Abstract
Overexpression of the brain and acute leukemia, cytoplasmic (BAALC) gene is implicated in myeloid leukemogenesis and associated with poor outcome in both acute myeloid leukemia (AML) and acute lymphoblastic leukemia patients. Additionally, high BAALC expression occurs in glioblastoma, melanoma, and childhood gastrointestinal stroma tumors, suggesting an oncogenic role for BAALC. However, the mechanisms underlying the deregulated expression are unknown. We hypothesized that a common heritable genetic feature located in cis might account for overexpression of BAALC in an allele-specific manner. By sequencing the genomic region of BAALC we identified nine informative single nucleotide polymorphisms (SNPs) and tested them for a possible association with BAALC expression levels. We show that BAALC overexpression occurs in the presence of the T allele of SNP rs62527607[GT], which creates a binding site for the activating RUNX1 transcription factor in the BAALC promoter region. The mechanism is demonstrated experimentally in vitro using luciferase reporter assays and electrophoretic mobility shift assay (EMSA) analysis. The association of high BAALC expression with the T allele and its correlations with RUNX1 expresser status are shown in vivo in a test set (n = 253) and validation set (n = 105) of samples from cytogenetically normal AML patients from different populations. Thus, we identify a heritable genomic feature predisposing to overexpression of an oncogene, thereby possibly leading to enhanced AML leukemogenesis. Our findings further suggest that genomic variants might become useful tools in the practice of personalized medicine.
Acute myeloid leukemia (AML) is a cytogenetically and molecularly heterogeneous disease characterized by clonal proliferation of myeloid precursors and maturation arrest of myeloid cells in the bone marrow. Despite cytogenetic and molecular-based stratification for risk-adapted therapies, only 40–45% of younger adult AML patients (<60 y) achieve long-term survival (1–3). Older AML patients fare even worse with a 2-y median overall survival (OS) of ∼7–15% (4–6). Predicting treatment response and outcome in patient subgroups has become an essential tool for treatment guidance. Numerous clinical, cytogenetic, and molecular variables are associated with AML outcome (7–30). Some variables are presently being exploited as therapeutic targets (e.g., internal tandem duplications of FLT3) (31).
The BAALC gene located on chromosome 8q22.3 was identified by cDNA-based representational difference analysis in leukemia patients (32). Its overexpression is a strong prognosticator associated with adverse outcome and its impact has been most extensively studied in the subgroup of cytogenetically normal (CN)-AML patients (33–37). Additionally, overexpression of BAALC has been described in acute lymphoblastic leukemia (38, 39), glioblastoma (32), melanoma (40), and childhood gastrointestinal stroma tumors (41). A functional role of BAALC overexpression in myeloid leukemogenesis has been recently reported, suggesting oncogenic potential of the BAALC gene (42). However, the mechanisms leading to up-regulation of the gene in leukemia blasts remain unknown.
On the basis of genome-wide mapping studies, heritable differences in the expression of genes are widespread and can be viewed as quantitative traits that are either regulated in trans (from elsewhere in the genome), or more often in cis (from within the locus itself) (43–45). Therefore, we hypothesized that a common heritable genetic feature located in cis might account for overexpression of BAALC in an allele-specific manner and lead to gene deregulation in AML patients.
Results and Discussion
rs6999622[CT] and rs62527607[GT] in the BAALC Promoter Region Associate with BAALC Expression.
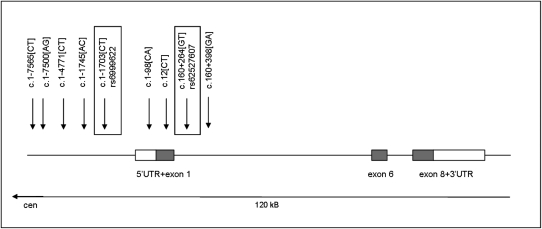
Using a sequence-based approach, we studied the genomic region of BAALC in 253 de novo CN-AML patients. Pre- and posttreatment peripheral blood (PB) or bone marrow (BM) samples were used for DNA resequencing and haplotyping. All exonic sequences of the main isoform of BAALC (consisting of exons 1, 6, and 8) including splice sites, promoter, and 5′- and 3′-untranslated regions (UTRs) were screened for mutations and SNPs. No mutations were detected, but the analysis revealed 30 sequence variants (SNPs). Nine of these 30 variants showed minor allele frequencies of >5% and were therefore selected as potential candidates for association with differential BAALC expression levels (Fig. 1).
Fig. 1.
BAALC genomic region with genotyped SNPs. Direct sequencing of the most common BAALC transcript variant 1-6-8 including 5′ and 3′ UTRs in 253 CN-AML patients revealed a total of nine informative SNPs (minor allele frequency >5%). SNPs were mainly located upstream of exon 1 and the 5′-UTR of BAALC. No informative SNPs were found in exons 6 and 8. Identified SNPs were genotyped in matched unaffected controls and in a CN-AML validation set with matched controls. Two of the nine SNPs were associated with high BAALC expresser status (rs6999622[CT] and rs62527607[GT], both highlighted in boxes). rs62527607[T] is predicted to create a RUNX1 TF binding site.
The nine SNPs were genotyped by SNaPShot analysis in 286 nonleukemic controls (Materials and Methods). Using the PHASE 2.0 program (46), 20 different haplotypes were generated using genotype data from both cases and controls (Table S1). No significant differences in the SNP and haplotype frequencies were observed in patients vs. controls. However, when comparing low and high BAALC expressers among the patients, two noncoding SNPs were significantly associated with high BAALC expresser status (rs6999622[CT]: genotype TT/CT vs. CC: P = 1.92 × 10−5, allele T vs. C: P = 1.54 × 10−4 and rs62527607[GT]: genotype TT/GT vs. GG: P = 2.01 × 10−4, allele T vs. G: P = 3.16 × 10−3) (Table 1), both assuming a dominant model for the genotype analyses.
Table 1.
Association of rs62527607[GT] genotype and BAALC expression in the test set and validation set and in the two sets combined
| Test set |
Validation set |
Combined sets |
||||||||
| BAALC expression | Low, n = 134 (%) | High, n = 119 (%) | *P value | Low, n = 53 (%) | High, n = 52 (%) | *P value | Low, n = 187 (%) | High, n = 171 (%) | **P value | |
| Genotype counts | GG | 108 (81) | 70 (59) | 43 (81) | 36 (69) | 151 (81) | 106 (62) | |||
| GT | 20 (15) | 46 (39) | 8 (15) | 16 (31) | 28 (15) | 62 (36) | ||||
| TT | 6 (5) | 3 (3) | 2 (4) | 0 (0) | 8 (4) | 3 (2) | ||||
| GT + TT | 26 (19) | 49 (41) | 2.01 × 10−4 | 10 (19) | 16 (31) | 0.16 | 36 (19) | 65 (38) | 9.30 × 10−5 | |
| OR (95% CI) | 2.91 (1.67, 5.16) | 1.91 (0.78, 4.86) | 2.59 (1.62, 4.22) | |||||||
| Allele count | G | 236 (88) | 186 (78) | 94 (89) | 88 (85) | 330 (88) | 274 (74) | |||
| T | 32 (12) | 52 (22) | 3.16 × 10−3 | 12 (11) | 16 (15) | 0.39 | 44 (12) | 68 (18) | 2.83 × 10−3 | |
| OR (95% CI) | 2.06 (1.28, 3.06) | 1.42 (0.64, 3.24) | 1.87 (1.25, 2.85) | |||||||
Test set, Cancer and Leukemia Group B; validation set, German-Austrian AML Study Group. All P values for genotypes compare GG with GT+TT and are obtained from *univariable or **multivariable logistic regression analyses. CI, confidence interval; OR, odds ratio.
The allele frequencies of the two SNPs were in agreement with those reported in populations of European ancestry in the dbSNP and the International Haplotype Mapping (HapMap) Project47 databases and their distributions were in Hardy-Weinberg equilibrium. The two SNPs are in almost complete linkage disequilibrium with each other [LD (rs6999622, rs62527607) D′= 0.97, R2 = 0.92, LOD = 136.35; derived from genotype data from cases and controls; Fig. S1]. Consequently, when comparing haplotype data of high and low BAALC CN-AML expressers, patients harboring at least one copy of the T allele of either SNP were more likely to belong to the high BAALC expressing group than those with the genotype CC of rs6999622 and the genotype GG of rs62527607 (P = 4.35 × 10−4; Table S2). This led us to hypothesize that these SNPs might affect BAALC expression either individually or in combination.
rs62527607[T] Creates a Binding Site for the Activating Transcription Factor RUNX1.
Because both SNPs are located in the BAALC promoter region (rs6999622 is located 1,703 bp upstream of the ATG of exon 1 and rs62527607 is located 264 bp into intron 1) (Fig. 1), we hypothesized that these SNPs might alter transcription factor (TF) binding sites. Indeed, using the TF-search algorithm (www.cbrc.jp/research/db/TFSEARCH.html), we predicted that rs62527607[T] creates a TF binding site for RUNX1. This led us to focus on rs62527607 as the most promising candidate marker to cause BAALC overexpression. RUNX1 is a known transcriptional activator that has been implicated in normal and malignant hematopoiesis (47).
RUNX1 Binds Preferentially to the T Allele of rs62527607[TG] and Promotes Its Transcriptional Activity.
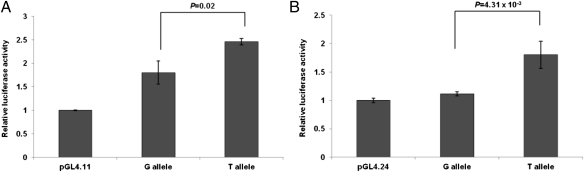
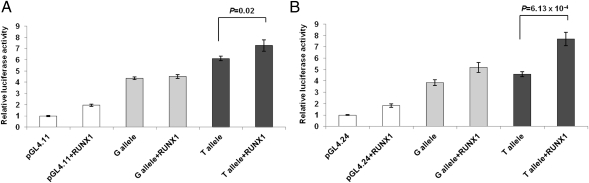
Luciferase reporter assays comparing the allelic activity of the respective SNP alleles, which were cloned into different vectors with their flanking genomic promoter sequence, demonstrated that rs62527607[T] increased transcription activity compared with rs62527607[G] [pGL4.11: P = 0.02 (Fig. 2A); pGL4.24: P = 4.31 × 10−3 (Fig. 2B) and compared with negative controls pGL4.11: P = 6.41 × 10−7 (Fig. 2A); pGL4.24: P = 2.17 × 10−3 (Fig. 2B)]. When testing the impact of RUNX1 on the regulatory potential of rs62527607, cotransfection of a RUNX1 expression construct with the luciferase gene reporters showed that with the promoterless vector pGL4.11 a significant increase could only be observed for rs62527607[T] (P = 0.02) and not for rs62527607[G] (P = 0.27, Fig. 3A). The minimal promoter vector pGL4.24 increased activity over baseline for both alleles of rs62527607[GT] (T allele: P = 6.13 × 10−4; G allele: P = 8.40 × 10−3; Fig. 3B). However, the RUNX1-induced increase of luciferase activity was stronger with the construct containing rs62527607[T] than with the construct containing rs62527607[G] (pGL4.11: 1.19-fold vs. 1.03-fold; pGL4.24: 1.67-fold vs. 1.35-fold).
Fig. 2.
(A and B) Luciferase reporter assays of rs62527607. Luciferase reporter constructs containing rs62527607 were transfected in triplicate into murine neuroblastoma cells (Neuro2a). Luciferase expression levels were normalized using a cotransfected Renilla construct (±SD). rs62527607 revealed activating potential compared with empty vectors without (pGL4.11) (A) and with promoter (pGL4.24) (B). The activating potential of rs62527607[T] is greater than that of rs62527607[G] [pGL4.11: P = 0.02 (A); pGL4.24: P = 4.31 × 10−3 (B)].
Fig. 3.
(A and B) Luciferase reporter assays of rs62527607 with cotransfected transcription factor RUNX1. Luciferase reporter constructs containing rs62527607 were transfected in triplicate into murine neuroblastoma cells (Neuro2a) and cotransfected with pIRES2-RUNX1-EGFP or empty expression construct as control. Luciferase expression levels were normalized using a cotransfected Renilla construct (±SD). Addition of the RUNX1 expression construct to the pGL4.11 system increased the activity of rs62527607[T] but not rs62527607[G] (T allele; P = 0.02; G allele: P = 0.27) (A). Using the minimal promoter system (pGL4.24) luciferase activity of both rs62527607 alleles increased (T allele: P = 6.13 × 10−4; G allele: P = 8.40 × 10−3), with rs62527607[T] being the more responsive allele (pGL4.11: 1.19-fold vs. 1.03-fold increase; pGL4.24: 1.67-fold vs. 1.35-fold increase) (B).
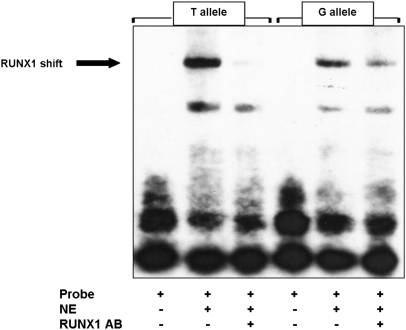
To confirm that the sequence containing rs62527607[T] is able to create a RUNX1 binding site, we performed an EMSA comparing the binding activities of the two alleles. The binding activity of RUNX1 to the sequence containing rs62527607[T] was stronger compared with rs62527607[G], thereby further supporting our hypothesis (Fig. 4).
Fig. 4.
Electrophoretic mobility shift assay (EMSA) of rs62527607[T] vs. rs62527607[G]. Nuclear extract of KG1a cells (NE) was added to a 20-bp biotin-labeled sequence (probe) containing either the T or G allele. For supershifting, RUNX1 antibody (AB) was added. RUNX1 bound to a greater extent to the sequence containing rs62527607[T], and the addition of the RUNX1-AB disrupted the binding.
RUNX1 Expresser Status Influences BAALC Expression Levels in the Presence of rs62527607[GT].
On the basis of the evidence that RUNX1 activated BAALC expression, we tested the correlation of RUNX1 and BAALC expression values in vivo in our test set of 253 CN-AML patients. Univariable analysis of the patient cohort revealed no significant correlation between RUNX1 and BAALC expression levels (P = 0.72). However, multivariable analysis revealed an interaction between genotypes (TT/GT vs. GG) and RUNX1 status (high/low) on BAALC expression (P = 0.05). When the analysis was restricted to patients with high RUNX1, the rs62527607 genotype (TT/GT vs. GG) revealed a positive correlation with BAALC levels (P = 1.18 × 10−4) in patients with genotype TT/GT, whereas no correlation of BAALC with the rs62527607 genotype was observed in the low RUNX1 expressing patient cohort (P = 0.21, Table 2).
Table 2.
Association of RUNX1 and BAALC expression levels depending on rs62527607[GT] genotype and RUNX1 expresser status in the test set and the validation set and in the two sets combined
| High RUNX1 | Test set |
Validation set |
Combined sets |
|||||||
| BAALC expression | Low, n = 74 (%) | High, n = 63 (%) | *P value | Low, n = 23 (%) | High, n = 29 (%) | *P value | Low, n = 97 (%) | High, n = 92 (%) | **P value | |
| Genotype counts | GG | 64 (86) | 35 (56) | 18 (78) | 18 (62) | 82 (85) | 53 (58) | |||
| GT | 8 (11) | 27 (43) | 4 (17) | 11 (38) | 12 (12) | 38 (41) | ||||
| TT | 2 (3) | 1 (2) | 1 (4) | 0 (0) | 3 (3) | 1 (1) | ||||
| GT + TT | 10 (14) | 28 (44) | 1.18 × 10−4 | 5 (22) | 11 (38) | 0.21 | 15 (15) | 39 (42) | 7.97 × 10−5 | |
| OR (95% CI) | 5.12 (2.29,12.24) | 2.20 (0.66, 8.19) | 4.02 (2.05, 8.21) | |||||||
| Low RUNX1 | ||||||||||
| BAALC expression | Low, n = 60 (%) | High, n = 56 (%) | Low, n = 30 (%) | High, n = 22 (%) | Low, n = 90 (%) | High, n = 78 (%) | ||||
| Genotype counts | GG | 44 (73) | 35 (62.5) | 25 (83) | 18 (82) | 69 (77) | 53 (68) | |||
| GT | 12 (20) | 19 (34) | 4 (13) | 4 (18) | 16 (18) | 23 (29) | ||||
| TT | 4 (7) | 2 (3.5) | 1 (3) | 0 (0) | 5 (6) | 2 (3) | ||||
| GT + TT | 16 (27) | 21 (37.5) | 0.21 | 5 (17) | 4 (18) | 0.89 | 21 (23) | 25 (32) | 0.24 | |
Test set, Cancer and Leukemia Group B; validation set, German-Austrian AML Study Group. All P values for genotypes compare GG with GT+TT and are obtained from *univariable or **multivariable logistic regression analyses. CI, confidence interval; OR, odds ratio.
Validation Set Supports the Association of rs62527607[GT] Genotype and BAALC Expression Levels.
For validation, we analyzed a set of 105 CN-AML patients from the German-Austrian AML Study Group (AMLSG, University of Ulm, Germany). The results supported the association of high BAALC expression with the T allele (genotypes TT/GT vs. GG, 31 vs. 19%, P = 0.16, Table 1), although it did not reach statistical significance. Also, when testing for the reported influence of RUNX1 on BAALC expression in patients with rs62527607 genotype TT/GT vs. GG in the high RUNX1 expressing group, patients with genotype TT/GT were more likely to belong to the BAALC high expressers (TT/GT vs. GG: 38 vs. 22%, P = 0.21, Table 2), whereas again no difference could be found in the low RUNX1 group (18 vs. 17%, P = 0.88).
Importantly, when the data from the test set and validation set were combined, the evidence for the association of genotypes TT/GT vs. GG with high BAALC expression became stronger [P = 9.30 × 10−5, odds ratio (OR) = 2.59 (1.62, 4.22; 95% confidence interval (CI)), Table 1]. The same was true for the in vivo influence of RUNX1 on BAALC expression levels in the presence of rs62527607 genotype TT/GT in patients belonging to the high RUNX1 group [P = 7.97 × 10−5, OR = 4.02 (2.05, 8.21; 95% CI), Table 2]. Taken together, these in vivo data suggest that not only the genotype but also the RUNX1 expresser status matters in the determination of BAALC expression levels in CN-AML patients.
Concluding Remarks
We report here on a SNP as a predisposing genetic factor for somatic events involved in a hematologic malignancy. In considering previous findings of a similar nature, the JAK2 gain-of-function mutation V617F is an illustrative example. It has been shown that a haplotype named 46/1 predisposes to the somatic mutation that apparently is a causative event in the development of myeloproliferative disorders (48–52). However, to the best of our knowledge the mechanism(s) by which the alleles predispose to the acquisition of V617F has not been explained. In contrast, rs62527607 does not predispose to an increased risk of leukemia, but to overexpression of the BAALC gene, which is implicated in leukemogenesis and associated with adverse outcome in CN-AML. We show that the mechanism through which BAALC overexpression occurs is related to the presence of the rs62527607[T] allele that creates a binding site for the activating RUNX1 transcription factor in the promoter region. This leads to constitutive BAALC overexpression compared with the rs62527607[G] allele. We are not suggesting that this mechanism is the only cause of increased expression of BAALC in leukemia, but we consider it as a major contributing factor for the activation of this oncogene. It is likely that BAALC acts as one component of a complex high-risk sequence of changes. One might even hypothesize that BAALC acts as a reporter without causative downstream effects. The analysis of this mechanism in other subtypes of leukemia, especially those with translocations involving the RUNX1 locus (e.g., t[8;21] [q22;q22]) or in patients harboring mutations of RUNX1 may be important in future research. In summary, we demonstrate that overexpression of BAALC in CN-AML is in part a heritable trait. This observation and its potential clinical implications are in line with the emerging concept of personalized medicine in leukemia.
Materials and Methods
Test Set.
Samples were obtained from 253 patients treated in Cancer and Leukemia Group B (CALGB) front-line clinical protocols [132 male, 121 female; median age: 56 (19–79) years]. At least 90% of the patients studied were of European ancestry. Patients with antecedent hematologic disorders were excluded. Cytogenetic analyses of pretreatment BM samples were performed by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion study, and centrally reviewed (53, 54). All patients were also enrolled in companion protocols CALGB 9665 (Leukemia Tissue Bank) and CALGB 20202 (molecular studies in AML) and gave informed consent for the research use of their specimens, in accordance with the Declaration of Helsinki. Sources for DNA samples were pre- or posttreatment PB (pretreatment: n = 139, posttreatment: n = 50) or BM (pretreatment: n = 27, posttreatment: n = 37); sample source for RNA isolation was pretreatment PB or BM mononuclear cells.
Preparation of pretreatment PB and BM samples was performed as previously described (6). In brief, mononuclear cells from pretreatment PB samples were enriched by Ficoll-hypaque gradient and frozen in liquid nitrogen. Total RNA was extracted from thawed samples (≥1 × 106 cells) using TRIzol reagent (Invitrogen) following the manufacturer's directions.
RNA from pretreatment PB or BM was used to determine expression levels of BAALC and RUNX1 using the Affymetrix U133 plus 2.0 array (Affymetrix) (SI Materials and Methods, Fig. S2). Determination of BAALC and RUNX1 expresser status (high/low) was performed using a median cutoff as previously described (6) (SI Materials and Methods).
Validation Set.
The validation set comprised pretreatment PB and BM samples of 105 CN-AML patients [51 male, 54 female; median age: 46 (18–60) years]. All patients were enrolled in AMLSG treatment protocols for younger adult patients (ages 16–60 y). Sample source for DNA samples was pretreatment PB (n = 104) or BM (n = 1); sample source for RNA isolation was pretreatment PB mononuclear cells. All patients gave informed consent for genetic analysis according to the Declaration of Helsinki. Approval was obtained from the institutional review boards of the participating AMLSG institutions.
For sample preparation, PB cells were enriched for mononuclear cells by Ficoll gradient and frozen at −80 °C. Total RNA was extracted from 1 × 107 cells with the AllPrep mini kit (Qiagen) according to standard protocol. A total of 2 μg of RNA was reverse transcribed into cDNA with the QuantiTect Reverse Transcription kit (Qiagen) in a 40-μL reaction mix according to the protocol.
Quantification of BAALC RNA expression in a set of 339 CN-AML patients was performed by RT-PCR in the laboratory of the University Hospital of Ulm as previously described (3). Determination of BAALC expresser status (high/low) was performed using a median cutoff. The 53 high- and 52 low-expressing samples used were chosen on the basis of tissue availability.
For determination of RUNX1 expression, RT-PCR amplification of RUNX1 and the house-keeping gene 18S was performed in triplicate at The Ohio State University. TaqMan Gene Expression primer-probe sets were obtained from ABI (Life Technologies/Applied Biosystems).
Controls.
DNA samples from 286 nonleukemic population-based controls with matching ancestry were obtained from the Human Cancer Genetics (HCG) Tissue Bank at The Ohio State University. Additionally, 190 nonleukemic population-based, ancestry-matched controls were provided by the University of Ulm.
Analysis of the BAALC Genomic Region.
We analyzed genomic DNA of 253 AML cases for mutations and polymorphisms by direct genomic sequencing of exons 1, 6, and 8 including splice sites, promoter, and 5′- and 3′- untranslated regions (UTRs) of BAALC. Primer sequences and PCR conditions are given in Table S3. Amplicons were bidirectionally sequenced using the Applied Biosystems 3730 DNA Analyzer (Life Technologies/Applied Biosystems). Sequencing traces were analyzed using SeqMan software.
Genotyping of the validation set and the control set for SNP rs62527607[GT] was performed using the ABI PRISM SNaPshot Multiplex kit (Life Technologies/Applied Biosystems).
Statistical Analyses.
Marker genotypes were evaluated for a potential association with overexpression of BAALC using univariable logistic regression analysis within the test and validation sets. Multivariable logistic regression was used to adjust for population differences of the test set and the validation set in the combined analysis. BAALC was analyzed as a dichotomized variable, using the median cut gene expression values. Results were obtained by assuming a dominant genotype model and multiplicative or allelic genotype models. Univariable and multivariable logistic regression analysis was performed to assess the association of BAALC expresser status and marker genotypes under the influence of RUNX1 expresser status. Haplotypes were derived from genotype data using the PHASE v2.1.1 (46) computer program. Haplotype and genotype frequency distributions between cases and controls or BAALC status (high/low) were compared using Fisher's exact test.
For analysis of the luciferase expression data the two-tailed homoscedastic Student's t test was applied to the normalized and log transformed luciferase expression data.
Luciferase Reporter Assays.
To study the regulatory potential of rs62527607 on BAALC gene expression, dual luciferase reporter constructs [pGL4.11 (promoterless), pGL4.24 (minimal promoter); Promega] were designed containing ∼500-bp genomic sequence flanking the respective variant. Primer sequences of the cloning primers were rs62527607 F: gcacgctagcGCCTGCACTCGGGTAAGTG (NheI) and rs62527607 R: gtgcctcgagCGCCTTGTGTATAAATCCA (XhoI). Neuro2a (murine neuroblastoma) cells (ATCC) were transfected in triplicate with reporter and control constructs. Relative expression was normalized using cotransfected Renilla luciferase (Promega).
To study the effect of RUNX1 on the regulatory potential of rs62527607, a CMV-driven expression construct pIRES2-RUNX1-EGFP (Clontech Laboratories/Takara Bio) was cotransfected with the luciferase reporter constructs. Primer sequences for cloning of the expression construct were RUNX1 F: gcaggctagcAGGAAGCGATGGCTTCAGAC (NheI) and RUNX1 R: cgtcgaattcCGCCTCAGTAGGGCCTCCAC (EcoRI). Empty expression vector pIRES2-EGFP was used as a control.
Electrophoretic Mobility Shift Assay (EMSA).
Nuclear proteins were extracted from KG1a cells (ATCC) using the Nuclear Extract kit (Active Motif) according to the manufacturer's instructions. In brief, cells were washed once in ice-cold PBS (PBS)/phosphatase inhibitors (PI), dislodged from the plates with a spatula, and centrifuged at 500 rpm for 5 min in 3 mL PBS/PI. The pellets were resuspended in 500 μL of hypotonic buffer for 10 min on ice. After addition of 25 μL detergent, the suspension was centrifuged for 30 s at 14,000 × g. The cytoplasmic fraction was removed and the pellet containing the nuclear fraction resuspended in 50 μL complete lysis buffer, vortexed, and incubated on ice on a rocking platform for 30 min. The lysate was centrifuged for 10 min at 14,000 × g and the supernatant containing the nuclear extract was immediately stored at −80 °C.
The 5′-biotinylated oligos 20 bp in length (25-nmol scale) were obtained from Integrated DNA Technologies. Oligo sequences were rs62527607[G] F: GCTTGCTCGCTGGTCGGG AG, rs62527607[G] R: CTCCCGACCAGCGAGCAAGC, rs62527607[T] F: GCTTGC TCTCTGGTCGGGAG, rs62527607[T] R: CTCCCGACCAGAGAGCAAGC. For annealing, concentrated complementary oligonucleotides were mixed at a 1:1 molar ratio and incubated at 95 °C for 5 min. The heat was then gradually reduced over hours until the oligonucleotides reached room temperature. Annealed oligos were diluted to a final concentration of 10 fmol. The LightShift Chemiluminescent EMSA kit (Pierce/Thermo Fisher Scientific) was used according to the manufacturer's instructions. Briefly, 20 fmol of each biotin-labeled oligonucleotide pair were incubated in EMSA binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT; pH 7.5) containing 2.5% (vol/vol) glycerol, 5 mM MgCl2, 50 ng/μL poly(dI-dC), 0.05% (vol/vol) Nonidet P-40, and 5 μg KG1a nuclear proteins for 20 min at room temperature. For achievement of a supershift, 400 ng RUNX1 antibody was added [AML1 (=RUNX1); Santa Cruz Biotechnology]. Complexes were resolved by electrophoresis on native 5% TBE Criterion Precast gels (Bio-Rad Laboratories) in 0.5× TBE buffer at 110 V. Gels were transferred to Biodyne B pre-cut modified nylon membranes (0.45 μM, Pierce/Thermo Fisher Scientific) using a Trans-Blot SD semi-dry transfer cell (Bio-Rad Laboratories). Membranes were cross-linked (UVC-508 UV Cross-linker, Ultra LUM) and visualized using the Chemiluminescent Nucleic Acid Detection system (Pierce/Thermo Fisher Scientific; 10-s exposure time).
Supplementary Material
Acknowledgments
We thank Jan Lockman, Ravi Patel, Somayeh S. Tarighat, and Cameron M. Beech for technical support; Deedra Nicolet for statistical analyses; Donna Bucci of the Cancer and Leukemia Group B (CALGB) Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center for sample processing and storage services; and The Ohio State University Comprehensive Cancer Center's Nucleic Acid Shared Resource and Microarray Shared Resource for technical support. This work was supported in part by the National Cancer Institute (Grants CA098933, CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657), the Coleman Leukemia Research Foundation, and the Pelotonia Fellowship Program (to A.-K.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203756109/-/DCSupplemental.
References
- 1.Farag SS, et al. Outcome of induction and postremission therapy in younger adults with acute myeloid leukemia with normal karyotype: A cancer and leukemia group B study. J Clin Oncol. 2005;23:482–493. doi: 10.1200/JCO.2005.06.090. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez HF, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361:1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnett AK, et al. Attempts to optimize induction and consolidation treatment in acute myeloid leukemia: Results of the MRC AML12 trial. J Clin Oncol. 2010;28:586–595. doi: 10.1200/JCO.2009.22.9088. [DOI] [PubMed] [Google Scholar]
- 4.Fröhling S, et al. Cytogenetics and age are major determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: Results from AMLSG trial AML HD98-B. Blood. 2006;108:3280–3288. doi: 10.1182/blood-2006-04-014324. [DOI] [PubMed] [Google Scholar]
- 5.Estey E. Acute myeloid leukemia and myelodysplastic syndromes in older patients. J Clin Oncol. 2007;25:1908–1915. doi: 10.1200/JCO.2006.10.2731. [DOI] [PubMed] [Google Scholar]
- 6.Burnett AK, et al. The addition of arsenic trioxide to low-dose Ara-C in older patients with AML does not improve outcome. Leukemia. 2011;25:1122–1127. doi: 10.1038/leu.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitman SP, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A cancer and leukemia group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 8.Thiede C, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 9.Fröhling S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 10.Falini B, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 11.Schnittger S, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 12.Döhner K, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 13.Farag SS, et al. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: Results from Cancer and Leukemia Group B 8461. Blood. 2006;108:63–73. doi: 10.1182/blood-2005-11-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuser M, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 15.Tonks A, et al. CD200 as a prognostic factor in acute myeloid leukaemia. Leukemia. 2007;21:566–568. doi: 10.1038/sj.leu.2404559. [DOI] [PubMed] [Google Scholar]
- 16.Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcucci G, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 18.Marcucci G, et al. Cancer and Leukemia Group B Study High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 19.Marcucci G, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paschka P, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschka P, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 23.Langer C, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2009;27:3198–3204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzeler KH, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: A comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 25.Metzeler KH, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker H, et al. Mutations of the Wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:788–792. doi: 10.1182/blood-2010-01-262543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker H, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwind S, et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood. 2011;118:4188–4198. doi: 10.1182/blood-2011-06-357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaidzik VI, et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 30.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: Still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 32.Tanner SM, et al. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci USA. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldus CD, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: A Cancer and Leukemia Group B study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 34.Bienz M, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 35.Santamaría C, et al. BAALC is an important predictor of refractoriness to chemotherapy and poor survival in intermediate-risk acute myeloid leukemia (AML) Ann Hematol. 2010;89:453–458. doi: 10.1007/s00277-009-0864-x. [DOI] [PubMed] [Google Scholar]
- 36.Eid MA, et al. BAALC and ERG expression in acute myeloid leukemia with normal karyotype: Impact on prognosis. Int J Lab Hematol. 2010;32:197–205. doi: 10.1111/j.1751-553X.2009.01168.x. [DOI] [PubMed] [Google Scholar]
- 37.Schwind S, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldus CD, et al. Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol. 2007;25:3739–3745. doi: 10.1200/JCO.2007.11.5253. [DOI] [PubMed] [Google Scholar]
- 39.Kühnl A, et al. High BAALC expression predicts chemoresistance in adult B-precursor acute lymphoblastic leukemia. Blood. 2010;115:3737–3744. doi: 10.1182/blood-2009-09-241943. [DOI] [PubMed] [Google Scholar]
- 40.Schrama D, et al. BRAFV600E mutations in malignant melanoma are associated with increased expressions of BAALC. J Carcinog. 2008;7(1) doi: 10.1186/1477-3163-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agaram NP, et al. Molecular characterization of pediatric gastrointestinal stromal tumors. Clin Cancer Res. 2008;14:3204–3215. doi: 10.1158/1078-0432.CCR-07-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heuser M, et al. Functional role of BAALC in leukemogenesis. Leukemia. 2011;26:532–536. doi: 10.1038/leu.2011.228. [DOI] [PubMed] [Google Scholar]
- 43.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 46.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman AD. Cell cycle and developmental control of hematopoiesis by Runx1. J Cell Physiol. 2009;219:520–524. doi: 10.1002/jcp.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 49.Jones AV, et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009;41:446–449. doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olcaydu D, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–454. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 52.Hermouet S, Vilaine M. The JAK2 46/1 haplotype: A marker of inappropriate myelomonocytic response to cytokine stimulation, leading to increased risk of inflammation, myeloid neoplasm, and impaired defense against infection? Haematologica. 2011;96:1575–1579. doi: 10.3324/haematol.2011.055392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Byrd JC, et al. Cancer and Leukemia Group B 8461 Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 54.Mrózek K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.