Background: CymA is the central menaquinol-7 dehydrogenase in anaerobic respiration of Shewanella sp.
Results: CymA uses menaquinone-7 as a cofactor.
Conclusion: CymA has one cofactor site that is specific for menaquinone-7 and one low affinity Q/QH2 site that is in equilibrium with the quinone pool.
Significance: The function of quinones needs to be reevaluated and crystallographically determined quinone binding pockets might not be the site of quinone conversion.
Keywords: Cytochromes, Dehydrogenase, Electrochemistry, Quinones, Respiration, Quartz-crystal Microbalance with Dissipation
Abstract
Little is known about enzymatic quinone-quinol interconversions in the lipid membrane when compared with our knowledge of substrate transformations by globular enzymes. Here, the smallest example of a quinol dehydrogenase in nature, CymA, has been studied. CymA is a monotopic membrane tetraheme c-type cytochrome belonging to the NapC/NirT family and central to anaerobic respiration in Shewanella sp. Using protein-film electrochemistry, it is shown that vesicle-bound menaquinone-7 is not only a substrate for this enzyme but is also required as a cofactor when converting other quinones. Here, we propose that the high concentration of quinones in the membrane negates the evolutionary pressure to create a high affinity active site. However, the instability and reactivity of reaction intermediate, semiquinone, might require a cofactor that functions to minimize damaging side reactions.
Introduction
Oxidoreductases that convert membrane-bound quinones play a key part in the metabolism of archaea, bacteria, and eukaryotes. Quinone oxidoreductases are involved in the electron transport chains of both oxidative phosphorylation and photosynthesis, where they catalyze the two-electron, two-proton conversion to drive the generation of transmembrane proton (ΔpH) and electrical gradients (ΔΨ). Generally, quinones are known as lipophilic electron and proton shuttles, but quinones may also function as cofactors. For instance, it is well known that the quinone in the QA site of the photosynthetic reaction center is a tightly bound cofactor and functions to transfer electrons on their way to reduce the quinone in the QB site (1, 2). In an ubiquinol oxidase from Escherichia coli, cytochrome bo3, a ubiquinone in the high affinity site, QH, also acts as a tightly bound cofactor (3, 4). The QH site accepts electrons from a ubiquinol at a low affinity site, QL, where the ubiquinone is thought to rapidly exchange with the quinone pool in the membrane. Like the QB in photosynthetic reaction centers, the QL site is seen to be the active site of cytochrome bo3. Although the QH is well characterized by spectroscopy and mutagenesis (e.g. (5)), the exact location of the QL-site remains elusive. No ubiquinone was found in the crystal structure, but close inspection of the structure revealed a possible quinone binding site with the properties expected for the QH site (6). Mutagenesis studies confirmed the proposed amino acid residues (Arg-71, Asp-75, His-98, Gln-101) did indeed form the QH binding pocket (6, 7). Still, no structural features were found consistent with the presence of a QL site, leading the crystallographers to speculate that there is either no QL site or that the QL site is only formed after a ubiquinone is bound to the QH site (6).
In cytochrome bc1, two ubiquinones have been shown to bind in a single quinone binding site (Qo) (8–12). The binding of both quinones seems to be cooperative as the binding affinity of both quinones is almost equally disrupted by mutagenesis of nearby residues or by binding of competitive inhibitors (8–12). Cytochrome bc1 operates via a unique Q cycle, which involves coupling of proton and electron transfers of ubiquinones at catalytic sites at opposite sides of the membrane (at the Qo and Qi sites). Proposals have been put forward that cytochrome bc1 binds multiple ubiquinones at the Qo site as a mechanism to prevent semiquinone-mediated short-circuiting, which would result in energy loss (for review, see Ref. 13). Similar to the case of cytochrome bo3, crystallography has been unable to detect any ubiquinone at the Qo site, and it remains unclear how two ubiquinones can bind at this site. Structures with inhibitors at the Qo site show only one inhibitor bound at a time, although distinct binding loci are found for the inhibitors stigmatellin and myxothiazol (13).
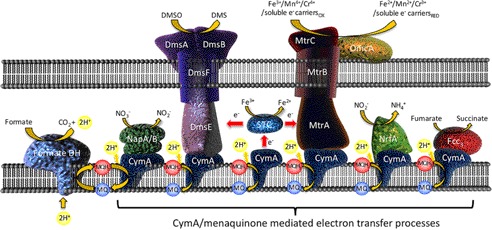
Here, we describe enzymatic studies of a relatively simple quinone converting enzyme, CymA, belonging to the NapC/NirT superfamily. In contrast to most other quinone oxidoreductases, members of this family are monotopic membrane proteins with one globular head domain facing the periplasm in Gram-negative bacteria. CymA is found in the inner membrane of the γ-proteobacterium Shewanella oneidensis MR-1, which is a facultative anaerobe and an important model organism for bioreactor, bioengineering, and bioremediation studies (14–18). Shewanella sp. are bestowed with a remarkably diverse multicomponent and branched electron transport chain (19–24). This respiratory plasticity allows S. oneidensis MR-1 to link proton translocation to the reduction of a wide variety of electron acceptors such as O2, fumarate, nitrate, arsenate, dimethyl sulfoxide, trimethylamine oxide, Fe(III), Mn(IV), Cr(VI), and V(V) oxides, and various forms of other carbonaceous and sulfur-based compounds (25–31). CymA plays a central role in this multibranched respiratory chain where it couples the oxidation of menaquinone-7 (MK-7) to a number of multi-heme cytochromes (19, 32–34), terminal reductases (35–38), and soluble extracellular electron carriers (21, 24, 39) (Fig. 1).
FIGURE 1.
Schematic of the role of CymA in S. oneidensis MR-1 anaerobic respiration. Inner membrane-bound dehydrogenases pass electrons through the lipophilic menaquinone pool to CymA where they are passed to four functionally distinct periplasmic reductases. CymA also plays the key role in shuttling electrons to terminal electron acceptors outside the cell though either outer-membrane-associated heme proteins directly such as DmsE or MtrA or via small tetraheme cytochrome (STC) as a periplasmic shuttle (46).
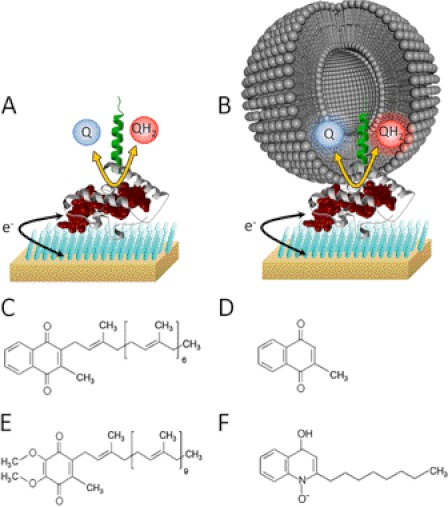
The only structure solved of the NapC/NirT family is NrfH (40, 41). Interestingly, NrfH has only been co-crystallized with NrfA, where NrfA contributes an axial ligand in the form of a lysine residue to one for the four heme groups. As far as we are aware, experiments to assess quinol oxidation by CymA have only been performed in the presence of redox partner proteins such as the periplasmic fumarate reductase Fcc3 (37, 38) or by chemical means (42), whereas characterization of the catalytic activity of the NapC/NirT superfamily has been restricted to intact cell studies, assays with membrane extracts, or soluble truncated mutants (23, 43–45). Recent characterization of CymA by electronic absorbance, magnetic circular dichroism, and electron paramagnetic resonance spectroscopy identifies three low-spin bishistidine coordinated hemes and one high-spin heme with His/H2O coordination.3 Binding the menaquinol analog, 2-heptyl-4-quinolinol-N-oxide (HQNO),4 to CymA identifies the high-spin heme as adjacent to the quinol binding site. The aim of this study was to characterize the enzyme kinetics and substrate specificity of this critical respiratory hub in Shewanella. We isolated full-length recombinant CymA expressed in S. oneidensis MR-1 and immobilized the enzyme on a gold electrode modified with a self-assembled monolayer. Protein-film electrochemistry was used to investigate menaquinone-7 (MQ-7) interactions with CymA in a lipophilic environment (Fig. 2), revealing that in this relatively small enzyme, menaquinone-7, is required as a cofactor.
FIGURE 2.
Schematics show a space-filling model of NrfH, a homologue of CymA, immobilized on a self-assembled monolayer-modified template-stripped ultra-flat gold electrode with menadione (MQ-0) in the solvent phase (A) and menaquinone-7 POPC vesicle with MQ-7 in the lipid-phase (B). The SAM is shown as blue bars, the CymA polypeptide is in white (globular head domain) or green (lipophilic tail domain), and the prosthetic heme groups is in red. C–F, structures of potential substrates and inhibitors of CymA used in this study are MQ-7 (C), MQ-0 (D), UQ-10 (E), and HQNO (F).
MATERIALS AND METHODS
Specialist Treatments of Chemicals
All solvents were HPLC grade (Fisher) and were used as received. MQ-7 (Wako) and ubiquinone-10 (UQ-10, Sigma) stock solutions were prepared at concentrations of 1 mg/ml in chloroform. Menadione (MQ-0, Sigma) was prepared as a stock at 4 mg/ml in 75:25 propanol:H2O and diluted 1:10 in 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) for use in electrochemical procedures. HQNO was purchased from Alexis Biochemicals, and a stock solution of 50 mm was prepared in chloroform. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; Avanti Polar Lipids, Inc., Alabaster, AL) was aliquoted to 5 mg and dried under an N2 stream before storage under an N2 atmosphere at −20 °C.
Expression and Purification of CymA
The cymA (locus tag SO4591) gene of S. oneidensis MR-1 (MR-1) was cloned according to a protocol described previously (47, 48). MR-1 cultures were grown microaerobically, and membranous CymA was isolated as described elsewhere.3 Full-length recombinant CymA was confirmed by mass spectrometry, N-terminal, sequencing, and immunoblotting. SDS-PAGE analysis identified ∼90% pure hemoprotein with a band at 24 kDa. Electronic absorption spectroscopy characterized a typical c-type cytochrome (A407 nm/A275 nm = 4) consistent with four inserted hemes per protein monomer. Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Sigma) with bovine serum albumin as the standard.
Solvent Phase Extraction of Quinones and HPLC
Quinones were isolated from CymA samples by solvent extraction (twice) with aqueous methanol:acetone (50/50 v/v) and petroleum ether (60/40 v/v) as described in previously (43). 0.5 ml of protein sample, typically 0.1–0.5 μm concentration, was stirred with 3 ml of acetone/methanol for 30 min then stirred with 1 ml of petroleum ether for 1 min and allowed to stand. The petroleum ether layer was pipetted off and evaporated under nitrogen gas. Quinone extracts were resuspended in 100 μl of ethanol/methanol (50/50 v/v). Identification and concentrations of MQ-7 were determined by high performance liquid chromatography analysis using a JASCO HPLC system with a Phenomenex reverse phase C18 (5 μm) column (dimensions 150 × 4.6 mm) with ethanol/methanol (50/50 v/v) as the mobile phase at 0.3 or 0.5 ml/min (49). Elution profiles were monitored by electronic absorbance (JASCO UV-visible multiwavelength detector) at 248, 270, and 290 nm to distinguish menaquinones (peak maxima at 248 and 270 nm) and ubiquinones (290-nm peak maximum). Concentrations of menaquinone-7 (Wako) standard samples were determined using an extinction coefficient ϵ248 nm = 18.9 (mm−1cm−1). Menaquinone-7 eluted at 18 min retention time (0.5 ml/min).
Vesicle Preparation
MQ-7, UQ-10, and HQNO were reconstituted into dried POPC lipids (Avanti) using a 50:50 chloroform/methanol mixture before being dried under a stream of nitrogen. The resulting film was resuspended by vortexing in 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) to a concentration of 5 mg/ml. A homogenous vesicle solution was prepared by extrusion through a 200-nm nucleopore track-etched polycarbonate membrane (Whatman) using a mini-extruder (Avanti) at 20 °C.
Reconstitution of CymA
On the day of reconstitution, dried POPC lipids containing quinone were prepared by vortexing in 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) to a concentration of 20 mg/ml. CymA was introduced into POPC liposomes by the method based on that of Carter and Gennis (50). Lipid vesicles, octylglucoside (OG), and CymA were mixed by inversion to final concentrations of 16.4 mg/ml lipid, 45 mm OG, and 0.16 mg/ml CymA. After 15 min of incubation on ice, the lipid/protein mixture was rapidly diluted 200-fold with 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) pre-cooled to 4 °C, and centrifuged at 142,000 × g for 1 h to pellet the proteoliposomes. The proteoliposomes were resuspended in the same volume of fresh cold 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) and centrifuged at 142,000 × g for 1 h to pellet the proteoliposomes. Proteoliposomes were resuspended in 1 ml of 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) and extruded through a 200-nm track-etched polycarbonate membrane using a mini-extruder (Avanti) to ensure unilamellar homogeneous vesicles.
Electrode Preparation and Modification
Routinely, experiments were carried out with ultra-flat template stripped gold (TSG) working electrodes prepared as described previously (51). 150 nm of 99.99% gold (Goodfellows) was evaporated on cleaned silicon wafers using an Edwards Auto 306 evaporator at <1–2 × 10−6 mbar. Cleaned 1.2-cm2 glass slides were glued to the gold layer with Epo-Tek 377 and cured for 2 h at 120 °C. Before use the glass slides were detached from the silicon wafers to expose the TSG surface. Self-assembled monolayers (SAMs) were prepared by incubating the TSG electrode with 1 mm concentrations of pure or mixed thiols: 8-mercaptooctanol (8-OH, Sigma), 8-amino-1-octanethiol (NBS Biologicals), 1-octanethiol (Sigma), or 8-mercaptooctanoic acid (Sigma) for 12 h. After incubation the excess thiol was gently washed away with isopropyl alcohol and methanol; the electrodes were then dried in a stream of N2 and incubated with 0.1 μm CymA (monomer concentration) for 20 min. After this the enzyme-modified electrode was 3× rinsed with water/buffer and used in the electrochemistry experiments, taking care the electrodes remained immersed in water/buffer at all times. MQ was routinely removed from the immobilized sample by washing the enzyme-modified electrode five times with 0.01% n-dodecyl β-d-maltoside (DDM; w/v) in buffer followed by rinsing three times with water/buffer to remove any residual detergent.
Cyclic Voltammetry (CV) and Electrochemical Impedance Spectroscopy
A bespoke glass spectroelectrochemical cell was used with a standard three-electrode setup (52). Typically, an ultra-flat TSG working electrode was embedded in a polytetrafluoroethylene (PTFE) holder with a rubber O-ring seal (A = 0.2 cm2); a platinum wire counter electrode and a saturated silver/silver chloride electrode (Ag/AgCl) completed the circuit in the buffer volume. All potentials are given versus the standard hydrogen electrode (SHE). Routinely, experiments were conducted in a 20 mm MOPS, 30 mm Na2SO4 (pH 7.4), buffer; however, for pH variation experiments, 20 mm MOPS was replaced by 6.7 mm MOPS, 6.7 mm MES, 6.7 mm Tris/HCl. The cell was used in a steel mesh Faraday cage to minimize electrical noise, and all experiments were conducted inside an N2 filled glovebox (MBraun MB 150 B-G) where the O2 levels were <1 ppm. All solutions were purged with N2 for 1 h and stored in the glovebox at least 24 h before use. Electrochemical measurements were recorded at 21 °C using an Autolab electrochemical analyzer with a PGSTAT30 potentiostat, SCANGEN and ADC750 modules, and FRA2 frequency analyzer (Ecochemie). To control SAM quality before electrode modification with CymA, electrochemical impedance spectroscopy impedance spectra were recorded on each SAM electrode before the addition of protein. Analog CVs were routinely recorded at a scan rate of 10 mV/s on SAMs and enzyme-modified electrodes before and after the addition of the substrate (MQ-7 and UQ-10 in 200 nm POPC vesicles, MQ-0 in 7.5% n-propyl alcohol, 92.5% 20 mm MOPS, 30 mm Na2SO4 (pH 7.4) (v/v)). Unless indicated otherwise, in all experiments using vesicles, non-directly associated vesicles were washed out after 30 min of incubation. Cyclic voltammetry experiments were routinely carried out by holding the potential at 0.3 V for 5 s before cycling at a scan rate of 10 mV/s in the potential window from 0.3 V to −0.6 mV (versus Ag/AgCl). The electroactive coverage of the enzyme was determined using the area of the base-line-subtracted signals from CVs measured at a scan rate (υ) of 10 mV/s of CymA on 8-OH modified gold in the absence of MQ-7 after washing with 0.01% DDM (w/v). The base line was subtracted using SOAS software, available from Dr. C. Léger (53), and the coverage was calculated with area/υ = nFAΓ, in which A is the surface area of the electrode, Γ is the enzyme coverage, F is the Faraday constant, and n is the number of electrons per CymA (n = 4).
Cyclic Voltammetry in Quartz Crystal Microbalance with Dissipation (QCM-D)
CV/QCM-D experiments were conducted with a Q-sense electrochemistry module (QEM 401) and a Ag/AgCl DRIREF-2SH electrode using a Q-Sense E4 (Q-Sense AB). Experiments were performed using gold sensor crystals at 22 °C, with the flow rate held at 70 μl/min before conducting procedures, and gold-modified QCM-D crystals were cleaned by bath-sonicating them with MilliQ water (30 min), 0.4% SDS (w/v) detergent (20 min), and again MilliQ water (20 min). After that, gold-covered crystals were treated for 20 min with UV/ozone (UV/ozone cleaning system, low pressure quartz-mercury vapor lamp emitting 254 and 185 nm UV) followed by 40 min of incubation in ethanol in a Soxlet extractor to reduce the gold oxides formed by the UV/ozone cleaning process. Immediately after reduction, the crystals were transferred into 1 mm thiol solutions and incubated for 16 h (as described previously). On graphs, changes in the dissipation (ΔD) and frequency (Δf) of the 7th overtone are presented, whereas 3rd, 5th, 9th, 11th, and 13th overtones were also recorded. The cumulative data were used during the modeling of viscoelastic properties of the adsorbed proteins. Modeling was performed using QTools 2 Qsense software under the assumptions of the Sauerbrey equation.
RESULTS
For interfacial electron transfer, electronic coupling between the electrode and the enzyme redox center is essential. Coupling depends on the orientation of the adsorbed protein on the electrode surface (54, 55), which in turn depends on several factors such as surface chemistry and topology of the electrode (54–56), pH, ionic strength, and viscosity of the buffer solution (57). Here, SAMs were used to control the physiochemical properties of a gold electrode. By changing the surface coverage of various thiol compounds, hydrophilicity (8-OH), hydrophobicity (1-octanethiol), and charge (8-mercaptooctanoic acid and 8-amino-1-octanethiol) were varied systematically on the electrode. The SAM layer composition was found to be crucial, and CymA electrochemistry was only detected on pure 8-OH or mixtures of 8-OH and 8-amino-1-octanethiol (up to 20% 8-amino-1-octanethiol in the thiol solution used to prepare the SAMs). The remainder of the work was carried out with SAMs of pure 8-OH as this was easiest to prepare.
Quartz-Crystal Microbalance
Using an electrochemical cell, QCM-D measured was combined with CV to examine the mass and viscoelastic properties of the adsorbed CymA under the electrochemical conditions described below. Immobilization of CymA (0.1 μm) was almost instantaneous, as manifested by the sharp drop in frequency to −15 Hz (Fig. 3). The frequency and dissipation remained constant irrespective of the addition of more CymA or removal of non-associated CymA from solution. Furthermore, the frequency and dissipation did not change significantly when conducting cyclic voltammetry, indicating that the CymA is not influenced by the application of electric fields within the potential window used in this work. The lack of significant changes in dissipation enables the use of the Sauerbrey equation to calculate the mass of the CymA film. A total protein mass of 0.24 μg/cm2 equates to ∼12 pmol/cm2 of CymA monomers when taking into account that 30% of the adsorbed mass is due to water associated with the protein film (58). Taking the geometric size of NrfH (40), a close homologue of CymA, this coverage is consistent with a close-packed monolayer of CymA on the electrode surface.
FIGURE 3.
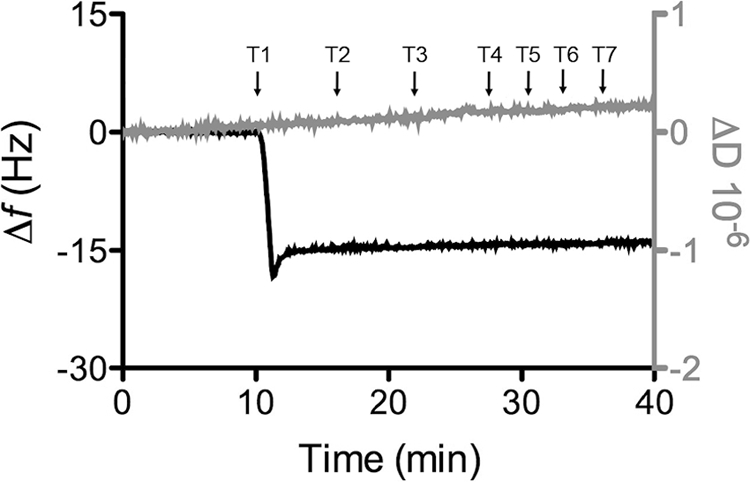
QCM-D results of a 8-OH- modified gold surface in buffer (20 mm MOPS, 30 mm Na2SO4 (pH 7.4)), plotting (black line, left axis) frequency and (gray line, right axis) dissipation against time. Time points (T1–7) of modifications to the procedure are indicated by arrows: T1, 0.1 μm CymA in buffer; T2, 0.01% DDM (w/v) in buffer; T3, buffer only. At T4, T5, T6, and T7, cyclic voltammograms (0.1 to −0.4 V versus SHE) were performed in buffer at 1, 10, 100, and 1000 mV/s, respectively. The plot shown is representative of triplicate experiments.
Non-catalytic Cyclic Voltammetry
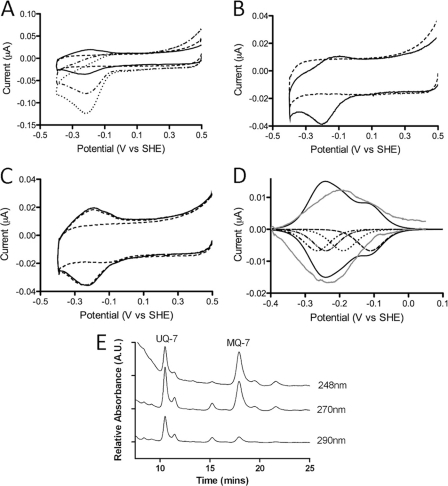
Contrary to expectations, cyclic voltammograms of CymA resembled a diffusion-controlled reductive wave independent on the presence of CymA in the electrolyte (dotted line, Fig. 4A). The current of the reduction wave decreased with successive scans (dot-dashed line, Fig. 4A), but clear non-turnover oxidation and reduction signals, like those previously reported for truncated mutants of CymA for which the N-terminal membrane-anchoring was removed (59), were never observed. We note again that QCM-D indicates that a stable monolayer of CymA is present on the electrode under these conditions. This suggests that the diffusion-controlled behavior is due to a small electroactive compound that co-purifies with CymA and co-immobilizes in the protein film. Solvent-phase extraction of the CymA followed by analysis with HPLC and electronic absorption spectroscopy revealed the presence of quinones with MQ-7 as the major component (Fig. 4E). The ratio MQ-7 to CymA was found to be ∼4 for the sample used to obtain Fig. 4A, although it was observed that this ratio varied between protein samples. We further note that the ratio on the electrode surface might not necessarily agree with the ratio in the protein sample, as either CymA or MQ-7 might preferentially adsorb in the electrode.
FIGURE 4.
CVs of 8-OH-modified TSG electrode before (blank) and after formation of a CymA film. All CVs were recorded in 20 mm MOPS, 30 mm Na2SO4 buffer (pH 7.4) at a 10 mV/s scan rate. A, CVs for as-isolated CymA are shown. The dotted and dash-dotted lines show the first and fifth scans of the same protein film (CVs were measured consecutively) compared with the blank (dashed line) and apoCymA redox peaks (solid line). See “Results” for more details and formation of the apoCymA film. B, CV of an as-isolated CymA sample after a DDM washing procedure on DEAE-Sepharose was included in the purification; see (solid line) compared to the blank (dashed line). See “Results” for more information on the preparation of the CymA sample. C, CV of immobilized apoCymA film is shown. Consecutive scans of CymA show redox peaks (solid and dash-dotted lines) compared with the blank (dashed line). D, base-line-subtracted CymA redox peaks (gray line, data taken from C) are compared with modeled data based on known reduction potentials of the four hemes; see “Results” for more information. The contribution of each heme is shown separately (dotted and dashed lines); the sum of these currents is shown as a solid black line. The modeling indicates electroactive CymA coverage of 3.3 pmol/cm2. E, representative HPLC analysis of solvent phase extract from purified CymA is shown. Menaquinone is characterized by absorption at 248 and 270 nm with little absorption at 290 nm. Identification of MQ-7 is by comparison to a standard. The peak at a retention time of 10.5 min is suggested to be UQ-7-based on comparison with literature and the fact that Shewanella sp. uses UQ-7 under aerobic growth.
As quinones could clearly contribute to the voltammetry by direct reaction at the electrode or with their redox transformation catalyzed by CymA, the sample of CymA was subject to an DDM wash while bound to DEAE-Sepharose. This procedure produced only minor perturbation of the electronic absorbance, magnetic circular dichroism, and electronic paramagnetic resonance spectra of CymA, indicating that this extra step did not drastically alter the heme environment. When this washed material was adsorbed on the electrode, the magnitude of the reductive wave is much reduced (for equal coverage of CymA on electrode) and there is evidence of discrete peaks for oxidation and reduction of CymA, i.e. non-turnover signals (Fig. 4B).
Further separation of CymA from co-purified materials was achieved by taking advantage of the high affinity of CymA for the gold electrode and high stability of the protein film. After five successive washes with 0.01% DDM (w/v) in situ, cyclic voltammograms of CymA films show non-turnover redox signals with peak maxima at −0.20 and −0.23 V versus SHE, indicating that it is possible to remove the co-purified quinones from CymA (solid line, Fig. 4A). QCM-D confirmed that the 0.01% DDM (w/v) washing procedure does not perturb the CymA film (Fig. 3). We refer to this DDM-washed CymA film as apoCymA. The area under the reduction peaks indicates a CymA coverage of 3–4 pmol/cm2 (assuming 4 electrons per monomer). It is difficult to unambiguously determine the reduction potential of the four heme groups in CymA, as they are unresolved in the broad redox peaks. Although multiple models can likely account for the observed redox peaks, the base-line-subtracted redox peaks can be adequately modeled using the four reduction potentials determined spectroelectrochemically (−110, −190, −240, and −265 mV versus SHE; Fig. 4D).3 This suggests that the CymA structure or the heme environments are not strongly perturbed by the immobilization procedures.
CVs of apoCymA up to 1000 V/s indicate that the interfacial electron-transfer rate is fast and not rate-limiting to catalysis. To characterize the interfacial electron-transfer kinetics further, peak potentials, Ep,red and Ep,ox, were plotted against scan rate on a logarithmic scale to construct a trumpet plot (Fig. 5A). Analysis of the trumpet plot according to Hirst and Armstrong (60) is used to determine an interfacial electron-transfer rate at zero over-potential (k0) of 1.5 × 103 s−1. This analysis assumes that the four hemes of CymA independently interact with the electrode. Although this assumption is likely to be incorrect, the high electron-transfer rate clearly illustrates that the reduction and oxidation kinetics of all four hemes is fast. Finally, CymA redox signals are moderately dependent on the pH of the aqueous phase (Fig. 5B).
FIGURE 5.

A, shown are experimental, Ep,red (open circles) and Ep,ox (closed circles) and fitted (line) peak positions as a function of scan rate in a 20 mm MOPS, 30 mm Na2SO4 buffer (pH 7. 4) of CymA 8-MO-modified TSG gold electrode. Ep,red and Ep,ox were fitted using the software Jellyfit, freely available from L. J. C. Jeuken (k0 is 1.5 × 103 s−1). A constant peak separation of 28 mV and a slope of 12 mV/log(scan rate) was added to the fit. B, peak potentials, Ep,red (open circles) and Ep,ox (closed circles), as a function of pH in a mixed 20 mm MES/MOPS/Tris, 30 mm Na2SO4 buffer for CymA immobilized on an 8-MO modified TSG gold electrode are shown. The values reported are the means of duplicate determinations, and the error associated with these values is less than 5%.
Catalytic Cyclic Voltammetry
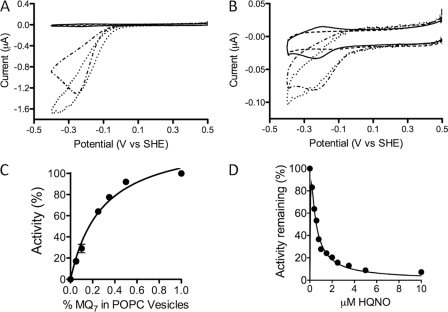
After the removal of the co-purified quinones from the CymA film on the electrode, creating an apoCymA film, catalytic properties were examined for the native lipophilic MQ-7 substrate. When 1 mg/ml of POPC vesicles containing 0.5% MQ-7 (w/w) were introduced to the electrolyte, diffusion-controlled reductive waves were observed (Fig. 6A). This signal can be blocked by inhibitors (see below), confirming the MQ-7 reduction is catalyzed by CymA. When the flux of vesicles toward the surface is increased by stirring the electrolyte in the electrochemical cell (see Ref. 52 for details of the electrochemical cell), the diffusion-limited behavior is transformed to a typical catalytic wave, confirming that MQ-7 reduction is diffusion-limited in the experiment without stirring. Either with or without stirring, it is striking that no MQ-7 oxidation is observed. Control experiments without CymA show that MQ-7-containing vesicles strongly adsorb on 8-OH-modified surfaces, giving rise to very small redox signals (Fig. 6B) that are invisible in the current scale used in Fig. 6A.
FIGURE 6.
A and B, shown are CVs of apoCymA adsorbed on an 8-MO-modified TSG gold electrode. Current is recorded in the absence (solid black line) and presence of 1 mg/ml POPC vesicles containing 0.5% (w/w) MQ-7 with (dotted black line) and without (dash-dotted black line) stirring at 100 rpm. CVs are shown before (A) and after (B) washing POPC vesicles out of the electrolyte. Blanks of 8-MO SAM before CymA adsorption are also shown (dashed black line). The gray voltammogram in B is of the control experiment (1 mg/ml POPC vesicles with 0.5% w/w MQ-7) without CymA adsorbed on the surface; the response is too small to resolve on the scale of Fig. 6. Relative current is measured as in B at −0.25 V versus SHE of the reductive scan with respect to MQ-7 concentration in the POPC vesicles (C) and relative current with respect to HQNO concentration in POPC vesicles containing 1% MQ-7 (w/w) (D). All CVs were recorded in 20 mm MOPS, 30 mm Na2SO4 buffer (pH 7.4) at a 10 mV/s scan rate.
These results suggest that POPC vesicles only loosely associate with CymA immobilized on the surface or that CymA-attached vesicles can easily be dislodged by other vesicles in the surroundings. Other explanations, like electron transfer between MQ-7 from different vesicles or diffusion limitation of MQ-7 within adsorbed vesicles, are unlikely because the control experiments (without CymA) do not show diffusion-controlled electrochemistry. It is possible that immobilized CymA on the surface is not able to reconstitute properly in a lipid membrane (i.e. insert its transmembrane α-helix into the lipid bilayer), resulting in a weaker and reversible interaction with the vesicles. However, when 8-OH-modified electrodes (without CymA) were incubated with CymA-reconstituted POPC/MQ-7 vesicles, the same pattern of diffusion-controlled reduction was observed, indicating that even if the CymA is properly inserted in the lipid vesicle, the interaction with lipid vesicle is weak. Whether the CymA-vesicle interaction is weakened by the electrode-CymA interaction is unclear at present.
To further investigate this effect, excess vesicles not directly associated with the CymA film were removed from the electrolyte by rinsing the cell with buffer solution. As before, cyclic voltammetry indicates diffusion-controlled MQ-7 reduction, although the catalytic currents are less than with MQ-7-containing vesicles in solution (Fig. 6B). The current of the reduction wave decreases with successive scans. This suggests that even though the lipid vesicles only loosely bind to the CymA film and can be replaced by other vesicles in solutions, a vesicle multilayer remains present on the CymA electrode after rinsing with buffer. We note that rinsing the CymA film with 0.01% DDM (w/v) returns the CV to the non-catalytic redox signals observed for the apoCymA film. Control experiments without CymA show that the vesicles bind strongly to the 8-OH-modified surface and are not removed when the electrolyte is exchanged with buffer (Fig. 6B). As for the control experiments where vesicles remain in solution, the redox signals do not display any diffusion-like limitations, and the direct electrochemical oxidation of the signals can be observed at ∼0.1 V versus SHE.
Examination of apoCymA with increasing amounts of MQ-7 in POPC vesicles revealed a Michaelis-Menten-like profile of MQ-7 reduction with an apparent Km ∼ 0.2% MQ-7 (w/w) in POPC vesicles (w/w) (Fig. 6C).
Many quinone oxidoreductases are specifically inhibited by HQNO (Fig. 2F), and in vivo studies have shown that HQNO is also an inhibitor for CymA (29, 61). Examination of an apoCymA film with increasing amounts of HQNO in the presence of 1.0% MQ-7 (w/w) revealed a typical inhibitory effect on the magnitude of the reductive wave as HQNO concentration increased. This yielded an apparent KI of 0.8 μm (Fig. 6D); dissociation constants of similar value have been measured for CymA using fluorescence-quench titrations on membrane extracts (45) and on purified enzyme.3
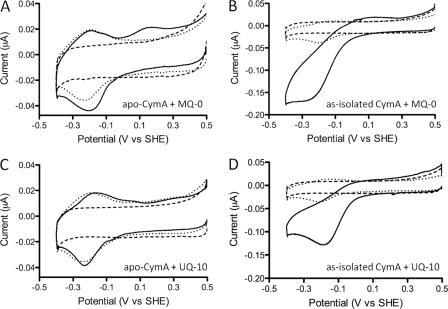
The true specificity of quinone oxidoreductases is difficult to discern in classical enzyme assays either due to the insolubility of the native substrate or due to the inability of native substrate to be removed from the enzyme without altering enzyme structure. Here, removing the co-purified quinones does not perturb the structure of CymA, as activity is restored upon the addition of MQ-7. This provides an opportunity to examine true substrate specificity of this class of enzyme. Substrate specificity was examined in the presence of menadione, a soluble analog of MQ-7 that lacks the isoprenoid chain, and ubiquinone-10 (MQ-0 and UQ-10, respectively; Fig. 2, D and E). Examination of activity with MQ-0 can identify potential function of the isoprenoid chain with regard to guiding substrate into a semihydrophobic pocket. Moreover, ubiquinone is structurally different in the head group from menaquinone, having a quinone head group instead of a napthoquinone (Fig. 2, E and C, respectively). Due to their hydrophilic nature, MQ-0 was added directly in solution (from an n-propyl alcohol stock), whereas UQ-10 was added in POPC vesicles as described for MQ-7. Regardless of concentration, MQ-0 and UQ-10 both failed to stimulate any catalytic turnover with apoCymA film (Fig. 7, A and C). Nonspecific, non-catalytic MQ-0 redox peaks can be seen in Fig. 7A. These MQ-0 signals are also observed in control experiments without CymA and appear superimposed on the CV of apoCymA, having no effect on the reduction potential of CymA (Fig. 7A). We note that HQNO, which is known to access the active site of CymA, does not induce an observable change in the redox signals of CymA. UQ-10 presented in vesicles also had no effect on catalysis (Fig. 7C). In contrast to MQ-0, vesicles containing UQ-10 are physically too large to access the electrode, which is shielded by a CymA monolayer, so no background ubiquinone redox signals are detected in the CV.
FIGURE 7.
CV from apoCymA (A and C) and as-isolated CymA (with MQ-7 bound; B and D) adsorbed on an 8-MO modified TSG gold electrode. CVs are recorded in the absence (dotted line) and presence (solid line) of either 1.25 μm MQ-0 (A and B) or 1 mg/ml POPC (C and D) vesicles containing 1% (w/w) UQ-10. CVs are shown after rinsing non-adsorbed POPC vesicles. All blanks of 8-MO SAM before CymA adsorption are shown (dashed black line). All CVs were recorded in 20 mm MOPS, 30 mm Na2SO4 buffer (pH 7.4) at a 10-mV/s scan rate.
Remarkably, when MQ-0 or UQ-10 is supplied to CymA films for which the tightly bound MQ-7 has not been removed by an in situ DDM wash, catalytic signals were observed (Fig. 7, B and D). For these experiments the CymA samples were used for which most of the co-purified quinone was removed by a DDM wash on the DEAE-Sepharose column before adsorption on 8-OH (see above).
These data indicate that the quinone co-purified with CymA is MQ-7 and that when either MQ-0 or UQ-10 is provided to CymA, MQ-7 remains bound. This is further confirmed by incubating apoCymA films with vesicles containing UQ-10 and small amounts of MQ-7 (0.05 or 0.1% (w/w)), which shows catalytic activity that is greater than for 0.05 or 0.1% MQ-7 alone. Given the requirement for MQ-7 presence, it seems unlikely that MQ-0 or UQ-10 access the quinone binding pocket directly during their “catalytic” turnover. It is, therefore, suggested that MQ-0 and UQ-10 reduction is mediated via quinone-quinone interactions and that MQ-7 may function as not just a substrate but also as a tightly bound cofactor.
DISCUSSION
CymA plays a central role in anaerobic respiration of Shewanella sp. where it transfers electrons from MQ-7, which is the dominant quinone under anaerobic conditions, to a variety of terminal oxidases. In complete contrast to this MQ-7 dehydrogenase function of CymA in vivo, the as-isolated CymA has a clear catalytic bias toward the reduction of MQ-7. Specific dose-dependent catalysis and specific inhibition by HQNO confirm that the dominant catalytic mode of as-isolated CymA is MQ-7 reduction. Although this is opposite to its proposed and demonstrated function in vivo, it has been demonstrated that CymA is indeed a reversible enzyme (62). Under aerobic conditions, when CymA is not expressed, the dominant quinones in Shewanella sp. are UQ-7 and UQ-8. In line with CymA function in anaerobic respiration, UQ-10 cannot be directly reduced by apoCymA (we note that oxidation of UQ-10 by CymA is thermodynamically unfeasible given the reduction potentials of CymA and UQ-10).
The purification of the CymA-MQ-7 complex and the catalytic behavior of CymA indicate that MQ-7 is tightly bound into a binding pocket of CymA, which we will name QMQ-7. The lack of activity demonstrated in the absence of MQ-7 also clearly indicates that CymA is highly specific for MQ-7 at the QMQ-7 site. This tightly bound MQ-7 facilitates the pass-off of electrons from the heme groups to MQ-0 in solution or UQ-10 in the membrane, which appear to bind to a less-specific and weaker “active-sit”' pocket, which we will name Qw. Despite the identity of quinone head groups between MQ-0 and MQ-7 (Fig. 2, C and D), MQ-0 is unable to occupy QMQ-7. However, MQ-0 can be reduced by CymA at the Qw site, during which MQ-7 remains tightly bound at the QMQ-7 site. This provides direct and clear evidence that the membrane environment and/or the isoprenoid tail play a critical role in facilitating the recognition or guidance of MQ-7 to the QMQ-7 site. Zargar and Saltikov have demonstrated that 2,3-dimethoxy-1,4-napthoquinol (DMNQH2) is able to reduce CymA in membrane preparations (45). However, based on the results shown here it is likely that in their membrane preparations CymA contains a tightly bound MQ-7. Elsewhere, we show that CymA used in this study is active in an MQ-0-based assay that couples NADH oxidation to fumarate reduction.3 This assay contains diaphorase to couple NADH oxidation to MQ-0 reduction and CymA to couple MQ-0 oxidation to the reduction of fumarate reductase (Fcc3), which reduces fumarate. It is striking that in this assay only 20–30% of the CymA is reduced by MQ-0,3 which we attribute to the fact that only a fraction of CymA has a MQ-7 bound (we note that the CymA used in this assay was purified according the protocol designed to remove bound MQ-7, resulting in the CV shown in Fig. 4B).
The experiments described here are a sobering warning to those who are trying to establish or “define” the specific activity and substrate specificity of oxidoquinone reductases. For CymA, it is ambiguous whether to classify its activity as MQ-7-specific or whether to classify MQ-7 as a specific cofactor of CymA. Either way, the results presented here show it is crucial to ascertain whether a quinone is co-purified when characterizing a quinone-converting enzyme, as even small amounts of lipophilic quinones can have a clear influence on the catalytic results obtained. Co-purification of lipids, including quinones, is a common feature of purifications of this type of metalloprotein, and similar problems are likely to occur for other enzymes. The premise of detergent-mediated removal of a quinone cofactor has been previously demonstrated in other enzymes like the QH site in cytochrome bo3, in which the tightly bound UQ-8 cofactor can be removed using harsher detergents (Triton X-100) (63, 64). The removal of the tightly bound UQ-8 does not damage cytochrome bo3, demonstrating the bound quinone is not critical to the native structure of this enzyme (63, 64).
If not for structural reasons, why would a quinone oxidoreductase use a quinone as a cofactor? Several properties of lipophilic quinones and quinone oxidoreductases make this class of membrane enzymes unique in nature. Localization of substrate and enzymes are restricted to the lipid membrane, where diffusion is an almost two-dimensional property. Combined with the fact that quinones typically have concentrations of several pmol/cm2 in the membrane, substrate-enzyme encounters will be much more frequent than typical encounters between globular enzymes and water-soluble substrates. We hypothesize that this has reduced the evolutionary pressure for a highly specific active site (Qw), which would only lower koff rates and reduce the turnover kinetics of the enzyme. It might be that a single hydrogen bond (via, for instance, a histidine or lysine side group) to one of the ketone groups of the quinone group provides sufficient binding energy for favorable binding in a membranous environment. The traditional function of an enzyme is to lower the transition-state activation energy and thereby increase the reaction rate. However, as has been noted before by Dutton and co-workers (65), this might not be required for quinone oxidation/reduction, which is an inherently fast reaction as long as a polar environment is provided to promote a rapid (de)protonation of the quinone. Neither of these arguments requires the presence of a quinone cofactor, as suggested here for MQ-7 in the QMQ-7 site. We hypothesize that the function of the quinone cofactors is to reduce the reactivity of the reaction intermediate, semiquinone. Semiquinones are generally believed to be amenable to side reactions, giving rise to damaging radical oxygen species. One way to reduce the reactivity of semiquinones is to stabilize the semiquinone in a protein binding pocket, reducing the free energy of any side reaction or sterically hinder access of compounds with which the semiquinone can react. However, this would require a specific quinone binding pocket, thereby lowering koff rates and reducing turnover numbers. By employing a quinone cofactor, in which the semiquinone is stabilized to minimize side reactions, a two-electron oxidoreduction reaction between the quinone substrate (at the Qw site) and quinone cofactor (at the QMQ-7 site) would provide a solution.
Acknowledgments
We thank Liang Shi, Jim Fredrickson, and John Zachara of Pacific Northwest National Laboratory for providing the strain of S. oneidensis MR-1 for overexpression of CymA.
Marritt, S. J., Lowe, T. G., Bye, J., McMillan, D. G. G., Shi, L., Fredrickson, J., Zaccharac, J., Richardson, D. J., Cheesman, M. R., Jeuken, L. J. C., and Butt, J. N. (2012) A functional description of CymA, the keystone for anaerobic respiratory flexibility in Shewanella. Biochem. J., DOI: 10.1042/BJ20120197.
- HQNO
- 2-heptyl-4-quinolinol-N-oxide
- MQ
- menaquinone
- UQ
- ubiquinone
- SAM
- self-assembled monolayer
- 8-OH
- 8-mercaptooctanol
- QCM-D
- quartz crystal microbalance with dissipation
- CV
- cyclic voltammetry
- DDM
- n-dodecyl β-d-maltoside
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- TSG
- template stripped gold.
REFERENCES
- 1. Remy A., Gerwert K. (2003) Coupling of light-induced electron transfer to proton uptake in photosynthesis. Nat. Struct. Biol. 10, 637–644 [DOI] [PubMed] [Google Scholar]
- 2. Okamura M. Y., Paddock M. L., Graige M. S., Feher G. (2000) Proton and electron transfer in bacterial reaction centers. Biochim. Biophys. Acta 1458, 148–163 [DOI] [PubMed] [Google Scholar]
- 3. Sato-Watanabe M., Mogi T., Ogura T., Kitagawa T., Miyoshi H., Iwamura H., Anraku Y. (1994) Identification of a novel quinone binding site in the cytochrome bo complex from Escherichia coli. J. Biol. Chem. 269, 28908–28912 [PubMed] [Google Scholar]
- 4. Sato-Watanabe M., Mogi T., Miyoshi H., Anraku Y. (1998) Characterization and functional role of the QH site of bo-type ubiquinol oxidase from Escherichia coli. Biochemistry 37, 5356–5361 [DOI] [PubMed] [Google Scholar]
- 5. Yap L. L., Samoilova R. I., Gennis R. B., Dikanov S. A. (2007) Characterization of mutants that change the hydrogen bonding of the semiquinone radical at the QH site of the cytochrome bo3 from Escherichia coli. J. Biol. Chem. 282, 8777–8785 [DOI] [PubMed] [Google Scholar]
- 6. Abramson J., Riistama S., Larsson G., Jasaitis A., Svensson-Ek M., Laakkonen L., Puustinen A., Iwata S., Wikström M. (2000) The structure of the ubiquinol oxidase from Escherichia coli and its ubiqionone binding site. Nat. Struct. Biol. 7, 910–917 [DOI] [PubMed] [Google Scholar]
- 7. Hellwig P., Yano T., Ohnishi T., Gennis R. B. (2002) Identification of the residues involved in stabilization of the semiquinone radical in the high affinity ubiquinone binding site in cytochrome bo3 from Escherichia coli by site-directed mutagenesis and EPR spectroscopy. Biochemistry 41, 10675–10679 [DOI] [PubMed] [Google Scholar]
- 8. Bartoschek S., Johansson M., Geierstanger B. H., Okun J. G., Lancaster C. R., Humpfer E., Yu L., Yu C. A., Griesinger C., Brandt U. (2001) Three molecules of ubiquinone bind specifically to mitochondrial cytochrome bc1 complex. J. Biol. Chem. 276, 35231–35234 [DOI] [PubMed] [Google Scholar]
- 9. Sharp R. E., Gibney B. R., Palmitessa A., White J. L., Dixon J. A., Moser C. C., Daldal F., Dutton P. L. (1999) Effect of inhibitors on the ubiquinone binding capacity of the primary energy conversion site in the Rhodobacter capsulatus cytochrome bc1 complex. Biochemistry 38, 14973–14980 [DOI] [PubMed] [Google Scholar]
- 10. Sharp R. E., Palmitessa A., Gibney B. R., White J. L., Moser C. C., Daldal F., Dutton P. L. (1999) Ubiquinone binding capacity of the Rhodobacter capsulatus cytochrome bc1 complex. Effect of diphenylamine, a weak binding Qo site inhibitor. Biochemistry 38, 3440–3446 [DOI] [PubMed] [Google Scholar]
- 11. Ding H., Moser C. C., Robertson D. E., Tokito M. K., Daldal F., Dutton P. L. (1995) Ubiquinone pair in the Qo site central to the primary energy conversion reactions of cytochrome bc1 complex. Biochemistry 34, 15979–15996 [DOI] [PubMed] [Google Scholar]
- 12. Ding H., Robertson D. E., Daldal F., Dutton P. L. (1992) Cytochrome bc1 complex (2Fe-2S) cluster and its interaction with ubiquinone and ubihydroquinone at the Qo site. A double occupancy Qo site model. Biochemistry 31, 3144–3158 [DOI] [PubMed] [Google Scholar]
- 13. Osyczka A., Moser C. C., Dutton P. L. (2005) Fixing the Q cycle. Trends Biochem. Sci. 30, 176–182 [DOI] [PubMed] [Google Scholar]
- 14. Biffinger J. C., Pietron J., Ray R., Little B., Ringeisen B. R. (2007) A biofilm-enhanced miniature microbial fuel cell using Shewanella oneidensis DSP10 and oxygen reduction cathodes. Biosens. Bioelectron. 22, 1672–1679 [DOI] [PubMed] [Google Scholar]
- 15. Fredrickson J. K., Romine M. F., Beliaev A. S., Auchtung J. M., Driscoll M. E., Gardner T. S., Nealson K. H., Osterman A. L., Pinchuk G., Reed J. L., Rodionov D. A., Rodrigues J. L., Saffarini D. A., Serres M. H., Spormann A. M., Zhulin I. B., Tiedje J. M. (2008) Toward environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603 [DOI] [PubMed] [Google Scholar]
- 16. Jensen H. M., Albers A. E., Malley K. R., Londer Y. Y., Cohen B. E., Helms B. A., Weigele P., Groves J. T., Ajo-Franklin C. M. (2010) Engineering of a synthetic electron conduit in living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 19213–19218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim H. J., Park H. S., Hyun M. S., Chang I. S., Kim M., Kim B. H. (2002) A mediator-less microbial fuel cell using a metal reducing bacterium, Shewanella putrefaciens. Enzyme Microb. Technol. 30, 145–152 [Google Scholar]
- 18. McLean J. S., Wanger G., Gorby Y. A., Wainstein M., McQuaid J., Ishii S. I., Bretschger O., Beyenal H., Nealson K. H. (2010) Quantification of electron transfer rates to a solid phase electron acceptor through the stages of biofilm formation from single cells to multicellular communities. Environ. Sci. Technol. 44, 2721–2727 [DOI] [PubMed] [Google Scholar]
- 19. Clarke T. A., Holley T., Hartshorne R. S., Fredrickson J. K., Zachara J. M., Shi L., Richardson D. J. (2008) The role of multiheme cytochromes in the respiration of nitrite in Escherichia coli and Fe(III) in Shewanella oneidensis. Biochem. Soc. Trans. 36, 1005–1010 [DOI] [PubMed] [Google Scholar]
- 20. Heidelberg J. F., Paulsen I. T., Nelson K. E., Gaidos E. J., Nelson W. C., Read T. D., Eisen J. A., Seshadri R., Ward N., Methe B., Clayton R. A., Meyer T., Tsapin A., Scott J., Beanan M., Brinkac L., Daugherty S., DeBoy R. T., Dodson R. J., Durkin A. S., Haft D. H., Kolonay J. F., Madupu R., Peterson J. D., Umayam L. A., White O., Wolf A. M., Vamathevan J., Weidman J., Impraim M., Lee K., Berry K., Lee C., Mueller J., Khouri H., Gill J., Utterback T. R., McDonald L. A., Feldblyum T. V., Smith H. O., Venter J. C., Nealson K. H., Fraser C. M. (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20, 1118–1123 [DOI] [PubMed] [Google Scholar]
- 21. Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., Bond D. R. (2008) Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Myers C. R., Myers J. M. (1993) Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108, 15–22 [Google Scholar]
- 23. Myers C. R., Myers J. M. (1993) Role of menaquinone in the reduction of fumarate, nitrate, iron(III), and manganese(IV) by Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 114, 215–222 [Google Scholar]
- 24. Newman D. K., Kolter R. (2000) A role for excreted quinones in extracellular electron transfer. Nature 405, 94–97 [DOI] [PubMed] [Google Scholar]
- 25. Burns J. L., DiChristina T. J. (2009) Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR-1 requires psrA, a homolog of the phsA gene of Salmonella enterica serovar typhimurium LT2. Appl. Environ. Microbiol. 75, 5209–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carpentier W., De Smet L., Van Beeumen J., Brigé A. (2005) Respiration and growth of Shewanella oneidensis MR-1 using vanadate as the sole electron acceptor. J. Bacteriol. 187, 3293–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gralnick J. A., Vali H., Lies D. P., Newman D. K. (2006) Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U.S.A 103, 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murphy J. N., Saltikov C. W. (2007) The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J. Bacteriol. 189, 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myers C. R., Nealson K. H. (1988) Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 30. Nealson K. H., Saffarini D. (1994) Iron and manganese in anaerobic respiration. Environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48, 311–343 [DOI] [PubMed] [Google Scholar]
- 31. Viamajala S., Peyton B. M., Apel W. A., Petersen J. N. (2002) Chromate reduction in Shewanella oneidensis MR-1 is an inducible process associated with anaerobic growth. Biotechnol. Prog. 18, 290–295 [DOI] [PubMed] [Google Scholar]
- 32. Field S. J., Dobbin P. S., Cheesman M. R., Watmough N. J., Thomson A. J., Richardson D. J. (2000) Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane multiheme c-type cytochromes from Shewanella frigidimarina NCIMB400. J. Biol. Chem. 275, 8515–8522 [DOI] [PubMed] [Google Scholar]
- 33. Myers C. R., Myers J. M. (1992) Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174, 3429–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pitts K. E., Dobbin P. S., Reyes-Ramirez F., Thomson A. J., Richardson D. J., Seward H. E. (2003) Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA. Expression in Escherichia coli confers the ability to reduce soluble Fe(III) chelates. J. Biol. Chem. 278, 27758–27765 [DOI] [PubMed] [Google Scholar]
- 35. Gao H., Yang Z. K., Barua S., Reed S. B., Romine M. F., Nealson K. H., Fredrickson J. K., Tiedje J. M., Zhou J. (2009) Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. Isme. J. 3, 966–976 [DOI] [PubMed] [Google Scholar]
- 36. Ross D. E., Ruebush S. S., Brantley S. L., Hartshorne R. S., Clarke T. A., Richardson D. J., Tien M. (2007) Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73, 5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwalb C., Chapman S. K., Reid G. A. (2002) The membrane-bound tetraheme c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella. Biochem. Soc. Trans. 30, 658–662 [DOI] [PubMed] [Google Scholar]
- 38. Schwalb C., Chapman S. K., Reid G. A. (2003) The tetraheme cytochrome cymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42, 9491–9497 [DOI] [PubMed] [Google Scholar]
- 39. Coursolle D., Baron D. B., Bond D. R., Gralnick J. A. (2010) The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192, 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodrigues M. L., Oliveira T. F., Pereira I. A., Archer M. (2006) X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel heme coordination. EMBO J. 25, 5951–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodrigues M. L., Scott K. A., Sansom M. S., Pereira I. A., Archer M. (2008) Quinol oxidation by c-type cytochromes. Structural characterization of the menaquinol binding site of NrfHA. J. Mol. Biol. 381, 341–350 [DOI] [PubMed] [Google Scholar]
- 42. Gescher J. S., Cordova C. D., Spormann A. M. (2008) Dissimilatory iron reduction in Escherichia coli. Identification of CymA of Shewanella oneidensis and NapC of E. coli as ferric reductases. Mol. Microbiol. 68, 706–719 [DOI] [PubMed] [Google Scholar]
- 43. Myers J. M., Myers C. R. (2000) Role of the tetraheme cytochrome cymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saffarini D. A., Blumerman S. L., Mansoorabadi K. J. (2002) Role of menaquinones in Fe(III) reduction by membrane fractions of Shewanella putrefaciens. J. Bacteriol. 184, 846–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zargar K., Saltikov C. W. (2009) Lysine 91 of the tetraheme c-type cytochrome CymA is essential for quinone interaction and arsenate respiration in Shewanella sp. strain ANA-3. Arch. Microbiol. 191, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qian Y., Paquete C. M., Louro R. O., Ross D. E., Labelle E., Bond D. R., Tien M. (2011) Mapping the iron binding site(s) on the small tetraheme cytochrome of Shewanella oneidensis MR-1. Biochemistry 50, 6217–6224 [DOI] [PubMed] [Google Scholar]
- 47. Shi L., Chen B., Wang Z., Elias D. A., Mayer M. U., Gorby Y. A., Ni S., Lower B. H., Kennedy D. W., Wunschel D. S., Mottaz H. M., Marshall M. J., Hill E. A., Beliaev A. S., Zachara J. M., Fredrickson J. K., Squier T. C. (2006) Isolation of a high affinity functional protein complex between omcA and mtrC. Two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi L., Lin J. T., Markillie L. M., Squier T. C., Hooker B. S. (2005) Overexpression of multi-heme c-type cytochromes. Biotechniques 38, 297–299 [DOI] [PubMed] [Google Scholar]
- 49. Bekker M., Kramer G., Hartog A. F., Wagner M. J., de Koster C. G., Hellingwerf K. J., de Mattos M. J. (2007) Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153, 1974–1980 [DOI] [PubMed] [Google Scholar]
- 50. Carter K., Gennis R. B. (1985) Reconstitution of the ubiquinone-dependent pyruvate oxidase system of Escherichia coli with the cytochrome o terminal oxidase complex. J. Biol. Chem. 260, 986–990 [PubMed] [Google Scholar]
- 51. Stamou D., Gourdon D., Liley M., Burnham N. A., Kulik A., Vogel H., Duschl C. (1997) Uniformly flat gold surfaces. Imaging the domain structure of organic monolayers using scanning force microscopy. Langmuir 13, 2425–2428 [Google Scholar]
- 52. Weiss S. A., Bushby R. J., Evans S. D., Henderson P. J., Jeuken L. J. (2009) Characterization of cytochrome bo3 activity in a native-like surface-tethered membrane. Biochem. J. 417, 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fourmond V., Hoke K., Heering H. A., Baffert C., Leroux F., Bertrand P., Léger C. (2009) SOAS. A free program to analyze electrochemical data and other one-dimensional signals. Bioelectrochemistry 76, 141–147 [DOI] [PubMed] [Google Scholar]
- 54. Chen X. X., Ferrigno R., Yang J., Whitesides G. A. (2002) Redox properties of cytochrome c adsorbed on self-assembled monolayers. A probe for protein conformation and orientation. Langmuir 18, 7009–7015 [Google Scholar]
- 55. El Kasmi A., Wallace J. M., Bowden E. F., Binet S. M., Linderman R. J. (1998) Controlling interfacial electron-transfer kinetics of cytochrome c with mixed self-assembled monolayers. J. Am. Chem. Soc. 120, 225–226 [Google Scholar]
- 56. Leopold M. C., Bowden E. F. (2002) Influence of gold substrate topography on the voltammetry of cytochrome c adsorbed on carboxylic acid terminated self-assembled monolayers. Langmuir 18, 2239–2245 [Google Scholar]
- 57. Höök F., Rodahl M., Kasemo B., Brzezinski P. (1998) Structural changes in hemoglobin during adsorption to solid surfaces. Effects of pH, ionic strength, and ligand binding. Proc. Natl. Acad. Sci. U.S.A. 95, 12271–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou C., Friedt J. M., Angelova A., Choi K. H., Laureyn W., Frederix F., Francis L. A., Campitelli A., Engelborghs Y., Borghs G. (2004) Human immunoglobulin adsorption investigated by means of quartz crystal microbalance dissipation, atomic force microscopy, surface acoustic wave, and surface plasmon resonance techniques. Langmuir 20, 5870–5878 [DOI] [PubMed] [Google Scholar]
- 59. Firer-Sherwood M., Pulcu G. S., Elliott S. J. (2008) Electrochemical interrogations of the mtr cytochromes from Shewanella. Opening a potential window. J. Biol. Inorg. Chem. 13, 849–854 [DOI] [PubMed] [Google Scholar]
- 60. Hirst J., Armstrong F. A. (1998) Fast-scan cyclic voltammetry of protein films on pyrolytic grapite edge electrodes. Characteristics of electron exchange. Anal. Chem. 70, 5062–5071 [DOI] [PubMed] [Google Scholar]
- 61. Myers C. R., Myers J. M. (1994) Ferric iron reduction-linked growth yields of Shewanella putrefaciens MR-1. J. Appl. Bacteriol. 76, 253–258 [DOI] [PubMed] [Google Scholar]
- 62. Ross D. E., Flynn J. M., Baron D. B., Gralnick J. A., Bond D. R. (2011) Toward electrosynthesis in Shewanella. Energetics of reversing the mtr pathway for reductive metabolism. PLOS One 6, e16649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rumbley J. N., Furlong Nickels E., Gennis R. B. (1997) One-step purification of histidine-tagged cytochrome bo3 from Escherichia coli and demonstration that associated quinone is not required for the structural integrity of the oxidase. Biochim. Biophys. Acta 1340, 131–142 [DOI] [PubMed] [Google Scholar]
- 64. Yap L. L., Lin M. T., Ouyang H., Samoilova R. I., Dikanov S. A., Gennis R. B. (2010) The quinone-binding sites of the cytochrome bo3 ubiquinol oxidase from Escherichia coli. Biochim. Biophys. Acta 1797, 1924–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Osyczka A., Zhang H., Mathé C., Rich P. R., Moser C. C., Dutton P. L. (2006) Role of the PEWY glutamate in hydroquinone-quinone oxidation-reduction catalysis in the Qo site of cytochrome bc1. Biochemistry 45, 10492–10503 [DOI] [PubMed] [Google Scholar]