Abstract
Poly(ADP-ribose) polymerase (PARP) inhibitors can generate synthetic lethality in cancer cells defective in homologous recombination. However, the mechanism(s) by which they affect DNA repair has not been established. Here we directly determined the effects of PARP inhibition and PARP1 depletion on the repair of ionizing radiation-induced single- and double-strand breaks (SSBs and DSBs) in human lymphoid cell lines. To do this, we developed an in vivo repair assay based on large endogenous Epstein–Barr virus (EBV) circular episomes. The EBV break assay provides the opportunity to assess quantitatively and simultaneously the induction and repair of SSBs and DSBs in human cells. Repair was efficient in G1 and G2 cells and was not dependent on functional p53. shRNA-mediated knockdown of PARP1 demonstrated that the PARP1 protein was not essential for SSB repair. Among 10 widely used PARP inhibitors, none affected DSB repair, although an inhibitor of DNA-dependent protein kinase was highly effective at reducing DSB repair. Only Olaparib and Iniparib, which are in clinical cancer therapy trials, as well as 4-AN inhibited SSB repair. However, a decrease in PARP1 expression reversed the ability of Iniparib to reduce SSB repair. Because Iniparib disrupts PARP1–DNA binding, the mechanism of inhibition does not appear to involve trapping PARP at SSBs.
Keywords: base excision repair, BRCA
Poly(ADP-ribose) polymerases (PARPs) are a family of enzymes that catalyze ADP ribosylation of a variety of cellular factors (1). Interest in PARPs, especially PARP1, was intensified by the discovery that PARP inhibition is toxic for cancer cells that are defective in the homologous recombination (HR) genes BRCA1 and BRCA2 (2, 3). These mutations are found in breast and ovarian cancer (4). Increased sensitivity to PARP inhibition has also been observed with cells defective in other DNA double-strand break (DSB) repair genes such as MRE11 (5) and ATM (6, 7). Despite these findings and the promising clinical utility of PARP inhibitors in treating HR-deficient cancers, the underlying mechanism(s) remains elusive. PARP1 and PARP2 are generally considered important enzymes in single-strand break (SSB) repair (8–10). Therefore, the synthetic lethality between PARP inhibition and HR defects has been attributed to accumulation of SSBs, which are subsequently converted to DSBs (11). However, unlike PARP inhibition, depletion of PARP1 resulted in only modest toxicity in BRCA2-deficient cells (2). Given the wide-ranging biological consequences expected from PARP inhibition, the synthetic lethality in cells with an HR defect might go beyond effects on just DNA repair. Elucidating the mechanisms of action is critical to addressing the efficacy of PARP inhibitors, understanding mechanisms of drug resistance, as well as extending PARP-related treatment to other types of cancers.
Because PARP participates in DNA repair processes, it is important to examine the effects of inhibitors on DNA damage and repair, especially SSBs and DSBs, which can lead to genome instability. Despite many repair studies in human cells, there is a lack of robust systems for accurate in vivo measurement of randomly induced SSBs and DSBs, thereby limiting opportunities to investigate underlying mechanisms of induction and repair, as well as the role of PARP. Currently, the most commonly used approach to detect random DSBs uses immunostaining of DSB-related biomarkers such as γ-H2AX (12). This approach is sensitive and has provided considerable understanding of proteins recruited to damage sites. However, detection of DSBs is indirect, and there are limitations with regard to knowing which proteins are directly required for repair and which are related cofactors that are part of downstream signaling or chromatin modification events. The comet assay, another commonly used approach, visualizes damage within the mass of nuclear DNA when DNA is subjected to an electric field. Comet assays are widely used to detect SSBs/DSBs as well as other lesions that can be converted into DNA strand breaks, such as alkali-labile sites (13, 14). However, the actual incidence of breaks cannot be determined directly, and there are limitations on assessing the specificity of the damage response (15, 16).
We have developed an approach for detecting SSBs and DSBs in human cells based on our previous findings with circular chromosomes in budding yeast. A single DSB dramatically changes the migration pattern of the chromosome during pulsed-field gel electrophoresis (PFGE) (17) because a DSB converts a circular chromosome into a unit-length linear form. We describe a human cell system based on Epstein–Barr virus (EBV) episomes. These minichromosomes are large circular molecules (165–180 kb) that have many human chromosome features, including nucleosomes with spacing typical of human chromatin (18, 19). Moreover, replication, which occurs only once per cell cycle, is controlled by host proteins (20), which makes this system ideal to study cell cycle-dependent DNA repair events. Here we show that the EBV break assay can assess directly and accurately the formation and repair of both SSBs and DSBs.
To address the impact of PARP and PARP inhibitors on DNA repair, we have used human lymphoblastoid cells containing the circular EBV episomal genome to assess ionizing radiation-induced SSB and DSB as well as repair. We evaluated the inhibitory properties of various widely used PARP inhibitors, including those showing clinical potential for cancer treatment such as Olaparib and Iniparib (21, 22). In addition, we confirmed the important difference between PARP knockdown and chemical inhibition of PARP on DNA strand break repair in vivo.
Results
Simultaneous, Quantitative Detection of SSBs and DSBs in Human Cells.
Yeast circular chromosomes have been used in combination with PFGE to monitor the formation and repair of DSBs (17, 23) as well as resection in vivo (24, 25). Large circular chromosomal DNA molecules (∼300 kb in Saccharomyces cerevisiae) are retained in the starting well during PFGE; however, a single random DSB generates a unit-length linear molecule detectable as a single band by Southern blot (Fig. 1). Previously, Johnson and Beerman (26) reported a similar approach to detect DSBs in EBV episomes from EBV-infected cells when the DNA was irradiated in plugs. We extended our yeast approach to EBV to quantitate radiation damage and repair within human cells. The EBV system provides the opportunity to monitor simultaneously the induction and repair in vivo of not only DSBs but also SSBs.
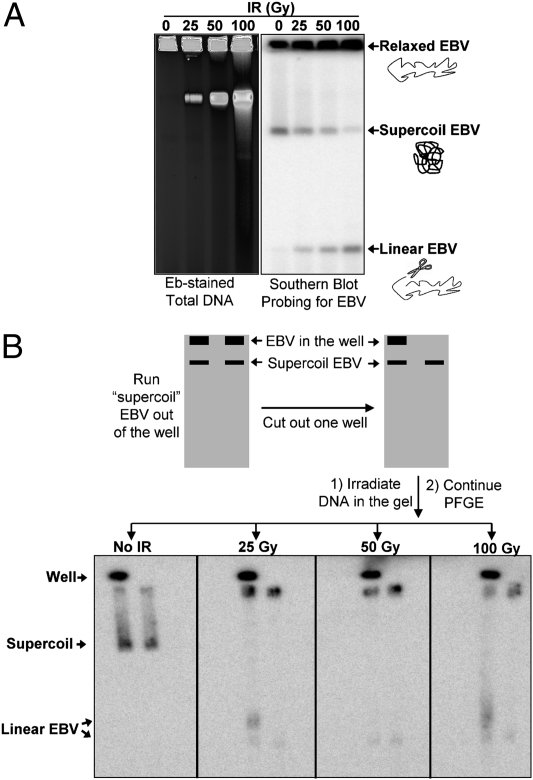
Fig. 1.
Detection of SSBs and DSBs in EBV. (A) IR changes the topology of EBV molecules in vivo in a dose-dependent manner. Following PFGE of irradiated cells, breaks in chromosomes were detected by ethidium bromide (Eb) staining (Left). The lower compaction band corresponds to broken high-molecular weight molecules. Southern blotting with an EBV-specific probe (Right) revealed three forms of EBV: relaxed circles (top band) that are not able to enter the gel, unit-size linear EBV molecules (bottom band) that were due to single DSBs in the EBV molecules, and supercoiled EBV (middle band). Note: The linear molecules arose primarily from DSBs in relaxed molecules that would have appeared in the well before IR. (B) Demonstration that the middle band is supercoiled EBV. Duplicate chromosomal DNA samples without damage were run on PFGE for 6 h to let the proposed “supercoil” DNA run out of the well. The well was then removed from one of the lanes to leave only the supercoiled DNA in the lane. Each gel slab containing the two lanes was irradiated, followed by PFGE for another 18 h. EBV was detected by Southern blotting. Without irradiation, the initial supercoiled band moves to a new position. However, because of nicking by the radiation the supercoiled DNA is relaxed, preventing further movement of the band. The radiation also generated broken molecules (i.e., induced DSBs) that would give rise to two linear EBV bands for the left lane (derived from DNA in the well and supercoil band) and only one for the right lane.
Without irradiation, two forms of EBV are detected in the DNA from Raji cancer cells (Burkitt lymphoma) or LCL35 EBV-transformed lymphoblast cells, as described in Fig. 1A. The majority of the molecules detected with the EBV-specific probe remained in the well. Following irradiation, there is loss of the upper band as well as the appearance of a much faster moving band that corresponds to linear EBV (based on a comparison with DNA markers) resulting from random single DSBs. Multiple DSBs result in a broad smear of DNA (17).
The mobility of the upper band and the loss at small doses suggested that this might be due to EBV supercoils, in which case SSBs would relax the material, preventing entry into the gel. If this were the case, then loss of the upper band would provide a direct measurement of SSBs. To demonstrate that the upper band is indeed supercoiled EBV, we examined the consequences of radiation on this band using the approach described in Fig. 1B, because SSBs would relax these molecules. Several plugs of unirradiated DNA were subjected to PFGE (6 h) to create lanes that had EBV DNA (presumably relaxed DNA) retained in the plug and a band of fast-running DNA (proposed supercoiled form). The gels were sliced so that each slice had two lanes; four slices with two lanes each were created. The four slices were irradiated with 0, 25, 50, or 100 Gy and subjected to further PFGE. If the upper band of DNA was actually due to supercoiled EBV, irradiation would lead to nicks and relaxation, preventing further movement upon the second round of PFGE. As shown in Fig. 1B, the EBV band in the unirradiated slab moved much farther during the subsequent PFGE, and there was no additional material contributed by the DNA in the well (compare left and right lanes after 0 dose to slab). However, irradiation with as little as 25 Gy essentially prevented any further migration of the band. We conclude that this is due to relaxation of the supercoiled form, which prevents further PFGE migration. In the right lane of each irradiated slab there was also a single higher-mobility band that was due to the linearization of molecules in the supercoiled band. In the left lane there were two fast-moving bands corresponding to linearized EBV from the plug and linearized DNA from the supercoiled band.
Thus, the mobility of supercoiled EBV provides a sensitive assay for monitoring SSB formation and repair in vivo. Because a single SSB would transform the EBV supercoil into a relaxed circle that remains trapped in the well, the efficiency of SSB induction can be determined by applying Poisson distributions to model the loss of material from the supercoiled band (SI Materials and Methods). The method does not allow an estimate of nicked and unnicked circles that remain in the well. For example, the incidence of SSBs resulting from 100 Gy is 11.3 SSBs/Mb, which would correspond to 680 SSBs/Gy in the human genome, as described in Table 1 (assuming 6 × 109 bp per diploid genome). Similar values have been obtained with less direct methods (27). The induction of DSBs can also be quantitated based on Poisson distribution by determining the appearance of material with a single DSB (i.e., the fast-moving linear band in Fig. 1; SI Materials and Methods). There was a near-linear increase in material corresponding to ∼0.016 DSBs⋅Mb−1⋅Gy−1 or ∼100 DSBs/Gy in the genome of human diploid cells (Table 1). This value corresponds to the range of 63–70 DSBs/Gy per cell from previous estimates (27) and nearly twice that measured in yeast with the same approach using changes in a circular chromosome (24). Comparable values of SSBs and DSBs were found for LCL35 cells, which have fivefold fewer copies of EBV than the Raji cell line (∼10 vs. ∼50), consistent with previous studies in which the initial number of DNA breaks is independent of cell type (28, 29).
Table 1.
Measurement of in vivo SSBs and DSBs induced by ionizing radiation
| IR (Gy) | SSBs/Mb | DSBs/Mb | SSBs/cell | DSBs/cell | SSB:DSB |
| 5 | 1.23 (1.10–1.67) | 0.09 (0.04–0.16) | 7,380 | 540 | 13.7 |
| 10 | 2.14 (1.72–2.60) | 0.16 (0.11–0.25) | 12,840 | 960 | 13.4 |
| 20 | 3.41 (2.84–3.71) | 0.33 (0.15–0.48) | 20,460 | 1,980 | 10.3 |
| 50 | 5.43 (3.95–5.56) | 0.59 (0.47–0.66) | 32,580 | 3,540 | 9.2 |
| 100 | 9.77 (9.24–10.95) | 1.15 (0.71–1.21) | 58,620 | 6,900 | 8.5 |
| 200 | 19.26 (18.3–22.9) | 2.33 (1.53–2.48) | 115,800 | 13,980 | 8.3 |
Number of SSBs and DSBs was calculated as described in SI Materials and Methods. Values presented are in the format of median and range (in parentheses) from five independent measurements. SSBs or DSBs per cell were calculated assuming a genome size of 6 × 109 bp in G2 phase (4N DNA content).
Using the EBV system, the ratio of SSB:DSB lesions induced in vivo by radiation was ∼10 over a 10–100 Gy dose range (Table 1), consistent with previous measurements with plasmids (30). The reduction in the SSB:DSB ratio with increasing doses also fits with previous data and proposed models (31, 32). Whereas a DSB could be generated if SSBs on opposite strands are closely spaced, we previously showed that for random SSBs generated during repair of methyl methanesulfonate (MMS) damage, only a few DSBs were produced following the generation of thousands of SSBs (17). The present ionizing radiation (IR) results are consistent with multiple free radicals being produced by single radiation events within a radius of only a few nanometers to generate DSBs (33).
Repair of IR-Induced DNA Breaks.
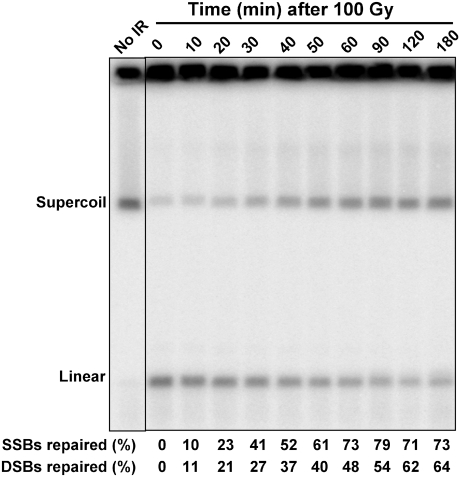
The EBV system provides a unique opportunity to quantitatively address repair of SSBs and DSBs. As shown in Fig. 2, repair of SSBs was first detected within several minutes after a dose of 100 Gy to a population of growing Raji cells (most of which are in G1). There was a small increase in supercoiled DNA by 10 min, and by 30 min about half the SSBs were repaired. Eventually, the amount of supercoiled DNA reached a level comparable to that before IR. The chromatin structure of the EBV minichromosomes is likely to be retained, including within the vicinity of breaks (34), so that repair of an SSB will result in the reappearance of supercoils when the DNA is displayed with PFGE. These results suggest that whereas before IR there is more relaxed EBV than supercoiled (compare DNA in the well vs. the supercoiled band), the reappearance of supercoiled molecules is largely due to repair of the IR-nicked supercoiled molecules. Included among the sources of preexisting relaxed DNA in the well are molecules that are replicating, gapped, or damaged.
Fig. 2.
Simultaneous detection of repair of SSBs and DSBs. Raji cells were γ-irradiated at a dose of 100 Gy and then incubated in complete medium at 37 °C to allow repair. Cells were collected at the indicated times and processed for PFGE and Southern blot analysis. The repair efficiencies of the SSBs and DSBs are indicated at the bottom.
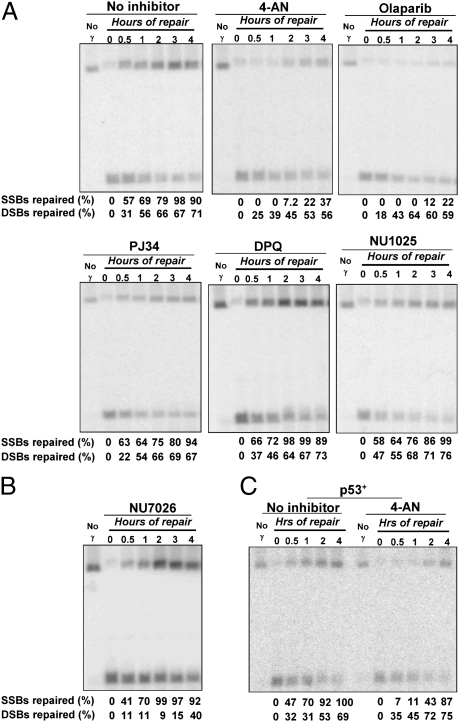
Similar to SSBs, ∼20% of the DSBs were repaired in the first 20 min; however, the subsequent reduction in broken molecules was less, with nearly half remaining at 60–90 min. To address a possible role for nonhomologous end joining (NHEJ), a key pathway for DSB repair in humans (35), we examined repair in cells exposed to NU7026, an inhibitor of DNA-dependent protein kinase (DNA-PK). Before irradiation, the population had been enriched for G2 cells using nocodazole to enhance the capability for recombinational repair. Because there was considerable reduction in DSB repair, with only ∼10% repair at 2 h compared with over 50% in mock-treated control cells, most of the early DSB repair was due to NHEJ (Fig. 3B). Repair of SSBs and DSBs appears to be independent of one another, because the NU7026 effect on DSB repair did not extend to SSBs. The subsequent limited repair could be due to homologous recombination. In a less quantitative analysis, ethidium bromide-stained gels also showed slow repair of the fragmented human genome in NU7026-treated cells compared with that of no inhibitor and the PARP inhibitor 4-amino-1,8-naphthalimide (4-AN) (Fig. S1).
Fig. 3.
Effect of PARP and NHEJ inhibitors on the repair of radiation-induced breaks. (A) Repair of IR-induced breaks (100 Gy) in nocodazole-enriched G2 Raji cells treated with PARP inhibitors: 20 μM DPQ; 50 μM NU1025; 10 μM PJ34; 10 μM 4-AN; 20 μM Olaparib. Cells were collected at the indicated times and processed for PFGE and Southern blot analysis. The repair efficiencies of SSBs and DSBs are indicated at the bottom of each panel. (B) Inhibition of DSB repair by the DNA-PK inhibitor NU7026 (10 μM). (C) Repair of IR-induced breaks in p53-competent cells (LCL35) in the absence or presence of 4-AN. LCL35 cells (p53+) enriched in G2 were exposed to 100 Gy followed by incubation with or without the PARP inhibitor 4-AN (10 μM) and then processed for PFGE and Southern blot analysis.
Inhibition of SSB Repair by PARP Inhibitors.
PARP inhibition has been used in cancer chemotherapy when combined with various drugs or in particular genetic backgrounds (11) where the degree of PARP inhibition appears related to clinical outcome (36, 37). The EBV system provides the opportunity to determine directly the effect of commonly used inhibitors on the ability of cells to repair IR-induced SSBs and DSBs.
Previously, the potencies of PARP inhibitors were mainly determined based on in vitro evaluation of NAD+ turnover or synthesis of poly(ADP ribose) chains in vivo (38). Although the impact of PARP inhibitors on DNA repair has been assessed using MMS sensitivity, comet assays, or H2AX foci formation, a side-by-side comparison of potential inhibitors on DNA repair of radiation damage has been lacking. Using the EBV system, we tested the following 10 inhibitors for effects on IR-induced SSB and DSB repair: 4-AN, NU1025, DPQ, PJ34, Olaparib, Iniparib, IQD, BYK204165, 3-AB, and DR2313 (See SI Materials and Methods for full chemical names). Cells were incubated with nocodazole before treatment with chemicals and IR to enrich for G2 cells and increase opportunities for HR repair. Most inhibitors (7/10; see DPQ, NU1025, and PJ34 in Fig. 3A) had little or no effect on SSB repair, with over 70% of SSBs repaired within 2 h following a dose of 100 Gy. However, there was inhibition of SSB repair by 4-AN (Fig. 3A), Olaparib (Fig. 3A), and Iniparib (Fig. 4D). Only 7% of SSBs were repaired in 4-AN–treated cells at 2 h, and Olaparib treatment resulted in almost no repair (Fig. 3A). The effect of Iniparib is weaker compared with 4-AN and Olaparib, resulting in 55% repair by 2 h (Fig. 4D). The efficiency of the inhibitors on PARP catalytic activity was examined with an in vitro poly(ADP ribosyl)ation (PARylation) assay. As shown in Fig. S2, all inhibitors used at the concentrations in the repair study, except Iniparib, efficiently inhibited PARylation. PJ32 and NU1025, which did not inhibit SSB repair as efficiently as 4-AN, caused over 95% inhibition of PARylation, which was greater than 4-AN. Thus, inhibition of SSB repair may not always reflect direct effects on PARP catalytic activity. The less effective inhibition of Iniparib might be due to a lack of direct targeting to the catalytic domain of PARP1. There was little or no effect on DSB repair by any of the PARP inhibitors (see examples in Fig. 3A), suggesting that under these conditions, PARP plays at most a minor role, even though it targets many genes involved in DSB repair.
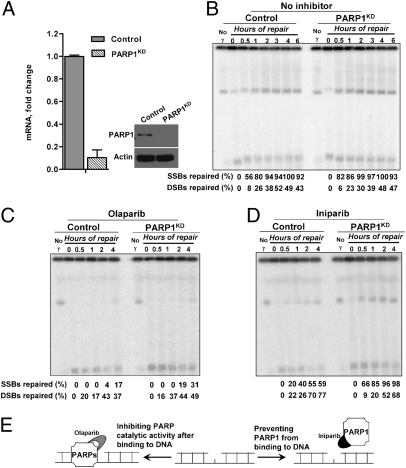
Fig. 4.
PARP1 depletion does not affect repair, but relieves the inhibition of repair by the PARP inhibitor Iniparib. (A) The level of PARP1 expression in control cells (scramble) or PARP1 knockdown LCL35 cells (PARP1KD) was determined by RT-PCR and Western blot analysis, as described in Materials and Methods. (B) Repair of SSBs and DSBs in control and PARP1KD cells following a dose of 100 Gy. The effect of PARP inhibitors, (C) Olaparib (20 μM) or (D) Iniparib (100 μM), on repair of SSBs and DSBSs in control and PARP1KD cells following a dose of 100 Gy. Cells were enriched for G2 by nocodazole. Following incubation, cells were collected and processed for PFGE and Southern blot analysis. (E) Description of how the different effects between Olaparib and Iniparib might arise. Olaparib inhibits the catalytic domain of PARPs and could trap a dysfunctional enzyme at the breaks, whereas Iniparib might prevent PARP1 binding but not other PARPs.
PARP1 Is Not Required for Repair of IR-Induced SSBs or DSBs.
To address further the possible role of PARP1 in DNA repair of IR-induced breaks, we created LCL35 cell lines expressing shRNA complementary to PARP1 (PARP1KD). The PARP1 mRNA levels were decreased by more than 80% compared with control cells expressing scrambled shRNA, and there was no detectable protein (Fig. 4A). As shown in Fig. 4B, PARP1 is not required to repair IR-induced SSBs or DSBs. Repair of both types of breaks following a dose of 100 Gy was comparable between the PARP1 knockdown and the PARP1-competent control cells.
The dramatic difference between PARP inhibition by some PARP inhibitors and PARP1 knockdown suggests that PARP1 protein per se is not required for SSB repair. Possibly it forms blocked repair intermediates or affects other repair-associated targets. To address the mechanism of PARP inhibition of SSB repair inhibition, we examined the effects of 4-AN, Olaparib, and Iniparib on repair in PARP1 knockdown cells. Control (scramble) and PARP1KD cells, enriched in G2 by nocodazole, were treated with 100 Gy and repair was determined. The knockdown of PARP1 did not prevent Olaparib inhibition, indicating that Olaparib may target other proteins involved in SSB repair, such as PARP2. Previous results with base-damaging agents suggested that inhibitors might trap PARP1 at DNA breaks, thereby uncoupling base excision repair (39). This hypothesis was tested using Iniparib. Unlike 4-AN and Olaparib, which mimic nicotinamide and compete for the catalytic domain of PARP1 and PARP2 (and possibly others), Iniparib is proposed to disrupt the interaction between PARP1 and DNA (40, 41) based on in vitro results. Iniparib inhibited SSB repair and the inhibition could be reversed by knockdown of PARP1 (Fig. 4D), suggesting that inhibition requires interaction with PARP1. Possibly Iniparib alters an interaction between PARP1 and repair components or even traps PARP1 at damage sites within cells, contrary to in vitro findings (Fig. 4E).
Repair of IR-Induced Breaks Is Not Affected by p53 or the Cell Cycle.
This study used two cell lines that contain EBV but differ in functional status of the tumor suppressor p53: Raji cells are mutated for p53 (Arg-213 and Thr-234), whereas LCL35 cells retain wild-type p53 function. Besides modulation of cell-cycle arrest in response to DNA damage, it has been suggested that p53 may play a direct role in at least some repair processes, including nucleotide excision repair (42, 43), base excision repair (44), and recombination repair (45). Here we tested whether p53 status would affect repair of SSBs and DSBs induced by IR. The Raji and LCL35 cells were treated with 100 Gy and subsequently with PARP inhibitors. As shown in Fig. 3C, repair of SSBs and DSBs was similar in both cell lines, suggesting that p53 was not required for the efficient repair of IR-induced breaks. Furthermore, the strong inhibition of SSB repair by the PARP inhibitor 4-AN was not related to p53 status at early times (Fig. 3C); however, there appeared to be reduced 4-AN inhibition in p53+ cells at later times.
We also compared repair of IR-induced DSBs and SSBs in G1 serum-deprived cells with the nocodazole-enriched G2 cells. There was no apparent difference in break repair in Raji cells (Fig. S3).
Discussion
The ability to detect three forms of EBV—linear, supercoiled, and relaxed—has led to the development of a sensitive system for direct and quantitative assessment of induction and repair of DSBs and SSBs that are randomly induced by IR. Because EBV has many chromosomal properties including chromatin organization, S phase-dependent replication, and much larger DNA size compared with plasmid-based assays, we propose that the EBV break assay is representative of damage and repair events in chromosomes. The precise measurements of breaks and repair are consistent with chromosomal results based on more indirect methods with chromosomal material including comet and gross PFGE measurements of fast-moving chromosomal materials (FAR analysis) (46).
The ability to simultaneously monitor SSBs and DSBs makes the EBV break assay a unique system for addressing in vivo repair. Supercoiled DNA of SV40 virus (which is much smaller, ∼5 kb) had previously been used to address SSBs and DSBs induced in vivo by γ radiation (47, 48). Cells containing recently transfected virus were irradiated with high doses (1,000 and 2,000 Gy); however, there was no repair. The levels of DSBs were similar to those we obtained with EBV minichromosomes as described here, and the ratio of 20 for SSBs:DSBs was within the range we describe for EBV. Some reports had indicated a higher ratio, but that might be due to methods that used high-temperature or alkaline conditions, which could lead to SSBs at sites of base damage.
We suggest that the SSB:DSB ratio can be used as an indicator of the incidence of clustered damage, where single events might give rise to localized, closely opposed SSBs. Based on our previous findings with MMS showing an SSB:DSB ratio in the range of ∼1,000 (17), the SSB:DSB ratio for IR-induced breaks cannot be attributed to random SSBs. Clustered DNA lesions are considered to be more destabilizing to the genome and require more complex repair systems than isolated single lesions (49–51).
The EBV break assay provides a direct measure of the ability of drugs/chemicals/environmental exposures along with genetic factors to affect the generation and repair of SSBs and DSBs. This could be extended to the impact of chromatin, compaction, and radiation quality on SSB and DSB induction as well as repair. Because the replication of individual EBV molecules is short relative to the genome in S phase, it should also be possible to address EBV changes during this phase. Based on results with circular chromosomes in yeast (52, 53), genetic factors that influence conversion of SSBs to DSBs can also be assessed. We found efficient repair of both IR-induced SSBs and DSBs at a dose of 100 Gy. The rate of DSB repair is roughly half that of SSB repair, although during the first 30 min they appear comparable.
Using the EBV break system, we were able to assess directly how p53 participates in repair of IR-induced breaks. Although there have been numerous studies suggesting p53 involvement in DNA repair, including interactions with RAD51 as well as the Bloom's (BLM) helicase at stalled replication forks (45, 54, 55), the studies typically rely on indirect analyses such as fluorescence-based detection of foci formation. Our direct measurements of DNA break formation and repair in p53 wild-type and p53 mutated cells demonstrate that p53 is not directly involved in repair of either SSBs or DSBs in the time frame investigated, although we do not exclude an effect on repair of residual breaks. This is not surprising, because initiation of DNA strand break repair, especially SSBs, is fairly quick, whereas up-regulation of genes targeted by p53 peaks at around 10 h following DNA damage and the stabilization of p53 itself is not seen until 30–60 min after treatment. However, we do not exclude the possibility that the mutated form of p53 in Raji cells might interact with repair proteins.
The EBV break system has also provided the opportunity to address the role of PARP and PARP inhibitors in repair of IR-induced breaks, which is relevant to chemotherapy because radiation is often included in treatment regimens. We did not detect an essential role for PARP1 in the repair of SSBs or DSBs, which is consistent with other studies of SSBs produced by other agents (39, 56). In addition, PARP1 is not required for recruitment of the major base excision repair proteins to sites of DNA damage (57). Depletion of PARP1 in cell extracts can even result in more rapid binding of repair proteins to DNA substrates (58). Therefore, if PARPs play a role in break repair, possibly other PARPs can substitute for PARP1, or the role of PARP1 might be greater during replication, which is not addressed here.
There was clearly an SSB-specific impact of the PARP inhibitors 4-AN, Olaparib, and Iniparib. These results are consistent with clinical trials for cancer treatment showing the effectiveness of Olaparib and Iniparib (21, 22, 59). Surprisingly, many inhibitors that can efficiently inhibit the catalytic activity of PARP (such as NU1025 and PJ32) failed to show an evident inhibition of SSB repair, raising the question of what property of the PARP inhibitors is relevant to in vivo inhibition of repair. Although PARP inhibition is considered to lead to DSB accumulation, which accounts for the lethal effect of PARP inhibitors in HR-deficient cells (2, 3, 60), none of the inhibitors resulted in elevated DSB levels or showed any apparent delay in the repair kinetics of IR-induced DSBs in G2 cells, suggesting SSBs are not converted into DSBs. However, SSBs might be converted to DSBs during replication (61–63), which would explain a requirement for BRCA1/2 and recombinational repair to prevent lethality that arises from PARP inhibitors.
The dramatic difference in SSB repair between PARP inhibition by some inhibitors and PARP1 knockdown suggests generation of blocked intermediates or inhibition of repair components. Because PARP1 molecules are quickly recruited to SSBs before other repair proteins (64), chemical inhibitors bound at the catalytic domain might trap the nonfunctional PARP1, preventing other repair proteins from being recruited. However, trapping probably does not account for PARP1-dependent inhibition of SSB repair by Iniparib, because it prevents PARP1 from binding to DNA (40, 41). Possibly the inhibitor leads to adverse interactions of PARP1 with other repair components that do not depend upon binding of PARP1 at SSBs.
In conclusion, the EBV break system provides a physical method to simultaneously monitor directly the in vivo formation and repair of both SSBs and DSBs in human cells. Using this system, we confirmed the dramatic difference between PARP inhibition by some PARP inhibitors and PARP1 knockdown as well as the inhibitory capacity of various widely used PARP inhibitors on DNA repair of radiation-induced breaks, providing insights into the mechanism of PARP inhibition on DNA repair. Beyond addressing many questions about break repair in human cells, the system is well-suited for use in studies that address broader issues of repair of a variety of DNA-damaging agents as well as applications in drug development and analysis.
Materials and Methods
Cell Lines, Plasmids, and Chemicals.
The EBV-containing Raji cell line derived from Burkitt’s lymphoma was obtained from the American Type Culture Collection (CCL-86); EBV-immortalized lymphoblastoid cell line LCL35 was from Micah Luftig (Duke University, Durham, NC). Depletion of PARP1 expression (PARP1KD) in LCL35 cells was effected using stably integrated PARP1 shRNA with MISSION shRNA lentiviral plasmids from Sigma-Aldrich. More information about cell culture, shRNA transfection, and chemicals is given in SI Materials and Methods.
Gene Expression by RT-PCR and Western Blot Analysis.
To determine PARP1 mRNA and protein levels in PARP1KD or control scramble-shRNA transfected LCL35 cells, real-time PCR and Western blot analysis were conducted as described in SI Materials and Methods.
Ionizing Radiation, Drug Treatment, and PFGE Analysis.
Cells kept on ice were irradiated in a 137Cs irradiator (J. L. Shepherd model 431, at a dose rate of 2.3 krad/min), and then treated with or without PARP/DNA-PK inhibitors for incubation repair. The collection of cells and preparation of agarose-DNA plugs for PFGE analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Norman Sharpless, Dr. Julie Horton, Dr. Michelle Heacock, Dr. Kin Chan, and Jim Westmoreland for critical reading of the manuscript, and Dr. Charles Romeo and Dr. Negin Martin at the National Institute of Environmental Health Sciences (NIEHS) Viral Vector Core Laboratory for generating LCL35 shRNA cell lines. This work was supported by the Intramural Research Program of the NIEHS (National Institutes of Health, Department of Health and Human Services) under Project 1 Z01 ES065073 (to M.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118078109/-/DCSupplemental.
References
- 1.Krishnakumar R, Kraus WL. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 3.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 5.Vilar E, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71:2632–2642. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weston VJ, et al. The PARP inhibitor Olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 7.Williamson CT, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Murcia JM, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 10.Wang ZQ, et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–369. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303(5654):92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 13.Collins AR. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 14.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 15.Collins AR, et al. The comet assay: Topical issues. Mutagenesis. 2008;23(3):143–151. doi: 10.1093/mutage/gem051. [DOI] [PubMed] [Google Scholar]
- 16.McKenna DJ, McKeown SR, McKelvey-Martin VJ. Potential use of the comet assay in the clinical management of cancer. Mutagenesis. 2008;23(3):183–190. doi: 10.1093/mutage/gem054. [DOI] [PubMed] [Google Scholar]
- 17.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson PJ, Farrell PJ. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 19.Shaw JE, Levinger LF, Carter CW., Jr Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates JL, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 22.O’Shaughnessy J, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 23.Game JC, Sitney KC, Cook VE, Mortimer RK. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westmoreland J, et al. RAD50 is required for efficient initiation of resection and recombinational repair at random, γ-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma W, Westmoreland J, Nakai W, Malkova A, Resnick MA. Characterizing resection at random and unique chromosome double-strand breaks and telomere ends. Methods Mol Biol. 2011;745(Pt 1):15–31. doi: 10.1007/978-1-61779-129-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson PG, Beerman TA. Damage induced in episomal EBV DNA in Raji cells by antitumor drugs as measured by pulsed field gel electrophoresis. Anal Biochem. 1994;220(1):103–114. doi: 10.1006/abio.1994.1305. [DOI] [PubMed] [Google Scholar]
- 27.Olive PL. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat Res. 1998;150(Suppl 5):S42–S51. [PubMed] [Google Scholar]
- 28.Banáth JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 29.Purschke M, Kasten-Pisula U, Brammer I, Dikomey E. Human and rodent cell lines showing no differences in the induction but differing in the repair kinetics of radiation-induced DNA base damage. Int J Radiat Biol. 2004;80(1):29–38. doi: 10.1080/09553000310001642885. [DOI] [PubMed] [Google Scholar]
- 30.Yokoya A, Cunniffe SM, O’Neill P. Effect of hydration on the induction of strand breaks and base lesions in plasmid DNA films by γ-radiation. J Am Chem Soc. 2002;124:8859–8866. doi: 10.1021/ja025744m. [DOI] [PubMed] [Google Scholar]
- 31.Shikazono N, Yokoya A, Urushibara A, Noguchi M, Fujii K. A model for analysis of the yield and the level of clustering of radiation-induced DNA-strand breaks in hydrated plasmids. Radiat Prot Dosimetry. 2011;143(2–4):181–185. doi: 10.1093/rpd/ncq528. [DOI] [PubMed] [Google Scholar]
- 32.Taucher-Scholz G, Kraft G. Influence of radiation quality on the yield of DNA strand breaks in SV40 DNA irradiated in solution. Radiat Res. 1999;151:595–604. [PubMed] [Google Scholar]
- 33.Blaisdell JO, Harrison L, Wallace SS. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat Prot Dosimetry. 2001;97(1):25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 34.Caldecott KW. Mammalian single-strand break repair: Mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor Olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 37.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor Olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 38.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 39.Ström CE, et al. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellisen LW. PARP inhibitors in cancer therapy: Promise, progress, and puzzles. Cancer Cell. 2011;19(2):165–167. doi: 10.1016/j.ccr.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 42.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 43.Smith ML, et al. p53-mediated DNA repair responses to UV radiation: Studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Offer H, et al. Direct involvement of p53 in the base excision repair pathway of the DNA repair machinery. FEBS Lett. 1999;450(3):197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 45.Akyüz N, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iliakis GE, Cicilioni O, Metzger L. Measurement of DNA double-strand breaks in CHO cells at various stages of the cell cycle using pulsed field gel electrophoresis: Calibration by means of 125I decay. Int J Radiat Biol. 1991;59:343–357. doi: 10.1080/09553009114550321. [DOI] [PubMed] [Google Scholar]
- 47.Krisch RE, Flick MB. Further studies of the induction and intracellular repair of DNA strand breaks using intranuclear SV40 as a test system. Radiat Res. 1988;116:462–471. [PubMed] [Google Scholar]
- 48.Krisch RE, Flick MB, Trumbore CN. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat Res. 1991;126:251–259. [PubMed] [Google Scholar]
- 49.Tounekti O, Kenani A, Foray N, Orlowski S, Mir LM. The ratio of single- to double-strand DNA breaks and their absolute values determine cell death pathway. Br J Cancer. 2001;84:1272–1279. doi: 10.1054/bjoc.2001.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shikazono N, O’Neill P. Biological consequences of potential repair intermediates of clustered base damage site in Escherichia coli. Mutat Res. 2009;669(1–2):162–168. doi: 10.1016/j.mrfmmm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat Res. 2011;711(1–2):123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma W, et al. The transition of closely opposed lesions to double-strand breaks during long-patch base excision repair is prevented by the coordinated action of DNA polymerase δ and Rad27/Fen1. Mol Cell Biol. 2009;29:1212–1221. doi: 10.1128/MCB.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma W, Westmoreland JW, Gordenin DA, Resnick MA. Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease. PLoS Genet. 2011;7:e1002059. doi: 10.1371/journal.pgen.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sengupta S, et al. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Süsse S, Janz C, Janus F, Deppert W, Wiesmüller L. Role of heteroduplex joints in the functional interactions between human Rad51 and wild-type p53. Oncogene. 2000;19:4500–4512. doi: 10.1038/sj.onc.1203809. [DOI] [PubMed] [Google Scholar]
- 56.Godon C, et al. PARP inhibition versus PARP-1 silencing: Different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodhouse BC, Dianova II, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair (Amst) 2008;7:932–940. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 58.Parsons JL, Dianova II, Allinson SL, Dianov GL. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J. 2005;272:2012–2021. doi: 10.1111/j.1742-4658.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 59.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bryant HE, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton JK, Stefanick DF, Zeng JY, Carrozza MJ, Wilson SH. Requirement for NBS1 in the S phase checkpoint response to DNA methylation combined with PARP inhibition. DNA Repair (Amst) 2011;10:225–234. doi: 10.1016/j.dnarep.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heacock ML, Stefanick DF, Horton JK, Wilson SH. Alkylation DNA damage in combination with PARP inhibition results in formation of S-phase-dependent double-strand breaks. DNA Repair (Amst) 2010;9:929–936. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.