Background: The three-dimensional structures of β-glucuronidase have been solved only for the GH2 enzymes.
Results: AcGlcA79A is composed of a (β/α)8-barrel domain and a β-domain.
Conclusion: The substrate binding site of AcGlcA79A is adapted for recognition of GlcA as a substrate.
Significance: This is the first report describing the crystal structure, mechanism, and catalytic residues of a GH79 enzyme.
Keywords: Crystal Structure, Enzyme Mechanisms, Enzyme Mutation, Enzyme Structure, Glycoside Hydrolases
Abstract
We present the first structure of a glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum, both as a product complex with β-d-glucuronic acid (GlcA) and as its trapped covalent 2-fluoroglucuronyl intermediate. This enzyme consists of a catalytic (β/α)8-barrel domain and a β-domain with irregular Greek key motifs that is of unknown function. The enzyme showed β-glucuronidase activity and trace levels of β-glucosidase and β-xylosidase activities. In conjunction with mutagenesis studies, these structures identify the catalytic residues as Glu173 (acid base) and Glu287 (nucleophile), consistent with the retaining mechanism demonstrated by 1H NMR analysis. Glu45, Tyr243, Tyr292–Gly294, and Tyr334 form the catalytic pocket and provide substrate discrimination. Consistent with this, the Y292A mutation, which affects the interaction between the main chains of Gln293 and Gly294 and the GlcA carboxyl group, resulted in significant loss of β-glucuronidase activity while retaining the side activities at wild-type levels. Likewise, although the β-glucuronidase activity of the Y334F mutant is ∼200-fold lower (kcat/Km) than that of the wild-type enzyme, the β-glucosidase activity is actually 3 times higher and the β-xylosidase activity is only 2.5-fold lower than the equivalent parameters for wild type, consistent with a role for Tyr334 in recognition of the C6 position of GlcA. The involvement of Glu45 in discriminating against binding of the O-methyl group at the C4 position of GlcA is revealed in the fact that the E45D mutant hydrolyzes PNP-β-GlcA approximately 300-fold slower (kcat/Km) than does the wild-type enzyme, whereas 4-O-methyl-GlcA-containing oligosaccharides are hydrolyzed only 7-fold slower.
Introduction
β-Glucuronidases hydrolyze β-glucuronic acid (GlcA)5-containing carbohydrates to release GlcA and are generally found in microorganisms, plants, and animals. GlcA is a component of proteoglycans, such as chondroitin sulfate proteoglycan, heparan sulfate proteoglycan, and hyaluronan from animals and arabinogalactan proteins from higher plants. Because β-glucuronidases are involved in the metabolism of proteoglycans, they are of biochemical, physiological, and medical interest. β-Glucuronidases have been used as reporter genes in many biological experiments in a manner similar to the use of green fluorescent protein and luciferase. β-Glucuronidases are classified into three glycoside hydrolase (GH) families, GH1, GH2, and GH79, according to their amino acid sequences in the Carbohydrate-Active EnZymes (CAZy) database (1, 2). The GH79 family includes heparanase (EC 3.2.1.166), baicalin-β-d-glucuronidase (EC 3.2.1.167), 4-O-methyl-β-glucuronidase, and β-glucuronidase (3–7). GH79, along with GH2, belongs to the GH-A clan (1, 2). The first reported crystal structure of a β-glucuronidase was of the human GH2 enzyme (8), which was followed by the report of the Escherichia coli GH2 β-glucuronidase structure (9). The GH2 family consists of only exo-acting enzymes, such as β-galactosidases, β-mannosidases, and β-glucuronidases, whereas the GH79 family consists of both exo-acting (β-glucuronidase) and endo-acting (heparanase) enzymes. Human heparanase plays a decisive role in disease-related processes, such as cell invasion, angiogenesis, and cancer metastasis. Several structural models of heparanase have been generated by computer programs using known clan GH-A structures as templates (10–12). However, to date, no experimental structure or even a preliminary crystallization report of a GH79 enzyme is available; the three-dimensional structure of a GH79 enzyme is eagerly anticipated.
For many years, bacterial β-glucuronidases were believed to be restricted to E. coli and closely related Enterobacteriaceae. Recently, however, the enzyme activity has been found among non-enterobacterial, anaerobic residents of the digestive tract, such as Bacteroides and Clostridium (13–15). Although these species exhibit lower β-glucuronidase activity than E. coli, they are ∼100-fold more abundant. Hence, it is suggested that they make a significant overall contribution to the enterohepatic circulation. Other bacterial genera with β-glucuronidase activity are Streptococcus, Staphylococcus, Corynebacterium, Alcaligenes, and many soil bacteria (16). In these bacteria, the enzyme is involved in carbohydrate and energy metabolism and may contribute to invasion during bacterial pathogenesis.
In contrast, GH79 β-glucuronidases from Arabidopsis thaliana, Aspergillus niger, and Neurospora crassa have been reported to act on plant proteoglycans (5–7). The A. niger and N. crassa enzymes have been shown to release both GlcA and 4-O-methyl-GlcA (MeGlcA) from arabinogalactan proteins (6, 7). No GH2 β-glucuronidases are known in plants, and no GH2 β-glucuronidase homologous sequences have been found in plants (17). Therefore, GH79 enzymes appear to play important roles in the metabolism of plant proteoglycans.
Thus, β-glucuronidases play an important role in plants, animals, and probably microorganisms. In this study, we present the three-dimensional structure of β-glucuronidase from Acidobacterium capsulatum (AcGlcA79A); this is the first structure of a GH79 enzyme. We also performed mutagenesis studies based on the structure of the enzyme-GlcA complex. Our results clearly demonstrate the structure-function relationship of GH79 β-glucuronidase and will help guide studies of other GH79 enzyme systems.
EXPERIMENTAL PROCEDURES
Substrates
p-Nitrophenyl (PNP)-glycosides including PNP-β-glucopyranoside (PNP-β-Glc), PNP-β-xylopyranoside (PNP-β-Xyl), and PNP-β-glucuronide (PNP-β-GlcA) were purchased from Sigma. MeGlcA-β-1,6-Gal-β-1,6-Gal (MeGlcA-β-1,6-Gal2) was prepared as described previously (6), and 2′,4′-dinitrophenyl 2-deoxy-2-fluoro-β-d-glucuronide (DNP-2FGlcA) was synthesized according to a method described previously (18).
Expression of AcGlcA79A and Mutant Generation
A. capsulatum NBRC15755 was obtained from the National Institute of Technology and Evaluation (Kazusa, Japan). The gene encoding a putative β-glucuronidase (ACP_2665; GenBankTM accession number ACO32043) was amplified from A. capsulatum genomic DNA by PCR using Phusion DNA polymerase (Finnzymes, Espoo, Finland) and the following primers: forward, 5′-CAT ATG GCT TTT GCC CGC GGC GGA CTG GCT-3′; reverse, 5′-AAG CTT AGC GAA TTC GAG CAA TGC GCC GGA-3′. The amplified DNA was cloned into pET30(+) (Novagen, Darmstadt, Germany) at NdeI and HindIII restriction enzyme sites (underlined). Recombinant enzymes were expressed using the T7 expression system in E. coli BL21-Gold(DE3) (Novagen) and purified with a C-terminal histidine tag (supplemental Fig. S1). Amino acid substitutions in AcGlcA79A were generated by inverse PCR using the expression vector pET30/AcGlcA79A as template DNA and the appropriate primers. Expression of mutants and their purification were performed in the same way as for wild-type AcGlcA79A. Further details can be found in the supplemental Experimental Procedures.
Crystallization, Data Collection, and Structural Determination
Initial crystallization screening of AcGlcA79A was conducted by the sitting drop, vapor diffusion method at 20 °C, mixing 0.3 μl of the protein solution (4.6 mg ml−1) and an equal volume of precipitant against 50 μl of reservoir solution using the following commercially available kits: JCSG+ Suite (Qiagen, Düsseldorf, Germany), Crystal Screen HT, and Index HT (Hampton Research, Aliso Viejo, CA). After refinement of the crystallization conditions, AcGlcA79A was crystallized by the sitting drop, vapor diffusion method with a precipitant solution composed of 2.0 m sodium phosphate monobasic monohydrate/potassium phosphate dibasic (0.5:9.5 (v/v), pH not adjusted) and with a protein concentration of 2.5 mg ml−1. Crystals with maximum dimensions of 200 × 200 × 500 μm were consistently obtained within a few days at 20 °C. Selenomethionine (Se-Met)-labeled AcGlcA79A was produced using the E. coli B834(DE3) methionine auxotroph and was crystallized under the same conditions as for the native enzyme. The GlcA complex was prepared by soaking the AcGlcA79A crystals in crystallization liquor containing GlcA powder for 5 min. The fluorinated glucuronide (2FGlcA) intermediate complex was prepared by adding the crystallization liquor containing 0.3% (w/v) DNP-2FGlcA into the AcGlcA79A crystallization drop and incubating for 5 min.
Diffraction experiments were conducted at the Photon Factory (PF) or the PF-Advanced Ring (PF-AR), High Energy Accelerator Research Organization, Tsukuba, Japan. The crystals were moved into the mother liquor containing 20% (v/v) glycerol as a cryoprotectant, and a single crystal was scooped in a nylon loop and flash frozen in a nitrogen gas stream at −178 °C. Diffraction data were collected with the Quantum 270 CCD detector (Area Detector Systems Corp., Poway, CA). Data were integrated and scaled using the programs DENZO and Scalepack in the HKL2000 program suite (19). AcGlcA79A crystals diffracted to ∼1.4-Å resolution (space group I4122). Assuming that the asymmetric unit of the crystal contained one AcGlcA79A molecule, the Matthews coefficient was calculated to be 2.7 Å3 Da−1 (20); this corresponded to the 55% solvent content of the crystals.
Structural analysis of AcGlcA79A was conducted through the multiwavelength anomalous dispersion method using Se-Met-labeled AcGlcA79A crystals. Seven selenium atom positions were determined, and initial phases were calculated using the AutoSol and AutoBuild wizards in the PHENIX program suite (21). The obtained initial model was further improved using the program ARP/wARP (22). Manual model rebuilding, introduction of water molecules, and molecular refinement were conducted using Coot and Refmac5 (23, 24). One phosphate ion and several glycerol molecules were added into the model. For the analyses of GlcA- and 2FGlcA-binding structures of AcGlcA79A, structural determination was conducted using the ligand-free structure as the starting model, and the bound ligand was observed in the difference electron density map. Refinement parameters for the ligands were generated by the GlycoBioChem PRODRG2 Server (25). Data collection and structure refinement statistics are given in Table 1. Stereochemistry of the models was analyzed with the program Rampage (26) in the CCP4 program suite (27). Structural drawings were prepared by the program PyMOL (Schrödinger, LLC, New York).
TABLE 1.
Data collection and structure refinement statistics of AcGlcA79A
Values in parentheses refer to the highest resolution shell. r.m.s.d., root mean square deviations.
| Data | Native | Se-Met |
GlcA complex | 2FGlcA complex | ||
|---|---|---|---|---|---|---|
| Peak | Edge | High remote | ||||
| Data collection | ||||||
| Space group | I4122 | I4122 | I4122 | I4122 | ||
| Unit cell parameters (Å) | a = b = 101.1, c = 217.9 | a = b = 101.2, c = 217.9 | a = b = 101.3, c = 217.8 | a = b = 101.5, c = 218.2 | ||
| Beam line | PF-AR NE-3A | PF-AR NE-3A | PF-AR NE-3A | PF BL-5A | ||
| Wavelength (Å) | 1.0000 | 0.97946 | 0.97967 | 0.96000 | 1.0000 | 1.0000 |
| Resolution (Å) | 50.0-1.50 (1.53-1.50) | 50.0-1.58 (1.61–1.58) | 50.0-1.60 (1.63-1.60) | 50.0-1.60 (1.63-1.60) | 50.0-1.80 (1.83-1.80) | 50.0-1.80 (1.86-1.80) |
| Rsyma | 0.062 (0.392) | 0.086 (0.373) | 0.053 (0.443) | 0.056 (0.656) | 0.061 (0.291) | 0.055 (0.579) |
| Completeness (%) | 100.0 (100.0) | 98.8 (97.2) | 98.2 (96.2) | 98.0 (92.2) | 99.9 (100.0) | 99.9 (99.9 |
| Multiplicity | 14.6 (14.4) | 29.5 (26.1) | 7.4 (6.8) | 7.3 (5.7) | 14.0 (14.0) | 24.0 (19.9) |
| Average I/σ(I) | 47.0 (6.8) | 52.0 (11.1) | 36.0 (3.8) | 32.9 (2.0) | 51.7 (10.4) | 33.1 (5.0) |
| Unique reflections | 90,146 (4,427) | 76,604 (3,725) | 73,731 (3,568) | 74,011 (3,416) | 52,642 (2,594) | 52,944 (5,219) |
| Observed reflections | 1,312,592 | 2,256,086 | 545,793 | 539,835 | 736,536 | 1,269,779 |
| Structure refinement | ||||||
| Resolution | 30.6-1.5 (1.54-1.50) | 38.4-1.80 (1.85-1.80) | 92.0-1.80 (1.85-1.80) | |||
| R factor | 0.172 (0.204) | 0.199 (0.262) | 0.214 (0.323) | |||
| Rfree factor | 0.190 (0.228) | 0.230 (0.296) | 0.248 (0.406) | |||
| r.m.s.d. from ideal value | ||||||
| Bond lengths (Å) | 0.007 | 0.011 | 0.014 | |||
| Bond angles (°) | 1.102 | 1.266 | 1.390 | |||
| No. of water molecules | 505 | 351 | 366 | |||
| Average B value | 16.5 | 22.1 | 24.5 | |||
| Ramachandran plot | ||||||
| Favored region (%) | 98.5 | 98.1 | 98.1 | |||
| Allowed region (%) | 1.3 | 1.7 | 1.9 | |||
| Outlier region (%) | 0.2 | 0.2 | 0.0 | |||
a Rsym = ΣhklΣi|Ii(hkl) − I(hkl)|/ΣhklΣiIi(hkl) where Ii(hkl) is the ith observation of reflection hkl and I(hkl) is the weighted average intensity for all observations i of reflection hkl.
Enzyme Assays
β-Glucuronidase activity was assayed using PNP-β-GlcA at 37 °C in McIlvaine buffer (pH 3.0) as described in the supplemental Experimental Procedures. The amount of PNP released was detected at A400 (extinction coefficient = 7284 m−1·cm−1). One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of PNP/min. The effects of pH and temperature on enzyme activity were investigated as described previously (28). The effect of pH on enzyme activity was examined with or without 90 mm formic acid or 90 mm acetic acid as described above.
The substrate specificity of AcGlcA79A toward various PNP-glycosides was determined. The assay method was identical to that described for PNP-β-GlcA. The kinetic parameters of the wild type and mutants of AcGlcA79A for PNP-β-GlcA, PNP-β-Glc, and PNP-β-Xyl were determined as follows. The reactions were performed in McIlvaine buffer (pH 3.5) containing 0.01–10 mm substrates, 0.1% (w/v) bovine serum albumin, and 0.9 nm-10.0 mm enzyme at 37 °C for up to 10 min. The amount of PNP released was determined from the A400 data. The kinetic parameters kcat and Km were determined by Lineweaver-Burk plot from three independent experiments and at five substrate concentrations. The substrate specificity and the catalytic efficiency of the wild type and mutants of AcGlcA79A were analyzed using MeGlcA-β-1,6-Gal2. Briefly, the enzyme (5 μm) was incubated with the substrate (10 μm) in McIlvaine buffer (pH 3.5) at 37 °C. At regular time intervals, the amount of degradation of each substrate was quantified by high performance anion exchange chromatography with pulsed amperometric detection (29). The assay was performed in duplicate.
Determination of Stereochemistry of Hydrolysis by 1H NMR
The substrates and recombinant enzymes were lyophilized twice from D2O before use. A solution of 15 mm PNP-β-GlcA (Koch-Light, UK) in 0.04 m deuterated sodium acetate buffer (pH 3.7) was incubated with 0.5 mg of AcGlcA79A. This enzyme concentration was needed to achieve a rapid substrate hydrolysis to minimize the effect of mutarotation on determination of the anomeric configuration of the primary product of hydrolysis. 1H NMR spectra were recorded at different time intervals on a Varian 400-MR (400-MHz) spectrometer.
RESULTS AND DISCUSSION
Expression and Characterization of AcGlcA79A
The cloning and expression of AcGlcA79A are described in the supplemental Experimental Procedures. Recombinant AcGlcA79A activities were tested using various PNP-glycosides as substrates. AcGlcA79A showed significant activity only for PNP-β-GlcA with negligibly weak or no activity for other substrates. Using PNP-β-GlcA as the substrate, maximal enzyme activity was detected at pH 3.0 and 50 °C. The enzyme was stable between pH 2.8 and 4.5 at 30 °C for 1 h and at pH 3.0 at <55 °C for 1 h. The Km and kcat values of AcGlcA79A for PNP-β-GlcA were 0.015 ± 0.001 mm and 34 ± 1 s−1, respectively (Tables 2 and 3). These values of AcGlcA79A are similar to the previously reported values of GH79 enzymes (7). The Km values of the A. niger and N. crassa recombinant GH79 enzymes were 0.030 and 0.038 mm, respectively, and the kcat values were 26.9 and 13.5 s−1, respectively (7). Because the A. niger and N. crassa enzymes showed 4-O-methyl-β-glucuronidase activity, the activity of AcGlcA79A for the MeGlcA-containing oligosaccharide MeGlcA-β-1,6-Gal2 was tested. AcGlcA79A barely hydrolyzed MeGlcA-β-1,6-Gal2, and its activity for MeGlcA-β-1,6-Gal2 was 103 times lower than that for PNP-β-GlcA (Table 4). These results suggest that AcGlcA79A is a genuine β-glucuronidase.
TABLE 2.
Activity of wild type (WT) and mutants of AcGlcA79A against PNP-β-GlcA
Each set of results is based on the average of three independent measurements. Results are means ± S.E.
| Km | kcat | kcat/Km | Relative to WT | |
|---|---|---|---|---|
| mm | s−1 | mm−1·s−1 | ||
| WT | 0.015 ± 0.001 | 34 ± 1 | 2303 | 1 |
| E173G | 0.5a | 2.0 × 10−4 | ||
| E173A | 0.1a | 5.6 × 10−5 | ||
| E287G | 0.2a | 9.1 × 10−5 | ||
| Y334F | 5.7 ± 0.5 | 67 ± 5 | 12 | 5.2 × 10−3 |
| Y243A | 0.55a | 2.4 × 10−4 | ||
| Y219A | 0.030 ± 0.001 | 31 ± 1 | 1026 | 0.5 |
| H327N | 5.5 ± 0.4 | 45 ± 4 | 8 | 3.5 × 10−3 |
| H327S | 0.57a | 2.5 × 10−4 | ||
| E45D | 4.8 ± 0.6 | 36 ± 3 | 8 | 3.3 × 10−3 |
a The enzyme (0.1 μm) was incubated with PNP-β-GlcA at a concentration of 1.5 μm. Kinetic parameters were not determined because mutants were too inactive to obtain individual kinetic constants. The activity of the rest of mutants such as E173Q, E287A, E287Q, Y334W, Y292A, H327K, H327T, and E45Q was not detected under this condition.
TABLE 3.
Activities of AcGlcA79A wild type and mutants for PNP-β-GlcA, PNP-β-Glc, and PNP-β-Xyl
Each set of results is based on the average of three independent measurements. Results are means ± S.E. ND, not detectable.
| Enzyme | Substrates |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PNP-β-GlcA |
PNP-β-Glc |
PNP-β-Xyl |
||||||||||||
| Km | kcat | kcat/Km | Relative to WT | Km | kcat | kcat/Km | Ratioa (Glc/GlcA) | Relative to WT | Km | kcat | kcat/Km | Ratiob (Xyl/GlcA) | Relative to WT | |
| mm | s−1 | mm−1·s−1 | mm | s−1 | mm−1·s−1 | mm | s−1 | mm−1·s−1 | ||||||
| WT | 0.015 ± 0.001 | 34 ± 1 | 2303 | 1 | 11 ± 0.4 | 3.1 × 10−4 ± 0.1 × 10−4 | 2.7 × 10−5 | 1.2 × 10−8 | 1 | 15 ± 1 | 1.5 × 10−3 ± 0.1 × 10−3 | 1.0 × 10−4 | 4.3 × 10−8 | 1 |
| Y334F | 5.7 ± 0.5 | 67 ± 5 | 12 | 5.2 × 10−3 | 9.6 ± 0.6 | 7.8 × 10−4 ± 0.5 × 10−4 | 8.2 × 10−5 | 6.8 × 10−6 | 567 | 11 ± 4 | 4.0 × 10−4 ± 0.8 × 10−4 | 3.7 × 10−5 | 3.1 × 10−6 | 72 |
| Y292A | NDc | 4.7 ± 0.5 | 8.6 × 10−5 ± 0.6 × 10−5 | 1.8 × 10−5 | 25 ± 3 | 9.8 × 10−4 ± 1.5 × 10−4 | 6.5 × 10−5 | |||||||
a (kcat/Km for PNP-β-Glc)/(kcat/Km for PNP-β-GlcA).
b (kcat/Km for PNP-β-Xyl)/(kcat/Km for PNP-β-GlcA).
c The enzyme (0.1 μm) was incubated with PNP-β-GlcA at a concentration of 1.5 μm.
TABLE 4.
Activity of AcGlcA79A wild type and E45D mutant for PNP-β-GlcA and MeGlcA-β-1,6-Gal2
The assay using MeGlcA-β-1,6-Gal2 was performed in duplicate. Results are means ± S.E.
|
kcat/Km |
Ratioa (MeGlcA/GlcA) | Relative to WT | ||
|---|---|---|---|---|
| PNP-β-GlcA | MeGlcA-β-1,6-Gal2 | |||
| mm−1·s−1 | ||||
| Wild type | 2,303 ± 355 | 7.7 × 10−3 ± 1.2 × 10−3 | 3.0 × 10−6 | 1 |
| E45D | 7.5 ± 1.7 | 1.1 × 10−3 ± 0.1 × 10−3 | 1.3 × 10−4 | 43 |
a (kcat/Km for MeGlcA-β-1,6-Gal2)/(kcat/Km for PNP-β-GlcA).
Three-dimensional Structure of AcGlcA79A
The crystal structure of AcGlcA79A was determined by the multiwavelength anomalous dispersion method using Se-Met derivative data. Native and GlcA complex structures were determined successively. Structure refinement statistics are summarized in Table 1. The quality and accuracy of the final structures were further demonstrated to show that all residues fall within the common regions of the Ramachandran stereochemistry plot. Recombinant AcGlcA79A is composed of a single polypeptide chain of 488 amino acids with the additional C-terminal residues (476KLAAALEHHHHHH488) derived from the expression vector and purification tag. Seventeen N-terminal residues (Met1–Ser17) and five C-terminal residues (His484–His488) were not identified because of a lack of electron density. Three cis-peptide bonds were found at Gly214-Pro215, Gly246-Pro247, and Ser457-Gly458. The final model consists of one AcGlcA79A molecule with one or two phosphate ions and one or four glycerol molecules. The overall structures of the ligand-free and GlcA complexes were almost the same, and the calculated root mean square difference was 0.2 Å for the Cα atom pairs.
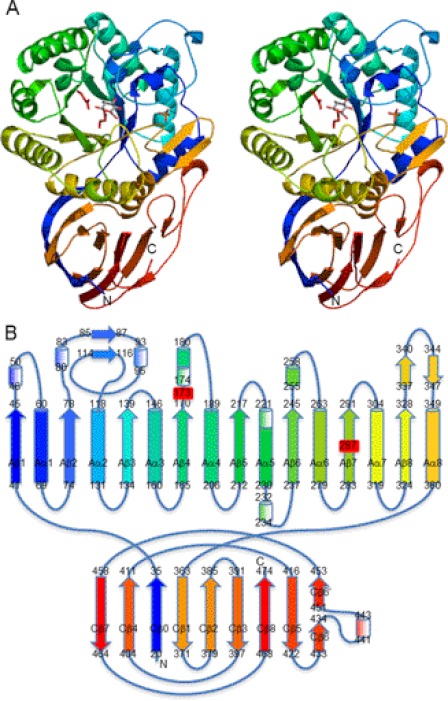
The protein consists of domains A and C (Fig. 1). The N-terminal β-strand (Val20–Ile35) is inserted into the domain C, and the peptide folds into the (β/α)8-barrel of domain A (Gly41–Ala360) and finally enfolds the β-domain of domain C. The secondary structure elements of AcGlcA79A are numbered in Fig. 1B.
FIGURE 1.
Structure of AcGlcA79A. A, stereoview of the ribbon model of AcGlcA79A-GlcA complex in rainbow-ordered colors. Two catalytic residues are displayed in red. The bound GlcA molecule and phosphate ion are shown as stick models. B, schematic topological diagram of AcGlcA79A. α-Helices are shown as cylinders, α310-helices are shown as shaded cylinders, and β-strands are shown as arrows. The colors correspond to those in A. Two catalytic residues are labeled in red.
Domain A consists of a (β/α)8-barrel, which is the catalytic domain in many glycoside hydrolases. The GlcA binding site is located at the C-terminal end of the central β-barrel and was designated as the catalytic pocket. GH79 and GH2 enzymes are classified into the same GH-A clan, and the overall folding of their structures shows certain similarities. The root mean square difference for the core Cα atoms between the catalytic domains of AcGlcA79A and E. coli GH2 β-glucuronidase was calculated to be 2.6 Å. In addition to the core secondary structures of the (β/α)8-barrel, several loops fold into some small substructures and contribute to the formation of the catalytic pocket (Fig. 1).
Domain C consists of nine β-strands with strands Cβ0 and Cβ4 aligned parallel and the others in interacting antiparallel configurations (Fig. 1B). The eight strands other than Cβ0 comprise a typical antiparallel β-domain structure containing Greek key motifs (Fig. 1). This C-terminal domain structure is observed in many glycoside hydrolases, although in many cases its function remains unknown.
Active Site and Catalytic Mechanism
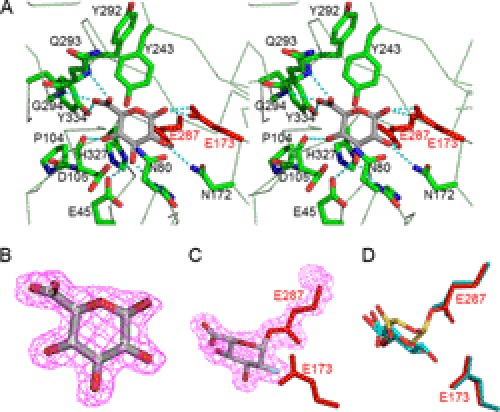
The active site of AcGlcA79A is located at the C-terminal end in the central (β/α)8-barrel and is represented by a pocket shape (Fig. 1). In the GlcA complex structure, Glu173 is located near the O1 atom of GlcA (2.3–2.4 Å), and Glu287 is located close to the C1 atom (3.0 Å). The distance between Glu173 and Glu287 of AcGlcA79A is 5.25 Å. The location of these acidic amino acids suggests that Glu173 and Glu287 are the catalytic residues of AcGlcA79A, functioning as acid/base and nucleophile, respectively, consistent with the expected retaining mechanism (Fig. 2). It has been reported that the distance between two catalytic residues is 7–10 Å in inverting glycosyl hydrolases and 5–6 Å in retaining glycosyl hydrolases (30). This is also apparent from the rule of the clan GH-A (4/7 superfamily) catalytic module in which the two catalytic residues are located posteriorly in Aβ4 and Aβ7 (31). The locations of Glu173 and Glu287 of AcGlcA79A are the same as those of the catalytic residues of GH2 enzymes and correspond to the 4/7 superfamily rule.
FIGURE 2.
Ligand-binding structure of AcGlcA79A. A, stereoview of the catalytic pocket of AcGlcA79A complexed with glucuronic acid. B, electron density for bound GlcA. Carbon atoms of GlcA are numbered. C, electron density for covalently bound 2FGlcA. D, superimposed model of the bound GlcA (yellow and red) and covalently bound 2FGlcA (cyan and red).
To verify the identities of the catalytic residues of AcGlcA79A, Gly, Ala, and Gln mutants of Glu173 and Glu287 were constructed. As expected, the activities of all mutants were at least 104 times lower than those of the wild-type enzyme (Table 2). The kcat/Km values of E173G, E173A, and E287G mutants were 0.5, 0.1, and 0.2 mm−1·s−1, respectively. Chemical rescue experiments were subsequently performed for E173G, E173A, E287G, and E287A using formic acid and acetic acid as exogenous nucleophiles (see supplemental Experimental Procedures). The β-glucuronidase activities of the mutants were measured in McIlvaine buffers (0.1 m citric acid and 0.2 m Na2HPO4) of various pH values with or without 90 mm formic acid and 90 mm acetic acid (supplemental Fig. S2). The pH activity profiles of E173G and E173A were the same with or without the chemicals, and the enzyme activities of the mutants remained significantly lower than that of the wild type. However, the pH activity profiles of E287G and E287A with formic acid were different in the pH range 2–5. The enzyme activity of E287G with formic acid near the optimum pH (2.6–3.5) recovered completely to the same level as that of the wild type. For E287A, 25% of the enzyme activity was recovered. In contrast, acetic acid did not affect the enzyme activities of E287G and E287A most likely because acetic acid is too large to enter the pocket created by the mutation. These data further suggest that Glu287 is the nucleophile.
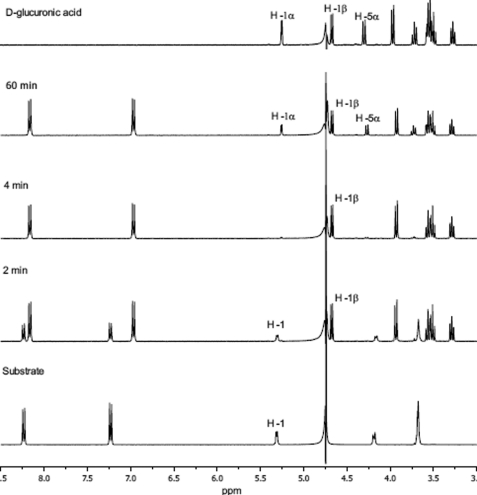
Because the catalytic mechanism of GH79 enzymes had not been verified experimentally, the anomeric configuration of the hydrolysis product was determined (Fig. 3). The hydrolysis of PNP-β-GlcA with AcGlcA79A in D2O was followed by 1H NMR analysis. The enzyme load was sufficient to complete the hydrolysis of 15 mm substrate in about 4 min. As shown in Fig. 3, the disappearance of the H1 signal of the glucuronide at 5.3 ppm was accompanied by parallel changes of the PNP aglycone signals into PNP signals (signals between 6.8 and 8.3 ppm). Free uronic acid was generated as the β-anomer as evidenced by the rapid appearance of the H1β resonance at 4.7 ppm. The α-anomer signal only appeared later as a result of mutarotation (Fig. 4 and supplemental Fig. S3). These results show that AcGlcA79A is a retaining enzyme utilizing a double displacement mechanism of hydrolysis. It is very probable that all enzymes belonging to this family will prove to be retaining. Consistent with this mechanism, we detected transglycosylation products generated by AcGlcA79A (data not shown). Similar transglycosylation reactions catalyzed by GH79 β-glucuronidase from A. niger have been reported previously (6, 7).
FIGURE 3.
Stereochemistry of hydrolysis by 1H NMR.
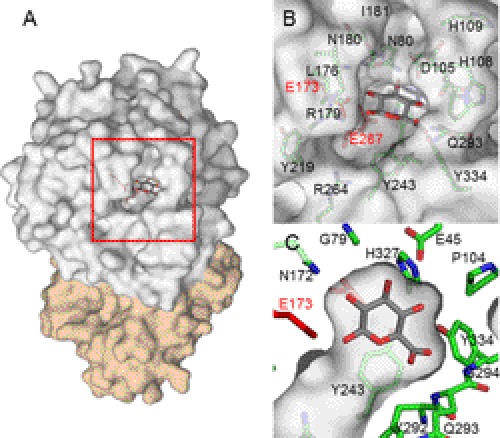
FIGURE 4.
Surface representation of catalytic pocket of AcGlcA79A. A, overall structure. B, close view around the catalytic pocket. C, cross-sectional view of the contact surface in the GlcA complex structure.
Ligand-binding Structure
The catalytic center of AcGlcA79A is pocket-shaped as is typically found in exo-acting enzymes (Figs. 2, A and B, and 4). In the GlcA complex structure, the aromatic ring of Tyr243 lies parallel to the pyranosyl ring of GlcA, providing stabilizing hydrophobic interactions. The O2 and O3 atoms of GlcA are recognized by hydrogen bonds, such as those between the O2 and Asn172-Nδ2 atoms and between the O3 and both Glu45-Oϵ2 and Asn80-N atoms. The O4 atom of GlcA is surrounded by Pro104 and His327 and forms a hydrogen bond with Asp105. Recognition of the GlcA carboxyl group is achieved by hydrogen bonds between O6A and Gln293-N and between O6B and both Gly294-N and Tyr334-Oη (Fig. 2). Fig. 4C shows a cross-sectional view of the active site pocket of the GlcA complex structure and clearly indicates that there is no extra space around the substrate. This was also indicated by a sugar soaking experiment in which no residual sugar was observed when the AcGlcA79A crystals were soaked with Glc, Xyl, or GalA.
2-Deoxy-2-fluoroglycosides, which act as mechanism-based inhibitors to form covalent intermediates, are powerful chemical tools for identifying the active site nucleophile in retaining glycosidases as reported by Withers and Aebersold (18). By adding DNP-2FGlcA to the AcGlcA79A crystal, the fluorinated glucuronide residue (2FGlcA) was observed to be covalently bound to Glu287 (Fig. 2C) through an α-configured linkage. The positions of the O2 and O3 atoms of 2FGlcA are almost the same as with GlcA, but the C1 atom is shifted by 1.3 Å toward Glu287 because of covalent bond formation (Fig. 2D).
Mutagenesis Study of AcGlcA79A
To investigate the basis of substrate specificity of AcGlcA79A, appropriate mutations were introduced at the residues that interact with GlcA, and the properties of the constructed mutants were characterized (Tables 2 and 3). Although GlcA and Glc are structurally different only at the C6 position (-COOH for GlcA and -CH2OH for Glc), AcGlcA79A discriminates greatly between GlcA and Glc. To address this discrimination, a Y334F mutant was constructed along with Y334W and Y292A mutants to alter the space around the C6 position of GlcA and change the position of the main chain of Gln293 and Gly294. Around the C6 position of GlcA, the main chain atoms of Gln293 and Gly294 and Tyr334-Oη interact with the carboxyl group of GlcA (Fig. 2).
As shown in Table 3, the catalytic efficiency of the Y334F mutant for hydrolysis of PNP-β-GlcA is lower than that of the wild type due to an increased Km value. In contrast, the activity for PNP-β-Glc is close to that of the wild type. Consequently, the activity ratio for PNP-β-Glc to PNP-β-GlcA (Glc/GlcA ratio) is 567 times higher than that of the wild type, suggesting that the para-OH of Tyr334 is necessary for recognizing the carboxyl group of GlcA. Tyr334 enables AcGlcA79A to interact predominantly with GlcA and exhibit higher β-glucuronidase activity. The Y334W mutant could not cleave PNP-β-GlcA at all presumably due to steric hindrance. When Tyr334 was substituted with tryptophan, the space around the C6 position of GlcA appeared to be filled with the aromatic ring of tryptophan so that GlcA could not possibly bind. The Y292A mutant also lost all activity for PNP-β-GlcA, whereas its catalytic efficiency with PNP-β-Glc was close to that of the wild-type enzyme. The positions of Gln293 and Gly294 may be influenced by the substitution of Tyr292 with alanine so that GlcA is unable to bind in the catalytic pocket. The catalytic efficiencies of Y334F and Y292A for PNP-β-Xyl were 0.37 and 0.65 times lower, respectively, than that of the wild type. However, the activity ratio of PNP-β-Xyl to PNP-β-GlcA (Xyl/GlcA ratio) was higher for both mutants. Y334F had a 72 times higher activity ratio than the wild-type enzyme. Although the mutants showed broader substrate specificity than the wild type, none of the activities for GlcA reached the level of the wild type. AcGlcA79A appears to discriminate GlcA through the residues around C6, including Tyr334, Tyr292, Gln293, and Gly294, while maintaining high enzyme activity toward GlcA.
Although some GH79 enzymes exhibit 4-O-methyl-β-glucuronidase activity, AcGlcA79A barely hydrolyzes the MeGlcA-containing oligosaccharide (Table 4). As shown in Fig. 4C, there is no extra space for a methyl group around the C4 position of GlcA in the GlcA complex structure. To investigate the discrimination mechanism for the 4-O-methyl group of MeGlcA, mutations at Glu45 and His327, which surround the C4 hydroxyl group of GlcA, were designed to make space for the 4-O-methyl group. The enzyme activities of His327 mutants for PNP-β-GlcA were eliminated or drastically reduced by 10−4–10−3 times relative to the wild-type AcGlcA79A with similar results being seen for Glu45 mutants (Table 2). In the case of the E45D mutation, the kcat and the Km values for PNP-β-GlcA were 36 ± 3 s−1 and 4.8 ± 0.6 mm, respectively, resulting in a 3.3 × 103 times reduction in activity relative to that of the wild-type enzyme (Tables 3 and 4). The carboxyl group of Glu45 is close to the O4 atom of GlcA at a distance of 3.2 Å and is hydrogen-bonded with the Thr81-Oγ1, Thr81-N, Asp85-N, and His327-Nϵ2 atoms. E45D might cause a structural distortion of the hydrogen-bonding network around the C4 position of GlcA and change the surrounding environment of C4. By contrast, the catalytic efficiency of the E45D mutant for MeGlcA-β-1,6-Gal2 was only 7 times lower than the wild type. As a result, the activity ratio for MeGlcA-β-1,6-Gal2 to PNP-β-GlcA (MeGlcA/GlcA ratio) of the E45D mutant was 43 times higher than that of the wild type, strongly suggesting that Glu45 is one of the key residues by which AcGlcA79A distinguishes the 4-O-methyl group of GlcA.
The substrate binding site of AcGlcA79A is specialized for recognition of GlcA as a substrate. A mutagenesis study revealed that Tyr334 and Tyr292 interact with the C6 position of GlcA and that Glu45 recognizes the C4 position of GlcA. Single amino acid mutations did not drastically change the substrate specificity because part of the substrate binding site was formed by the main chain atoms of the protein. These results will help guide studies of other GH79 enzyme systems.
Supplementary Material
Acknowledgments
We thank the beamline researchers at Photon Factory for assistance with synchrotron radiation. We also thank Dr. Iveta Uhliariková from the Institute of Slovak Academy of Sciences for assistance with the NMR experiment and Yuan Liu from the Tokyo University of Agriculture and Technology for assistance with the constructions of expression vectors for the mutants.

This article contains supplemental Experimental Procedures and Figs. S1–S3.
The atomic coordinates and structure factors (codes 3VNY, 3VNZ, and 3VO0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GlcA
- glucuronic acid
- AcGlcA79A
- β-glucuronidase from A. capsulatum
- GH
- glycoside hydrolase
- MeGlcA
- 4-O-methyl-GlcA
- PNP
- p-nitrophenol
- PNP-β-Glc
- PNP-β-glucopyranoside
- PNP-β-Xyl
- PNP-β-xylopyranoside
- PNP-β-GlcA
- PNP-β-glucuronide
- MeGlcA-β-1,6-Gal2
- MeGlcA-β-1,6-Gal-β-1,6-Gal
- DNP-2FGlcA
- 2′,4′-dinitrophenyl 2-deoxy-2-fluoro-β-d-glucuronide.
REFERENCES
- 1. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 3. Parish C. R., Freeman C., Hulett M. D. (2001) Heparanase: a key enzyme involved in cell invasion. Biochim. Biophys. Acta 1471, M99–M108 [DOI] [PubMed] [Google Scholar]
- 4. Sasaki K., Taura F., Shoyama Y., Morimoto S. (2000) Molecular characterization of a novel β-glucuronidase from Scutellaria baicalensis Georgi. J. Biol. Chem. 275, 27466–27472 [DOI] [PubMed] [Google Scholar]
- 5. Eudes A., Mouille G., Thévenin J., Goyallon A., Minic Z., Jouanin L. (2008) Purification, cloning and functional characterization of an endogenous β-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 49, 1331–1341 [DOI] [PubMed] [Google Scholar]
- 6. Kuroyama H., Tsutsui N., Hashimoto Y., Tsumuraya Y. (2001) Purification and characterization of a β-glucuronidase from Aspergillus niger. Carbohydr. Res. 333, 27–39 [DOI] [PubMed] [Google Scholar]
- 7. Konishi T., Kotake T., Soraya D., Matsuoka K., Koyama T., Kaneko S., Igarashi K., Samejima M., Tsumuraya Y. (2008) Properties of family 79 β-glucuronidases that hydrolyze β-glucuronosyl and 4-O-methyl-β-glucuronosyl residues of arabinogalactan-protein. Carbohydr. Res. 343, 1191–1201 [DOI] [PubMed] [Google Scholar]
- 8. Jain S., Drendel W. B., Chen Z. W., Mathews F. S., Sly W. S., Grubb J. H. (1996) Structure of human β-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 3, 375–381 [DOI] [PubMed] [Google Scholar]
- 9. Wallace B. D., Wang H., Lane K. T., Scott J. E., Orans J., Koo J. S., Venkatesh M., Jobin C., Yeh L. A., Mani S., Redinbo M. R. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hulett M. D., Hornby J. R., Ohms S. J., Zuegg J., Freeman C., Gready J. E., Parish C. R. (2000) Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry 39, 15659–15667 [DOI] [PubMed] [Google Scholar]
- 11. Sapay N., Cabannes E., Petitou M., Imberty A. (2012) Molecular model of human heparanase with proposed binding mode of a heparan sulfate oligosaccharide and catalytic amino acids. Biopolymers 97, 21–34 [DOI] [PubMed] [Google Scholar]
- 12. Gandhi N. S., Freeman C., Parish C. R., Mancera R. L. (2012) Computational analyses of the catalytic and heparin-binding sites and their interactions with glycosaminoglycans in glycoside hydrolase family 79 endo-β-d-glucuronidase (heparanase). Glycobiology 22, 35–55 [DOI] [PubMed] [Google Scholar]
- 13. McBain A. J., Macfarlane G. T. (1998) Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J. Med. Microbiol. 47, 407–416 [DOI] [PubMed] [Google Scholar]
- 14. Nakamura J., Kubota Y., Miyaoka M., Saitoh T., Mizuno F., Benno Y. (2002) Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol. Immunol. 46, 487–490 [DOI] [PubMed] [Google Scholar]
- 15. Dabek M., McCrae S. I., Stevens V. J., Duncan S. H., Louis P. (2008) Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 66, 487–495 [DOI] [PubMed] [Google Scholar]
- 16. Ritz K., Dighton J., Giller K. E. (1994) Beyond the Biomass: Compositional and Functional Analysis of Soil Microbial Communities, pp. 149–156, John Wiley and Sons Ltd., Chichester, UK [Google Scholar]
- 17. Arul L., Benita G., Sudhakar D., Thayumanavan B., Balasubramanian P. (2008) β-Glucuronidase of family-2 glycosyl hydrolase: a missing member in plants. Bioinformation 3, 194–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Withers S. G., Aebersold R. (1995) Approaches to labeling and identification of active site residues in glycosidases. Protein Sci. 4, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 21. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen S. X., Morris R. J., Fernandez F. J., Ben Jelloul M., Kakaris M., Parthasarathy V., Lamzin V. S., Kleywegt G. J., Perrakis A. (2004) Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60, 2222–2229 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 26. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 27. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ichinose H., Yoshida M., Fujimoto Z., Kaneko S. (2008) Characterization of a modular enzyme of exo-1,5-α-l-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl. Microbiol. Biotechnol. 80, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ichinose H., Yoshida M., Kotake T., Kuno A., Igarashi K., Tsumuraya Y., Samejima M., Hirabayashi J., Kobayashi H., Kaneko S. (2005) An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280, 25820–25829 [DOI] [PubMed] [Google Scholar]
- 30. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 31. Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) β-Glucosidase, β-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxyl-terminal ends of β-strands four and seven. FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.