in DNA replication by DNA polymerase I: A computer simulation analysis. Structure. 2010;18:83–93. doi: 10.1016/j.str.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: Control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 34.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′–5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 36.Sydow JF, et al. Structural basis of transcription: Mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 39.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 40.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Palangat M, et al. Efficient reconstitution of transcription elongation complexes for single-molecule studies of eukaryotic RNA polymerase II. Transcription. 2012 doi: 10.4161/trns.20269. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.