Abstract
Eukaryotic cells contain hundreds of metalloproteins, and ensuring that each protein receives the correct metal ion is a critical task for cells. Recent work in budding yeast and mammalian cells has uncovered a system of iron delivery operating in the cytosolic compartment that involves monothiol glutaredoxins, which bind iron in the form of iron-sulfur clusters, and poly(rC)-binding proteins, which bind Fe(II) directly. In yeast cells, cytosolic monothiol glutaredoxins are required for the formation of heme and iron-sulfur clusters and the metallation of some non-heme iron enzymes. Poly(rC)-binding proteins can act as iron chaperones, delivering iron to target non-heme enzymes through direct protein-protein interactions. Although the molecular details have yet to be explored, these proteins, acting independently or together, may represent the basic cellular machinery for intracellular iron delivery.
Keywords: Iron Metabolism, Iron-Sulfur Protein, Metalloenzymes, Metalloproteins, Protein-Metal Ion Interaction, Frataxin, Glutaredoxin, Iron Chaperone, Poly(rC)-binding Protein
Introduction
Metalloproteins are very abundant in all types of cells, where they perform a plethora of enzymatic and regulatory functions. A variety of transition metal ions contribute to the metalloproteome of a cell, with iron and zinc being the most abundant (1). Estimates of the number of cellular proteins containing metal ions vary substantially, and evidence from bacteria suggests that many of the proteins that contain metal ions have not yet been annotated as metalloproteins (2). One method of estimating the prevalence of metalloproteins is to examine structural databases. A survey of enzymes for which three-dimensional structures have been determined indicated that 9% contained zinc, 8% iron, 6% manganese (in many cases, the biologically relevant metal is magnesium), and 1% copper (3). A second approach involves the identification of metalloproteins based on the homology of metal-binding domains and metal-binding sites of known metalloproteins to protein sequences derived from sequenced genomes. This type of bioinformatics approach indicates that zinc proteins constitute ∼10% of the eukaryotic proteome, non-heme iron proteins account for ∼1% of the proteome, and copper proteins are <1% of the proteome (4).
Given the presence of so many different metalloproteins, the cell faces several obstacles in ensuring that apoproteins receive the correct metal ion. First, although the primary coordination spheres of metal-binding sites can readily exclude ions based on charge, coordination geometry, and polarity, these sites may not have the capacity to discriminate between divalent metal cations and may even bind a non-cognate metal more tightly than the correct one (1). Second, some metal ions, such as iron and copper, are redox-active and, in the presence of oxygen, can catalyze the formation of dangerous reactive oxygen species. Thus, cells must tightly regulate the uptake and distribution of these metals to use them while avoiding the twin toxicities of mismetallation and oxidative damage. One cellular strategy is to maintain the pools of “free” or unliganded metals at exceedingly low levels. This appears to be especially true for the tightest binding metals, zinc and copper (5). A second, complementary strategy involves the use of metallochaperones.
Metallochaperone is the term coined to describe a protein that specifically binds metal ions and delivers them to target apoproteins through direct protein-protein interactions (6). Metallochaperones are thought to enhance the efficiency of metal transfer to target apoproteins and to prevent metal transfer to non-cognate sites. Nickel and copper chaperones were identified more than a decade ago. Structural and biochemical studies of these proteins suggest that metal delivery occurs by a ligand exchange reaction that depends on both the specificity of the protein-protein interaction and the affinities for the metal ligand. Iron chaperones have been identified more recently, and the structural and biochemical details of metal delivery for these proteins are not yet clear. Candidate iron chaperones include frataxin, poly(rC)-binding proteins (PCBPs),2 and the Grx3-type monothiol glutaredoxins.
Frataxin
The human disease Friedreich ataxia is an inherited neurodegenerative disorder usually caused by a reduction in the cellular levels of the protein frataxin (7). Frataxin is expressed in the mitochondria of all eukaryotic cells, and a large amount of data support a role for frataxin in the mitochondrial assembly of iron-sulfur clusters (8). Frataxin binds iron through a patch of carboxylic amino acid residues located on the distal surface of helix 1 and β-sheet 1. Iron binding is relatively low affinity (Kd = 2.5–55 μm), and the stoichiometry of binding differs when measured in frataxins from different species. Iron binding is important for in vivo activity, however, as substituting alanine residues for the clusters of acidic residues results in both loss of iron binding and a reduced capacity to support Fe-S cluster assembly. The core Fe-S cluster assembly complex in mitochondria consists of the cysteine desulfurase Nfs1, an accessory protein (Isd11), and a scaffold protein (Isu1 or Isu2) (nomenclature from budding yeast) (9). Frataxin interacts directly with Isu in an iron-dependent manner and facilitates the transfer of iron to Isu during Fe-S cluster assembly. Because of the capacity of frataxin to bind iron and transfer it to Isu via a direct protein-protein interaction, frataxin has been considered a mitochondrial iron chaperone. Some studies indicate that frataxin can directly transfer iron to or stabilize iron in other Fe-S proteins, such as mitochondrial aconitase (10). Recent in vitro studies using purified human proteins suggest, however, that the role of frataxin in Fe-S cluster assembly may be more complex.
Rather than simply providing iron to Isu for cluster assembly, frataxin and frataxin-bound iron also stimulate the activity of the cysteine desulfurase. Purified human Nfs1, Isd11, Isu, and frataxin form a stable complex in vitro that is not dependent on the presence of iron (11). The addition of frataxin alone to a complex of Nfs1, Isd11, and Isu results in a 316-fold increase in the catalytic efficiency of the cysteine desulfurase, which is due to an increase in both the kcat and the affinity for cysteine. The addition of frataxin and Fe(II) together results in a further increase in catalytic efficiency. Frataxin addition also increases the efficiency of Fe-S cluster production by the Fe-S core complex. Naturally occurring missense mutations in frataxin that are associated with disease progression lead to both reduced binding to the Nfs1-Isd11-Isu complex and reduced stimulation of cysteine desulfurase activity (12).
Yeast expressing a mutant form of Isu1 containing an M107I substitution can partially bypass the requirement for frataxin in Fe-S cluster assembly (13). This substitution of Met-107 for a branched chain amino acid occurs naturally in the forms of IscU expressed in prokaryotes, which are much less dependent on frataxin for Fe-S cluster assembly (14). The mechanism through which yeast cells expressing Isu1 M107I bypass the requirement for frataxin is not clear, but could involve iron delivery, cysteine desulfurase activation, or Fe-S cluster transfer to downstream recipient proteins. These studies suggest that Nfs1, Isd11, Isu, and frataxin together form the core Fe-S assembly complex in mitochondria and that, in eukaryotes, frataxin acts as an allosteric activator of the cysteine desulfurase, in addition to its role as an iron donor in Fe-S cluster assembly.
PCBPs
PCBP1 was recently identified as a cytosolic iron chaperone that delivers iron for incorporation into ferritin (15). Ferritin is a highly conserved iron storage protein expressed in bacteria, plants, and higher eukaryotes (16). In metazoans, ferritin is a heteropolymer composed of 24 subunits of heavy (H) and light (L) chains that form a hollow sphere into which iron is deposited. H-ferritin (but not L-ferritin) contains a ferroxidase center, located on the interior surface of the heteropolymer, which is structurally similar to the catalytic sites of oxo-bridged diiron monooxygenases. Fe(II) gains access to the ferroxidase center through funnel-shaped pores lined with carboxylic amino acid residues.
Fungi differ from other organisms in that they do not express ferritins. Human H- and L-ferritins can be expressed in yeast, where they assemble into functional heteropolymers, but contain relatively little iron. PCBP1 was identified in a genetic screen for human genes that could increase the amount of iron stored in human ferritin when the proteins were expressed together in yeast cells. Depletion of PCBP1 in cultured human cells inhibits the incorporation of iron into ferritin, which also leads to an increase in the labile iron pool and an increase in the iron-mediated degradation of IRP2 (iron regulatory protein 2). When ferritin was immunoprecipitated from yeast cells expressing both ferritin and PCBP1, PCBP1 was detected in a complex with ferritin, but only when the immune complexes were isolated in buffers containing ferrous iron. In vitro studies of PCBP1 supported the observations in yeast and human cells. Purified human PCBP1 was found to bind ferrous iron at a 3:1 iron/PCBP1 ratio with an affinity of 0.9–5.8 μm using isothermal titration calorimetry. Purified PCBP1 increased the incorporation of iron into apoferritin in vitro, indicating that PCBP1 could directly bind and donate iron to ferritin.
PCBP1 (also called α-CP1 or heterogeneous nuclear ribonucleoprotein (hnRNP) E1) was originally identified as an RNA-binding protein that binds to C-rich motifs in the 3′-UTR of α-globin mRNA (17–19). PCBP1 is one member of a family of four homologous proteins, each of which contains three hnRNP K homology (KH) domains. KH domains are conserved, single-stranded nucleic acid-binding motifs, and PCBP family members function in the processing, stability, and translation of host and viral RNAs. PCBPs also function in transcriptional regulation and protein-protein interactions. PCBP1 is encoded by an intronless gene found only in mammals and was likely formed via retrotransposition of a minor splice variant of PCBP2 mRNA. PCBP1 and PCBP2 (including its alternatively spliced isoforms) are very highly expressed in essentially all mammalian cells, whereas PCBP3 and PCBP4 are generally expressed at much lower levels. PCBP1 and PCBP2 localize to both the cytosol and nucleus. They have been reported to self-associate and to form complexes with other PCBPs and hnRNPs.
Two major questions emerged from the initial studies of PCBP1 and ferritin. 1) Are other cytosolic iron enzymes dependent on PCBP1 for metallation, and 2) are other PCBP family members acting as iron chaperones? Recent data indicate that the answer to both questions is yes. PCBP1 and PCBP2 play a role in the metallation of the iron-dependent prolyl hydroxylases (PHDs) that regulate hypoxia-inducible factor (HIF) 1α (20). HIF is a heterodimeric transcription factor that activates the expression of genes involved in the response to hypoxia (21–23). HIF1 and its paralog HIF2 are regulated by multiple mechanisms, but the primary mechanism is through the oxygen-dependent degradation of the α-subunit. Under hypoxic conditions, HIFα (HIF1α or HIF2α) accumulates in cells, binds to the β-subunit (HIFβ or ARNT), and activates transcription. Under normoxic or hyperoxic conditions, HIFα is hydroxylated on two conserved proline residues. The hydroxyproline residues are recognized and bound by the von Hippel-Lindau tumor suppressor protein, which then recruits components of the ubiquitination machinery and targets HIFα for degradation in the proteasome. This pathway has received considerable attention because defects in this degradation pathway lead to the formation of cancers in humans, especially clear cell renal carcinomas and hemangioblastomas.
Three HIF PHDs mediate the hydroxylation of HIFα: PHD1, PHD2, and PHD3 (also called HPH3, HPH2, and HPH1 or EGLN2, EGLN1, and EGLN3, respectively) (24, 25). These enzymes are members of a large conserved class of Fe(II)- and 2-oxoglutarate-dependent dioxygenases. PHDs and other enzymes of this class coordinate a single Fe(II) ion deep in a solvent-accessible active site through an HX(D/E)XnH triad. The iron cofactor is relatively labile in vivo, as treatment of cells with iron chelators can inactivate the enzyme. Because PHD activity is regulated by the availability of oxygen and 2-oxoglutarate, PHD has been proposed to function directly as an oxygen sensor. PHD2 accounts for the majority of HIF PHD activity in cells (26). Similar to almost all non-heme iron enzymes, the mechanism by which PHDs receive iron in their active site was unknown.
Depletion of PCBP1 in mammalian cells using siRNA was associated with a loss of PHD activity (20). Cells lacking PCBP1 exhibited loss of iron incorporation into PHD2, reduced hydroxylation of proline residues on HIF1α, and decreased degradation and accumulation of HIF1α. The effects of PCBP1 depletion were greatly increased when cells were treated briefly with an iron chelator. These observations pointed to a role for PCBP1 in the delivery of iron to PHDs and, more specifically, indicated that PCBP1 was important for the remetallation of PHD when iron deficiency increased the loss of metal from the active site of the enzyme.
Depletion of PCBP2 in mammalian cells led to similar, but not identical, effects on PHD as did depletion of PCBP1. Depletion of PCBP2 was also associated with loss of iron incorporation into PHD2, reduced prolyl hydroxylation of HIF1α, and accumulation of HIF1α protein. Although PCBP2 depletion had no effect on HIF1α accumulation and HIF1α transcriptional activity in iron-sufficient cells, the effects of PCBP2 depletion were similar to the effects of PCBP1 depletion in cells transiently subjected to iron deficiency. The results suggested that cells needed both PCBP1 and PCBP2 for the iron-dependent activation of PHDs.
Confirmation that both PCBP1 and PCBP2 were involved in iron delivery to PHDs came from in vitro studies of PHD activity from cells lacking PCBP1 or PCBP2. Lysates lacking PCBPs were nearly devoid of PHD activity, which could be fully restored with high concentrations of iron, confirming that both PCBP1 and PCBP2 were necessary for PHD activity and that the loss of PHD activity was attributable to loss of the iron cofactor. The addition of purified PCBP1 in complex with Fe(II) to lysates lacking PCBP1 could activate PHD, but the addition of the same PCBP1-Fe(II) complex to lysates lacking PCBP2 had no effect. These studies confirmed that PCBP1 and PCBP2 had non-redundant functions in the delivery of iron to PHD.
The PCBP-mediated delivery of iron to PHD may occur via a direct protein-protein interaction. PCBP1 and PHD2 are detected together in immune complexes isolated from cells expressing endogenous levels of both proteins. The complex containing PCBP1 and PHD2 is detectable in iron-deficient as well as iron-sufficient cells. Whether iron changes the composition or stability of the complex remains to be determined. PCBP2 is also detected in a complex with PCBP1, but whether PCBP1, PCBP2, and PHD2 form a single complex also remains to be determined.
FIH1 (factor inhibiting HIF1) is an asparagyl hydroxylase (similar to the PHDs) that is also involved in the oxygen- and iron-dependent regulation of HIF1α (27–29). The in vivo activity of FIH1 is reduced in cells lacking PCBP1, and FIH1 is also present in a complex with PCBP1. Thus, the iron chaperone activity of PCBP1 and PCBP2 is likely involved in the metallation of multiple members of this enzyme family. The iron- and 2-oxoglutarate-dependent dioxygenases are a large group of enzymes, and the human genome encodes >60 predicted members (22, 23). In addition to the amino acid hydroxylases, this enzymes class includes the Jumonji-type histone demethylases and the AlkB-type DNA and RNA demethylases. Thus, PCBP iron chaperones may be involved in the activation of a large group of cellular enzymes affecting a diversity of cellular processes.
Monothiol Glutaredoxins
Glutaredoxins are a large, evolutionarily conserved family of enzymes generally involved in redox reactions as protein thiol reductases, using glutathione as a source of reducing equivalents (30). Monothiol glutaredoxins represent a subclass of the glutaredoxin family and typically do not have oxidoreductase activity. Instead, they bind Fe-S clusters and participate in Fe-S cluster biogenesis and iron homeostasis. Recent work also suggests that Grx3-type monothiol glutaredoxins may also function as iron chaperones. The Grx3-type monothiol glutaredoxins have a modular structure that consists of an N -terminal thioredoxin domain and C-terminal glutaredoxin domains. Budding yeast cells contain two Grx3-type monothiol glutaredoxins, Grx3 and Grx4, which are expressed at similar levels, are 63% identical, and appear to be functionally equivalent (31–33). Mammals express a single Grx3-type glutaredoxin, which contains two C-terminal glutaredoxin domains in addition to the N-terminal thioredoxin domain. Each of the monothiol glutaredoxin domains contains an active site with the conserved CGFS motif. Both human and yeast Grx3 can form homodimers containing a bridging 2Fe-2S cluster that is coordinated through the active site cysteines and two molecules of glutathione (34, 35). In vitro, the Fe-S cluster bound by the Grx3 homodimer is labile and lost in the presence of oxygen or reductant.
Grx3-type glutaredoxins from several species have been found to interact with BolA-like proteins (30). BolA-like proteins are conserved and widely expressed in eukaryotes and prokaryotes, where their genes are frequently found in operons containing monothiol glutaredoxins. The BolA-like protein from yeast is Fra2, and Fra2 has been found, using multiple experimental approaches, to form a complex with Grx3/4 (35, 36). Grx3/4 and Fra2 form a heterodimer that also contains a bridging 2Fe-2S cluster. This cluster differs from that of the Grx3/4 homodimer in that it is resistant to degradation by both oxygen and reductant and is coordinated by the nitrogen ligand of His-103 in Fra2, rather than by a cysteine residue.
Grx3/4 and Fra2 are required for iron sensing by the major iron-dependent transcription factor in budding yeast, Aft1 (31, 32, 36). Under iron-deficient conditions, Aft1 activates the transcription of genes involved in the response to iron deficiency (37). Under iron-sufficient conditions, Aft1 is inactive and is exported from the nucleus to the cytosol. Grx3/4, Fra2, and another uncharacterized protein (Fra1) are found together in a complex with Aft1 (36). Yeast strains without Grx3 and Grx4, without Fra2, or without Fra1 are unable to inactivate Aft1 in the setting of iron sufficiency. One model for the iron-dependent inactivation of Aft1 involves the direct transfer of the 2Fe-2S cluster from Grx3/4-Fra2 to Aft1, with the result being the export of Aft1 from the nucleus and the loss of Aft1 transcriptional activity.
Grx3-type monothiol glutaredoxins have roles in iron transfer beyond those that regulate the activity of Aft1. Yeast strains lacking both Grx3 and Grx4 exhibit defects in the metallation of a variety of iron-dependent enzymes expressed in both the cytosol and mitochondria (31, 38). Yeast cells lacking Grx3 and Grx4 have defects in the biosynthesis of Fe-S clusters and heme, two mitochondrial processes that depend on the import of iron from the cytosol into the mitochondria. These defects are detectable as a loss of activity of mitochondrial and cytosolic Fe-S cluster and heme enzymes. This loss of activity is due to the absence of Fe-S clusters and heme cofactors in the respective enzymes. Unlike yeast strains with primary defects in the mitochondrial Fe-S cluster assembly machinery, strains lacking Grx3 and Grx4 exhibit depletion (rather than accumulation) of mitochondrial iron pools and accumulation of cytosolic iron pools. These phenotypes would be most consistent with a defect in mitochondrial iron delivery. However, ribonucleotide reductase, a cytosolic oxo-bridged diiron protein, also exhibits a lower level of iron binding and activity in the Grx-depleted strain, suggesting that Grx3/4 is also involved in the metallation of at least one cytosolic diiron enzyme. Whether Grx3/4 acts as an iron chaperone and can directly transfer iron to the iron-binding subunit of ribonucleotide reductase is not yet clear, nor is it clear whether Grx3/4 can directly transfer iron to the mitochondria or whether Fra2 participates in any of these metal delivery processes. However, Grx4 does require the cysteine residue that coordinates the Fe-S cluster in the glutaredoxin active site to support the activity of the heme and Fe-S enzymes. Of note, not all investigators studying the phenotypes of a strain carrying deletions of both GRX3 and GRX4 have observed the slow growth and enzymatic defects that would be associated with profound reductions in the metallation of all iron-dependent enzymes (33).
Recent studies in fission yeast indicate that a monothiol glutaredoxin, Grx4, is required for iron sensing by its iron-dependent transcription factors, Fep1 and Php4 (39, 40). Although these transcription factors have no similarity to Aft1, Grx4 can be found in a complex with each transcription factor in this fungal species, but it is not known whether direct transfer of an iron cofactor occurs. Whether mammalian Grx3 has similar roles in iron cofactor assembly and delivery is not known. Similarly, whether Grx3 interacts genetically or physically with PCBPs in mammalian cells remains to be determined.
Conclusion
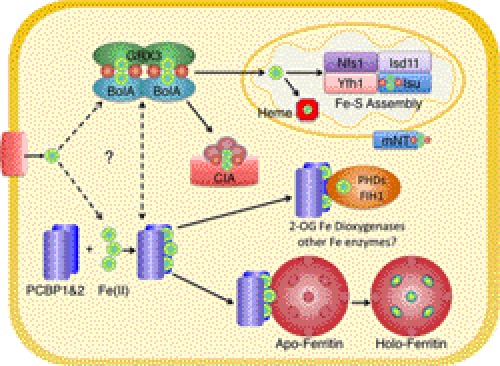
The iron chaperone field is in its infancy, with many more questions unanswered than resolved (Fig. 1). Studies implicating the monothiol glutaredoxins in iron delivery have largely been performed in budding yeast cells, which do not contain clear PCBP orthologs. Two proteins in yeast, Pbp2 and Hek2, contain three KH domains and exhibit homology to hnRNP K and, to a lesser extent, PCBPs (41–43). Both yeast proteins have been characterized as RNA-binding proteins, but deletion of either gene was not associated with an iron-related phenotype.3 Perhaps it is not surprising to discover that yeast and human cells may manage their intracellular iron pools in a different way, given that yeast cells do not rely on ferritins for cytosolic iron storage. One possibility is that yeast cells rely directly on Grx3/4 and Fra2 to deliver iron, bypassing a PCBP-like iron chaperone. Interestingly, yeast Grx3 and Grx4 each contain a sequence motif with 12–14 carboxylic amino acid residues, which are located between the thioredoxin and glutaredoxin domains.4 Frataxin contains a similar acidic patch that is involved in iron binding (8), and one might speculate that the acidic motif in Grx3/4 could have a similar function. This acidic motif is not present in human Grx3. Also of note is that BolA-like proteins from bacteria and mice exhibit a KH domain fold similar to the individual KH domains of PCBPs (44). Thus, the KH domains of PCBPs could interact with Grx3 in a manner similar to that of the BolA-like proteins. Whether other proteins assist in the PCBP1-PCBP2 iron chaperone complex in mammalian cells remains to be determined.
FIGURE 1.
Model of iron delivery in mammalian cells. PCBP1 and PCBP2 bind cytosolic Fe(II) and directly deliver it to ferritin, 2-oxoglutarate dioxygenases (such as PHDs and FIH1), and possibly other non-heme iron enzymes (such as oxo-bridged diiron enzymes). Although mammalian Grx3 has not been characterized, based on the functions of Grx3/4 in yeast, it likely associates with mammalian BolA2, with each glutaredoxin domain coordinating a 2Fe-2S cluster with one BolA2. In a speculative model of iron chaperone function, PCBPs may interact with Grx3-BolA in a way that effects the transfer of iron from one complex to the other. Grx3-BolA may deliver iron to mitochondria (perhaps via mitoNEET (mNT)) for heme and Fe-S cluster biosynthesis. Yfh1 (frataxin), along with Nfs1, Isd11, and Isu (yeast nomenclature), forms the core Fe-S cluster assembly complex in mitochondria. Grx3-BolA may also interact with the cytosolic Fe-S cluster assembly machinery (CIA).
Other proteins may be involved in the delivery of iron to intracellular organelles. Miner1 and mitoNEET are two paralogous proteins expressed on the cytoplasmic surface of the endoplasmic reticulum and the outer membrane of mitochondria, respectively (45, 46). A loss-of-function mutation in Miner1 is associated with the autosomal recessive disorder Wolfram syndrome 2, in which affected individuals present with multiple symptoms, including neurodegeneration and diabetes mellitus (47). Both proteins can coordinate a labile 2Fe-2S cluster. In the case of mitoNEET, the Fe-S cluster can be directly transferred to apoferredoxin in vitro, and in cultured cells, iron from mitoNEET can be taken up by mitochondria. These observations raise the possibility that Miner1 or mitoNEET can deliver iron to transporters in their respective organelles. Further studies will be needed to either support or refute this model.
Acknowledgment
I thank T. L. Stemmler for helpful discussions.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of NIDDK. This is the second article in the Thematic Minireview Series on Metals in Biology 2012.
C. C. Philpott and H. Shi, unpublished data.
C. C. Philpott, unpublished data.
- PCBP
- poly(rC)-binding protein
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- KH
- hnRNP K homology
- PHD
- prolyl hydroxylase
- HIF
- hypoxia-inducible factor.
REFERENCES
- 1. Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 2. Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole F. L., 2nd, Jenney F. E., Jr., Lancaster W. A., Praissman J. L., Shanmukh S., Vaccaro B. J., Trauger S. A., Kalisiak E., Apon J. V., Siuzdak G., Yannone S. M., Tainer J. A., Adams M. W. (2010) Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782 [DOI] [PubMed] [Google Scholar]
- 3. Andreini C., Bertini I., Cavallaro G., Holliday G. L., Thornton J. M. (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218 [DOI] [PubMed] [Google Scholar]
- 4. Andreini C., Bertini I., Rosato A. (2009) Metalloproteomes: a bioinformatic approach. Acc. Chem. Res. 42, 1471–1479 [DOI] [PubMed] [Google Scholar]
- 5. Outten C. E., O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 6. Rosenzweig A. C. (2002) Metallochaperones: bind and deliver. Chem. Biol. 9, 673–677 [DOI] [PubMed] [Google Scholar]
- 7. Pandolfo M. (2009) Friedreich ataxia: the clinical picture. J. Neurol. 256, Suppl. 1, 3–8 [DOI] [PubMed] [Google Scholar]
- 8. Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010) Frataxin and mitochondrial Fe-S cluster biogenesis. J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. (2010) Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulteau A. L., O'Neill H. A., Kennedy M. C., Ikeda-Saito M., Isaya G., Szweda L. I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245 [DOI] [PubMed] [Google Scholar]
- 11. Tsai C. L., Barondeau D. P. (2010) Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139 [DOI] [PubMed] [Google Scholar]
- 12. Tsai C. L., Bridwell-Rabb J., Barondeau D. P. (2011) Friedreich ataxia variants I154F and W155R diminish frataxin-based activation of the iron-sulfur cluster assembly complex. Biochemistry 50, 6478–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoon H., Golla R., Lesuisse E., Pain J., Donald J., Lyver E. R., Pain D., Dancis A. (2012) Mutation in the Fe-S scaffold protein Isu bypasses frataxin deletion. Biochem. J. 441, 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pohl T., Walter J., Stolpe S., Soufo J. H., Grauman P. L., Friedrich T. (2007) Effects of the deletion of the Escherichia coli frataxin homolog CyaY on the respiratory NADH:ubiquinone oxidoreductase. BMC Biochem. 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hintze K. J., Theil E. C. (2006) Cellular regulation and molecular interactions of the ferritins. Cell. Mol. Life Sci. 63, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaudhury A., Chander P., Howe P. H. (2010) Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16, 1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Makeyev A. V., Liebhaber S. A. (2002) The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostareck-Lederer A., Ostareck D. H. (2004) Control of mRNA translation and stability in hematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell 96, 407–411 [DOI] [PubMed] [Google Scholar]
- 20. Nandal A., Ruiz J. C., Subramanian P., Ghimire-Rijal S., Sinnamon R. A., Stemmler T. L., Bruick R. K., Philpott C. C. (2011) Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 14, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 22. Loenarz C., Schofield C. J. (2008) Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4, 152–156 [DOI] [PubMed] [Google Scholar]
- 23. Ozer A., Bruick R. K. (2007) Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3, 144–153 [DOI] [PubMed] [Google Scholar]
- 24. Bruick R. K., McKnight S. L. (2001) A conserved family of prolyl 4-hydroxylases that modify HIF. Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 25. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 26. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) HIF prolyl hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF1α in normoxia. EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., Ratcliffe P. J., Pugh C. W., Schofield C. J. (2002) Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 28. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 30. Rouhier N., Couturier J., Johnson M. K., Jacquot J. P. (2010) Glutaredoxins: roles in iron homeostasis. Trends Biochem. Sci. 35, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ojeda L., Keller G., Muhlenhoff U., Rutherford J. C., Lill R., Winge D. R. (2006) Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281, 17661–17669 [DOI] [PubMed] [Google Scholar]
- 32. Pujol-Carrion N., Belli G., Herrero E., Nogues A., de la Torre-Ruiz M. A. (2006) Glutaredoxins Grx3 and Grx4 regulate nuclear localization of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 119, 4554–4564 [DOI] [PubMed] [Google Scholar]
- 33. Rodríguez-Manzaneque M. T., Ros J., Cabiscol E., Sorribas A., Herrero E. (1999) Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8180–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haunhorst P., Berndt C., Eitner S., Godoy J. R., Lillig C. H. (2010) Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem. Biophys. Res. Commun. 394, 372–376 [DOI] [PubMed] [Google Scholar]
- 35. Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., Riggs-Gelasco P. J., Huynh B. H., Johnson M. K., Outten C. E. (2009) The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumánovics A., Chen O. S., Li L., Bagley D., Adkins E. M., Lin H., Dingra N. N., Outten C. E., Keller G., Winge D., Ward D. M., Kaplan J. (2008) Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 283, 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Philpott C. C., Protchenko O. (2008) Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E., Lillig C. H., Lill R. (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jbel M., Mercier A., Labbé S. (2011) Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1. Eukaryot. Cell 10, 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mercier A., Labbé S. (2009) Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J. Biol. Chem. 284, 20249–20262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Denisenko O., Bomsztyk K. (2002) Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol. Cell. Biol. 22, 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Irie K., Tadauchi T., Takizawa P. A., Vale R. D., Matsumoto K., Herskowitz I. (2002) The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mangus D. A., Amrani N., Jacobson A. (1998) Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18, 7383–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kasai T., Inoue M., Koshiba S., Yabuki T., Aoki M., Nunokawa E., Seki E., Matsuda T., Matsuda N., Tomo Y., Shirouzu M., Terada T., Obayashi N., Hamana H., Shinya N., Tatsuguchi A., Yasuda S., Yoshida M., Hirota H., Matsuo Y., Tani K., Suzuki H., Arakawa T., Carninci P., Kawai J., Hayashizaki Y., Kigawa T., Yokoyama S. (2004) Solution structure of a BolA-like protein from Mus musculus. Protein Sci. 13, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conlan A. R., Axelrod H. L., Cohen A. E., Abresch E. C., Zuris J., Yee D., Nechushtai R., Jennings P. A., Paddock M. L. (2009) Crystal structure of Miner1: the redox-active 2Fe-2S protein causative in Wolfram syndrome 2. J. Mol. Biol. 392, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zuris J. A., Harir Y., Conlan A. R., Shvartsman M., Michaeli D., Tamir S., Paddock M. L., Onuchic J. N., Mittler R., Cabantchik Z. I., Jennings P. A., Nechushtai R. (2011) Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc. Natl. Acad. Sci. U.S.A. 108, 13047–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Amr S., Heisey C., Zhang M., Xia X. J., Shows K. H., Ajlouni K., Pandya A., Satin L. S., El-Shanti H., Shiang R. (2007) A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am. J. Hum. Genet. 81, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]