Background: trans-Encoded HLA-DQ molecules are biologically interesting, but no structures of such molecules exist.
Results: X-ray crystal structure of the trans-encoded DQ2.3 (DQA1*03:01/DQB1*02:01) was determined. Structural data are presented together with functional T-cell data.
Conclusion: DQ2.3 has preference for negative charged anchors at P1 and P4.
Significance: This work helps to understand why DQ2.3 is associated with a particular risk for type 1 diabetes.
Keywords: Autoimmunity, Diabetes, Immunology, Major Histocompatibility Complex (MHC), X-ray Crystallography, Celiac Disease, Gluten, Type 1 Diabetes
Abstract
MHC class II molecules are composed of one α-chain and one β-chain whose membrane distal interface forms the peptide binding groove. Most of the existing knowledge on MHC class II molecules comes from the cis-encoded variants where the α- and β-chain are encoded on the same chromosome. However, trans-encoded class II MHC molecules, where the α- and β-chain are encoded on opposite chromosomes, can also be expressed. We have studied the trans-encoded class II HLA molecule DQ2.3 (DQA1*03:01/DQB1*02:01) that has received particular attention as it may explain the increased risk of certain individuals to type 1 diabetes. We report the x-ray crystal structure of this HLA molecule complexed with a gluten epitope at 3.05 Å resolution. The gluten epitope, which is the only known HLA-DQ2.3-restricted epitope, is preferentially recognized in the context of the DQ2.3 molecule by T-cell clones of a DQ8/DQ2.5 heterozygous celiac disease patient. This preferential recognition can be explained by improved HLA binding as the epitope combines the peptide-binding motif of DQ2.5 (negative charge at P4) and DQ8 (negative charge at P1). The analysis of the structure of DQ2.3 together with all other available DQ crystal structures and sequences led us to categorize DQA1 and DQB1 genes into two groups where any α-chain and β-chain belonging to the same group are expected to form a stable heterodimer.
Introduction
Major histocompatibility complex (MHC) proteins play a critical role in immune recognition by displaying antigens to T-cell receptors (TCRs)5 within the context of MHC-peptide complex to elicit a T-cell-mediated immune response. In humans, MHC proteins are encoded by the human leukocyte antigen (HLA) genes found on chromosome 6. HLA is the single most important genetic factor that predisposes to most autoimmune diseases (1) and contributes 35–50% of the genetic disease association in type 1 diabetes and celiac disease (2, 3). CD4+ T-cells recognize antigens in the context of MHC class II molecules that are heterodimers of α- and β-chains. In humans, there are three isotypes of MHC class II molecules: HLA-DR, HLA-DQ, and HLA-DP. In HLA-DR, the polymorphic variation is provided by the β-chain alone as the α-chain is monomorphic. However, in DQ and DP, both the α-chains and the β-chains are polymorphic. As a result, unique DQ and DP molecules can be formed with α- and β-chains encoded on the same chromosome (i.e. encoded in cis) or on opposite chromosomes (i.e. encoded in trans). The occurrence of trans-encoded HLA class II molecules is well documented in the literature (4). However, evidence suggests that not every α- and β-chain pairing will form a stable heterodimer (5, 6). Hence, it is generally considered that alleles of DQα- and DQβ-chains pair up predominantly in cis rather than in trans (5, 7). Nevertheless, studies on type 1 diabetes indicate that trans-encoded HLA molecules may play a role in pathogenesis (8). It has been observed that individuals who are heterozygous for DQ2.5 (DQA1*05:01/DQB1*02:01) and DQ8 (DQA1*03:01/DQB1*03:02) are susceptible to type 1 diabetes with an almost 5-fold higher risk than those who are homozygous for either of the DQ variants (1, 9, 10). This phenomenon can be explained by the formation of trans-encoded molecules DQ8.5 (DQA1*05:01/DQB1*03:02) and DQ2.3 (DQA1*03:01/DQB1*02:01), which could present one or a few specific diabetogenic epitopes to CD4+ T-cells, possibly inducing an immune response that leads to destruction of insulin-producing pancreatic β-islet cells (8). A strong argument for involvement of the DQ2.3 heterodimer in type 1 diabetes comes from trans-racial gene mapping studies that have found that this heterodimer, which is typically found in the trans-configuration among Caucasian subjects, exists and is overrepresented in the cis-configuration among type 1 diabetes patients of African origin (11, 12). The increased diabetes risk of the African DQ2.3 (DQA1*03:01/DQB1*02) carrying DR7 haplotype is contrasted by a protecting effect of the DQ2.2 (DQA1*03:01/DQB1*02) carrying DR7 haplotype of European origin, speaking to the functional importance of α-chain in the DQ2.3 molecule (12).
Celiac disease patients mount T-cell responses to gluten (consisting of the α-, β-, and γ-gliadins as well as glutenin components) in the context of the celiac disease-associated DQ2.5 and DQ8 molecules (13). This human disease with in vivo expansion of T-cell clones specific for naturally selected epitopes offers a unique system to study the structure-function relationship of peptide-MHC complexes. Here, we have used this model system, offering a natural T-cell epitope restricted by HLA-DQ2.3, to study the x-ray crystal structure and function of a trans-encoded HLA-DQ molecule.
EXPERIMENTAL PROCEDURES
Gluten-specific T-cells and T-cell Proliferation Assay
T-cell reagents were established from intestinal biopsies of celiac disease patients. Separate, single biopsy specimens were challenged with chymotrypsin-treated gluten (0.2 mg/ml) or the overlapping M36999 γ-gliadin peptides (10 μm of each peptide) (14). T-cell cloning and T-cell proliferation assays were performed as described previously (14, 15). T-cell reactivity was tested in 3-day proliferative restimulation assays by the incorporation of [3H]thymidine during the final 24 h. T-cell lines that showed reactivity for TG2-treated gluten presented via DQ2.3 were isolated from five out of six DQ8/DQ2.5 heterozygote patients. We obtained three T-cell clones from three individual T-cell lines of one patient that were reactive to variants of the DQ2.5-glia-γ-4c epitope (PQPEQPEQPFPQPQ or PQTEQPEQPFPQPQ) in the context of DQ2.3. These were TCC548.3.5.6 (TRAV9-2*02/TRBV7-2*01, presented in detail in this study), TCC548.3.5.3 (TRAV12-3*01/TRBV15*02), and TCC548.1.8.5 (TRAV19*01/TRBV7-7*01). In addition, the T-cell clone TCC387.19 (TRAV8-1*01/TRBV4-3*01) from a DQ2.5-positive and DQ8-negative celiac disease patient was used for comparison. B-lymphoblastoid cells were used as antigen-presenting cells (supplemental Fig. 1). DQ2.5-mediated presentation was tested with the line CD114 (DQA1*05:01/DQB1*02:01 homozygous). DQ8-mediated presentation was tested with the cell line IHW9092 (DQA1*03:01/DQB1*03:02 homozygous). Presentation by DQ2.3 was tested with the cell line IHW9102 (DQA1*03:03/DQB1*02:02 homozygous), and presentation by DQ8.5 was tested with an HLA class II-negative bare lymphocyte syndrome cell line transfected with DQA1*05:01 and DQB1*03:02 (BLS WK; transfected by William Kwok) (16, 17).
Peptide Binding Assay
Flat-bottom plates (Nunc) were coated with the HLA-DQ-specific mAb SPV-L3 (2 μg/well). Lysates from EBV-transformed B-cell lines DQ2.5 (DQA1*05:01/DQB1*02:01) or DQ2.3 (DQA1*03:03/DQB1*02:02), 2 × 105 and 4 × 105 cells respectively, were added to each well. The captured HLA-DQ molecules were incubated with biotin-labeled P418 indicator peptide (EPRAPWIEQEGPEYW; MHC class I-derived) (18) at 0.5 μm, and synthetic peptides of several concentrations were added to the wells in a competitive binding assay as described elsewhere (19). The bound biotinylated indicator peptide was detected with europium-conjugated streptavidin (PerkinElmer Life Sciences), and the signals were measured as time-resolved fluorescence in a 1420 Victor3 multilabel counter (PerkinElmer Life Sciences).
Production of DQ2.3 Heterodimer
Constructs for production of the HLA α-chain (DQA1*03:01 made from cDNA of the DQA1*03:01/DQB1*03:02 homozygous lymphoblastoid B-cell IHW9092) and β-chain (DQB1*02:01 modified from pAcAB3-DQB1*02:01) (20) were introduced into the expression plasmid pRmHa3. The sequence coding for the transmembrane region of each chain was excluded. A nucleotide segment was introduced so that the peptide RDSGPQPEQPEQPFPQPQ (underlined sequence representing residues 68–81 of a γ-gliadin; National Center for Biotechnology Information (NCBI) accession number AAK84776) with Gln-to-Glu substitutions at two positions (P1 and P4) followed by a 15-residue thrombin-cleavable linker GAGSLVPR↓GSGGGGS (arrow indicates cleavage site) was linked to the N-terminal part of the DQβ-chain (21). The C-terminal part of the α-chain contained a Cys followed by a human rhinovirus 3C protease cleavage site, an acidic leucine zipper, and a His6 tag. The C-terminal part of the β-chain also contained a Cys residue, an HRV 3C protease cleavage site, a basic leucine zipper, a FLAG tag, and a BirA biotinylation site. An interchain disulfide bond was formed by the C-terminally introduced Cys residues and the leucine zipper pair promoted MHC dimer formation, whereas the His6 and FLAG tags facilitated protein purification. Drosophila Schneider 2 (S2; Invitrogen) cells were co-transfected with a pair of DNA plasmids encoding the α- and β-chain separately as well as with a blasticidin resistance gene (pCoBlast, Invitrogen) that ensured survival of the transfected cells. Expression of DQ2.3 was induced with CuSO4. The recombinant protein was isolated from culture supernatant by anti-FLAG tag affinity chromatography (anti-FLAG M2 affinity gel, Sigma) followed by gel filtration chromatography (Superdex 200, GE Healthcare). Gel filtration revealed three peaks. The first peak was a high molecular weight material, i.e. aggregated DQ2.3 molecules. MS analysis showed that the second peak contained the DQ2.3 molecule and a protein identified as Drosophila super coiling factor (FlyBase database; FlyBaseID: FBgn0025682) (22), likely competing with the linked gluten peptide for binding to the peptide groove. The third peak contained the DQ2.3 molecule and was further purified before crystallization.
Functional Testing of Water-soluble DQ2.3 Molecules
Purified molecules were site-specifically biotinylated using the biotin-protein ligase BirA (Avidity). Biotinylated molecules were bound to streptavidin-conjugated plates (Roche Applied Science) overnight. T-cell clones were added to the plate, and recognition of HLA molecules was measured as [3H]thymidine incorporation in a proliferative assay. Biotinylated DQ2.3 molecules were conjugated to multimers with streptavidin-phycoerythrin (Invitrogen). Staining of DQ2.3 reactive T-cell clones was analyzed on a FACSCalibur flow cytometer (BD Biosciences).
X-ray Crystallography
For crystallization, leucine zipper portion of the DQ2.3 heterodimer was removed by human rhinovirus 3C protease digestion for 16 h at 4 °C. The resulting mixture was purified by anion-exchange chromatography using a Resource Q column (GE Healthcare) followed by size exclusion chromatography using a Shodex KW-803 column (Showa Denko K.K.). Crystals were grown using the hanging drop vapor diffusion method at 18 °C. Protein concentration was 4 mg/ml, and the crystallization solution was 0.2 m Li2SO4, 0.1 m Tris, pH 8.5, 30% (w/v) PEG4000, 8% glycerol. Crystals grew to full size in 1 week. Crystals were dehydrated by the addition of 5 μl 0.2 m Li2SO4, 0.1 m Tris, pH 8.5, 40% (w/v) PEG4000, 8% glycerol to the hanging drop and 1 ml of the same buffer to the well and incubating at 18 °C for 12 h. The DQ2.3 crystal belonged to the space group C2 with cell dimensions a = 74.9 Å, b = 114.9 Å, c = 138.0 Å, and β = 103.4°. The initial diffraction data set was collected at beam line BL13B1 of the Taiwan National Synchrotron Radiation Research Center, and the final data set was collected at beam line 9-3 of the Stanford Synchrotron Radiation Laboratory. X-ray diffraction data were indexed and integrated using HKL2000 (23). Structure was solved by molecular replacement using Phaser (CCP4) (24). The α-chain of DQ8 (1JK8) and β-chain of DQ2.5 (1S9V) were used as search models. Structure was refined by Refmac (CCP4) (25), Coot (26), Buster (27), and PHENIX (28). The final Rwork and Rfree values are 0.210 and 0.283, respectively. The quality of the final model was verified by PROCHECK (29). Crystallographic parameters, data collection, and refinement statistics are given in supplemental Table 1.
RESULTS
T-cell Recognition of DQ2.5-Glia-γ-4c/DQ8-Glia-γ-1a Gluten Epitope Presented via cis- or trans-Encoded Heterodimers
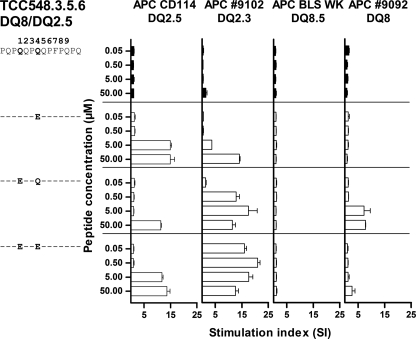
We describe three T-cell clones (TCC548.3.5.6, TCC548.3.5.3, and TCC548.1.8.5) derived from a DQ8/DQ2.5 heterozygous celiac disease patient that are specific for variants of the DQ2.5-glia-γ-4c epitope that harbor deamidated residues (PQPEQPEQPFPQPQ or PQTEQPEQPFPQPQ). These T-cell clones, in contrast to similar T-cell clones described earlier (14), have undergone maturation in thymus in the presence of the DQ2.3 molecule. Also of note, the epitope studied is recognized in the same register by DQ8-restricted T-cells (then named DQ8-glia-γ-1a). The preference for the positioning of the Glu residues introduced by deamidation is different for the DQ2.5- and DQ8-restricted T-cells, DQ2.5-restricted T-cells being sensitive to deamidation at position P4 and DQ8-restricted T-cells being sensitive to deamidations at P1 (14). The three T-cell clones carried different TCRs, but they showed similar reactivity patterns when tested against variants of the DQ2.5-glia-γ-4c epitope in the context of different antigen-presenting cells (APCs). Importantly, we found that a peptide with deamidations at both P1 and P4 was recognized much better when presented by the trans-encoded DQ2.3 molecule than the cis-encoded DQ2.5 molecule (Fig. 1). The same three T-cell clones were tested for their ability to recognize peptides with Gln-to-Glu substitutions at P1 and/or P4 in the context of various cis- or trans-encoded HLA molecules, and they gave similar response patterns. Substitution at P1 had an effect in the context of DQ2.3 and DQ8, whereas substitution at P4 had an effect in the context of both DQ2.3 and DQ2.5 (Fig. 1).
FIGURE 1.
T-cell recognition of cis- and trans-encoded DQ heterodimers. The T-cell clone TCC548.3.5.6 was derived from a DQ8/DQ2.5 heterozygote celiac disease patient and tested against peptides with Gln to Glu substitutions (indicated by single-letter codes) at position P1 and/or P1 and P4 with four different APCs: DQ2.5 (CD114), DQ2.3 (IHW9102), DQ8.5 bare lymphocyte syndrome (BLS WK), and DQ8 (IHW9092). An increased response is seen when the peptide with Gln-to-Glu substitutions at both P1 and P4 is presented in the context of trans-encoded DQ2.3 to the T-cell clone from the heterozygote DQ8/DQ2.5 patient.
Peptide Binding to cis- or trans-Encoded Heterodimers
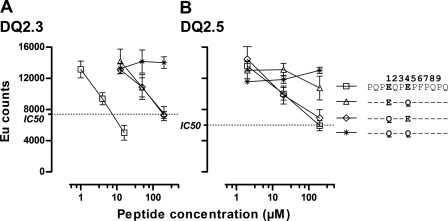
Peptide binding assays showed that the peptide variant with Glu at both P1 and P4 bound stronger to the DQ2.3 molecule (IC50 = 7.50 μm) when compared with peptide variants with Gln-to-Glu substitutions at either P1 or at P4 alone (both IC50 ≥ 200 μm) (Fig. 2A). This correlated with the T-cell response pattern described above, suggesting that the improved T-cell recognition (Fig. 1) could be explained by improved HLA binding. Similar peptide binding analysis for DQ2.5 showed that Gln-to-Glu substitution at P4 exerted the biggest effect as the variants with Gln-to-Glu substitutions at both P1 and P4 or only at P4 bound better than the variants with no substitution or substitution only at P1 (Fig. 2B). Thus, enhanced HLA binding could also explain the improved T-cell recognition of the P4 Glu-substituted peptide in the context of DQ2.5 (Fig. 1).
FIGURE 2.
Peptides with Gln to Glu substitutions (indicated by single-letter codes) at position P1 and P4, P1, or P4 were tested in a competitive peptide binding assay for binding to HLA DQ2.3 (A)and DQ2.5 molecules (B). The peptide with the Gln-to-Glu substitution at both P1 and P4 binds the strongest to the trans-encoded DQ2.3 molecule. Error bars indicate mean of triplicate ± S.D. EU counts, Europium photon counts per second.
Determining T-cell Recognition by Ala and Lys Scans
Single position Ala- or Lys-substituted variants of PQPEQPEQPFPQPQ were next tested in proliferative T-cell assays with the T-cell clones TCC387.19 and TCC548.3.5.6 using APCs expressing DQ2.5 or DQ2.3 (supplemental Fig. 2). The most interesting observation in this experiment related to recognition of the peptide with Ala substitution at P1. Both TCC548.3.5.6 and TCC387.19 showed impaired recognition of the P1 Ala peptide when compared with the P1 Glu peptide in the context of DQ2.3, whereas in the context of DQ2.5, both T-cell clones recognized the P1 Ala peptide best (Fig. 3). As this effect was seen with both T-cell clones, it may suggest that the P1 pocket of DQ2.5 is better at accommodating Ala than the P1 pocket of DQ2.3.
FIGURE 3.
T-cell recognition of peptides substituted with Ala in position P1 and Glu in position P4. Synthetic peptides were tested in a T-cell proliferative assay with the T-cell clones DQ2 TCC387.19 (A) and DQ2.3 TCC548.3.5.6 (B) using the DQ2.5 APC or the DQ2.3 APC. For both T-cell clones, the response is the highest to the Ala in P1-substituted peptide in the context of DQ2.5, whereas the response against the DQ2.3 is the highest when the bound peptide harbors a negative charge in P1. Error bars indicate mean of triplicate ± S.D. Ala and Glu are indicated by single-letter codes.
Functional Testing of Water-soluble trans-Encoded DQ2.3 Molecules
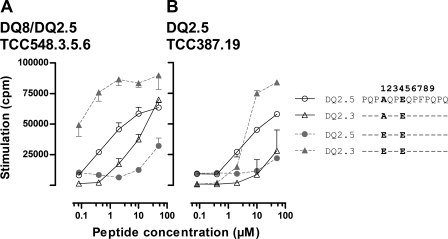
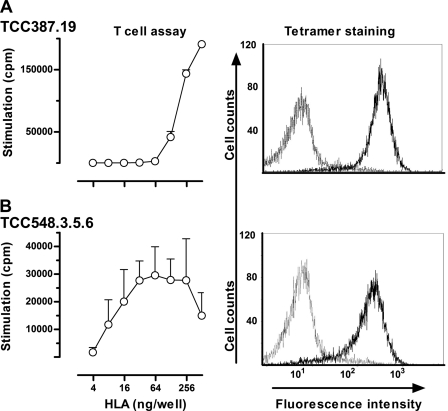
We produced water-soluble DQ2.3 molecules in S2 cells displaying the PQPEQPEQPFPQPQ peptide. Purified monomeric DQ2.3 molecules were biotinylated and added to streptavidin-coated plastic wells to test whether the recombinant molecules were recognized by antigen-specific T-cell clones. Both TCC387.19 and TCC548.3.5.6 recognized the recombinant molecules with the tethered peptide (Fig. 4). Moreover the clones were specifically stained with multimerized HLA/peptide molecules obtained by coupling biotinylated DQ2.3 HLA molecules to phycoerythrin-labeled streptavidin. As evident in Fig. 4, both TCC387.19 and TCC548.3.5.6 were effectively stained, indicating that the water-soluble HLA molecules were indeed correctly folded and were properly recognized by the T-cells. Once the integrity of the trans-encoded DQ2.3 molecule was verified, we went on to determine the x-ray crystal structure of DQ2.3 to gain structural insight into trans-encoded heterodimer formation and epitope recognition.
FIGURE 4.
T-cell recognition of recombinant water-soluble DQ2.3 trans-encoded molecules. Purified and BirA-biotinylated DQ8/2 molecules were bound to streptavidin-coated microtiter plates and tested for recognition by T-cell clones TCC387.19 (A) and TCC548.3.5.6 (B) in a proliferative T-cell assay (mean of triplicate ± S.D.). The T-cells recognize the water-soluble DQ2.3 molecule in a dose-dependent manner. In addition, the same T-cell clones were stained with biotinylated DQ2.3 coupled to phycoerythrin-labeled streptavidin and analyzed by flow cytometry. The DQ2-αI tetramer was used for negative control staining. The T-cell clones stain positively with the DQ2.3 tetramer.
Structure of DQ2.3 in Complex with PQPEQPEQPFPQPQ
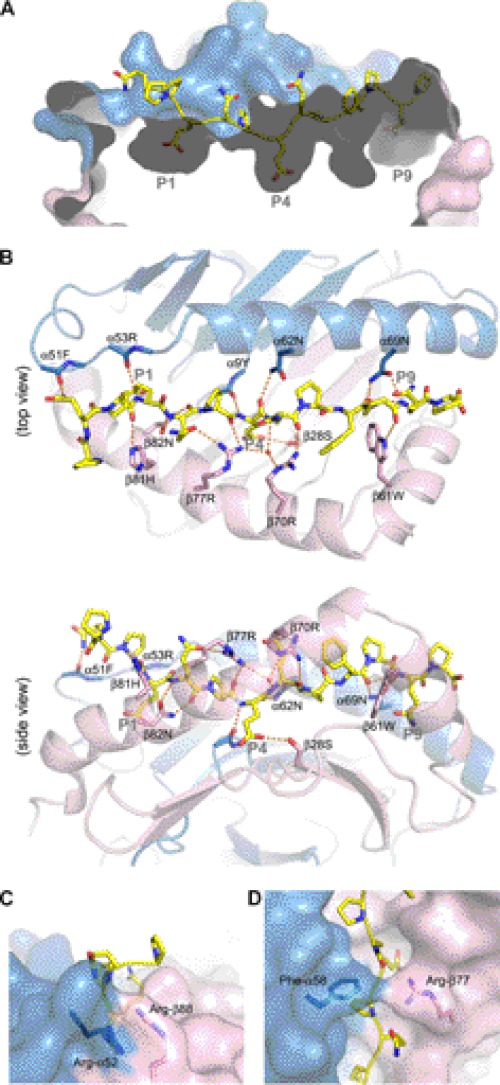
The structure of DQ2.3:DQ2.5-glia-γ-4c complex was determined to 3.05 Å resolution using x-ray crystallography. Statistics for data collection and structure refinement are given in supplemental Table 1, and a representative electron density map is shown in supplemental Fig. 3. Asn-β33 is the only residue that is in the disallowed region of the Ramachandran plot. The loop structure at this particular position adopts an unusual type II β-turn (30), which places the φ and ψ angles of the residue at the i+1 position of the turn in otherwise disallowed configurations. The overall structure of DQ2.3 is highly similar to previously reported DQ crystal structures: DQ2.5, DQ8, and DQ6.2.
Thirteen residues of the tethered peptide (PQPEQPEQPFPQP) are visible in the electron density map, occupying the P–3 to P10 binding register. The apparent anchoring residues are P1 Glu, P4 Glu, and P9 Gln, whose side chains are buried inside individual pockets found along the DQ2.3 peptide-binding groove (Fig. 5A). There are a total of 15 hydrogen bonds between DQ2.3 and peptide, of which 12 are mediated by the peptide main chain and three are mediated by the peptide side chain groups (Fig. 5B). Despite having three Pro residues within the nine-core HLA-binding sequence, only one conserved main chain hydrogen bond is found missing: P6 nitrogen to Asn-α62 Oδ1. The DQ2.3 residues involved in hydrogen bonding to the DQ2.5-glia-γ-4c epitope are Tyr-α9, Phe-α51, Arg-α53, Asn-α69, Ser-β28, Trp-β61, Arg-β70, Arg-β77, His-β81, and Asn-β82. Interestingly, all of these residues are also present in the cis-encoded DQ2.5 (31).
FIGURE 5.
Crystal structure of DQ2.3:DQ2.5-glia-γ-4c complex (PDB ID: 4D8P). DQ2.3 α-chain and β-chain are colored in blue and pink, respectively. The gliadin peptide is drawn as a stick model (yellow, carbon; blue, nitrogen; red, oxygen). A, surface representation of the peptide-binding groove in DQ2.3. B, putative hydrogen-bond interactions between DQ2.3 and the gliadin peptide are shown as orange dashes. C, the gate structure found adjacent to the P1 pocket of DQ2.3. D, the roof structure found above the P4 pocket of DQ2.3.
Two unusual structural features are present in DQ2.3 that are not seen in other MHC class II molecules. The first unique structure is the “gate” formed by Arg-α52 and Arg-β88 (Fig. 5C), which is located at the P1 end of the peptide-binding groove. The guanidinium group of Arg-α52 and Arg-β88 is stacked in a face-to-face manner with a Cζ to Cζ separation distance of 3.6 Å. Although Arg-Arg pairing is not common in protein structures, a survey of the Protein Data Bank by Magalhaes et al. (32) found 41 analogous Arg-Arg interactions. This gate creates a shallow barrier at the P1 end of the peptide-binding groove. Related to this, the P–3 to P–1 segment of the peptide is protruding out at a roughly 90° angle relative to the rest of the peptide sitting in the MHC-binding groove. The second unique structure is the “roof” formed by Phe-α58 and Arg-β77, which lies over the P3 and P4 section of the bound peptide (Fig. 5D). Arg-β77 side chain forms a hydrogen bond with the P2 Gln side chain and the P3 main chain carbonyl oxygen. The occurrence of hydrogen-bonding interaction between Arg-β77 and the nonanchoring P2 side chain and P3 backbone is unusual. Extensive shielding of the bound peptide by MHC side chains is not seen in any of the previous class II MHC crystal structures. Incidentally, α58 and β77 are involved in TCR recognition, particularly with the CDR3 loops, as observed in different class II MHC-TCR complex structures determined to date (33).
DISCUSSION
Our results demonstrate that the DQ2.3 molecule combines the peptide binding signatures of the DQ2.5 and DQ8 molecules. This results in a binding motif with preference for negatively charged anchor residues at both the P1 and the P4 positions. In this way, some epitopes can be presented even more effectively in the context of the trans-encoded DQ2.3 molecule. This has relevance for understanding how the trans-encoded DQ2.3 molecule is predisposing to type 1 diabetes.
We found that the responses of the T-cell clones in the context of the DQ2.3 molecule were improved by Gln-to-Glu substitutions in the peptide at the P1 and P4 positions (Fig. 1). Results from peptide binding assays suggest that the enhanced T-cell recognition of the deamidated peptides, at least in part, is explained by improved HLA binding (34). There are multiple positively charged residues near the P1 pocket (Arg-α52 and His-α24) and the P4 pocket (Arg-β77, Arg-β70, Lys-β71) that may establish long range electrostatic interaction with the P1 or P4 Glu of the gliadin peptide or establish hydrogen bonds with the peptide through water molecules, which in the current crystal structure are not visible due to insufficient resolution.
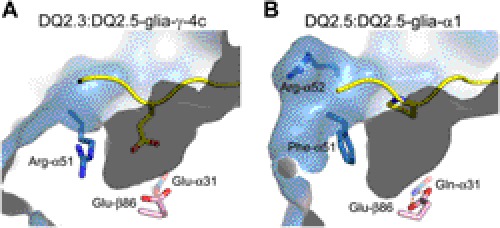
Differential recognition of the P1 Ala-substituted PQPEQPEQPFPQPQ peptide by a T-cell clone in the context of DQ2.5 versus DQ2.3 points to structural differences between the two HLA molecules around the P1 pocket (Fig. 3). Although the overall shape and size of the P1 pocket in DQ2.3 and DQ2.5 are highly similar, the polarity of the residues found at the surface of the respective P1 pockets is dissimilar (Fig. 6). In DQ2.5, Gln-α31 and Glu-β86 are found at the bottom of the P1 pocket and form a bidentate hydrogen bond to one another (donor-acceptor distances are 2.8 and 3.0 Å). In contrast, Glu-α31 and Glu-β86 are found at the bottom of the P1 pocket in DQ2.3 (carboxylate oxygen to carboxylate oxygen distances are 2.9 and 3.4 Å). Furthermore, DQ2.3 has Arg-α51 on the side of the P1 pocket, whereas DQ2.5 has Phe-α51. Binding of the P1 Ala-substituted peptide to either DQ2.3 or DQ2.5 will result in a mostly vacant P1 pocket, which may lead to conformational shift of the above mentioned residues as well as introduction of solvent molecules inside the P1 pocket. Although the details of such rearrangement need to be assessed in a future study, we predict that the DQ2.5 will form a more stable complex with an aliphatic amino acid such as Ala in P1 in contrast to the DQ2.3 molecule that prefers a negatively charged amino acid in this position.
FIGURE 6.
P1 pocket of DQ2.3 (PDB ID: 4D8P) (A) and DQ2.5 (PDB ID: 1S9V) (B). Selected α-chain residues (light blue, carbon; dark blue, nitrogen; red, oxygen) and β-chain residues (pink, carbon; red, oxygen) are shown as a stick model. For both structures, the bound peptide is represented as a yellow ribbon with the P1 residue side chain shown as a stick model (yellow, carbon; dark blue, nitrogen; red, oxygen).
It is generally considered that alleles encoded in cis (i.e. on a single chromosome) have been evolutionarily selected to form a stable heterodimer, whereas trans-encoded heterodimers may not necessarily be able to form a stable or functional MHC (35). Regardless, a number of studies have shown that αβ heterodimers can form among a subset of different MHC class II molecules, including pairing of mixed haplotypes (e.g. mouse MHC class II AαbAβk (35)), mixed isotypes (e.g. human MHC class II DRαDQβ (36) or mouse MHC class II EαdAβd (37)), and mixed species (e.g. human and mouse MHC class II DRαAβd (38)). Some structural explanations for the bias for cis-encoded heterodimer formation have been given for mouse MHC class II I-A molecules, where a subset of specific residues has been identified that can dictate which of the α- and β-chain alleles can form a stable heterodimer (39). However, limited knowledge exists as to the rules that determine formation of MHC class II heterodimers.
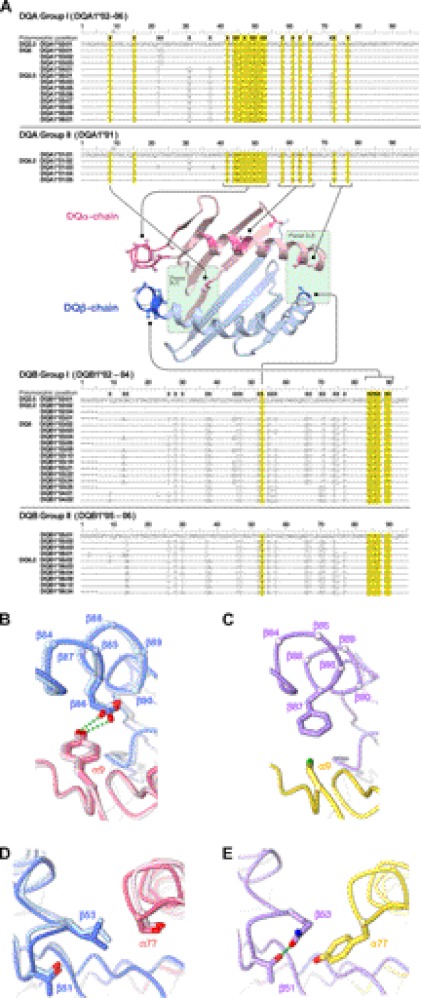
In DQ molecules, polymorphic residues at the αβ dimerization interface are concentrated at the two ends of the peptide-binding groove. On the P1 side, α44–α54 and β84–β90, two clusters in the α and β chains where polymorphism occurs most extensively, come together to form the α/β interface. On the P9 side, polymorphic residues α73/α77 and β53 contact each other at the α/β interface. Sequence distribution at these polymorphic positions reveals that DQ alleles can be divided into two major groups. Group I consists of DQA1*02 through DQA1*06 and DQB1*02 through DQB1*04, whereas group II is composed of DQA1*01 and DQB1*05 and DQB1*06 (Fig. 7A). Interestingly, according to the dbMHC database (40), only 10 out of 4,233 reported HLA-DQ haplotypes carry a group I-group II mixed pair, indicating potential evolutionary selection pressure against the cross-group pairing. A survey of available DQ crystal structures revealed some key structural features on the αβ dimerization interface that may be dictating the observed allele-pairing bias.
FIGURE 7.
Polymorphic residues at α-chain/β-chain dimerization interface. A, amino acid sequence alignment of known DQA1 and DQB1 alleles. Polymorphic positions are indicated by X. Polymorphic positions at the α/β dimerization interface are highlighted in yellow. Their locations are also indicated by a darker shade in the ribbon diagram of a representative DQ structure (DQ8, PDB ID: 2NNA). For non-glycine residues, their Cα–Cβ bonds are shown in stick-and-ball representation to indicate the general orientation of the side chain (dark blue, nitrogen; red, oxygen; green, sulfur). B, hydrogen-bonding interactions between α9 and β86 side chain near the P1 pocket. The α- and β-chains are shown in pink and blue, respectively. The two structures of group I molecules are shown with DQ2.5 (1S9V) in a lighter shade than DQ8 (2NNA). C, same position as in B for a group II molecule DQ6.2 (1UVQ). The α- and β-chains are shown in yellow and purple, respectively. D, in the two group I molecules, DQ2.5 and DQ8, polymorphic residues α77 and β53 contact each other in the P9 pocket at the α–β interface. The same molecules and color-coding are used as in B. E, the same position as in D for the group II molecule DQ6.2.
On the P1 side of the molecule in DQ2.5 and DQ8 (group I), absolutely conserved Tyr-9α and Glu-86β side chains form an H-bond to each other (Fig. 7B). In DQ6 (group II), however, β87 is found at the position where β86 occurs in DQ2.5 and in DQ8 (both group I) (Fig. 7C). Cys-9α is absolutely conserved, and β87 carries either a Tyr or a Phe residue, allowing the pair to have a good steric fit. However, mixing of group I and group II alleles will result either in the loss of an H-bond or the introduction of a steric clash. Furthermore, although the α44–α54 and β84–β90 polymorphic clusters both assume a helical structure, their local conformations are different in groups I and II. In DQ6 (group II), the backbone conformation near Phe-51α draws the bulky side chain close to the β-chain at Ala-86β and Gly-89β. In group I, β86 and β89 are conserved for Glu and Thr, respectively. Simple overlay of DQ6 and DQ8 structures shows steric incompatibility between Phe-51α and Thr-89β side chains. Similarly, in DQ8 (group I), the difference in the helical structure shifts the β-chain-contacting residue from Phe-51α to Arg-52α, where the Arg-52α side chain is buried deep inside the αβ interface and forms an H-bond with the previously mentioned Glu-86β. Again, overlay of DQ6 and DQ8 structures indicates that the Arg-52α will clash against Ala-86β. At the other end of the peptide-binding groove, polymorphic α77 and β53 residues contact each other. In group I, α77 and β53 are conserved for Ser and Leu, respectively (Fig. 7D). In group II, α77 and β53 are conserved for Tyr and Gln (Fig. 7E). Structural overlay of DQ2.5, DQ8, and DQ6 shows that pairing of the group II α-chain and the group I β-chain will likely suffer from steric crash between the Tyr-77α and Leu-53β side chains. Taken together, mixed chain pairing (group I α-chain paired with group II β-chain or group II α-chain paired with group I β-chain) will lead to loss of interchain hydrogen-bond interaction or result in suboptimal packing at the heterodimer interface.
Our crystal structure shows that the P1 pocket in DQ2.3 is significantly different from that of DQ2.5 due to the polymorphic MHC residues found in this region. Additionally, we have demonstrated that DQ2.3 presents a gluten epitope to T-cells much more efficiently than DQ2.5. Therefore, trans-encoded MHCs have the potential to drastically alter an individual's immune response toward a particular antigen. Sequence analysis of the HLA-DQ gene products has revealed that certain haplotype pairings are more likely to produce a structurally stable trans-encoded αβ heterodimer than others, which is also reflected in the high frequency of compatible pairings seen in the NCBI dbMHC database.
Supplementary Material
Acknowledgments
We thank Marit Sandvik for technical assistance and Elizabeth Mellins for providing the pRmHa3 plasmid. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and NIGMS.
This work was supported by grants from the Novo Nordisk Foundation, the Juvenile Diabetes Research Foundation, and the Research Council of Norway (to L. M. S.) and by Singapore Biomedical Research Council Grant 07/1/21/19/546 (to C. Y. K.).

This article contains supplemental Figs. 1–3 and Table 1.
The atomic coordinates and structure factors (code 4D8P) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TCR
- T-cell receptor
- TCC
- T-cell clone
- APC
- antigen-presenting cell.
REFERENCES
- 1. Thorsby E. (1997) Invited anniversary review: HLA-associated diseases. Hum. Immunol. 53, 1–11 [DOI] [PubMed] [Google Scholar]
- 2. Todd J. A., Walker N. M., Cooper J. D., Smyth D. J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S. F., Payne F., Lowe C. E., Szeszko J. S., Hafler J. P., Zeitels L., Yang J. H., Vella A., Nutland S., Stevens H. E., Schuilenburg H., Coleman G., Maisuria M., Meadows W., Smink L. J., Healy B., Burren O. S., Lam A. A., Ovington N. R., Allen J., Adlem E., Leung H. T., Wallace C., Howson J. M., Guja C., Ionescu-Tîrgovişte C., Simmonds M. J., Heward J. M., Gough S. C., Dunger D. B., Wicker L. S., Clayton D. G. (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Heel D. A., Franke L., Hunt K. A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M. C., Barnardo M. C., Bethel G., Holmes G. K., Feighery C., Jewell D., Kelleher D., Kumar P., Travis S., Walters J. R., Sanders D. S., Howdle P., Swift J., Playford R. J., McLaren W. M., Mearin M. L., Mulder C. J., McManus R., McGinnis R., Cardon L. R., Deloukas P., Wijmenga C. (2007) A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 39, 827–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charron D. J., Lotteau V., Turmel P. (1984) Hybrid HLA-DC antigens provide molecular evidence for gene trans-complementation. Nature 312, 157–159 [DOI] [PubMed] [Google Scholar]
- 5. Kwok W. W., Nepom G. T. (1991) Structural and functional constraints on HLA class II dimers implicated in susceptibility to insulin dependent diabetes mellitus. Baillieres Clin. Endocrinol. Metab. 5, 375–393 [DOI] [PubMed] [Google Scholar]
- 6. Kwok W. W., Kovats S., Thurtle P., Nepom G. T. (1993) HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J. Immunol. 150, 2263–2272 [PubMed] [Google Scholar]
- 7. McFarland B. J., Beeson C. (2002) Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med. Res. Rev. 22, 168–203 [DOI] [PubMed] [Google Scholar]
- 8. Nepom G. T., Erlich H. (1991) MHC class II molecules and autoimmunity. Annu. Rev. Immunol. 9, 493–525 [DOI] [PubMed] [Google Scholar]
- 9. Koeleman B. P., Lie B. A., Undlien D. E., Dudbridge F., Thorsby E., de Vries R. R., Cucca F., Roep B. O., Giphart M. J., Todd J. A. (2004) Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans-dimer associations with disease. Genes Immun. 5, 381–388 [DOI] [PubMed] [Google Scholar]
- 10. Erlich H., Valdes A. M., Noble J., Carlson J. A., Varney M., Concannon P., Mychaleckyj J. C., Todd J. A., Bonella P., Fear A. L., Lavant E., Louey A., Moonsamy P. (2008) HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 57, 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Todd J. A., Mijovic C., Fletcher J., Jenkins D., Bradwell A. R., Barnett A. H. (1989) Identification of susceptibility loci for insulin-dependent diabetes mellitus by trans-racial gene mapping. Nature 338, 587–589 [DOI] [PubMed] [Google Scholar]
- 12. Noble J. A., Johnson J., Lane J. A., Valdes A. M. (2011) Race-specific type 1 diabetes risk of HLA-DR7 haplotypes. Tissue Antigens 78, 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sollid L. M. (2002) Celiac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2, 647–655 [DOI] [PubMed] [Google Scholar]
- 14. Tollefsen S., Arentz-Hansen H., Fleckenstein B., Molberg O., Ráki M., Kwok W. W., Jung G., Lundin K. E., Sollid L. M. (2006) HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Invest. 116, 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Molberg Ø., Solheim Flaete N., Jensen T., Lundin K. E., Arentz-Hansen H., Anderson O. D., Kjersti Uhlen A., Sollid L. M. (2003) Intestinal T-cell responses to high molecular weight glutenins in celiac disease. Gastroenterology 125, 337–344 [DOI] [PubMed] [Google Scholar]
- 16. Kwok W. W., Schwarz D., Nepom B. S., Hock R. A., Thurtle P. S., Nepom G. T. (1988) HLA-DQ molecules form α-β heterodimers of mixed allotype. J. Immunol. 141, 3123–3127 [PubMed] [Google Scholar]
- 17. Reichstetter S., Kwok W. W., Nepom G. T. (1999) Impaired binding of a DQ2- and DQ8-binding HSV VP16 peptide to a DQA1*0501/DQB1*0302 trans-class II heterodimer. Tissue Antigens 53, 101–105 [DOI] [PubMed] [Google Scholar]
- 18. Vartdal F., Johansen B. H., Friede T., Thorpe C. J., Stevanović S., Eriksen J. E., Sletten K., Thorsby E., Rammensee H. G., Sollid L. M. (1996) The peptide-binding motif of the disease associated HLA-DQ (α 1* 0501, β 1* 0201) molecule. Eur. J. Immunol. 26, 2764–2772 [DOI] [PubMed] [Google Scholar]
- 19. Vader W., Stepniak D., Kooy Y., Mearin L., Thompson A., van Rood J. J., Spaenij L., Koning F. (2003) The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc. Natl. Acad. Sci. U.S.A. 100, 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quarsten H., McAdam S. N., Jensen T., Arentz-Hansen H., Molberg Ø., Lundin K. E., Sollid L. M. (2001) Staining of celiac disease-relevant T cells by peptide-DQ2 multimers. J. Immunol. 167, 4861–4868 [DOI] [PubMed] [Google Scholar]
- 21. Kozono H., White J., Clements J., Marrack P., Kappler J. (1994) Production of soluble MHC class II proteins with covalently bound single peptides. Nature 369, 151–154 [DOI] [PubMed] [Google Scholar]
- 22. Jüse U., Fleckenstein B., Bergseng E., Sollid L. M. (2009) Soluble HLA-DQ2 expressed in S2 cells copurifies with a high affinity insect cell-derived protein. Immunogenetics 61, 81–89 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D. Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. Wilmot C. M., Thornton J. M. (1988) Analysis and prediction of the different types of β-turn in proteins. J. Mol. Biol. 203, 221–232 [DOI] [PubMed] [Google Scholar]
- 31. Kim C. Y., Quarsten H., Bergseng E., Khosla C., Sollid L. M. (2004) Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 101, 4175–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Magalhaes A., Maigret B., Hoflack J., Gomes J. N., Scheraga H. A. (1994) Contribution of unusual arginine-arginine short-range interactions to stabilization and recognition in proteins. J. Protein Chem. 13, 195–215 [DOI] [PubMed] [Google Scholar]
- 33. Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 34. Fallang L. E., Bergseng E., Hotta K., Berg-Larsen A., Kim C. Y., Sollid L. M. (2009) Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat. Immunol. 10, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 35. Braunstein N. S., Germain R. N. (1987) Allele-specific control of Ia molecule surface expression and conformation: implications for a general model of Ia structure-function relationships. Proc. Natl. Acad. Sci. U.S.A. 84, 2921–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lotteau V., Teyton L., Burroughs D., Charron D. (1987) A novel HLA class II molecule (DRα-DQβ) created by mismatched isotype pairing. Nature 329, 339–341 [DOI] [PubMed] [Google Scholar]
- 37. Spencer J. S., Freed J. H., Kubo R. T. (1993) Expression and function of mixed isotype MHC class II molecules in normal mice. J. Immunol. 151, 6822–6832 [PubMed] [Google Scholar]
- 38. Lechler R. I., Sant A. J., Braunstein N. S., Sekaly R., Long E., Germain R. N. (1990) Cell surface expression of hybrid murine/human MHC class II β-α dimers. Key influence of residues in the amino-terminal portion of the β 1 domain. J. Immunol. 144, 329–333 [PubMed] [Google Scholar]
- 39. Zhu Y., Rudensky A. Y., Corper A. L., Teyton L., Wilson I. A. (2003) Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. J. Mol. Biol. 326, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 40. Helmberg W., Dunivin R., Feolo M. (2004) The sequencing-based typing tool of dbMHC: typing highly polymorphic gene sequences. Nucleic Acids Res. 32, W173–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.