Background: Abasic sites are the most frequent DNA lesions and are often bypassed by incorporating an adenosine opposite that lesion.
Results: We determined structures of DNA polymerase in complex with different nucleotides opposite an abasic site.
Conclusion: Interaction of the incoming nucleotide with a single amino acid governs nucleotide selection opposite abasic sites.
Significance: This work furthers the understanding of the bypass of a mutagenic lesion by DNA polymerases.
Keywords: DNA Damage, DNA Enzymes, DNA Polymerase, DNA Repair, X-ray Crystallography, Abasic Sites
Abstract
Cleavage of the N-glycosidic bond that connects the nucleobase to the backbone in DNA leads to abasic sites, the most frequent lesion under physiological conditions. Several DNA polymerases preferentially incorporate an A opposite this lesion, a phenomenon termed “A-rule.” Accordingly, KlenTaq, the large fragment of Thermus aquaticus DNA polymerase I, incorporates a nucleotide opposite an abasic site with efficiencies of A > G > T > C. Here we provide structural insights into constraints of the active site during nucleotide selection opposite an abasic site. It appears that these confines govern the nucleotide selection mainly by interaction of the incoming nucleotide with Tyr-671. Depending on the nucleobase, the nucleotides are differently positioned opposite Tyr-671 resulting in different alignments of the functional groups that are required for bond formation. The distances between the α-phosphate and the 3′-primer terminus increases in the order A < G < T, which follows the order of incorporation efficiency. Additionally, a binary KlenTaq structure bound to DNA containing an abasic site indicates that binding of the nucleotide triggers a remarkable rearrangement of enzyme and DNA template. The ability to resolve the stacking arrangement might be dependent on the intrinsic properties of the respective nucleotide contributing to nucleotide selection. Furthermore, we studied the incorporation of a non-natural nucleotide opposite an abasic site. The nucleotide was often used in studying stacking effects in DNA polymerization. Here, no interaction with Tyr-761 as found for the natural nucleotides is observed, indicating a different reaction path for this non-natural nucleotide.
Introduction
The most common DNA damage under physiological conditions consists of abasic sites resulting from spontaneous hydrolysis of the N-glycosidic bond between the sugar moiety and the nucleobase in DNA (1). Because the genetic information is lost by the cleavage of the nucleobase, abasic sites bear a high mutagenic potential (2–4). In most cases, the lesion is removed by DNA repair systems using the sister strand to guide for incorporation of the right nucleotide. However, undetected lesions, or those formed during S phase, pose a challenge to DNA polymerases and block replication (5, 6). Several studies indicated the mutagenic potential of these lesions in translesion synthesis, which is more pronounced in animal as compared with bacterial cells presumably due to higher translesion synthesis in eukaryotes (4, 7, 8). Interestingly, it has been shown that in human cells, an adenosine (A) is preferentially incorporated opposite abasic sites (4). Further in vitro and in vivo studies of DNA polymerases from family A (including human DNA polymerases γ and θ) and B (including human DNA polymerases α, ϵ, and δ) in the presence of the stabilized tetrahydrofuran abasic site analog F (supplemental Fig. S1) have shown that purines, in particular adenosine, and to a lesser extent guanosine, are most frequently incorporated opposite the lesion. The strong preference for adenosine has been termed “A-rule” (7–20).
A set of studies concerning the behavior of DNA polymerases from different sequence families showed that there are multiple mechanisms to overcome an abasic site. Most translesion DNA polymerases follow various loop-out mechanisms (11, 21–25). Thereby, the nucleotide selection is influenced by the following upstream templating bases resulting in deletions and complex mutation spectra. Recently, an amino acid templating mechanism was found for the “error-free” bypass of an abasic site by the yeast Rev1 DNA polymerase member of family Y (26). Because guanine is cleaved most frequently (2), the preference of Rev1 for dCMP incorporation opposite an abasic site represents the “best guess.”
However, the determinants of the A-rule are still controversially discussed. Structural and functional studies have added significantly to our understanding of the basic mechanisms of translesion synthesis by DNA polymerases (27, 28). Superior stacking as well as solvation properties of adenine have been discussed as the driving force behind adenine selection (17, 29–31). However, previously reported structures of KlenTaq, the large fragment of Thermus aquaticus (Taq) DNA polymerase I, which is a member of the sequence family A, suggests that this enzyme follows the A-rule by applying an amino acid templating mechanism (19, 20). Thereby, interaction with Tyr-671 seems to be the basis of the preference of purines. It facilitates nucleotide incorporation by mimicking a pyrimidine nucleobase directing for purine incorporation opposite abasic sites due to the enhanced geometric fit to the active site (32, 33). The crucial role of this tyrosine in translesion synthesis, which is highly conserved throughout evolution in DNA polymerase family A from bacteria to humans (34), was further probed by analysis of site-directed mutations (19). Interestingly, when the six-membered ring of tyrosine was mutated to the bicyclic indole of tryptophan, the A-rule was shifted to a “C/T-rule,” further supporting geometric factors as determinants of nucleotide selection opposite an abasic site.
Still, several aspects remain to be clarified to fully understand the mechanism of nucleotide selection opposite an abasic site by DNA polymerases from the family A. These aspects include the overall conformation of the enzyme-DNA complex in the binary state prior to nucleotide binding. Furthermore, the order of nucleotide incorporation efficiency opposite abasic sites by DNA polymerases such as KlenTaq is A, G, T, and C. The mechanistic understanding for this observation is sparse due to the lack of structural data. Here, we report a structure of a binary complex of KlenTaq DNA polymerase bound to the primer/template bearing an abasic site lesion. Additionally, we present several ternary structures of KlenTaq caught incorporating different nucleotides opposite an abasic site, providing insights into the preference order of nucleotide incorporation efficiency opposite this lesion.
EXPERIMENTAL PROCEDURES
Protein and Oligonucleotides
Protein expression and purification were conducted as described (35). In brief, an Escherichia coli codon-optimized KlenTaq gene (amino acids 293–832 of Taq gene; purchased from GeneArt) was cloned into a pET-21b vector without any purification tags and expressed in E. coli strain BL21 (DE3). Of note, codon optimization resulted in changes within the gene sequence without affecting the amino acid sequence. After heat denaturation and ultracentrifugation, a PEI precipitation was performed. The resulting material was purified by anion exchange (Q Sepharose) chromatography followed by size-exclusion chromatography (Superdex 75) in 20 mm Tris HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm β-mercaptoethanol.
Oligonucleotides were purchased from Metabion or Thermo Scientific. The dideoxy cytosine modified primer was synthesized on an Applied Biosystems 392 DNA/RNA synthesizer using 5′-dimethoxytrityl-2′,3′-dideoxy cytosine modified N-succinoyl-long chain alkylamino controlled pore glass, which was purchased from Glen Research. The synthesized oligonucleotide was purified by preparative PAGE on a 12% polyacrylamide gel containing 8 m urea (DMT-OFF). The nucleotide dNITP3 was synthesized starting from dNI purchased from Berry & Associates Inc., according to published procedures (36).
Crystallization and Structure Determination
KlenTaq (buffer: 20 mm Tris HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm β-mercaptoethanol) was incubated in the presence of DNA primer (5′-d(GAC CAC GGC GC)-3′), an abasic site F containing template (5′-d(AAA FNG CGC CGT GGT C)-3′); N represents the templating base directing the processing of ddGTP, ddTTP, or ddCTP.
KlenTaqF-G-I
The crystallization was set up using purified KlenTaq (11 mg/ml), DNA template/primer duplex, and ddGTP in a molar ratio of 1:3:50 and in the presence of 20 mm MgCl2. The crystallization solution was mixed in a 1:1 ratio with the reservoir solution containing 0.05 m sodium cacodylate (pH 6.5), 0.2 m NH4OAc, 0.01 m Mg(OAc)2, and 25% PEG 8000.
KlenTaqF-G-II
The crystallization was set up as for KlenTaqF-G-I except using KlenTaq (11 mg/ml):DNA template/primer duplex:ddGTP = 1:3:50 in the presence of 20 mm MgCl2. The reservoir solution contained 0.05 m sodium cacodylate (pH 6.5), 0.2 m NH4OAc, 0.01 m Mg(OAc)2, and 28% PEG 8000.
KlenTaqF-T
The crystallization was set up as for KlenTaqF-G-I except using KlenTaq (11 mg/ml):DNA template/primer duplex:ddTTP = 1:3:50 in the presence of 20 mm MgCl2. The reservoir solution contained 0.05 m sodium cacodylate (pH 6.5), 0.2 m NH4OAc, 0.01 m Mg(OAc)2, and 28% PEG 8000.
KlenTaqF-binary-II
The crystallization was set up as for KlenTaqF-G-I except using KlenTaq (11 mg/ml):DNA template/primer duplex:ddCTP = 1:1.5:60 in the presence of 20 mm MgCl2. The reservoir solution contained 0.1 m Tris HCl (pH 8.0), 0.2 m Mg(formiate)2, and 15% PEG 8000.
KlenTaqF-binary
The crystallization was set up using purified KlenTaq (8 mg/ml), a template (5′-d(AAA FGG CGC CGT GGT C)-3′), and a previously synthesized primer with a dideoxy cytosine at its 3′-end, in a molar ratio of 1:1.4 and in the presence of 20 mm MgCl2. The crystallization solution was mixed in 1:1 ratio with the reservoir solution containing 0.1 m Tris HCl (pH 7.5), 0.2 m Mg(formiate)2, and 12% PEG 8000.
KlenTaqF-NI
The crystallization was set up as for KlenTaqF-binary except using KlenTaq (8 mg/ml):DNA template/primer duplex = 1:1.4 and in the presence of 20 mm MgCl2. The reservoir solution contained 0.1 m Tris HCl (pH 7.0), 0.2 m Mg(formiate)2, and 18% PEG 8000. The crystals were soaked overnight by adding 0.5 μl of dNITP (20 mm) solution. The occupancy of the bound dNITP was refined to 0.77.
Crystals were produced by the hanging drop vapor diffusion method by equilibrating against 1 ml of the reservoir solution for 5 days at 18 °C. The crystals were frozen in liquid nitrogen. Datasets were collected at beamline PXI (X06SA) of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland), at a wavelength of 1.000 or 0.900 Å using a PILATUS 6M detector. Data reduction was performed with the XDS package (37, 38). The structures were solved by difference Fourier techniques using KlenTaq wild type (Protein Data Bank (PDB) 3LWL) as model. Refinement was performed with PHENIX (39), and model rebuilding was done with COOT (40). Figures were made with PyMOL (41).
Primer Extension Assays
Primer extension was performed as described (19). In brief, for incorporation opposite F, 20 μl of the KlenTaq reactions contained 100 nm primer (5′-d(CGT TGG TCC TGA AGG AGG ATA GG)-3′), 130 nm template (5′-d(AAA TCA FCC TAT CCT CCT TCA GGA CCA ACG TAC)-3′), 100 μm dNTPs in buffer (20 mm Tris HCl, pH 7.5, 50 mm NaCl, and 2 mm MgCl2), and 500 nm of the respective KlenTaq polymerase. Reaction mixtures were incubated at 37 °C. Incubation times are provided in the respective figure legends. Primer was labeled using [γ-32P]ATP according to standard techniques. Reactions were stopped by the addition of 45 μl of stop solution (80% (v/v) formamide, 20 mm EDTA, 0.25% (w/v) bromphenol blue, 0.25% (w/v) xylene cyanol) and analyzed by 12% denaturing PAGE. Visualization was performed by phosphorimaging.
Enzyme Kinetics
The rates of single turnovers in pre-steady-state kinetics were determined as described (19). In brief, 15 μl of radiolabeled primer/template complex (200 nm) and DNA polymerase (2 mm) in reaction buffer (see primer extension assay) were rapidly mixed with 15 μl of a dNTP solution in reaction buffer at 37 °C. Quenching was achieved by adding previously described stop solution. For reaction times longer than 5 s, a manual quench was performed. The analysis of dNTP incorporation opposite to the abasic site primer (for sequences, see “Primer Extension Assays”) and templates (for sequences, see “Primer Extension Assays”) were applied. Quenched samples were analyzed on a 12% denaturing PAGE followed by phosphorimaging. For kinetic analysis, experimental data were fit by nonlinear regression using the program GraphPad Prism 4. The data were fit to a single exponential equation: (conversion) = A × (1 − exp(−kobs t)). The observed catalytic rates (kobs) were then plotted against the dNTP concentrations used, and the data were fit to a hyperbolic equation to determine the Kd of the incoming nucleotide. The incorporation efficiency is given by kpol/Kd.
RESULTS
Overall Structure of KlenTaq Complexes in Presence of an Abasic Site
All KlenTaq crystals grew in the same space group and very similar cell parameters (supplemental Table S1) as those reported earlier (19, 20, 35, 42–45). Thus, significant differences in crystal packing forces acting on the active site should be negligible. Nevertheless, parts of the finger domain differ in their mobility (as measured by temperature factors) between the different structures. This is a consequence of the structural differences, which were induced in the active site assembly by the primer/template complexes and respective nucleotides that were investigated. In the following, the active sites for the various complexes are shown and described in detail.
Structure of KlenTaq in Binary Complex with DNA Duplex Containing an Abasic Site (KlenTaqF-binary)
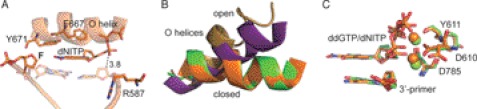
To address the issue, if the overall conformation of the enzyme-DNA complex is affected by an abasic site prior to nucleotide binding, we crystallized KlenTaq bound to primer/template duplex containing an abasic site following a similar strategy as reported recently (19, 20, 35, 43, 46). The structure was solved by difference Fourier techniques at a resolution of 1.9 Å (supplemental Table S1). KlenTaqF-binary is very similar to one reported for KlenTaq in a binary complex bound to undamaged DNA duplex (PDB 4KTQ; KlenTaqbinary) (35) as reflected by an r.m.s.d. for Cα atoms of 0.60 Å (Fig. 1). However, remarkable structural changes were observed for the single-stranded 5′-overhang of the DNA template, which is rotated around the helical axis. Thereby, the lesion is flipped out of a developing DNA duplex (Fig. 1A). This conformation is stabilized by stacking of the 5′-upstream nucleobases of the template strand on the top of the primer/template duplex (Fig. 1, B and C) and a distinct hydrogen-bonding network of amino acid residues Glu-615 and Tyr-671 with the DNA template (Fig. 1D). In contrast, in KlenTaqbinary, the backbone rotates 5′ to the DNA duplex, redirecting the remainder of the single-stranded template out of the DNA polymerase active site (Fig. 1E).
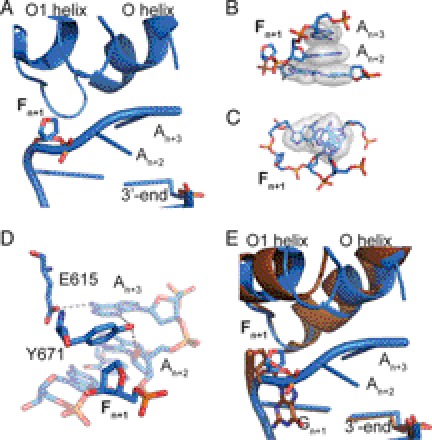
FIGURE 1.
Structure of KlenTaqF-binary (blue). A, template stacking assembly of An+2 and An+3 in KlenTaqF-binary. The abasic site analog Fn+1 is located extrahelically. B, the stick and surface depiction highlights the template stacking arrangement. C, top view of the primer/template stacking arrangement. D, hydrogen-bonding network of the amino acid side chains Tyr-671 and Glu-615 with the template strand. E, same arrangement as in A for the superimposed structures of KlenTaqF-binary (blue) and KlenTaqbinary (brown).
KlenTaq with ddGTP Opposite an Abasic Site (KlenTaqF-G-I, KlenTaqF-G-II)
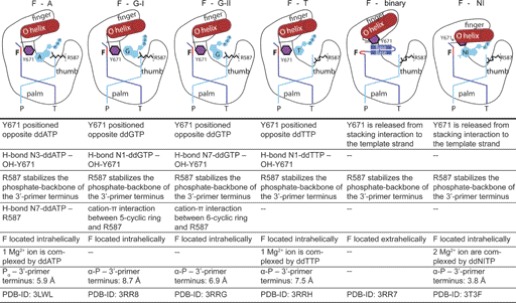
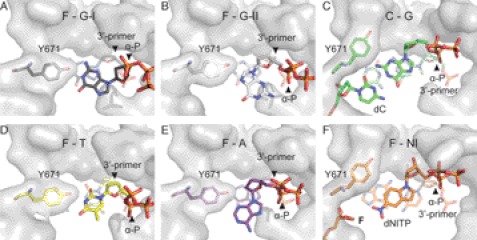
To gain insights into the structural basis for the preference of adenosine over guanosine of KlenTaq in bypassing an abasic site, we crystallized KlenTaq as a ternary complex bound to a primer/template duplex and ddGTP opposite the abasic site. Two structures were reproducibly found in several crystallization trials and solved by difference Fourier techniques at resolutions of 2.3–2.4 Å (KlenTaqF-G-I and KlenTaqF-G-II) (supplemental Table S1). The obtained structures show conformational heterogeneity in the active site region. In particular, we found two different orientations of the incoming ddGTP (Fig. 2, A and B, and supplemental Fig. S2, A and B). However, the overall conformation of the two ddGTP-trapped structures is very similar to the ternary complex of KlenTaq harboring ddATP opposite an abasic site (PDB 3LWL; KlenTaqF-A; r.m.s.d. for Cα of 0.42 Å) (19), showing the same remarkable structural changes as compared with the undamaged case (KlenTaq bound to an undamaged primer/template duplex processing a ddGTP; PDB 1QSS; KlenTaqC-G) (42). Thus, the conformation of the O helix leaves the active site more open, similar to the one in KlenTaqF-A, and somehow between the open (PDB 2KTQ; KlenTaqopen) (35) and closed (KlenTaqC-G) conformations in the reported ternary complexes (Fig. 2E). Like in KlenTaqF-A, we found that the abasic site is intrahelically located and that Tyr-671 is positioned opposite the incoming ddGTP at the place that is usually occupied by the templating nucleobase (Fig. 2A). However, remarkable differences between KlenTaqF-A and KlenTaqF-G-I appear in the positioning and interaction pattern of the incoming triphosphates (Fig. 2A and supplemental Fig. S3, A and B). In KlenTaqF-G-I, Tyr-671 stabilizes ddGTP via hydrogen bonds between the Tyr-671 hydroxyl group and the N1 of guanine, whereas in KlenTaqF-A, the N3 of adenine interacts with Tyr-671 (Fig. 2A and supplemental Fig. S3A). Furthermore, an important stabilizing factor of the incoming nucleotide is Arg-587. In KlenTaqF-A, Arg-587 forms a hydrogen bond to the N7 of adenine (supplemental Fig. S3A). However, in KlenTaqF-G-I, ddGTP is stabilized by cation-π interaction with Arg-587. Because this type of interaction strongly depends on the distribution of the electron density in the aromatic ring system, there is in the case of purines a clear preference for arginine to position itself below or above the imidazole five-atom ring system (47). Indeed, this is observed in KlenTaqF-G-I for Arg-587 (Fig. 2, A and C). The binding arrangement of ddGTP causes a misalignment of the α-phosphate, resulting in a large distance of the α-phosphate to the 3′-primer terminus (8.7 Å). Hence, KlenTaqF-G-I might represent an initial binding event of the incoming triphosphate (Fig. 2A). In comparison, the guanine base in KlenTaqF-G-II is rotated around the N-glycosidic bond, and the N7 of guanine points to the hydroxyl group of Tyr-671 (Fig. 2, B and D, and supplemental Fig. S2B). Thereby, the sugar moiety is realigned, resulting in a shortened distance between the α-phosphate and 3′-primer terminus of about 6.9 Å. However, the reorientation of the nucleobase positions the six-membered heterocycle above Arg-587 (Fig. 2, B and D), a less favored arrangement observed only in rare cases (47). In summary, the distance between 3′-OH and the α-phosphate increases in the following direction KlenTaqF-A < KlenTaqF-G-II < KlenTaqF-G-I that are all longer than the one observed in the canonical case (Fig. 2G).
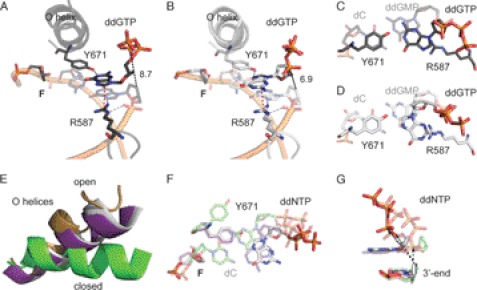
FIGURE 2.
Structure of KlenTaqF-G-I (black) and KlenTaqF-G-II (gray). A, stabilization network of ddGTP in KlenTaqF-G-I. The amino acid side chains Arg-587 and Tyr-671 are labeled. Gray, red, and black dashed lines indicate hydrogen-bonding interactions, cation-π interaction, and distance (Å), respectively. B, the same as in A for KlenTaqF-G-II. C, top view of the nascent base pair opposite F (KlenTaqF-G-I). In the front, the incoming ddGTP opposite Tyr-671 is depicted. The first nucleobase pair of the primer/template terminus is shown as transparent. D, same view as in C for KlenTaqF-G-II. E, the O helices from the superimposition of KlenTaqF-G-II (gray), KlenTaqF-A (purple), KlenTaqC-G (green), and KlenTaqopen (gold) are highlighted. F, comparison of the nascent base pairs of KlenTaqF-G-II (gray), KlenTaqF-A (purple), and KlenTaqC-G (green). G, the incoming ddNTPs of KlenTaqF-G-II (gray), KlenTaqF-A (purple), and KlenTaqC-G (green) and the respective 3′-primer terminus are shown. The arrow indicates the displacement of the α-phosphate regarding to the 3′-primer terminus.
Pyrimidines Opposite an Abasic Site
Pyrimidine nucleotides are incorporated opposite an abasic site by KlenTaq with poor efficiencies (19). Furthermore, we demonstrated that the purine preference could be switched to pyrimidine preference by a single site-directed mutagenesis of Tyr-671 to Trp (19). This suggests that the low incorporation efficiencies of pyrimidines relies on a specific interaction with this amino acid residue. To get structural insights, we crystallized KlenTaq in the presence of a primer/template duplex and ddTTP. The structure was solved by difference Fourier techniques at a resolution of 1.8 Å (KlenTaqF-T) (supplemental Table S1). Although kinetic studies elucidated that thymidine incorporation opposite an abasic site is unfavorable, we observed a bound thymidine triphosphate in the active site (Fig. 3A and supplemental Fig. S2C). Once more we obtained a semiclosed enzyme conformation as can be seen from the superimposition of this structure with KlenTaqA-T (PDB 1QTM; processing a canonical base pair with an incoming ddTTP) (42), KlenTaqF-A, and KlenTaqopen (Fig. 3B). The O helix does not pack against the nascent base pair, allowing Tyr-671 to position itself between the incoming ddTTP and the abasic site as is observed for Tyr-671 in KlenTaqF-A (Fig. 3A and supplemental Fig. S3A). The ddTTP is stabilized via H-bonding between the N1 of thymidine and the hydroxyl group of Tyr-671 as well as π-stacking interactions to Phe-667. In contrast to KlenTaqF-A in KlenTaqF-T, Arg-587 is released from the stabilization pattern of the incoming triphosphate, as observed for the purine-trapped structures, while still interacting with the phosphate backbone of the 3′-primer terminus. The resulting thymidine tyrosine pair is accommodated in the active site in a way that results in a misalignment of the α-phosphate and thereby positions the sugar moiety on top of the 3′-primer terminus (Fig. 3C).
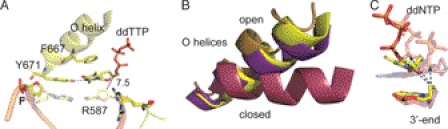
FIGURE 3.
Structure of KlenTaqF-T (yellow). A, stabilization network of ddTTP. The amino acid side chains Arg-587, Phe-667, and Tyr-671 are labeled. Gray and black dashed lines indicate hydrogen-bonding interactions and distance (Å), respectively. B, the O helices from the superimposition of KlenTaqF-T (yellow), KlenTaqF-A (purple), KlenTaqA-T (magenta), and KlenTaqbinary (gold) are highlighted. C, the incoming ddNTPs of KlenTaqF-T (yellow), KlenTaqF-A (purple), and KlenTaqA-T (berry) and the respective 3′-primer terminus are shown. The arrow indicates the displacement of the α-phosphate regarding the 3′-primer terminus.
The same approach as described before was used to co-crystallize KlenTaq in the presence of a primer/template duplex and ddCTP (KlenTaqF-binary-II). However, in all conducted approaches, only the formations of binary complexes of KlenTaq and the primer/template were obtained. These structures are very similar to the binary structure KlenTaqF-binary (supplemental Fig. S4).
Nucleotide Analog dNITP Incorporation Opposite an Abasic Site
Base stacking capability was suggested to play a decisive role in influencing the incorporation efficiency opposite an abasic site (17, 31, 48–50). Intriguingly, nucleotides that bear nucleobase surrogates with strong stacking but lacking hydrogen-bonding capability such as dNITP (Fig. 5A) are incorporated opposite abasic sites with higher efficiency as their natural counterparts. Therefore, non-natural nucleotides such as dNITP were used to investigate abasic site bypass as a probe for stacking. To get insights into how dNITP is processed opposite an abasic site by KlenTaq, we performed soaking experiments with crystals of a binary complex of KlenTaq in the presence of an abasic site and dNITP. The structure was solved by difference Fourier techniques at a resolution of 1.9 Å (supplemental Table S1 and supplemental Fig. S2D). In the presence of dNITP, the enzyme is accumulated in a closed and productive complex, very similar to reported canonical cases (e.g. PDB 1QTM; KlenTaqA-T; r.m.s.d. for Cα of 0.53 Å) and in contrast to KlenTaqF-A (Fig. 4, A and B). In disparity with the structures of KlenTaq complexed with natural nucleotides opposite an abasic site, Tyr-671 is released from its stacking interaction to the template strand and provides space for the incoming dNITP (Fig. 4A). The hydrophobic nucleotide analog perfectly stacks on the developing DNA duplex, resulting in a proper alignment of the α-phosphate and recruitment of two catalytically essential magnesium ions (Fig. 4C and supplemental Fig. S5).
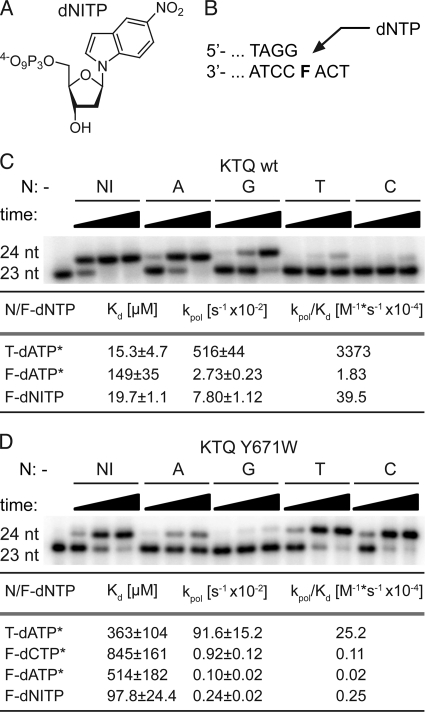
FIGURE 5.
Nucleotide incorporation opposite an abasic site analog F. A, structure of nucleotide analog dNITP. B, partial primer/template sequence used in primer extension experiments. C, single nucleotide (nt) incorporation of KlenTaq wild type opposite F for 1, 10, or 60 min, respectively. The respective dNTP is indicated. Transient kinetic analysis of nucleotide incorporation opposite T/F by KlenTaq wild type was performed. *, kinetic data were first reported in Ref. 19. D, single nucleotide incorporation of KlenTaq Y671W mutant opposite F for 10, 60, or 120 min, respectively. The respective dNTP is indicated. Transient kinetic analysis was performed as in C for KlenTaq Y671W.
FIGURE 4.
Structure of KlenTaqF-NI (orange). A, stabilization network of dNITP. The amino acid side chains Arg-587, Phe-667, and Tyr-671 are labeled. Black dashed lines indicate distance (Å). B, the O helices from the superimposition of KlenTaqF-NI (cyan), KlenTaqF-A (purple), KlenTaqC-G (green), and KlenTaqbinary (gold) are highlighted. C, the incoming ddNTPs of KlenTaqF-NI (cyan) and KlenTaqC-G (green), the respective Mg2+ ions, and the respective catalytically relevant residues Asp-610, Tyr-611, and Asp-785 are shown.
Furthermore, kinetics show that dNIMP is incorporated more efficiently than any other natural nucleotide by KlenTaq opposite an abasic site resulting in the following order of incorporation efficiencies (kpol/Kd): NI ≫ A > G ≫ T > C (Fig. 5C and supplemental Fig. S6A). Due to the decrease in Kd and simultaneous increase in kpol, dNIMP shows a 22-fold increase in incorporation efficiency as compared with dAMP. Interestingly, if KlenTaq mutant Y671W is used, we obtained the following incorporation efficiency (kpol/Kd) order: NI > C > T ≫ A > G (Fig. 5D and supplemental Fig. S6B). Therefore, one can assume that Tyr-671 does not influence the incorporation of dNIMP, whereas it clearly directs the incorporation of the natural nucleotides.
DISCUSSION
General Aspects
Although the bypass of a non-instructive lesion by DNA polymerases is extensively studied, the mechanisms are not fully understood yet. Besides initial binding of the nucleotide and the chemical reaction, noncovalent intermediates with energetic minima are crucial for the reaction pathway. These intermediates are believed to be responsible for preselection of nucleotides and represent kinetic checkpoints explaining the overall accuracy and fidelity of these enzymes (51–53). The present structural study provides further insights into how KlenTaq DNA polymerase, a member of sequence family A DNA polymerases, is able to bypass abasic sites (Table 1). Independent of the incoming natural nucleotide opposite the abasic site, the obtained ternary complexes show two noticeable alterations as compared with the canonical cases. Firstly, it is always observed that Tyr-671 is placed opposite the incoming nucleotides, and secondly, the enzyme adopts a semiclosed conformation. Such a conformation was also associated with mismatch incorporation (54). However, FRET studies of Klenow fragment DNA polymerase in the presence of mismatches indicate that along with the open form, the closed state is also partially occupied (55). Because crystallographic data only provide single snapshots, it is likely that in solution, there is also a distribution of the populations between the open and closed state even in the presence of an abasic site. Similarly to nucleotide discrimination between matched and mismatched nucleotides, DNA polymerases might also undergo several conformational changes as preselection steps before preceding the phosphoryl transfer in the presence of an abasic site. Therefore, the observed conformations might represent various local energy minima along the reaction coordinate. Along these lines, computer simulations of the fidelity of T7 DNA polymerase support the hypothesis that the formation of mismatches may also occur with high energy barriers from a partially open protein conformation (56). In general, there is an ongoing discussion whether the chemical step demands similar conformational states processing a correct or incorrect nucleotide (52–54, 57). However, the incoming nucleotides opposite an abasic site in the herein depicted structures are not properly aligned for the phosphoryl transfer. This suggests either that a rearrangement to the closed conformation has to occur or that the incorporation step proceeds from the semiclosed conformation after appropriate conformational changes of the nucleotide. Both scenarios support the observed overall poor incorporation efficiencies opposite abasic sites (19).
TABLE 1.
Summary of KlenTaq structures bound to an abasic site analog F
Structure of Binary Complex Containing Abasic Site and KlenTaq
In the binary complex of KlenTaq bound to an abasic site, we observed the 5′-upstream template strand stacking on the primer/template duplex. Nucleotide binding forces the release of the template strand from its stacking arrangement and triggers a remarkable rearrangement of the bound DNA template rather than a reorganization of the enzyme (supplemental Fig. S7). Furthermore, the ability to resolve the stacking arrangement might be dependent on the intrinsic properties of the respective nucleotide, such as the stacking and solvation ability of the nucleobase. It remains to be elucidated whether this arrangement is sequence-specific.
Nucleotide Selection Opposite Abasic Sites of KlenTaq, Adenosines versus Guanosines
The structural data offer valuable clues how nucleotide selection is performed opposite a non-instructive lesion and moreover support the previous kinetic analysis of abasic site bypass by KlenTaq (19). In the case of an incoming guanosine, we obtained several crystal structures showing the incoming triphosphate in two different orientations. This heterogeneity was not observed in the presence of the other nucleotides. In KlenTaqF-G-I and KlenTaqF-G-II, Tyr-671 is filling the space of the vacant nucleobase of the abasic site and is located opposite the incoming ddGTP and thereby roughly mimics the geometry of a nascent nucleobase pair. Of note, such a selection of purines mediated by Tyr-671 is also observed for KlenTaqF-A. However, in KlenTaqF-G-I and KlenTaqF-G-II, the interaction with Tyr-671 causes misalignment of the α-phosphate resulting in enlarged distances to the 3′-primer terminus (Figs. 2F and 6, A and B). The different steric constraints of the purine structures account for the preference of adenosine over guanosine. In detail, the exocyclic C2-NH2 group of guanine prevents the same arrangement of the nucleotide, as was found for adenosine (Table 1). Due to the steric restriction of the active site, the assembly of the sterically more demanding guanosine results in an alignment with an enlarged distance between the α-phosphate and the 3′-primer terminus in comparison with adenosine. Interestingly, the complex showing the stronger cation-π interaction between the Arg-587 and the five-membered ring (KlenTaqF-G-I) results in an arrangement of the ddGTP obviating a possible attack at the α-phosphate. The closer orientation of the nucleotide toward the 3′-primer terminus suggests that the structure of KlenTaqF-G-II forming a less stable cation-π interaction (47) may represent one step ahead on the reaction coordinate in comparison with KlenTaqF-G-I. In conclusion, we find that the distance between the α-phosphates and the 3′-primer terminus (see Table 1, 8.7–6.9 Å for guanosines versus 5.9 Å for adenosine) correlates well with the measured incorporation efficiencies. This indicates that active misalignment of the incoming guanosines governs their unfavorable processing by KlenTaq in comparison with adenosines.
FIGURE 6.
Geometry fit to active site. A, active site assembly in the presence of an abasic site analog F. The nascent base pair of Tyr-671 and ddGTP (KlenTaqF-G-I) is depicted. The surface of the surrounding active site residues is shown in gray. B, same as in A for KlenTaqF-G-II. C, active site assembly in the case of an undamaged template showing the nascent base pair of dC and ddGTP (KlenTaqC-G). D, same as in A for KlenTaqF-T. E, same as in A for KlenTaqF-A. F, same as in C for KlenTaqF-NI showing the nascent base pair of F and dNITP. A–F, the arrows indicate the positioning of the α-phosphate and the 3′-primer terminus, respectively.
Nucleotide Selection Opposite Abasic Sites by KlenTaq, Purines versus Pyrimidines
To shine light on the incorporation mechanisms of the less favored pyrimidines, we investigated a structure bearing an incoming ddTTP opposite an abasic site. The DNA polymerase interacts with the incoming nucleotide by hydrogen bonding of Tyr-671 with the nucleobase. This results in a nucleoside triphosphate conformation where the sugar moiety is positioned above the 3′-primer terminus instead of the α-phosphate (Figs. 3C and 6D). In this scenario, all components of the active site are assembled and organized in a topological and geometrical arrangement that does not allow the enzyme to proceed with the chemical step, explaining the very low incorporation efficiency. In contrast to purines, ddTTP shows low B-factors (supplemental Table S1), which indicates that it is well stabilized opposite Tyr-671. Thus, the tightly bound thymidine probably stalls the DNA polymerase in a “nonproductive” complex by active misalignment, whereas the purine nucleobases enhance the geometric fit to the active site, resulting in an arrangement with shortened distances between the α-phosphate and the 3′-primer terminus as compared with the pyrimidine base (Figs. 2G and 6, B and E).
Nucleobase Stacking and Abasic Site Bypass
Superior stacking and solvation properties of adenine have been discussed as a driving force behind adenine selection opposite abasic sites (17, 29–31). The nucleotide dNITP contains a nucleobase surrogate lacking hydrogen-bonding capability but increased stacking ability in comparison with their structural congener purines. These properties build the basis for the motivation to employ the nucleotide in abasic site bypass (17, 31, 48–50). We find, in contrast to the results obtained with nucleotides containing the natural nucleobases, that binding of the nucleotide analog dNITP readily allows the enzyme to change its active site conformation in a closed, productive complex that is similar to those found for undamaged DNA substrates. The accumulation of the enzyme in a productive conformation is clearly supported by the kinetic data, which show a significant increase in incorporation efficiency of dNIMP as compared with dAMP opposite the abasic site. Furthermore, the circumstance that dNITP is processed with higher efficiency than the favored nucleotides of either KlenTaq wild type or KlenTaq mutant Y671W shows that Tyr-671 apparently lost its selection criteria because both the dNITP and the Tyr-671 residues are not compatible with the geometric constraints of the active site. Our results suggest that due to its increased stacking ability, the nucleobase surrogate imposes active site conformations that differ significantly from those induced by the natural nucleobases. Consequently, results that are obtained by usage of dNITP instead of the natural nucleotides should be interpreted with caution toward their significance in abasic site bypass by natural nucleotides. It is noteworthy that dNITP is known as a chain terminator (59, 60) as well as universal base (61) and has shown no preference for incorporation opposite one of the four natural nucleobases. Again, enforcement of aberrant enzyme and DNA conformations due to the strong stacking ability of dNITP might be the origin of the observed properties.
Taken together, the present structural study of KlenTaq in complex with different nucleoside triphosphates opposite an abasic site reveals that natural nucleotides are bound, due to interaction with Tyr-671 and the geometric confines of the active site, in positions that require additional conformational changes for proper alignment to facilitate the chemical step. The degree of misalignment, measured as distance of the α-phosphate to the 3′-primer terminus, parallels the order of incorporation efficiency of KlenTaq opposite an abasic site. These conformational changes might be accompanied by barriers of varied energy explaining the observed decline in nucleotide incorporation efficiency from A and G to T and C as well as the strong block of the abasic site lesion. The resulting pausing of DNA synthesis might allow the DNA polymerase to be replaced by repair systems that use the sister strand for error-free repair.
The human DNA polymerase θ and γ are members of the sequence family A as KlenTaq. Interestingly, the human DNA polymerase θ also obeys the A-rule (18), and its intrinsically error-prone abasic site bypass might contribute to somatic hypermutation of Ig genes (62). Furthermore, the role of Tyr-671 in nucleotide selection opposite an abasic site is highlighted. Tyr-671 is highly conserved in family A DNA polymerases (19, 34) and is known to be involved in the discrimination process between canonical and noncanonical base pairs (63–65). Along these lines, Leob and co-workers (63) demonstrated that Tyr-671 is essential for maintaining Taq DNA polymerase I activity. Mutations at that position are hardly tolerated and compromise activity with the exception of phenylalanine. Nevertheless, mutations of the homolog residue Tyr-766 of the Klenow fragment from E. coli DNA polymerase I to serine or alanine result in significant decrease in fidelity (64). Interestingly, mutation of the corresponding Tyr-955 in human DNA polymerase γ has been attributed to progressive external ophthalmoplegia, stressing its importance in accurate function of DNA polymerases (58, 66).
Supplementary Material
Acknowledgments
We thank the beamline staff of the Swiss Light Source (SLS) and Konstanz Research School Chemical Biology for support.
This work was supported by the Deutsche Forschungsgemeinschaft through a grant in SPP 1170.

This article contains supplemental Figs. S1–S7 and Table S1.
- dNITP
- 5-nitroindolyl-2′-deoxyriboside 5′-triphosphate
- ddTTP
- dideoxythymidine 5′-triphosphate
- r.m.s.d.
- root mean square deviation
- dNIMP
- 5-nitroindoyl-2′-deoxyriboside 5′-monophosphate
- ddGTP
- dideoxyguanosine 5′-triphosphate
- ddCTP
- dideoxycytidine 5′-triphosphate
- ddATP
- dideoxyadenosine 5′-triphosphate
- ddNTP
- dideoxynucleoside 5′-triphosphate.
REFERENCES
- 1. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2. Loeb L. A., Preston B. D. (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 20, 201–230 [DOI] [PubMed] [Google Scholar]
- 3. Hoeijmakers J. H. (2001) Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 [DOI] [PubMed] [Google Scholar]
- 4. Avkin S., Adar S., Blander G., Livneh Z. (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 99, 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodman M. F., Cai H., Bloom L. B., Eritja R. (1994) Nucleotide insertion and primer extension at abasic template sites in different sequence contexts. Ann. N.Y. Acad. Sci. 726, 132–142 [DOI] [PubMed] [Google Scholar]
- 6. Hubscher U., Maga G., Spadari S. (2002) Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71, 133–163 [DOI] [PubMed] [Google Scholar]
- 7. Pagès V., Johnson R. E., Prakash L., Prakash S. (2008) Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl. Acad. Sci. U.S.A. 105, 1170–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaaper R. M., Kunkel T. A., Loeb L. A. (1983) Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc. Natl. Acad. Sci. U.S.A. 80, 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sagher D., Strauss B. (1983) Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry 22, 4518–4526 [DOI] [PubMed] [Google Scholar]
- 10. Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. (1987) Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem. 262, 6864–6870 [PubMed] [Google Scholar]
- 11. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1997) Abasic translesion synthesis by DNA polymerase β violates the “A-rule”: novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem. 272, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 12. Shibutani S., Takeshita M., Grollman A. P. (1997) Translesional synthesis on DNA templates containing a single abasic site: a mechanistic study of the “A-rule”. J. Biol. Chem. 272, 13916–13922 [DOI] [PubMed] [Google Scholar]
- 13. Strauss B. S. (2002) The “A-rule” revisited: polymerases as determinants of mutational specificity. DNA Repair 1, 125–135 [DOI] [PubMed] [Google Scholar]
- 14. Taylor J. S. (2002) New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat. Res. 510, 55–70 [DOI] [PubMed] [Google Scholar]
- 15. Freisinger E., Grollman A. P., Miller H., Kisker C. (2004) Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 23, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hogg M., Wallace S. S., Doublié S. (2004) Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 23, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reineks E. Z., Berdis A. J. (2004) Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry 43, 393–404 [DOI] [PubMed] [Google Scholar]
- 18. Seki M., Masutani C., Yang L. W., Schuffert A., Iwai S., Bahar I., Wood R. D. (2004) High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23, 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Obeid S., Blatter N., Kranaster R., Schnur A., Diederichs K., Welte W., Marx A. (2010) Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 29, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Obeid S., Schnur A., Gloeckner C., Blatter N., Welte W., Diederichs K., Marx A. (2011) Learning from directed evolution: Thermus aquaticus DNA polymerase mutants with translesion synthesis activity. ChemBioChem 12, 1574–1580 [DOI] [PubMed] [Google Scholar]
- 21. Ling H., Boudsocq F., Woodgate R., Yang W. (2004) Snapshots of replication through an abasic lesion: structural basis for base substitutions and frameshifts. Mol. Cell 13, 751–762 [DOI] [PubMed] [Google Scholar]
- 22. Fiala K. A., Hypes C. D., Suo Z. (2007) Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 282, 8188–8198 [DOI] [PubMed] [Google Scholar]
- 23. Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2009) DNA synthesis across an abasic lesion by human DNA polymerase ι. Structure 17, 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi J. Y., Lim S., Kim E. J., Jo A., Guengerich F. P. (2010) Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 404, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherrer S. M., Fiala K. A., Fowler J. D., Newmister S. A., Pryor J. M., Suo Z. (2011) Quantitative analysis of the efficiency and mutagenic spectra of abasic lesion bypass catalyzed by human Y-family DNA polymerases. Nucleic Acids Res. 39, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2011) DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J. Mol. Biol. 406, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang W., Woodgate R. (2007) What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zahn K. E., Wallace S. S., Doublié S. (2011) DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Curr. Opin. Struct. Biol. 21, 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matray T. J., Kool E. T. (1999) A specific partner for abasic damage in DNA. Nature 399, 704–708 [DOI] [PubMed] [Google Scholar]
- 30. Hwang H., Taylor J. S. (2004) Role of base stacking and sequence context in the inhibition of yeast DNA polymerase η by pyrene nucleotide. Biochemistry 43, 14612–14623 [DOI] [PubMed] [Google Scholar]
- 31. Zahn K. E., Belrhali H., Wallace S. S., Doublié S. (2007) Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry 46, 10551–10561 [DOI] [PubMed] [Google Scholar]
- 32. Goodman M. F. (1997) Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. U.S.A. 94, 10493–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kool E. T. (2002) Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 71, 191–219 [DOI] [PubMed] [Google Scholar]
- 34. Delarue M., Poch O., Tordo N., Moras D., Argos P. (1990) An attempt to unify the structure of polymerases. Protein Eng. 3, 461–467 [DOI] [PubMed] [Google Scholar]
- 35. Li Y., Korolev S., Waksman G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Di Pasquale F., Fischer D., Grohmann D., Restle T., Geyer A., Marx A. (2008) Opposed steric constraints in human DNA polymerase β and E. coli DNA polymerase I. J. Am. Chem. Soc. 130, 10748–10757 [DOI] [PubMed] [Google Scholar]
- 37. Kabsch W. (2010) XDS. Acta Crystallogr. D 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabsch W. (2010) Integration, scaling, space-group assignment, and post-refinement. Acta Crystallogr. D 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 42. Li Y., Mitaxov V., Waksman G. (1999) Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad. Sci. U.S.A. 96, 9491–9496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y., Waksman G. (2001) Crystal structures of a ddATP-, ddTTP-, ddCTP, and ddGTP- trapped ternary complex of Klentaq1: insights into nucleotide incorporation and selectivity. Protein Sci. 10, 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Obeid S., Baccaro A., Welte W., Diederichs K., Marx A. (2010) Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 21327–21331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Betz K., Streckenbach F., Schnur A., Exner T., Welte W., Diederichs K., Marx A. (2010) Structures of DNA polymerases caught processing size-augmented nucleotide probes. Angew. Chem., Int. Ed. Engl. 49, 5181–5184 [DOI] [PubMed] [Google Scholar]
- 46. Korolev S., Nayal M., Barnes W. M., Di Cera E., Waksman G. (1995) Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-Å resolution: structural basis for thermostability. Proc. Natl. Acad. Sci. U.S.A. 92, 9264–9268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wintjens R., Liévin J., Rooman M., Buisine E. (2000) Contribution of cation-π interactions to the stability of protein-DNA complexes. J. Mol. Biol. 302, 395–410 [DOI] [PubMed] [Google Scholar]
- 48. Vineyard D., Zhang X., Donnelly A., Lee I., Berdis A. J. (2007) Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org. Biomol. Chem. 5, 3623–3630 [DOI] [PubMed] [Google Scholar]
- 49. Sheriff A., Motea E., Lee I., Berdis A. J. (2008) Mechanism and dynamics of translesion DNA synthesis catalyzed by the Escherichia coli Klenow fragment. Biochemistry 47, 8527–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Motea E. A., Lee I., Berdis A. J. (2011) Quantifying the energetic contributions of desolvation and π-electron density during translesion DNA synthesis. Nucleic Acids Res. 39, 1623–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Joyce C. M., Benkovic S. J. (2004) DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324 [DOI] [PubMed] [Google Scholar]
- 52. Johnson K. A. (2010) The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta 1804, 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Joyce C. M. (2010) Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta 1804, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu E. Y., Beese L. S. (2011) The structure of a high-fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 286, 19758–19767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Santoso Y., Joyce C. M., Potapova O., Le Reste L., Hohlbein J., Torella J. P., Grindley N. D., Kapanidis A. N. (2010) Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 107, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Florián J., Goodman M. F., Warshel A. (2005) Computer simulations of protein functions: searching for the molecular origin of the replication fidelity of DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 102, 6819–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Structures of DNA polymerase β with active site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Graziewicz M. A., Longley M. J., Bienstock R. J., Zeviani M., Copeland W. C. (2004) Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat. Struct. Mol. Biol. 11, 770–776 [DOI] [PubMed] [Google Scholar]
- 59. Smith C. L., Simmonds A. C., Felix I. R., Hamilton A. L., Kumar S., Nampalli S., Loakes D., Hill F., Brown D. M. (1998) DNA polymerase incorporation of universal base triphosphates. Nucleosides Nucleotides 17, 541–554 [Google Scholar]
- 60. Loakes D. (2001) Survey and summary: the applications of universal DNA base analogues. Nucleic Acids Res. 29, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loakes D., Brown D. M. (1994) 5-Nitroindole as a universal base analogue. Nucleic Acids Res. 22, 4039–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masuda K., Ouchida R., Hikida M., Kurosaki T. (2007) DNA polymerases η and θ function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 282, 17387–17394 [DOI] [PubMed] [Google Scholar]
- 63. Suzuki M., Baskin D., Hood L., Loeb L. A. (1996) Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 93, 9670–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bell J. B., Eckert K. A., Joyce C. M., Kunkel T. A. (1997) Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J. Biol. Chem. 272, 7345–7351 [DOI] [PubMed] [Google Scholar]
- 65. Minnick D. T., Bebenek K., Osheroff W. P., Turner R. M., Jr., Astatke M., Liu L., Kunkel T. A., Joyce C. M. (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J. Biol. Chem. 274, 3067–3075 [DOI] [PubMed] [Google Scholar]
- 66. Copeland W. C., Ponamarev M. V., Nguyen D., Kunkel T. A., Longley M. J. (2003) Mutations in DNA polymerase γ cause error-prone DNA synthesis in human mitochondrial disorders. Acta Biochim. Pol. 50, 155–167 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.