Background: TssL is a core component of the T6SS and has homologue in the T4bSS.
Results: The TssL cytoplasmic domain adopts a globular α-helical domain and forms dimers.
Conclusion: Dimer formation involves the trans-membrane segment, but contacts mediated by the cytoplasmic domain are important for TssL function.
Significance: The structural and functional characterization of TssL leads to a better understanding of T6SS and T4bSS assembly.
Keywords: Bacterial Pathogenesis, Membrane Proteins, Membrane Trafficking, Protein Secretion, Structural Biology, DotU, IcmH, TssL, Type IV Secretion, Type VI Secretion
Abstract
The Type VI secretion system (T6SS) is a macromolecular system distributed in Gram-negative bacteria, responsible for the secretion of effector proteins into target cells. The T6SS has a broad versatility as it can target both eukaryotic and prokaryotic cells. It is therefore involved in host pathogenesis or killing neighboring bacterial cells to colonize a new niche. At the architecture level, the T6SS core apparatus is composed of 13 proteins, which assemble in two subcomplexes. One of these subcomplexes, composed of subunits that share structural similarities with bacteriophage tail and baseplate components, is anchored to the cell envelope by the membrane subcomplex. This latter is constituted of at least three proteins, TssL, TssM, and TssJ. The crystal structure of the TssJ outer membrane lipoprotein and its interaction with the inner membrane TssM protein have been recently reported. TssL and TssM share sequence homology and characteristics with two components of the Type IVb secretion system (T4bSS), IcmH/DotU and IcmF, respectively. In this study, we report the crystal structure of the cytoplasmic domain of the TssL inner membrane protein from the enteroaggregative Escherichia coli Sci-1 T6SS. It folds as a hook-like structure composed of two three-helix bundles. Two TssL molecules associate to form a functional complex. Although the TssL trans-membrane segment is the main determinant of self-interaction, contacts between the cytoplasmic domains are required for TssL function. Based on sequence homology and secondary structure prediction, we propose that the TssL structure is the prototype for the members of the TssL and IcmH/DotU families.
Introduction
The Type VI secretion system (T6SS)5 is a macromolecular system encoded within the genomes of Gram-negative bacteria. This pathway has been recently described to translocate protein substrates into both eukaryotic and prokaryotic cells (1–6). T6SS-dependent translocation of proteins into eukaryotic cells leads to cytoskeleton rearrangements and cytotoxicity (7–9). More recently, the translocation of canonical protein effectors degrading the peptidoglycan layer into the periplasm of recipient bacterial cells has been demonstrated (10, 11). The T6SS has also been shown to be involved in stress sensing or biofilm formation, although the mechanistic bases for these phenotypes have not been described yet (12–14). The T6SS is therefore a versatile machine that adapts to the specific needs of each bacterium.
The T6SS is composed of 13 core components named Tss (Type six secretion (15)), but several additional subunits might be encoded within T6SS gene clusters (1, 16–18). Two proteins, Hcp (hemolysin-coregulated protein) and VgrG (valine-glycine repeat protein), are released in the culture supernatant and are suggested to form the extracellular portion of the T6SS machine (16). Interestingly, Hcp and VgrG are structurally related to two components of the bacteriophage T4, the tail protein and the needle-like structure, respectively. Hcp forms hexameric rings, a structure that is superimposable to the major tail protein gpV (19, 20). VgrG assembles as trimers resembling the bacteriophage gp27/gp5 complex (7, 21, 22). Recently, electron microscopy studies have proposed that the TssB and TssC proteins form structures resembling the bacteriophage sheath (21, 23, 24) From these observations and the structural similarities of TssE with the bacteriophage baseplate component gp25 (1), a model on how the T6SS is structurally arranged and how it functions has recently emerged (25, 26). It has been proposed that the T6SS forms an inverted bacteriophage-like structure that will puncture the target cell to deliver the protein effectors.
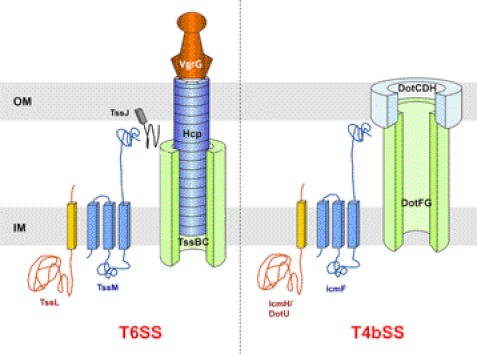
Aside from bacteriophage-related components, a number of membrane-embedded proteins are associated with the T6SS. At least three proteins, TssJ, TssL, and TssM, form a trans-envelope complex that may be augmented by TagL, an additional protein containing a peptidoglycan-binding domain (27, 28). TssJ is an outer membrane lipoprotein (12) whose structure has been reported recently (29, 30). TssJ interacts with the inner membrane TssM protein (29, 31). TssM forms a stable complex with TssL (31, 32). TssL and TssM are closely related to two proteins associated with the Type IVb secretion system (T4bSS), IcmH/DotU and IcmF, respectively (Fig. 1). These two components are required for the optimal function of the T4bSS in Legionella pneumophila and Coxiella burnetii (33, 34). The TssL protein and its T4bSS IcmH/DotU counterpart are inner membrane proteins (32, 35–38). The membrane topology of the TssL has been determined; TssL has a single trans-membrane segment (TMS) located at the C terminus of the protein (36). However, in some cases, the TMS can be followed by an additional domain protruding into the periplasm and carrying a peptidoglycan-binding motif (28, 32). The topology of the IcmH/DotU protein is not known, but the absence of labeling with the membrane-impermeant sulfo-NHS-biotin suggests that, as shown for TssL, the bulk of the protein locates in the cytoplasm (38). In L. pneumophila, IcmH/DotU stabilizes IcmF, whereas IcmF is absolutely required for IcmH/DotU production (38). Regarding the T6SS homologues, de Bruin et al. (35) reported that TssL stabilizes TssM. Taken together, these data suggest that the TssL-TssM pair is closely related to the IcmH/DotU-IcmF pair in terms of localization, topology, interaction, and stabilization. However, although the primary sequences of the TssL and IcmH/DotU proteins are not conserved, both proteins belong to the DUF2077 family (Pfam PF09850). This and the observation that TssL and IcmH/DotU hold similar secondary structure predictions (supplemental Fig. S1) suggest that these two proteins share a common fold. In this study, we report the crystal structure of the cytoplasmic domain of the enteroaggregative Escherichia coli (EAEC) TssL protein. TssL is formed by the association of two helix bundles each composed of three helices. This globular structure is connected to the TMS through a linker of ∼20 residues. Analysis of the crystal packing and mutagenesis studies further showed that TssL form dimers. Although the main dimer interface involves the trans-membrane segment, functional contacts occur between the cytoplasmic helices 1 of each protomer. Based on the similarities between the T6SS TssL and T4bSS IcmH/DotU protein, we propose that the EAEC TssL structure represents the prototypic fold for the DUF2077 family.
FIGURE 1.
Schematic models of bacterial Type VI (left) and Type IVb (right) secretion systems. The localizations and topologies of the T6SS-associated TssL (orange) and TssM (blue) proteins, as well as those of their T4bSS homologues, IcmH/DotU and IcmF, respectively, are schematized. TssM interacts with the TssJ outer membrane lipoprotein (29, 31), whereas IcmF stabilizes the outer membrane-associated DotH protein (37). OM, outer membrane; IM, inner membrane.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Medium, Growth Conditions, and Chemicals
E. coli K12 DH5α was used for cloning procedures. The enteroaggregative E. coli strain 17-2 (kindly provided by Arlette Darfeuille-Michaud, University of Auvergne, Clermont-Ferrand, France) and its ΔtssL derivative (ΔsciP (27)) were used for this study. EAEC strains were routinely grown in LB medium at 37 °C with shaking. Expression of tssL (from plasmid pIBA-TssL (36)) was obtained by the addition of anhydrotetracycline. For the Hcp release assay, the sci-1 gene cluster was induced by the addition of the iron chelator 2,2′-dipyridyl (125 μm final concentration), 30 min prior to harvesting the cells (39). Plasmids were maintained by the addition of ampicillin (100 μg/ml for E. coli K12, 200 μg/ml for EAEC). Anhydrotetracycline, used at 0.2 μg/ml throughout the study, was purchased from IBA. The anti-TolB polyclonal antibodies are from the laboratory collection, whereas the anti-HA (3F10 clone, Roche Applied Science) and anti-FLAG (M2 clone, Sigma-Aldrich) monoclonal antibodies are commercially available.
Constructions for in Vivo Studies
The plasmids used for this study are listed in supplemental Table S1. Polymerase chain reactions (PCRs) were performed with a Biometra thermocycler, using the Phusion DNA polymerase (Thermo scientific). Custom oligonucleotides were synthesized by Sigma-Aldrich and are listed in supplemental Table S1. For in vivo complementations, the pIBA-TssL plasmid, encoding an N-terminally FLAG epitope-tagged TssL protein (36), has been used. For bacterial two-hybrid assay, the fragment encoding the TssL cytoplasmic domain has been cloned downstream the T18 or T25 sequence in vectors pEB354 and pEB355 (40) to yield pTssL-T18 and pTssL-T25, respectively. These plasmids have been constructed by a double PCR technique, allowing the amplification of the gene of interest flanked by extensions annealing to the target vector (41). The TssL variants have been constructed by site-directed mutagenesis using pIBA-TssL, pTssL-T18, or pTssL-T25 as template and complementary oligonucleotides introducing the required mismatch(es) (supplemental Table S1). All constructs have been verified by restriction analyses and DNA sequencing (GATC).
Hcp Release Assay
Supernatant and cell fractions have been separated as described previously (36). Briefly, 2 × 109 cells producing hemagglutinin (HA) epitope-tagged Hcp (from plasmid pOK-HcpHA (27)) were harvested and collected by centrifugation at 2,000 × g for 5 min. The supernatant fraction was then subjected to a second low-speed centrifugation and then at 16,000 × g for 15 min. The supernatant was filtered on sterile polyester membranes with a pore size of 0.2 μm (Membrex 25 PET, membraPure GmbH) before precipitation with 15% trichloroacetic acid (TCA). Cells and precipitated supernatants were resuspended in loading buffer and analyzed by SDS-PAGE and immunoblotting with the anti-HA antibody. As control for cell lysis, Western blots were probed with antibodies raised against the periplasmic TolB protein.
Biofilm Assay
The adherence was tested on cover-glasses after incubation at 30 °C without agitation for 20 h. Attached bacteria were stained with 1% crystal violet for 15 min and washed four times with water. For quantification, the ring of stained bacteria was collected with 500 μl of 95% ethanol and diluted in the same volume of water. The absorbance of the suspension was then measured at 590 nm. This value was divided by the absorbance of the bacterial culture (measured at 600 nm), and data are reported relative to the wild-type strain.
Bacterial Two-hybrid Assay
We used the adenylate cyclase-based two-hybrid technique (42) using previously published protocols (40). Briefly, pairs of proteins to be tested were fused to the two catalytic domains T18 and T25 of the Bordetella adenylate cyclase. After co-transformation of the BTH101 strain with the two plasmids producing the fusion proteins, plates were incubated at 30 °C for 2 days. 600 μl of LB medium supplemented with ampicillin, kanamycin, and 0.5 mm isopropyl-β-thio-galactoside were inoculated with independent colonies. Cells were grown at 30 °C overnight, and the β-galactosidase activity was determined as described by Miller (43). The values presented are the mean of three independent assays, each in triplicate.
Constructions for in Vitro Studies and Purification Procedures
The sequence encoding the tssL cytoplasmic domain (residues 1–183) of enteroaggregative E. coli strain 17-2 was cloned into the pETG-20A expression vector (a kind gift from Dr. Arie Geerlof, European Molecular Biology Laboratory (EMBL), Hamburg, Germany) according to standard GatewayTM protocols. The final construct allows the production of the soluble cytoplasmic TssL fragment (TssLC) fused to an N-terminal hexahistidine-tagged thioredoxin followed by a tobacco etch virus protease cleavage site. The plasmid was transformed into the E. coli T7 Iq pLysS expression strain (New England Biolabs). Cells were grown at 37 °C in terrific broth until the A600 reached 0.9, and tssL expression was induced with 0.5 mm isopropyl-β-thio-galactoside overnight at 17 °C. After cell harvesting, cells were resuspended in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.25 mg/ml lysozyme and broken by sonication. The soluble proteins were separated from inclusion bodies and cell debris by centrifugation 30 min at 20,000 × g. The TssL fusion was purified using Ni2+ affinity chromatography (5-ml HisTrap column, GE Healthcare, on an ÄKTA FPLC system) and eluted with a step gradient of imidazole. The TssL fusion protein was digested overnight at 4 °C by a hexahistidine-tagged tobacco etch virus protease using a 1:10 (w/w) protease:protein ratio. The tobacco etch virus protease and contaminants were bound on a second Ni2+ affinity chromatography, and the native TssL cytoplasmic fragment was collected in the flow-through and separated on a preparative Superdex 75 gel filtration column (GE Healthcare) equilibrated in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% glycerol. The final concentration of the TssLC solution was 23 mg/ml.
The seleno-methionine (Se-Met) derivative of TssL was produced in minimal medium supplemented with 100 mg/liter lysine, phenylalanine, and threonine and 50 mg/liter isoleucine, leucine, valine, and seleno-methionine. Protein production was performed overnight at 17 °C after the addition of 0.5 mm isopropyl-β-thio-galactoside, and the Se-Met-TssLC protein was purified as described above.
Crystallization and Structure Determination
TssLC crystallization trials were carried out by the sitting-drop vapor diffusion method in 96-well Greiner crystallization plates at 20 °C, using a nanodrop-dispensing robot (Cartesian Inc.) (44). Crystals grew in a few days after mixing 300 nl of the TssL protein at 23 mg/ml with 100 nl of 4.3 m NaCl, 0.1 m Bicine, pH 10.6. Crystals were cryoprotected with mother liquor supplemented with 10% glycerol and flash-frozen in liquid nitrogen.
Two single anomalous dispersion datasets were collected on the same crystal at the selenium edge wavelength. The first dataset was collected at the European Synchrotron Radiation Facility (ESRF) ID14-4 beamline (Grenoble, France), and the second was collected at the SOLEIL Proxima 1 beamline (Saint-Aubin, Gif-sur-Yvette, France) with a 30° offset reorientation of the crystal using the kappa axis of the Proxima 1 goniometer. After processing the two datasets using the XDS (45) program, the scaling was done using XSCALE (see Table 1). The selenium sites were determined using SHELXD (46), and the phase was calculated using PHASER (47). These phases were then improved by solvent flattening and histogram matching using the PARROT program (48), assuming 49% solvent content. This results in interpretable maps from which an initial model comprising more than 90% of the expected amino acids was built using BUCCANEER (49). Structure refinement was performed with autoBUSTER, alternated with rebuilding using COOT (50), using translation/libration/screw parameters.
TABLE 1.
Data collection (ID14–4 at ESRF and PROXIMA 1 at SOLEIL) and refinement statistics
| Proxima-1 Soleil | IDA4–4 ESRF | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | ||
| Resolution limitsa (Å) | 50.00–2.74 (2.81–2.74) | 45.0–2.63 (2.70–2.63) |
| Rmergea,b (%) | 4.6 (57.0) | 5.6 (68) |
| No. of observationsa | 35,674 (2550) | 65,807 (4720) |
| No. of unique reflectionsa | 10,304 (745) | 11,601 (853) |
| Mean ((I)/sd(I))a,c | 15.6 (2.1) | 20.2 (2.5) |
| Completenessa (%) | 99.4 (96.9) | 99.4 (99.8) |
| Multiplicitya | 3.5 | 5.7 |
| Refinement | ||
| Resolutiona (Å) | 43.7–2.63 (2.94–2.63) | |
| No. of reflectionsa | 6278 (1576) | |
| No. of protein/water/glycerol atoms | 1288/31/6 | |
| No. of test set reflections | 604 (167) | |
| Rwork/Rfreea,d | 0.235/0.257 | |
| r.m.s.d.e bonds (Å)/angles (°) | 0.09/1.07 | |
| B-wilson/B-average | 70.2/69.8 | |
| Coot's Ramachandran Preferred/allowed % | 97.6/2.4 | |
a Numbers in brackets refer to the highest resolution bin.
b Σ (| Ih,i − Ih|))/Σ (Ih,i).
c Σi = 1,n Ih,i/n.
d Σ (|Fobs − Fcalc|)/Σ (|Fobs|).
e r.m.s.d., root mean square deviation.
Cloning, Expression, and Purification of Full-length TssL
Full-length TssL Production
The full-length tssL gene of enteroaggregative E. coli strain 17-2 was cloned into the pETG-20A expression vector according to standard GatewayTM protocols. The plasmid was transformed into the E. coli C43(DE3) expression strain (51). Cells were grown at 37 °C in terrific broth to an A600 of 0.9 prior to induction of the tssL expression with 0.3 mm isopropyl-β-thio-galactoside overnight at 17 °C.
Membrane Isolation and Detergent Solubilization
All steps were performed at 4 °C. Cells were resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5% glycerol (v/v), 0.5 mg/ml lysozyme, 20 μg/ml DNase, 20 mm MgSO4, 1 mm phenylmethylsulfonyl fluoride (PMSF)) supplemented with protease inhibitors (Complete EDTA-free, Roche Applied Science). Upon lysis by sonication, cell debris and inclusion bodies were removed by centrifugation at 20,000 × g for 20 min. Membranes were collected from the cleared suspension by ultracentrifugation for 1.5 h at 125,000 × g (Ti45; Beckman Coulter). The membrane pellet was resuspended in buffer A (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, 5% glycerol (v/v), PMSF, and Complete (Roche Applied Science)) supplemented with 1% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace SOLgrade) by gentle stirring for 1 h at 4 °C. The insoluble material was removed by ultracentrifugation for 30 min at 125,000 × g.
Purification
The full-length TssL protein was purified from the detergent-solubilized membrane proteins using a protocol similar to the soluble TssL fragment except that all buffers were supplemented with 0.05% DDM (Anatrace ANAgrade). For chemical cross-linking studies, proteins were dialyzed against 10 mm sodium phosphate buffer (Na2HPO4/NaH2PO4) (pH 6.3), 150 mm NaCl supplemented (full-length TssL) or not (TssL cytoplasmic domain) with 0.05% DDM.
Analytical Gel Filtration
2 mg of each protein standards (ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and ribonuclease (13.7 kDa) (GE Healthcare)) and 1 mg of the soluble or full-length TssL protein were run separately on an analytical Superdex 75 10/30 column (GE Healthcare) equilibrated in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% (v/v) glycerol, 0.05% DDM (Anatrace ANAgrade).
Chemical Cross-linking
Chemical cross-linking experiments were performed with para-formaldehyde (PFA, Merck) essentially as described previously (52). For in vivo studies, cells were washed and resuspended in 10 mm sodium phosphate buffer (Na2HPO4/NaH2PO4) (pH 6.3), 150 mm NaCl prior to sonication. Cleared cell lysates were treated with PFA (0.2 or 1% final concentration). For in vitro studies, 10 μg of protein were mixed in a final volume of 20 μl with 1% PFA. After incubation for 25 min at 25 °C, the cross-linking reaction was quenched by the addition of 50 mm Tris-HCl, pH 6.8, and samples were analyzed by SDS-PAGE and immunodetection or Coomassie Blue staining.
RESULTS
We recently reported that the TssL protein is anchored into the inner membrane through a C-terminal trans-membrane segment (36). The bulk of the protein (amino acids 1–183) resides in the cytoplasm. To gain insight into its biological function, we purified the soluble, cytoplasmic fragment of the TssL protein encoded within the sci-1 T6SS gene cluster of EAEC (GenBankTM accession number CBG37349; locus tag EC042_4527). This fragment was fused in the Gateway vector pETG20A to an N-terminal hexahistidine-tagged thioredoxin for purification (53). A Se-Met TssLC derivative was produced in minimal medium and purified by affinity chromatography and gel filtration. The native TssLC protein was obtained upon fusion and tag cleavage using the tobacco etch virus protease.
Crystal Structure of EAEC TssLC Domain
TssLC crystallized readily with 4.3 m NaCl as precipitant. We collected a dataset from a unique Se-Met crystal. The structure was solved from a 2.7 Å resolution single anomalous dispersion map calculated from a combined dataset collected with a unique crystal exposed at beamlines ID14-4 (ESRF) and Proxima 1 (Soleil synchrotron). The structure was further refined with the ID14-4 dataset at 2.63 Å resolution (Table 1).
A unique TssLC molecule is contained in the asymmetric unit, and the polypeptide chain could be traced from residue Ile-6 to Arg-178, with a gap between Glu-79 and Trp-85 due to scarce electron density. TssLC is an eight-α-helix protein constituted of two three-helix bundles connected by two short helices that presents, overall, a hook shape (Fig. 2). The three first long helices (h1–3) form a tight bundle and are followed by a disordered loop of 4 polar residues (loop 3–4). The first helix is bended, due to the presence of a proline residue at position 15, which induces a helix kink. Two short helices (h4 and h5) and loops connect the two bundles. The second helix bundle is formed by three helices (h6–8), shorter than those of the first bundle. Of particular interest, a long stretch of 10 residues protrudes between helices 7 and 8 (amino acids 138–147), formed by two elongated structures separated by a loop. The central part of this loop (amino acids 141–146) is mainly composed of charged residues (RSQDDE). The last helix ends at residue 157 and is followed by an elongated unstructured segment (residues 158–178), which links the TssL cytoplasmic core domain to the TMS. This linker segment is ordered in the crystal due to packing contacts with a symmetry-related protein.
FIGURE 2.
Structure of enteroaggregative E. coli T6SS TssL cytoplasmic domain. A, stereo view of the EAEC TssL cytoplasmic domain in ribbon representation and rainbow coloring from blue (N terminus (Nt)) to red (C terminus (Ct)); the figure was made with PyMOL (57). IM, inner membrane. B, top view of the TssL cytoplasmic domain, with the same coloring as in A (k, kink). C, amino acid sequence of TssL. The amino acids are colored according to the same rainbow code. D, topology graphic of TssL (same coloring as in A) The three-helix bundles (hb1 and hb2) and the h3-h4 and h7-h8 loops are indicated.
Searching the Protein Data Bank for structurally related proteins with Dali (54) returned hits in the twilight zone, with the best z scores comprised between 5 and 6. However, examination of the aligned structures reveals that only three-helix bundle proteins have been selected, which superimpose on the TssL first helix bundle with root mean square deviations comprised between 3.2 and 5.6 Å (data not shown). These hits are therefore probably not significant to suggest a molecular function for TssL. Therefore, the TssLC structure defines a new fold.
Examination of TssLC surface and calculations of contact electrostatic potential did not highlight any hydrophobic or positively/negatively charged surface patches, but indicated a scattered and balanced charge distribution over the whole surface (supplemental Fig. S2). In particular, we did not observe any patch of positively charged residues that might be candidates for interactions with the phosphate groups of the inner membrane lipids nor patches of hydrophobic residues that might interact through their hydrophobic lateral chains.
Analysis of the surface cavities using the CASTp software (55) returned three spots of open crevices. The first cavity involves the portion of space between the base of the linker and the core (1000 Å3; supplemental Fig. S2, indicated by an arrow). The second and third small cavities (280 Å3) are located at either side of the domain between the helix bundles.
The conserved residues of TssL identified in supplemental Fig. S1 were mapped onto the TssLC structure (supplemental Fig. S3). Although several of these residues are structural determinants of the helix association within the bundles, a large conserved area is located on the core domain facing the 1000 Å cavity. This groove forms at the interface between the two helix bundles. This conserved area must be structurally important to maintain the interaction between the two-helix bundles or to accommodate a protein partner.
TssL Structure-Function Analysis
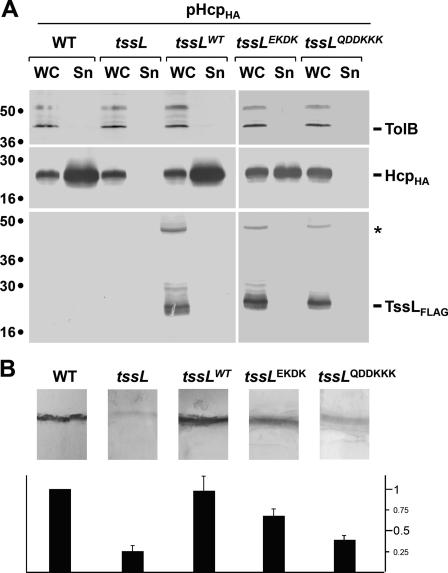
To identify functional regions of the TssL protein, we introduced substitutions to inverse the charge of loops connecting h3 and h4 (E79K/D82K, called EKDK) and h7 and h8 (Q143K/D144K/D145K, called QDDKKK). Loop h3-h4 is disordered in the crystal and probably subjected to flexibility. Loop h7-h8 has an outgoing plug shape protruding from the core domain and might be involved in contacting a protein partner. Substitutions were introduced into plasmid pTssL, which encodes a functional N-terminally FLAG-tagged TssL protein (36), and the constructs were assayed for complementation of a nonpolar tssL mutation. Complementation was tested by using the biofilm and Hcp release assays as reporters, two assays previously shown to reflect the functionality of the EAEC Sci-1 T6SS (12). Both mutant TssL proteins were produced at levels comparable with the WT TssL protein (Fig. 3). Although mutations of the charged residues of the h3-h4 loop caused a strong defect in T6SS assembly without abrogating it, mutations within the h7-h8 loop completely abolished TssL function (Fig. 3). These results suggest that the plug-shaped h7-h8 loop is critical for TssL function and might be required for TssL activity or to functionally associate with an interacting partner. It is worthy to note that this loop is not conserved among the TssL proteins.
FIGURE 3.
Structure-function analysis of negatively charged loops of TssL. A, in vivo Hcp release assay. HcpHA release was assessed by separating whole cells (WC) and supernatant (Sn) fractions from wild-type (WT) or tssL cells carrying the empty vector (tssL), the plasmid encoding wild-type TssL (tssLWT), or plasmids encoding the TssL-E79K-D82K (tssLEKDK) or TssL-Q144K-D145K-D146K (tssLQDDKKK) variants. 2 × 108 cells and the TCA-precipitated material of the supernatant from 5 × 108 cells were subjected to 12.5% acrylamide SDS-PAGE and immunodetection using the anti-TolB polyclonal antibodies (lysis control; upper panel) and the anti-HA (Hcp detection, middle panel) and anti-FLAG monoclonal antibodies (TssL detection, lower panel). The proteins are indicated on the right, and the molecular mass markers are indicated on the left (in kDa). The asterisk indicates the dimeric form of TssL observable on SDS denaturing gels. B, biofilm formation. Biofilms formed in static cultures of WT, tssL, tssLWT, tssLEKDK, and tssLQDDKKK cells were visualized on cover-glass by crystal violet staining (upper panel) and quantified using the ethanol solubilization procedure, relative to the WT EAEC strain (lower graph). Error bars indicate S.D.
TssL Dimer Formation
Although a unique molecule of TssLC is present in the asymmetric unit, Western blot analyses showed that a TssL dimer can be detected upon denaturation in SDS-containing loading buffer and boiling (Fig. 3, indicated by asterisk in anti-FLAG blots). The crystal structure was analyzed for probable packing interactions. Packing interactions extracted with the PISA server (56) revealed putative interactions (supplemental Fig. S4). The packings involving three or four TssL molecules (supplemental Fig. S4, D and E) can be ruled out because they will prevent proper insertion of the TssL TMS in the inner membrane. In the 1190 Å2 configuration (supplemental Fig. S4A), the largest contact area involves residues of h8 and of the linker (amino acids 148–177). In the 700 Å2 configuration (supplemental Fig. S4B), the main contacts involve helix h1 (residues 11–30). Finally, the 690 Å2 symmetry-related pair (supplemental Fig. S4C) involves contacts between core residues 27–41 and 77–79 and the linker.
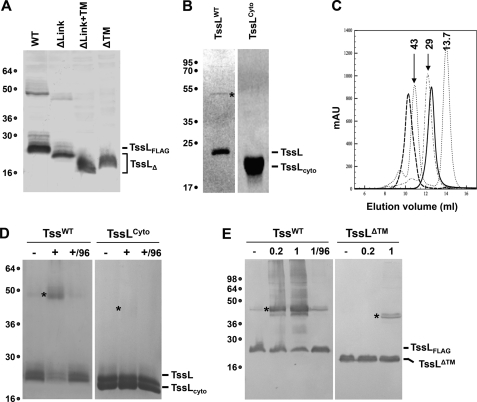
We therefore designed substitution mutations targeting the putative dimerization interfaces and tested these TssL variants for dimer formation. Because two of the hypotheses involve the linker, we first constructed a TssL derivative lacking amino acids 165–178 (TssL-Δlink). The second hypothesis was tested by targeting residues of h1 facing the exterior of the core domain. Substitutions of Leu-18 (L18S) and Ser-21 (S21E) were constructed to provoke a large clash with the symmetrical molecule through charge repulsion. Deletion of the linker region led to a defect in T6SS function as shown by the absence of Hcp release in culture supernatant (Fig. 4A) and lower levels of biofilm (Fig. 4B). Similarly, substitutions L18R, L18S, S21E, and S21K completely abolished TssL function. However, dimers were still detectable upon Western blot analyses of all these TssL variants (Fig. 4A). One of the elements present in the full-length TssL protein but absent in the structure is the TMS. To test whether the TMS is required for dimer formation, we introduced a stop codon at position 165 or 184, leading to a cytoplasmic soluble TssL derivative lacking both the linker and the TMS (165stop; TssL-Δlink+TM) or only the TMS (184stop; TssL-ΔTM). Although these derivatives were produced at levels comparable with the wild-type protein, dimers were not observed upon Western blot analyses (Fig. 5A). To confirm this result, the full-length TssL protein was produced and purified from detergent-solubilized membrane extracts. As shown in Fig. 5B, the full-length protein, but not the soluble derivative, forms oligomers detectable on Coomassie Blue-stained SDS-PAGE. The presence of only TssL in the oligomeric bands was confirmed by mass spectrometry analyses (data not shown). The full-length TssL and the isolated cytoplasmic domain were further analyzed using gel filtration. Although the isolated cytoplasmic domain was monomeric in solution, a unique peak corresponding to the molecular weight of two molecules was obtained with full-length TssL (Fig. 5C). To confirm these data, in vivo and in vitro cross-linking experiments were performed with para-formaldehyde. In vitro, a weak band corresponding to a dimer of the cytoplasmic domain of TssL can be observed upon Coomassie Blue staining, whereas most of the full-length TssL protein assembles as dimer (Fig. 5D). In vivo, 0.2% of formaldehyde was sufficient to efficiently achieve full-length TssL dimer formation (Fig. 5E), whereas a faint dimerization of the TssL cytoplasmic domain was observed with 1% formaldehyde. The TssL protein is therefore dimeric, and dimer formation is mainly controlled by the TMS.
FIGURE 4.
Mutational analysis of probable TssL dimer interfaces. A, in vivo Hcp release assay. HcpHA release was assessed by separating whole cells (WC) and supernatant (Sn) fractions from tssL cells carrying the empty vector (tssL), the plasmid encoding wild-type TssL (tssLWT) or plasmids encoding the TssL-Δlinker (tssLΔlink), the TssL-L18S (tssLL18S), the TssL-S21E (tssLS21E), or the TssL-L18S-S21E (tssLL18S-S21E) variants. The asterisk indicates the dimeric form of TssL observable on SDS denaturing gels. B, biofilm formation. Biofilms formed in static cultures of tssL, tssLWT, tssLΔlink, tssLL18S, tssLS21E, or tssLL18S-S21E cells were visualized on cover-glass by crystal violet staining (upper panel) and quantified using the ethanol solubilization procedure, relative to the WT EAEC strain (lower graph). Error bars indicate S.D.
FIGURE 5.
The TssL trans-membrane segment is required for dimer formation. A, cell extracts of tssL strains producing the wild-type TssL protein (WT) or TssL deleted of the linker (Δlink), of the linker and trans-membrane segment (Δlink+TM), or of the TMS (ΔTM) were subjected to 12.5% acrylamide SDS-PAGE and immunodetection using the anti-FLAG monoclonal antibody. The proteins are indicated on the right, and the molecular mass markers are indicated on the left (in kDa). The asterisks indicate the dimeric forms of the TssL variants. B, the purified detergent-solubilized full-length TssL protein (TssLWT, theoretical mass of the monomer: 24 kDa) and the purified cytoplasmic domain of TssL (TssLcyto, theoretical mass of the monomer: 20 kDa) were subjected to 12.5% acrylamide SDS-PAGE and detected by Coomassie Blue staining. The proteins are indicated on the right, and the molecular mass markers are indicated on the left(in kDa). The asterisk indicates the dimeric form of TssL. C, analytical gel filtration analysis of TssLWT (dashed thick line) and TssLcyto (continuous thick line) on a Superdex 75 column, calibrated with molecular size markers. The sizes of the markers are indicated on the top of each peak (in kDa). mAU, milliabsorbance units. D, in vitro chemical cross-linking using para-formaldehyde. The detergent-solubilized full-length TssL protein (TssLWT) and the cytoplasmic domain of TssL (TssLcyto) were treated (+) or not (−) with 1% PFA and subjected to 12.5% acrylamide SDS-PAGE and detected by Coomassie Blue staining. As control, a PFA-treated sample was heated 20 min at 96 °C (+/96) to break the cross-links. E, in vivo chemical cross-linking using para-formaldehyde. Lysates from tssL cells producing the wild-type TssL protein (TssLWT) or TssL deleted of the trans-membrane segment (TssLΔTM) were treated or not with PFA (0.2 or 1% as indicated) and subjected to 12.5% acrylamide SDS-PAGE and immunodetection using the anti-FLAG monoclonal antibody. The proteins are indicated on the right, and the molecular mass markers are indicated on the left(in kDa). The asterisks indicate the dimeric form of the TssL variants.
However, when tested in a bacterial two-hybrid assay, the soluble cytoplasmic domain gave a reproducible weak but significant positive signal (Fig. 6), suggesting that the soluble domain is able to dimerize in the in vivo conditions. The L18S and S21E substitutions were introduced into the bacterial two-hybrid constructs. As shown in Fig. 6, the L18S and S21E mutations drastically decrease the β-galactosidase activity, suggesting that dimerization in the cytoplasm occurs through contacts between the h1 of two adjacent TssL molecules. To validate this hypothesis, we took advantage of the observation that a smaller serine residue at position 18 (L18S) provides more space to a glutamate residue at position 21 (S21E) to fit properly. In this configuration, the serine residue can also form a hydrogen bond with the lateral chain of the glutamate of the adjacent TssL molecule to restore a favorable dimer interface (supplemental Fig. S5). In the bacterial two-hybrid assay, the TssL-L18S/S21E variant dimerizes (Fig. 6), demonstrating that contacts between the h1 helices of two adjacent TssL molecules can be restored by suppressive mutations. Taken together, these results support a model in which TssL self-interaction occurs mainly through the TMS, but in which contacts between helices 1 of the soluble domain contribute to the stability of the TssL dimer (see model in Fig. 7). However, cytoplasmic contacts are critical for TssL function; although the single L18S and S21E TssL derivatives were unable to complement the biofilm and Hcp release defects of the tssL mutant, production of a TssL protein carrying both substitutions was fully functional (Fig. 4).
FIGURE 6.
Bacterial two-hybrid analysis of TssL cytoplasmic domain. Pairs of plasmids corresponding to proteins fused to the T18 or T25 domains of the Bordetella adenylate cyclase (as indicated) were transformed into the BTH101 reporter strain, and the β-galactosidase activity is reported in Miller units. Error bars indicate S.D.
FIGURE 7.
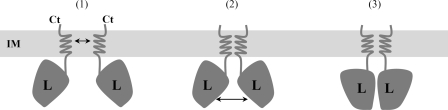
Schematic model for TssL self-interaction. TssL dimerization through the inner membrane-anchored trans-membrane segment (i) induces contacts between the cytoplasmic domains (ii), leading to a functional TssL dimer (iii).
DISCUSSION
The T6SS is an important determinant of bacterial pathogenesis. It can act as a direct virulence factor by producing and translocating effector domains into eukaryotic host cells such as the Vibrio cholerae or the Aeromonas hydrophila T6SS (7–9). Several T6SS have been found to be required for bacterial targeting, and the Pseudomonas aeruginosa HSI-1 T6SS has been shown to export canonical substrates into bacterial recipient cells (2–5, 10, 11). The overall architecture of the T6SS is not known, but the current data converge to the idea that the T6SS is composed of two subcomplexes. We recently reported co-immunoprecipitation data that identify four components, TssL, TssM, TssJ, and TagL, constituting the membrane-associated subcomplex (27). Although TssJ is an outer membrane lipoprotein (12), the three other components are anchored in the inner membrane. TssM interacts with TssJ, and this interaction has been characterized through different approaches (29, 31). TssM and TssJ associate onto a 1:1 complex with a Kd in the micromolar range (29). The TssJ protein has been structurally characterized and folds as a β-sandwich characteristic of the transthyretin family (29, 30). One of the TssJ loops has been shown to specifically contact TssM (29).
Here, we report the structural characterization of the cytoplasmic domain of the TssL subunit. We recently showed that TssL is a membrane protein anchored to the inner membrane protein through a C-terminal trans-membrane segment (36). The cytoplasmic domain (amino acids 1–184) was produced and crystallized. The TssL cytoplasmic domain is composed of two bundles of three helices, each of which are combined to form a globular structure. Although TssL is an inner membrane protein, its cytoplasmic domain behaves as a classical soluble protein; (i) it possesses numerous charged residues that are scattered on the whole domain surface, and (ii) it does not exhibit any hydrophobic patches. This compact domain is connected to the inner membrane by a ∼20-amino acid linker. The absence of catalytic residues or of a putative catalytic site suggests that TssL is a structural component of the T6SS.
The overall TssLC fold resembles a hook, and one may hypothesize that TssL recruits other subunits during the assembly of the apparatus or the protein effectors prior to their translocation. Indeed, TssL interacts with the TssM inner membrane protein (31, 32). A yeast two-hybrid approach showed that the cytoplasmic domains of the Agrobacterium tumefaciens TssL and TssM proteins are required for their association (32). An additional support for this interaction is the observation that TssL stabilizes TssM in Francisella tularensis (35). In the TssLC structure, several elements might be required to mediate the contacts with TssM. First, the plug-shaped h7-h8 loop, which has been shown to be required for TssL function (Fig. 3), might be necessary for this interaction. Interestingly, this loop is not conserved among the TssL homologues. However, the TssM-TssJ interaction is mediated by a nonconserved loop, which has been proposed to serve as a specificity determinant during the T6SS assembly, especially when several T6SS are encoded within the genome (29). A similar role for the TssL h7-h8 loop can be suggested. Another TssL structural element that can accommodate the cytoplasmic domain of TssM is the highly conserved groove located on the core domain and facing the linker (see supplemental Fig. S3). Experiments currently underway are dedicated to define the contribution of these elements for formation of the TssL-TssM complex.
In this study, we also showed that the TssL protein oligomerizes. The detergent-solubilized, membrane-extracted full-length protein is dimeric in solution, as shown upon gel filtration (Fig. 5). This dimer is sufficiently stable to be partly resistant to boiling in SDS-loading buffer as a band corresponding to a dimer can be observed upon SDS-PAGE, both from purified full-length protein and from cell extracts. Dimer formation is strengthened by chemical cross-linking both in vivo and in vitro. The dimerization mainly involves the trans-membrane segment, which has been previously shown to be important for TssL function (36). A purified TssL protein deleted of the TMS is monomeric in solution. However, dimerization of the cytoplasmic domains is visualized upon chemical cross-linking or in bacterial two-hybrid experiments. These results demonstrate that contacts between the cytoplasmic domains are established, albeit at low affinity, suggesting that this interaction is transient or weak. How the cytoplasmic domains self-interact has been tested by analyzing the possible packings. Although the most probable packing (interface of 1190 Å) involves contacts between the linker of two adjacent molecules, our in vivo data do not support this mode of self-interaction; a TssL protein deleted of the linker is still able to form dimers. The data rather support a model in which contacts are mediated by the h1 helices of the two monomers. Mutagenesis of 2 residues located at the interface, Leu-18 and Ser-21, abrogates TssL cytoplasmic dimerization in the bacterial two-hybrid assay. Interestingly, the combination of the L18S and S21E substitutions, which reconstitute a favorable dimer interface, restores the interaction in the bacterial two-hybrid assay, as well as the phenotypes associated with the tssL mutation. It is also worthy to note that although the S21E TssL variant is unstable, the L18S/S21E TssL variant is as stable as the WT protein (Fig. 4), suggesting that reconstituting the cytoplasmic contacts within the TssL dimer has a stabilizing effect. These results demonstrate that (i) two TssL proteins contact each other through the cytoplasmic domains and (ii) cytoplasmic contacts are functionally relevant for TssL function. Interestingly, dimerization of the Serratia marcescens TssL cytoplasmic domain has also been evidenced in a bacterial two-hybrid assay,6 strengthening the functional role of this interface. As noted above, this packing is not predicted to be the more probable based on the contact area (∼700 Å). However, in this packing, the linkers are arranged in a parallel manner, allowing the two TMS to be brought together. The contacts mediated by the TMS therefore significantly increase the interface between the two monomers.
From these data, we propose a zipper-like model in which the TssL TMS is responsible for the dimerization of the protein. Formation of the contacts in the membrane might increase the local concentration, sufficiently to favor the association between the cytoplasmic domains (Fig. 7). The TssL dimer being the functional form of TssL, it would be interesting to test whether dimer formation triggers the recruitment of a protein partner, such as TssM.
The sequence alignments provided in this study show that although the TssL homologues are not highly conserved, the predicted secondary structures are identical. We can therefore consider the Sci-1 EAEC TssLC three-dimensional structure as a prototype for all the T6SS-associated TssL homologues. More interestingly, the T6SS TssL proteins and the T4bSS-associated IcmH/DotU proteins also share conservation of the secondary structures, and we therefore propose that the TssL structure might also represent a prototype for all the DUF2077 family members. Our results therefore extend the list of characteristics shared by the TssL-TssM and IcmH/DotU-IcmF pairs, highlighting the homology between these complexes associated with two different secretion systems. What is the function of these pairs in the T6 and T4bSS and how these pairs have been exchanged during the evolution are two questions that remain to be answered.
Supplementary Material
Acknowledgments
We thank the members of the Cascales, Lloubès, Bouveret, and Sturgis research groups for discussions, Emmanuelle Bouveret for use of the TECAN microplate reader, for plasmids, and for advice regarding the bacterial two-hybrid technique, Sarah Coulthurst (University of Dundee, Scotland, United Kingdom) for helpful comments and sharing unpublished results, and the two anonymous reviewers for constructive comments. We gratefully acknowledge the help of Pierre Legrand (Proxima 1) for data collection, the ESRF and Soleil synchrotrons for beamtime allocation, and Eric Azaraille for encouragements.
This work was supported by the CNRS and funded by a grant from the Agence Nationale de la Recherche (Grant ANR-10-JCJC-1303-03) (to E. C) and by the CNRS, the Université de la Méditerranée, and grants from the Marseille-Nice Génopole, IBiSA, and the Fondation de la Recherche Médicale (Grant FRM DEQ2011-0421282) (to C. C.).

This article contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (code 3U66) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
S. Coulthurst, personal communication.
- T6SS
- Type VI secretion system
- T4bSS
- Type IVb secretion system
- Tss
- Type VI secretion subunit
- Hcp
- hemolysin-co-regulated protein
- VgrG
- valine-glycine repeat protein
- EAEC
- enteroaggregative E. coli
- SAD
- single anomalous dispersion
- DDM
- n-dodecyl-β-d-maltopyranoside
- PFA
- para-formaldehyde
- TMS
- trans-membrane segment(s)
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 2. Schwarz S., Hood R. D., Mougous J. D. (2010) What is Type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., Rohmer L., Tolker-Nielsen T., Skerrett S. J., Mougous J. D. (2010) Burkholderia Type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6, e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S. (2010) The Vibrio cholerae Type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murdoch S. L., Trunk K., English G., Fritsch M. J., Pourkarimi E., Coulthurst S. J. (2011) The opportunistic pathogen Serratia marcescens utilizes Type VI secretion to target bacterial competitors. J Bacteriol. 193, 6057–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The Type VI secretion system: translocation of effectors and effector domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 7. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J.J. (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Translocation of a Vibrio cholerae Type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010) A Type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP-ribosylation of actin. J Bacteriol. 192, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A Type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell A. B., Hood R.D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aschtgen M. S., Bernard C.S., De Bentzmann S., Lloubès R., Cascales E. (2008) SciN is an outer membrane lipoprotein required for Type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190, 7523–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weber B., Hasic M., Chen C., Wai S. N., Milton D. L. (2009) Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11, 3018–3028 [DOI] [PubMed] [Google Scholar]
- 14. de Pace F., Nakazato G., Pacheco A., de Paiva J. B., Sperandio V., da Silveira W. D. (2010) The Type VI secretion system plays a role in Type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun. 78, 4990–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shalom G., Shaw J. G., Thomas M. S. (2007) In vivo expression technology identifies a Type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 [DOI] [PubMed] [Google Scholar]
- 16. Cascales E. (2008) The Type VI secretion toolkit. EMBO Rep. 9, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009) Dissecting the bacterial Type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filloux A., Hachani A., Bleves S. (2008) The bacterial Type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 19. Mougous J. D., Cuff M.E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) The phage λ major tail protein structure reveals a common evolution for long-tailed phages and the Type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V.V., Arisaka F., Rossmann M. G. (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 [DOI] [PubMed] [Google Scholar]
- 23. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009) Remodeling of VipA/VipB tubules by ClpV-mediated threading is crucial for Type VI protein secretion. EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cascales E., Cambillau C. (2012) Structural biology of Type VI secretion systems. Philos. Trans. R Soc. Lond. B. Biol. Sci. 367, 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanamaru S. (2009) Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. U.S.A. 106, 4067–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Records A. R. (2011) The Type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 24, 751–757 [DOI] [PubMed] [Google Scholar]
- 27. Aschtgen M. S., Gavioli M., Dessen A., Lloubès R., Cascales E. (2010) The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol. Microbiol. 75, 886–899 [DOI] [PubMed] [Google Scholar]
- 28. Aschtgen M. S., Thomas M. S., Cascales E. (2010) Anchoring the Type VI secretion system to the peptidoglycan: TssL, TagL, TagP … what else? Virulence 1, 535–540 [DOI] [PubMed] [Google Scholar]
- 29. Felisberto-Rodrigues C., Durand E., Aschtgen M.S., Blangy S., Ortiz-Lombardia M., Douzi B., Cambillau C., Cascales E. (2011) Towards a structural comprehension of bacterial Type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7, e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rao V. A., Shepherd S. M., English G., Coulthurst S. J., Hunter W. N. (2011) The structure of Serratia marcescens Lip, a membrane-bound component of the Type VI secretion system. Acta Crystallogr. D Biol. Crystallogr. 67, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng J., Leung K. Y. (2007) Dissection of a Type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206 [DOI] [PubMed] [Google Scholar]
- 32. Ma L. S., Lin J. S., Lai E. M. (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker A motif is required for Type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Segal G., Feldman M., Zusman T. (2005) The Icm/Dot Type IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29, 65–81 [DOI] [PubMed] [Google Scholar]
- 34. Nagai H., Kubori T. (2011) Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol. 2, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Bruin O. M., Duplantis B. N., Ludu J. S., Hare R. F., Nix E. B., Schmerk C. L., Robb C. S., Boraston A. B., Hueffer K., Nano F. E. (2011) The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB, and DotU suggest roles in Type VI secretion. Microbiology 157, 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aschtgen M. S., Zoued A., Lloubès R., Journet L., Cascales E. (2012) The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC. MicrobiologyOpen. 1, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sexton J. A., Miller J. L., Yoneda A., Kehl-Fie T. E., Vogel J. P. (2004) Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun. 72, 5983–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. VanRheenen S. M., Duménil G., Isberg R. R. (2004) IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun. 72, 5972–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brunet Y. R., Bernard C.S., Gavioli M., Lloubès R., Cascales E. (2011) An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a Type VI secretion gene cluster. PLoS Genetics 7, e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Battesti A., Bouveret E. (2008) Improvement of bacterial two-hybrid vectors for detection of fusion proteins and transfer to pBAD-tandem affinity purification, calmodulin-binding peptide, or 6-histidine tag vectors. Proteomics 8, 4768–4771 [DOI] [PubMed] [Google Scholar]
- 41. van den Ent F., Löwe J. (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem. Biophys Methods 67, 67–74 [DOI] [PubMed] [Google Scholar]
- 42. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller J. H. (ed) (1992) A Short Course in Bacterial Genetics: A Laboratory Manual for Escherichia coli and Related Bacteria, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Sulzenbacher G., Gruez A., Roig-Zamboni V., Spinelli S., Valencia C., Pagot F., Vincentelli R., Bignon C., Salomoni A., Grisel S., Maurin D., Huyghe C., Johansson K., Grassick A., Roussel A., Bourne Y., Perrier S., Miallau L., Cantau P., Blanc E., Genevois M., Grossi A., Zenatti A., Campanacci V., Cambillau C. (2002) A medium-throughput crystallization approach. Acta Crystallogr. D. Biol. Crystallogr. 58, 2109–2115 [DOI] [PubMed] [Google Scholar]
- 45. Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A. 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 47. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J Appl Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cowtan K. (2010) Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 50. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 51. Miroux B., Walker J. E. (1996) Overproduction of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 52. Cascales E., Bernadac A., Gavioli M., Lazzaroni J. C., Lloubes R. (2002) Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184, 754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vincentelli R., Canaan S., Offant J., Cambillau C., Bignon C. (2005) Automated expression and solubility screening of His-tagged proteins in 96-well format. Anal. Biochem. 346, 77–84 [DOI] [PubMed] [Google Scholar]
- 54. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 57. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.