Background: GRIN is an effector of Go/i known to regulate neurite extension.
Results: GRIN modulates MAPK by sequestering Sprouty, an inhibitor of the MAPK pathway, and this is reversed by active Gαo binding.
Conclusion: Activation of Gαo can inhibit the MAPK pathway by releasing Sprouty from GRIN binding.
Significance: This is a novel mechanism of GPCR regulation of the MAPK pathway.
Keywords: Cannabinoid Receptors, Cell Signaling, G Protein-coupled Receptors (GPCR), G Proteins, MAP Kinases (MAPKs)
Abstract
Gαo/i interacts directly with GRIN (G protein-regulated inducer of neurite outgrowth). Using the yeast two-hybrid system, we identified Sprouty2 as an interacting partner of GRIN. Gαo and Sprouty2 bind to overlapping regions of GRIN, thus competing for GRIN binding. Imaging experiments demonstrated that Gαo expression promoted GRIN translocation to the plasma membrane, whereas Sprouty2 expression failed to do so. Given the role of Sprouty2 in the regulation of growth factor-mediated MAPK activation, we examined the contribution of the GRIN-Sprouty2 interaction to CB1 cannabinoid receptor regulation of FGF receptor signaling. In Neuro-2A cells, a system that expresses all of the components endogenously, modulation of GRIN levels led to regulation of MAPK activation. Overexpression of GRIN potentiated FGF activation of MAPK and decreased tyrosine phosphorylation of Sprouty2. Pretreatment with Go/i-coupled CB1 receptor agonist attenuated subsequent FGF activation of MAPK. Decreased expression of GRIN both diminished FGF activation of MAPK and blocked CB1R attenuation of MAPK activation. These observations indicate that Gαo interacts with GRIN and outcompetes GRIN from bound Sprouty. Free Sprouty then in turn inhibits growth factor signaling. Thus, here we present a novel mechanism of how Go/i-coupled receptors can inhibit growth factor signaling to MAPK.
Introduction
Gαo, the most abundant G protein in the mammalian brain, belongs to the Gαo/i family, containing ∼80% sequence homology to Gαi1 (1). Gαo is also expressed in adenohypophysis, heart, kidney, medulla, olfactory epithelium, and chromaffin cells (2). Despite its discovery over two decades ago, the direct signaling partners of Gαo and effectors are not fully characterized. Studies, using yeast two-hybrid screens and cDNA expression cloning, have yielded novel direct effectors of Gαo. They include the GTPase-activating protein for the small G protein Rap (RapGAP),3 Gαz-GAP (also known as RGS20), the putative tyrosine phosphatase paladin, the regulator of G protein signaling 17 (RGS-17), the Purkinje cell protein-2 (Pcp2), and the G protein-regulated inducer of neurite outgrowth (GRIN) (3–5). The GRIN protein family, consisting of three gene products, GRIN1–3 (GPRIN1–3), is highly expressed in the brain and binds selectively to activated forms of Gαo, Gαz, and Gαi (5). GRIN isoforms co-localize with Gαo at the growth cone of neuronal cells and promote neurite extension in Neuro-2A cells when co-expressed with the constitutively active mutant Gαo Q205L (6). Gαo-GRIN1 modulates Cdc42 activation and neuronal morphology (6). We have previously identified GRIN3 (originally named IP6) as a direct interacting protein of Gαo, in a yeast two-hybrid study using the constitutively active form Q205L-Gαo as bait (3). This family member shows sequence homology to KIAA0514 (amino acids 205–459), which was later renamed GRIN2 (3, 5).
In the present study, we used the yeast two-hybrid system to identify potential downstream partners of the Gαo-GRIN2 pathway. GRIN2 was used as the bait to screen a human brain cDNA library, and among the candidate proteins hits was Sprouty2, a known regulator of receptor tyrosine kinase signaling. Sprouty2 has been shown to inhibit both neuronal differentiation and survival through a negative-feedback loop that down-regulates neurotrophic-driven signals (7, 8). Our results show that Sprouty2 and GRIN isoforms interact in exogenously expressed and endogenous systems. We have characterized the interaction and functional consequence of GRIN in communicating Gαo signals to Sprouty2. In a fully endogenous system, activation of Go/1-coupled receptor can inhibit subsequent stimulation of MAPK1,2 by receptor tyrosine kinase, and thus the GRIN-Sprouty interaction can serve as a temporal bridge to connect heterotrimeric receptor G protein pathway regulation of receptor tyrosine kinase signaling to MAPK1,2.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK-293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, and Neuro-2A cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 1 mm sodium pyruvate, and 1% penicillin-streptomycin. Neuro-2A cells were obtained from ATCC (CCl-131). Lipofectamine 2000 (Invitrogen) was used for introduction of exogenous DNA into HEK-293T and Neuro-2A cells according to the manufacturer's instructions. For HEK-293T cells, 1.8 × 106 cells were plated on 60-mm dishes, and 24 h later, the cells were transfected with 5 μg of DNA, or 5.5 × 106 cells were plated on 100-mm dishes, and 24 h later, the cells were transfected with 10 μg of DNA. For imaging experiments, Neuro-2A cells (1.2 × 105) were plated on 35-mm MatTek® plates and transfected using Lipofectamine 2000 the following day. Neuro-2A cells were serum-starved in serum-free media (0.1% BSA modified Eagle's medium).
Treatment with Growth Factors or Agonists
Cells were treated with various growth factors or agonists in culture using serum-free media. Basic fibroblast growth factor (bFGF, Invitrogen) was dissolved in PBS with 0.1% BSA to a stock concentration of 100 ng/ml and stored at −20 °C until use. Epidermal growth factor (EGF, Millipore) was dissolved in PBS to a stock concentration of 50 ng/ml and stored at −20 °C. Glial-derived neurotrophic factor (Promega) was dissolved in PBS at a stock concentration of 100 ng/ml and stored at −20 °C. HU-210 (Sigma) was dissolved in DMSO at a stock concentration of 10 mm and stored at −80 °C. Carbachol (Tocris) was dissolved in DMSO.
DNA Constructs
The cDNA clone KIAA0514 was identified in public databases and was generously supplied by Dr. T. Nagase (Kazusa DNA Research Institute). This cDNA was subcloned into various vectors (pPC86 (SalI-NotI), pBk FLAG (SalI-NotI), pcDNA3.1-HA (EcoRI-NotI), and pGEX4T2 (SalI-NotI)). Mouse GRIN2 was cloned by one-step RT-PCR (Qiagen) from mouse whole brain mRNA using the following primers: 5′-ATG AGT TCTAGC CAC CCT GAA-3′ (forward) and 5′-TCA TTC TGG AGC CGC TCC AGA-3′(reverse). The PCR fragment was then purified and subcloned into various vectors (pcDNA3.1 using restriction sites HindIII-EcoRI; pCMV FLAG using restriction sites HindIII-EcoRI; pcDNA3.1-HA using restriction sites HindIII-NotI; pEGFP N1 using restriction sites BglII-HindIII; and pEGFP C1 using restriction sites BglII-KpnI). Mouse GRIN2 truncation constructs resulting in 1–100, 1–200, and 1–300 amino acids (aa) were subcloned into pcDNA3.1-HA using the restriction sites HindIII-NotI. The constructs pcDNA3 Sprouty2, pcDNA3 FLAG mouse Sprouty1, Sprouty2, Sprouty3, and Sprouty4 were described previously (9). Sprouty2 full-length and truncation constructs (1–295, 1–279, 1–250, 1–238, and 1–209 aa) were subcloned into pcDNA3.1-HA using the restriction sites HindIII-NotI, whereas Sprouty2 truncations (1–173 and 174–315 aa) were subcloned into pDsRed-monomer-N1 using the restriction sites NheI-HindIII. pEGFP N1 Sprouty2 was cloned using the BglII-HindIII restriction sites. For GST pulldown constructs, full-length (human) hGRIN2, hGRIN2 1–204, and hGRIN2 205–461 were cloned into the vector pGEX4T2 (Amersham Biosciences) using the restrictions sites SalI-NotI. Clones were verified by automated DNA sequencing.
Yeast Two-hybrid Screen
For the yeast two-hybrid study, a GRIN2 1–360-aa truncation construct was cloned into the GAL4 DNA activation domain plasmid pPC86 using SalI/NotI restriction sites and used as the bait to screen a human brain cDNA library for potential interacting proteins (3).
GST Pulldown
Bacterially expressed purified protein was bound to glutathione-Sepharose and incubated with HEK-293T cell lysates expressing pcDNA3 FLAG or pcDNA3 FLAG human Sprouty2 followed by SDS-PAGE and immunoblotting with M2-FLAG antibodies.
Co-immunoprecipitation
HEK-293T and Neuro-2A cells were transfected with the indicated constructs using Lipofectamine 2000. Cells were harvested 48 h later using ice-cold lysis buffer (1% Triton X-100, 50 mm Tris-HCl, pH 7.5, 10 mm EDTA, pH 8.0, 150 mm NaCl) supplemented with 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm sodium orthovanadate, 5 μg/ml benzamidine, 5 mm sodium fluoride, 1 mm β-glycerophosphate, and 1 mm phenylmethylsulfonyl fluoride). One mg of whole protein was precleared with protein G-agarose (Roche Applied Science) at 4 °C for 3 h. Immunoprecipitations were performed overnight with 4 μg of M2-FLAG (Sigma) monoclonal antibody at 4 °C or 4 μg of anti-HA monoclonal antibody followed by a 4-h incubation with 30 μl of protein-G agarose. Samples were washed three times with lysis buffer containing protease inhibitors and then resolved with SDS-PAGE. After transfer to nitrocellulose, the membranes were blocked with 5% nonfat dry milk and immunoblotted with M2-FLAG, anti-Gαo, or anti-HA antibodies followed by either rabbit or mouse TrueBlot (eBioscience) or routine rabbit or mouse (Pierce) HRP-conjugated secondary antibodies. Whole cell lysates were resolved on SDS-PAGE, transferred, blocked, and immunoblotted with anti-M2-FLAG, anti-Gαo, or anti-HA antibodies followed by either rabbit or mouse HRP-conjugated secondary antibodies (Pierce). Bands were visualized using the ECL detection method.
RT-PCR
Neuro-2A cells were grown on 100-mm plates to 70% confluency. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's protocol. One-step RT-PCR (Qiagen) was used to run PCRs for mouse GRIN2, mouse GRIN1, mouse Sprouty1,Sprouty2, Sprouty3, and Sprouty4 from Neuro-2A total RNA. The primers used were as follows: mSprouty1, 5′-TACGATTCTGTCCCTAGACC-3′ (forward) and 5′-TATTCCACCATGCTCTCC-3′ (reverse); mSprouty2, 5′-GATCAGATCAGAGCCATCC-3′ (forward) and 5′-ACACAGCACACACAAGTCC-3′ (reverse); mSprouty3, 5′-GCAATGATTATGTGGAACG-3′ (forward) and 5′-AGGCAGGGTAAGAAAAGG-3′ (reverse); mSprouty4, 5′-ATTGACCAGATGAAGACC-3′ (forward) and 5′-AGTGATAGAAGATACCTTGC-3′ (reverse). Each reaction contained 0.6 μm of each gene-specific primer, reaction mix, and 2 μg of total RNA. The annealing temperature was 54 °C, and the extension time was 40 s.
Real-time RT-PCR
Total RNA was isolated from cells using TRIZOL reagent (Invitrogen), and real-time RT-PCR was performed on a LightCycler (Roche Applied Science) using the QuantiTect SYBR Green RT-PCR system (Qiagen) according to the manufacturer's instructions. Mouse GRIN1 (QT00262136) and tubulin (QT00251664) primer pairs were purchased from Qiagen from the QuantiTect primer assay collection. Data were analyzed using the LightCycler data analysis software. Crossing points were calculated using the second derivative maximum method. All reactions were done in triplicate, and data for GRIN were normalized against tubulin RNA levels for each sample.
Antibodies
Antibodies specific for GRIN were made by immunizing rabbits with a keyhole limpet hemocyanin-conjugated peptide specific for GRIN2 (Sigma Genosys). The sequence for the synthesized peptide for GRIN2 was N-DPEVLGVAIQKHLEMQFE-C. Antibodies were affinity-purified on peptide-conjugated columns (Pierce).
Imaging
Neuro-2A cells were trypsinized and counted 1 day prior to transfection. 5.0 × 104 cells were plated onto 35-mm MatTek® dishes. Cells were transfected with the appropriate GFP constructs using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-four hours after transfection, living Neuro-2A cells were imaged using a Zeiss LSM 510 META microscope in an inverted configuration with a pinhole setting of 1.0 using a UV Plan-Apochromat 63 × 1.4 NA oil objective and an optical zoom 2× in one x-y focal plane with the appropriate stage adapter configured for 35-mm MatTek® dishes. Multiple (6–8) 2–5-μm Z section slices and single images were obtained for each sample and condition. Representative images are shown.
siRNA Knockdown Experiments
Small interfering RNA smart pool reagents designed for mGRIN1 (catalog number M-046253-00) and siCONTROL Non-Targeting siRNA pool (catalog number D-001206-13-05) were purchased from Dharmacon. Neuro-2A cells were trypsinized and counted 1 day prior to transfection. Then, 2.5 × 105 Neuro-2A cells were plated onto 35-mm dishes. Twenty-four hours later, cells were transfected with the appropriate siRNA oligonucleotide (final concentration 50 nm) using DharmaFECT 2 transfection reagent, according to the manufacturer's instructions. After 48 h, one set of cells was trypsinized, split, and plated onto four 35-mm plates at a density of 3.5 × 104 cells. RNA was harvested from a parallel set of transfected cells using the TRIzol method. Six to eight hours after plating, cells were serum-starved with 0.1% BSA modified Eagle's medium for 16 h. The next day, cells were stimulated with the appropriate agonist and/or growth factors for the times indicated. Protein levels of phospho-p44/42, total p44/42, and tubulin for each sample were determined by harvesting total cellular lysates followed by separation using SDS-PAGE and immunoblotting as preciously described.
RESULTS
GRIN Proteins Interact with Gαo
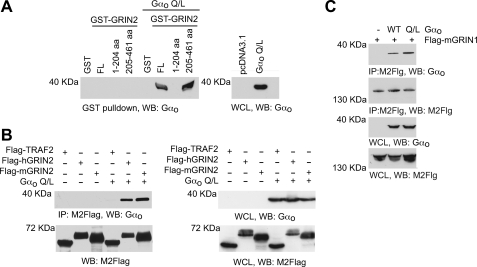
Our laboratory and others have found that GRIN isoforms can interact with Go/i (3, 5, 6). We characterized the nature of the interaction by GST pulldowns and co-immunoprecipitation assays. To identify the region on GRIN involved in the interaction with Gαo, GST fusion constructs were made with human GRIN2 (hGRIN2) full-length sequence and with truncated N-terminal region of hGRIN2 (containing 1–204 aa) and truncated C-terminal region of hGRIN2 (containing 205–461 aa). The various bacteria-expressed GST-tagged proteins were then incubated with HEK-293T cell lysates expressing an empty vector (pcDNA3.1) control or the constitutively active mutant Gαo Q205A. Only the GST-hGRIN2 full-length sequence and the GST-hGRIN2 205–461-aa truncation were able to pull down Gαo Q205A (Fig. 1A, left panel), suggesting that the C-terminal region of hGRIN2 is required for the interaction with Gαo. The interaction of hGRIN2 and Gαo was confirmed by co-immunoprecipitation of overexpressed FLAG-tagged hGRIN2 and Gαo Q205L (Fig. 1B). FLAG-TRAF2, a non-Gαo interacting protein, failed to immunoprecipitate Gαo (Fig. 1B). Whole cell lysates showed that FLAG-TRAF-2, FLAG-GRIN2, and Gαo were expressed accordingly (Fig. 1B, right panel). A GenBankTM search uncovered hGRIN mouse homologs mGRIN1 (long isoform) and mGRIN2 (short isoform). Although these two isoforms share only 48% homology at the C-terminal region, both FLAG-mGRIN1 and FLAG-mGRIN2 interact with Gαo, as shown by immunoprecipitation (Fig. 1, B and C).
FIGURE 1.
GRIN proteins interact with Gαo. A, GST pulldown of GRIN2 and Gαo. Human GRIN2 1–461 (FL), 1–204, and 205–461 were expressed as GST fusion proteins in bacteria and purified (left panel). GST fusion proteins were incubated with cell lysates from 293T cells exogenously expressing empty vector (pcDNA3.1) or the constitutively activated Gαo Q205L (Q/L) mutant and then immunoblotted with Gαo specific antibodies. Whole cell lysates (WCL) were immunoblotted for Gαo expression (right panel). Representative immunoblots (WB) from one of three experiments are shown. B, co-immunoprecipitation of GRIN2 and Gαo. HEK-293T lysates were immunoprecipitated (IP) with the M2-FLAG antibody. Immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-Gαo antibody and M2-FLAG antibody (left panel). Whole cell lysate was also resolved using SDS-PAGE and immunoblotted with anti-Gαo antibody and M2-FLAG antibody (right panel). Representative immunoblots from one of three independent experiments are shown. C, co-immunoprecipitation of GRIN1 and Gαo. HEK-293T lysate was immunoprecipitated with the M2-FLAG antibody and resolved using SDS-PAGE. Immunoprecipitates were immunoblotted with anti-Gαo antibody (panel 1) or M2-FLAG antibody (panel 2). Whole cell lysate was also resolved using SDS-PAGE and immunoblotted with the anti-Gαo antibody (panel 3) and M2-FLAG antibody (panel 4). Representative immunoblots from one of two independent experiments are shown. WT, wild type.
Sprouty2 Interacts with GRIN
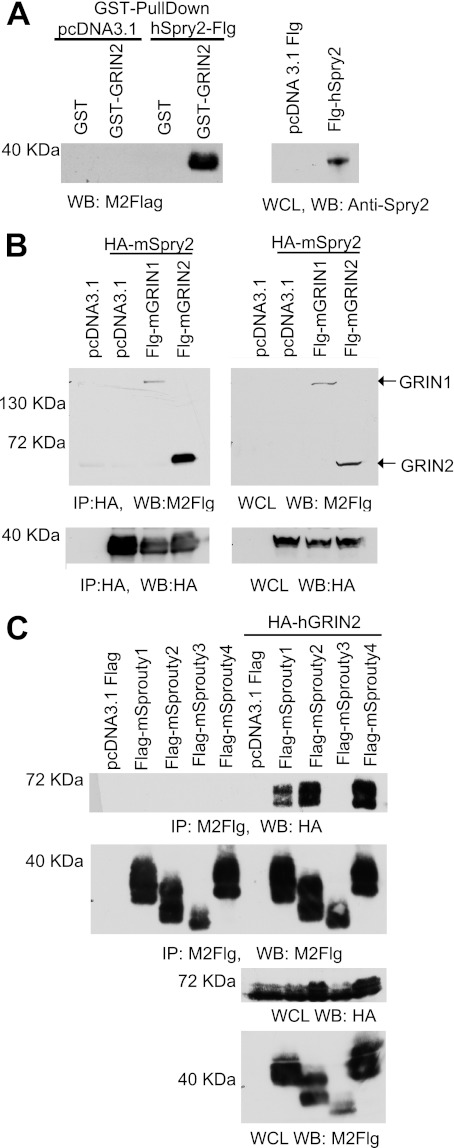
To identify potential interactors of GRIN, a yeast two-hybrid screen was performed using full-length GRIN2 as bait. This resulted in the induction of reporter genes in the absence of interacting proteins, suggesting that full-length GRIN2 contains a transcriptional activation domain (supplemental Fig. S1). To overcome this, we made truncations of GRIN2 and found that the truncation GRIN2 1–360 was the construct that lacked the transcriptional activation domain, and thus, we used it as bait in the yeast two-hybrid study. We screened a human brain cDNA library and found a number of potential GRIN2 interactors (Table 1). These interactors were confirmed both in growth (on plates lacking leucine, tryptophan, and histidine) and in β-gal assay (β-galactosidase activity). The first protein identified was E2F4, a known transcription repressor that has been shown to function in growth suppression and differentiation (10). E2F4 represses genes during quiescence, heterodimerizes with p130 after cell cycle exit, and thereby induces differentiation in developing neurons (11). A second interactor candidate identified was neural epidermal growth factor-like like 2 (Nell2). Nell2 is a neuron-specific thrombospondin-1-like extracellular protein containing six epidermal growth factor-like domains that is highly expressed in the hippocampus (12, 13). Nell2 was found to act within the CNS and peripheral nervous system progenitors to promote neuronal differentiation. Nell2−/− mice have defects in spatial learning (14, 15). The Na+/H+ exchanger (NHEI) was also identified to directly interact with hGRIN2. NHEI has been shown to play a major role in intracellular pH homeostasis and cell volume regulation. Stimulation of NHEI is required for cell proliferation, and its activity can be modulated by pH, tumor promoters, growth factors, and cell surface receptor signaling (16–19). Sprouty2, another direct interacting protein, belongs to a family of four mammalian proteins with a conserved C-terminal cysteine-rich domain that has been shown to regulate tyrosine kinase receptor signaling (20, 21). hSprouty2 and hGRIN2 direct interaction was confirmed by GST pulldown (Fig. 2A). Co-transfection of HA-mSprouty2 with mGRIN1 or mGRIN2 in HEK-293T cells followed by immunoprecipitation showed that both GRIN isoforms interact with Sprouty2 (Fig. 2B). Of note, Sprouty2 expression appears as a doublet in whole cell lysate gels, and Sprouty2 expression has been shown by other groups to exist as a doublet and sometimes triplet due to serine phosphorylation (22–24). Given their sequence homology, other members of the Sprouty family can potentially interact with GRIN2 proteins. We overexpressed FLAG-tagged mouse Sprouty1, Sprouty2, Sprouty3, and Sprouty4 in conjunction with control vector (pcDNA3.1) or HA-GRIN2 in 293T cells. After immunoprecipitating Sprouty proteins and immunoblotting for GRIN2, we found that Sprouty1, Sprouty2, and Sprouty4 were able to interact with GRIN2 whereas Sprouty3 could not (Fig. 2C). Whole cell lysates showed that Sprouty proteins and GRIN2 proteins were expressed in 293T cells (Fig. 2C, lower panel).
TABLE 1.
Results of hGRIN2 (1–360-aa) two-hybrid screen
FIGURE 2.
GRIN proteins associate with Sprouty. A, GST pulldown of Sprouty2 and GRIN2. Human GRIN2 1–461 full-length and GST alone were expressed as GST fusion proteins in bacteria and purified. GST fusion proteins were bound to glutathione beads and incubated with cell lysates from HEK-293T cells exogenously expressing empty vector (pcDNA3.1 FLAG (pcDNA3.1 Flg)) or FLAG-hSprouty2 (Flg-hSprouty2). GST-hGRIN2-associated proteins (left panel) were immunoblotted with M2-FLAG antibodies, whereas HEK-293T whole cell lysates (WCL, right panel) were immunoblotted with anti-Sprouty2 antibodies. Representative immunoblots (WB) from one of three experiments are shown. B, co-immunoprecipitation (IP) of GRIN1 and GRIN2 with Sprouty2. HEK-293T lysate was immunoprecipitated with anti-HA (12C25) antibody and resolved using SDS-PAGE. Immunoprecipitates were immunoblotted with M2-FLAG (top left panel) and an anti-HA antibody (bottom left panel). Whole cell lysate was resolved using SDS-PAGE, immunoblotted with the M2-FLAG antibody (top right panel), and further blotted with the anti-HA antibody (bottom right panel). Representative immunoblots from one of three experiments are shown. Flg, FLAG. C, co-immunoprecipitation of GRIN2 with Sprouty proteins. HEK-293T cells were transfected with the indicated expression constructs, and total cell lysates were immunoprecipitated with the M2-FLAG antibody and resolved using SDS-PAGE. M2-FLAG immunoprecipitates were immunoblotted with an anti-HA antibody (panel 1) and with M2-FLAG antibody (panel 2). Whole cell lysates were immunoblotted with the anti-HA antibody and M2-FLAG antibody. Representative immunoblots from one of two experiments are shown.
The Conserved C-terminal Domain of Sprouty2 Is Important for Interaction with GRIN2
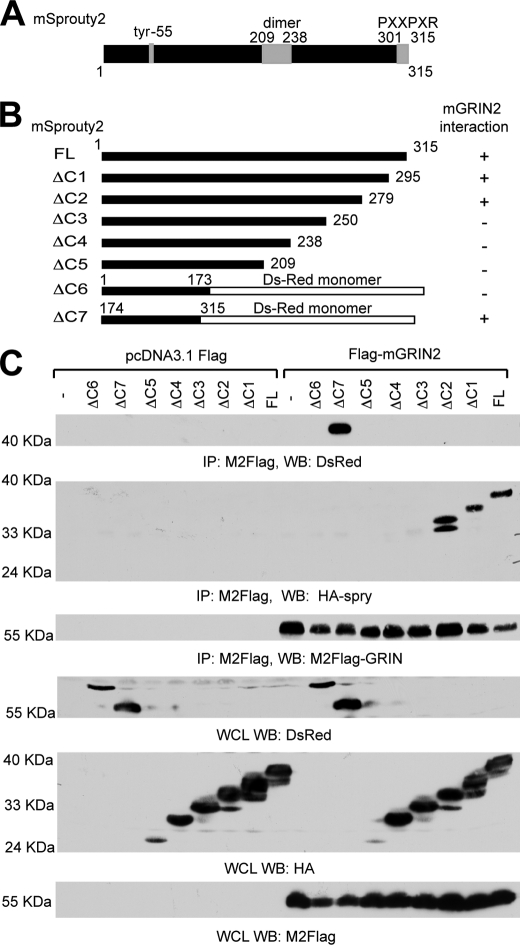
To further characterize the Sprouty2-GRIN interaction, we sought to determine the regions of Sprouty2 responsible for GRIN2 binding. The ability of Sprouty to inhibit the MAPK signaling pathway is dependent on its C terminus (region 295–315 aa) (25). The C terminus also contains a dimerization domain (209–238 aa, Fig. 3A). Based on this, we made several C-terminal truncations of mSprouty2 tagged with hemagglutinin (HA) or DsRed monomer as follows: HA-Sprouty2 1–315 full-length, HA-Sprouty2 1–295 (Δ1), HA-Sprouty2 1–279 (Δ2), HA-Sprouty2 1–250 (Δ3), HA-Sprouty2 1–239 (Δ4), HA-Sprouty2 1–209 (Δ5), DsRed monomer N1-Sprouty2 1–73 (Δ6), and DsRed monomer N1 Sprouty2 174–315 (Δ7) (Fig. 3B). We co-expressed these truncations with FLAG-mGRIN2 in HEK-293T cells and immunoprecipitated with M2-FLAG antibody. The only mSprouty2 truncations detected after mGRIN2 immunoprecipitation were those containing region 250–279 (Fig. 3C). This suggests that the region of Sprouty2 that interacts with GRIN2 lies in the C-terminal conserved cysteine region and resides between amino acids 250–279.
FIGURE 3.
The conserved C-terminal domain of Sprouty2 is essential for GRIN interaction. A, schematic diagram of the Sprouty2. B, constructs used in these studies and summary of results. ΔC1 to ΔC5 mSprouty2 truncations were tagged with an HA epitope. Mouse Sprouty2 truncations ΔC6 and ΔC7 were made into fusion proteins with the pDsRed. C, Sprouty2 regions essential for GRIN2 binding. HEK-293T cells were co-transfected with the indicated plasmids (ΔC1-ΔC7 and FLAG-mGRIN2) and immunoprecipitated (IP) with the M2-FLAG antibody. Immunoprecipitates were immunoblotted (WB) with anti-DsRed (panel 1), anti-HA (panel 2), and M2-FLAG antibodies (panel 3), respectively. Whole cell lysates (WCL) were also immunoblotted with anti-DsRed antibody (panel 4), anti-HA antibody (panel 5), and M2-FLAG antibody (panel 6). Representative immunoblots from one of three independent experiments are shown.
Sprouty2 and Gαo Compete for GRIN2 Binding
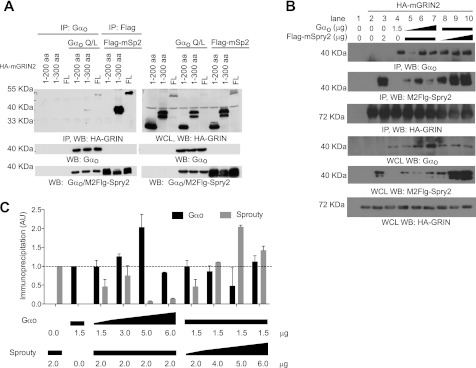
We next set out to map the regions of GRIN2 necessary for Sprouty2 and Gαo binding. We made truncated HA-tagged mGRIN2 variants: HA-mGRIN2 and HA-mGRIN2 1–300 (Fig. 4A). A full-length tagged construct was also made (1–455 aa). The truncations and the full-length construct were co-expressed with either Q205L-Gαo or FLAG-mSprouty2 in HEK-293T cells followed by immunoprecipitation with anti-HA antibody. The immunoprecipitate was tested for the presence of Gαo or FLAG-mSprouty2. Only full-length HA-mGRIN2 and HA-mGRIN2 1–300 were able to co-immunoprecipitate Gαo or FLAG-mSprouty2. This suggests that the 200–300-aa region of mGRIN2 is required for Gαo and Sprouty2 binding.
FIGURE 4.
Sprouty2 and Gαo compete for GRIN2 binding. A, the region of 200–300 aa of GRIN is essential for Sprouty2 and Gαo binding. Left panel, HEK-293T cells were co-transfected with the indicated expression constructs, and total cell lysates were prepared as described previously. Lysates were immunoprecipitated (IP) with the anti-Gαo antibody or with the M2-FLAG antibody and immunoblotted (WB) with the anti-HA antibody (panel 1), with the anti-Gαo antibody (panel 2), and with the M2-FLAG antibody (panel 3). Panel 3 contains bands for Gαo (single band) and FLAG-Sprouty2 (doublet bands). Right panel, whole cell lysates (WCL) were immunoblotted with the anti-HA antibody (panel 1), with the anti-Gαo antibody (panel 2), and with the M2-FLAG antibody (panel 3). Panel 3 contains bands for Gαo (single band) and FLAG-Sprouty2 (doublet bands). Representative immunoblots from one of two independent experiments are shown. mSp2, mouse Sprouty2; mG2, mouse GRIN2. B, Sprouty2 and Gαo compete for GRIN2 binding. HEK-293T cells were co-transfected with the indicated plasmids: Gαo Q205L (Q/L) mutant (1.5, 3.0, or 6.0 μg), FLAG-mSprouty2 (2.0, 4.0, or 6.0 μg), and HA-mGRIN2 (4 μg). Vector pcDNA3.1 was co-transfected to make the total amount of DNA transfected equal. Lysates were prepared as described previously and immunoprecipitated with anti-HA antibody. Immunoprecipitates were immunoblotted with anti-Gαo (panel 1), M2-FLAG (panel 2), or anti-HA (panel 3) specific antibodies. The triangle indicates increasing concentration of DNA transfected, and the rectangle indicates the same concentration of DNA transfected. A representative blot from three independent experiments is shown. Lower panel, whole cell lysates were immunoblotted with anti-Gαo (panel 1), M2-FLAG (panel 2), or anti-HA (panel 3) specific antibodies. Representative immunoblots from one of three independent experiments are shown. mSpry2; mouse Sprouty2. C, corresponding densitometry for the average of 3–6 independent experiments testing the following DAN concentrations: Gαo Q205L (Q/L) mutant (1.5, 3.0, 5.0, or 6.0 μg), FLAG-mSprouty2 (2.0, 4.0, 5.0 or 6.0 μg), and HA-mGRIN2 (4 μg). AU, arbitrary units. Error bars are S.E.
Because the same region on mGRIN2 is implicated in binding to Gαo and Sprouty2, it is possible that these two proteins compete for GRIN2 binding. To determine whether this was the case, we set up competitive co-immunoprecipitation experiments. The amount of Gαo cDNA transfected in Neuro-2A cells was varied (from 1.5 to 6.0 μg), whereas the amount of FLAG-Sprouty2 (2.0 μg) and HA-mGRIN2 cDNA (4.0 μg) was kept constant. GRIN2 was immunoprecipitated and tested for the presence of Gαo and FLAG-mSprouty2. Increasing Gαo expression led to a decrease in the amount of FLAG-Sprouty2 immunoprecipitated (Fig. 4, B and C). In parallel, we varied the amount of FLAG-mSprouty2 cDNA transfected (from 2 to 6 μg), whereas the amount of Gαo (1.5 μg) and HA-mGRIN2 (4.0 μg) was kept constant. Similarly to Gαo, as the expression of FLAG-mSprouty2 increased, the amount of Gαo co-immunoprecipitated decreased (Fig. 4, B and C), suggesting competition for GRIN binding.
Tyrosine Phosphorylation Modulates Sprouty2-GRIN2 Interaction
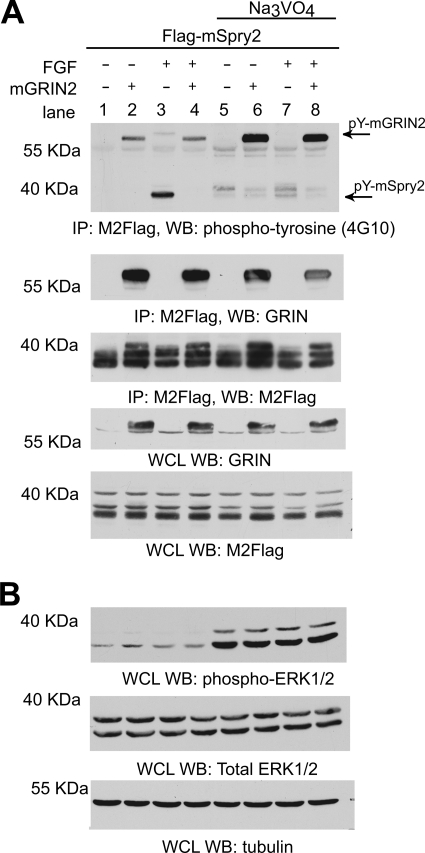
Sprouty2 has been shown to be tyrosine-phosphorylated, and this phosphorylation is important for its inhibitory activity of the MAPK pathway (23, 25–27). This tyrosine phosphorylation, at Tyr-53, is induced by bFGF signaling (25). We wanted to determine whether the interaction with GRIN2 affected the tyrosine phosphorylation state of Sprouty2. To do so, we co-transfected Neuro-2A cells with FLAG-mSprouty2 with vector or mGRIN2. Transfected cells were serum-starved and then stimulated for 30 min with bFGF in the presence or absence of a 1-h pretreatment with sodium orthovanadate to inhibit protein phosphotyrosyl-phosphatases. FLAG-tagged mSprouty2 was immunoprecipitated with mGRIN2 (Fig. 5A). Immunoblotting was then performed using anti-phospho-Tyr antibody (4G10) followed by anti-GRIN and M2-FLAG antibodies. mSprouty is tyrosine-phosphorylated upon bFGF treatment (lanes 1 and 3). mGRIN2 is also tyrosine-phosphorylated in the presence or absence of bFGF. Pretreatment with sodium orthovanadate enhanced the tyrosine phosphorylation of mGRIN2 (Fig. 5A, row 1, compare lanes 2 and 6 with lanes 4 and 8). Tyrosine-phosphorylated mGRIN2 co-immunoprecipitated with FLAG-mSprouty2 irrespective of bFGF stimulation. On the other hand, when mGRIN2 was co-expressed with mSprouty2, the amount of tyrosine-phosphorylated mSprouty2 co-immunoprecipitated decreased (lanes 3 and 4), suggesting that mGRIN2 co-expression prevents the tyrosine phosphorylation of mSprouty2. Pretreatment with sodium orthovanadate shifts up the motility of mSprouty2 band upward, suggesting further modifications (lanes 3 and 7). However, the amount of tyrosine-phosphorylated mSprouty2 co-immunoprecipitated decreased in the presence of mGRIN2 even when the cells were treated with sodium orthovanadate (lanes 7 and 8). Whole cells lysates show that FLAG-mSprouty2 and GRIN2 proteins were expressed similarly (Fig. 5A, lower panels). Cells treated with bFGF and/or sodium orthovanadate have higher phosphoERK1/2 levels, indicating that the stimulation and treatment worked as expected (Fig. 5B). The fibroblast growth factor (FGF) stimulation of MAPK is damped due to overexpression of Sprouty.
FIGURE 5.
Expression of mGRIN2 inhibits tyrosine phosphorylation of mSprouty2. A, co-immunoprecipitation of tyrosine-phosphorylated (pY) GRIN and Sprouty. Neuro-2A cells were transfected with the indicated expression constructs. Cells were serum-starved overnight. One set of cells was treated with sodium orthovanadate (1 μm) for 1 h prior to stimulation with 40 ng/ml FGF for 30 min as indicated. Total cell lysates were immunoprecipitated (IP) with M2-FLAG antibody and immunoblotted (WB) with anti-phospho-tyrosine (G410) (top panel); anti-GRIN antibody (middle panel); and M2-FLAG antibody (bottom panel). In the whole cell lysate (WCL) panels, whole cell lysate was immunoblotted with anti-GRIN antibody (panel 1) and M2-FLAG antibody (panel 2). Of note, the M2-FLAG antibody detected some nonspecific bands (panel 2), and the third band is in fact Sprouty2. mSpry2, mouse Sprouty2. Na3VO4, sodium orthovanadate. B, whole cell lysates from A were immunoblotted with anti-phospho-p44/42 ERK1/2 antibody (panel 1), anti-total p44/42 ERK1/2 antibody (panel 2), and anti-tubulin antibody (panel 3). Representative immunoblots from one of three independent experiments are shown.
GRIN2-GFP Fusion Proteins Localize to Plasma Membrane in Gαo-expressing Cells but Not Sprouty2-expressing Cells
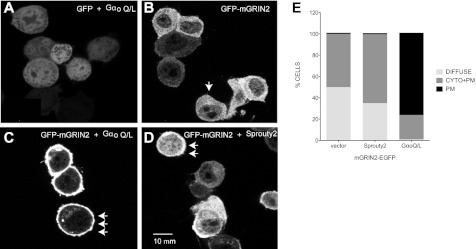
To determine whether GRIN2 interaction with Gαo or Sprouty2 had any effect on GRIN2 localization, we made GRIN2-GFP fusion proteins. GRIN2 was placed in the N terminus of GFP, and co-immunoprecipitation experiments showed that this fusion protein retains the ability to interact with Gαo and Sprouty2 (data not shown). We determined the localization of the GRIN2-GFP construct when co-expressed with Gαo-Q205L, Sprouty2, or empty vector expression in Neuro-2A cells. When overexpressed along with empty vector, GRIN2-GFP localized to the cytoplasm (Fig. 6, B and E, DIFFUSE). However, upon expression of Gαo Q205L, GRIN2-GFP primarily localized to the plasma membrane (Fig. 6, C and E, PM). Gαo Q205L expression had no effect on enhanced green fluorescent protein (EGFP) localization (Fig. 6A). This plasma membrane enrichment was less pronounced with Sprouty2 co-expression (Fig. 6, D and E, CYTO+PM). Sprouty2 localization was not affected by expression of Gαo Q205L (supplemental Fig. S2).
FIGURE 6.
Gαo expression induces translocation of GRIN2 to plasma membrane. A–D, Neuro-2A cells were transiently transfected with expression plasmids for pEGFP and Gαo Q205L (Q/L) mutant (A); GFP-mGRIN2 (B); GFP-mGRIN2 and Gαo Q205L (C); and GFP-mGRIN2 and Sprouty2 (D). After 24 h, live cells were imaged in a laser scanning microscope 510-META. Scale bar, 10 μm. Representative images from one of four experiments are shown. E, subcellular distribution of GRIN2-GFP coexpressed with vector (n = 137), Gαo Q205L (n = 203), or Sprouty2 (n = 53) was scored in Neuro-2A cells as diffuse, cytoplasmic and plasma membrane-bound (CYTO+PM) or mostly plasma membrane-bound (PM). GRIN2 distribution was considered diffuse as shown in the cell marked by a single arrow in panel B; GRIN2 distribution was considered cytoplasmic and plasma membrane-bound as shown in the cells marked by the double arrow in panel D; and GRIN2 distribution was considered mostly plasma membrane-bound as shown by the triple arrow in panel C.
Active Gαo Signaling Attenuates FGF Activation of MAPK
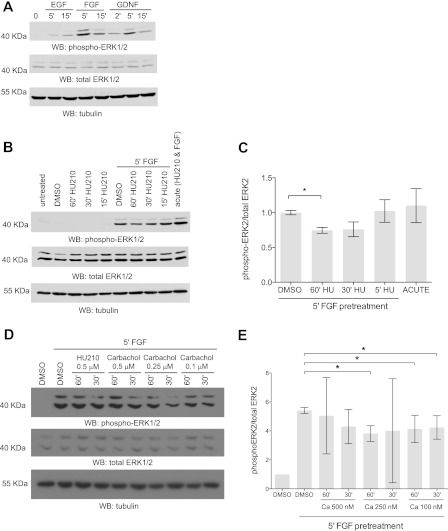
The previous experiments were performed using exogenous expression systems. We sought to find a cell line expressing all three proteins: GRIN, Sprouty2, and Gαo. Neuro-2A cells endogenously express GRIN1 and Gαo (5, 6). Sprouty2 is also endogenously present in Neuro-2A cells (data not shown). Sprouty2 has been shown to be regulate growth factor activation of ERK1/2 (25). The growth factors EGF, FGF, and glial-derived neurotrophic factor activate MAPK in Neuro-2A cells (Fig. 7A). Because the GRIN-Sprouty2 interaction can be outcompeted by Gαo signaling, we determined the functional significance of this regulation in the FGF-dependent activation of MAPK. We used HU-210, a CB1 receptor agonist, to examine the effects of Gαo signaling on FGF stimulation of MAPK pathway in Neuro-2A cells. Serum-starved Neuro-2A cells were pretreated with HU-210 (500 nm) for various time periods (15, 30, and 60 min) followed by FGF stimulation for 5 min. HU-210 pretreatment for 30 or 60 min was able to decrease FGF activation of MAPK (Fig. 7, B and C), whereas a 15-min pretreatment with HU-210 had no effect. Simultaneous co-treatment with HU-210 and FGF enhanced MAPK activation (Fig. 7B, acute). This is expected as Gβγ subunits from Go/i can activate MAPK (28–32). We sought to test whether FGF activation of MAPK could be repressed by other Go/i-coupled receptors. Pretreatment by carbachol, a muscarinic agonist, also decreased FGF activation of MAPK (Fig. 7, D and E), suggesting that this is a general mechanism not limited to CB1R signaling.
FIGURE 7.
Active Gαo signaling attenuates FGF signaling. A, Neuro-2A cells express functional FGF receptors. Neuro-2A cells were serum-starved overnight and then treated with 50 ng/ml EGF, 20 ng/ml FGF, or 20 ng/ml glial-derived neurotrophic factor (GDNF) for various periods of time (0, 2, 5, or 15 min). Western blots (WB) were performed with monoclonal anti-dual phospho-p44/42 MAPK (top panel), polyclonal anti-dual total p44/42 MAPK (middle panel), and anti-tubulin antibodies (bottom panel). Representative immunoblots from one of three experiments are shown. B, CB1R prestimulation decreases FGF signaling. Neuro-2A cells were serum-starved overnight and pretreated with 500 nm HU-210 at different time points (15, 30, and 60 min) followed by stimulation with FGF for 5 min. Total cell lysate was resolved by immunoblotted with monoclonal anti-dual phospho-p44/42 ERK1/2 (top panel), polyclonal anti-dual total p44/42 ERK1/2 (middle panel), and anti-tubulin antibody (bottom panel). Representative immunoblots from one of three independent experiments are shown. C, corresponding densitometric data for the average of three independent experiments. Error bars indicate S.E. D, muscarinic receptor prestimulation decreases FGF signaling. Neuro-2A cells were serum-starved overnight and pretreated with different concentrations of carbachol (500, 250, or 100 nm) at different time points (30 and 60 min) followed by stimulation with FGF for 5 min. Total cell lysate was immunoblotted with monoclonal anti-dual phospho-p44/42 ERK1/2 (top panel), polyclonal anti-dual total p44/42 ERK1/2 (middle panel), and anti-β tubulin antibody (bottom panel). Representative immunoblots from one of three independent experiments are shown. E, corresponding densitometric data for the average of three independent carbachol experiments. *, p < 0.05. Error bars indicate S.E.
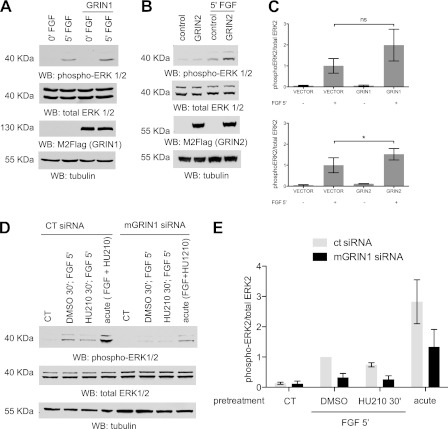
GRIN Potentiates FGF Activation of MAPK
Because Sprouty2 negatively regulates activation of MAPK, we determined whether exogenous mGRIN expression could modulate FGF-mediated MAPK activation. Neuro-2A cells were transfected with vector or FLAG-mGRIN1 or GRIN2. Cells were then serum-starved for 16 h and stimulated with 20 ng/ml FGF for 5 min. Expression of either mGRIN1 or mGRIN2 potentiated FGF activation of MAPK (Fig. 8, A–C). To implicate GRIN in the HU-210-dependent attenuation of MAPK activation by FGF, we abolished endogenous mGRIN1 expression using small interfering RNAs. We hypothesized that by decreasing GRIN1 levels, we would attenuate FGF activation of MAPK even further because without GRIN expression, sequestration and inhibition of Sprouty2 would decrease, and thus, Sprouty2 inhibition of the MAPK pathway would be enhanced. Neuro-2A cells were transfected with negative control pool siRNA or mGRIN1 pool siRNA. GRIN1 mRNA was decreased by 80% after 48 h of transfection with the GRIN1 pool siRNA (supplemental Fig. S3). siRNA transfected cells were serum-starved and then pretreated with DMSO or HU-210 (500 nm) for 30 min followed by FGF stimulation for 5 min. In cells exposed to mGRIN1 siRNA, pretreatment with HU-210 produced no difference from DMSO control after 5 min of FGF stimulation (Fig. 8, D and E). In addition, the levels of phospho-p44/42 for the GRIN1 siRNA pool decreased when compared with the negative control pool siRNA. This suggests that decreasing mGRIN1 expression attenuates FGF activation of ERK1,2.
FIGURE 8.
GRIN expression levels modulate MAPK activation by FGF. A, overexpression of GRIN2 enhances FGF stimulation of MAPK. Neuro-2A cells were transfected with either pcDNA3.1 (vector control) or pcDNA3.1 mGRIN2. Eight hours after transfection, cells were serum-starved overnight. They were then treated with FGF (20 ng/ml) for 5 min as indicated. Total cell lysate was immunoblotted (WB) with monoclonal anti-dual phospho-p44/42 ERK1/2 (top panel), polyclonal anti-dual total p44/42 ERK1/2 (second panel), anti-GRIN (third panel) antibody, and anti-tubulin antibody (bottom panel). Representative immunoblots from one of three experiments are shown. B, overexpression of GRIN1 enhances FGF stimulation of MAPK. Neuro-2A cells were transfected with either pcDNA3.1 FLAG (vector control) or pcDNA3.1 FLAG-mGRIN1. Eight hours after transfection, cells were serum-starved with 0.5% FBS modified Eagle's medium for 16 h before stimulation. They were then treated with FGF (40 ng/ml) for 5 min as indicated. Cells were harvested, and 60 μg of total cell lysate was resolved by SDS-PAGE and immunoblotted with monoclonal anti-dual phospho-p44/42 ERK1/2 (top panel), polyclonal anti-dual total p44/42 ERK1/2 (second panel), M2-FLAG (third panel) antibody, and anti-tubulin antibody (bottom panel). Representative immunoblots from one of three experiments are shown. C, densitometric data for the average of three independent experiments of panel B. Error bars indicate S.E. ns, not significant. D, GRIN1 knockdown attenuates FGF stimulation of MAPK. Negative pool siRNA and GRIN1-specific siRNA (Dharmacon) were transfected into Neuro-2A cells. Cells were serum-starved overnight and pretreated with DMSO or HU-210 (500 nm) for 30 min followed by stimulation with FGF (20 ng/ml) for 5 min as indicated. For the acute stimulation, HU-210 (500 nm) and FGF (20 ng/ml) were added simultaneously for 5 min. Whole cell lysate was immunoblotted with monoclonal anti-dual phospho-p44/42 ERK1/2 (top panel), polyclonal anti-dual total p44/42 ERK1/2 (middle panel), and anti-tubulin antibody (bottom panel). Representative immunoblots from one of three independent experiments are shown. CT, control. E, the corresponding densitometric data for the average of three experimental trials are shown. *, p < 0.05. Error bars indicate S.E.
DISCUSSION
Our laboratory and others have found GRIN to be a direct interactor of Gαo (3, 5, 33). GRIN preferentially interacts with activated Gαo over the wild-type form (Fig. 1C), suggesting that GRIN proteins may be direct downstream effectors for Gαo signaling. GRIN family proteins contain conserved motifs that are important for Gαo binding and for Sprouty2 binding. A Gαo binding region lies within the semiconserved 200–300-amino acid region of GRIN2 where Sprouty2 binding occurs as well. Within this region, there are 15 conserved residues between GRIN1 and GRIN2, suggesting that these residues are potential contact sites for Sprouty2 and Gαo binding. Mutational analysis and perhaps further truncations within the GRIN2 200–300-aa region can further define the area of interaction on GRIN2 that binds Sprouty2 and Gαo. Similar to GRIN2, GRIN1 has two established C-terminal binding regions (821–851 and 902–932 aa) for Gαo (6). Gαo expression induces the translocation of GRIN2 from the cytoplasm to the plasma membrane, and the C-terminal region of GRIN2 is critical for this localization (data not shown). Similarly, GRIN1 has been shown to be membrane-bound, and the C-terminal region of GRIN1 (region 772–827) is required for its localization to the plasma membrane (6). In fact, two conserved C-terminal cysteine residues, found in GRIN proteins, are required for GRIN1 membrane localization. These two cysteine residues (923–924 aa) are putative palmitoylation sites (6) and appear to be conserved in GRIN2. Thus, it is possible that Gαo may induce the membrane recruitment of GRIN2 by inducing the addition of palmitoyl moieties. Palmitoylation, a reversible modification, has been shown to affect protein activity as well as subcellular localization by changing the proximity of the protein to its activators and/or targets (34–37).
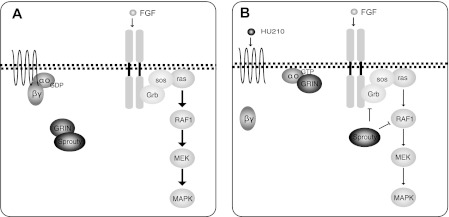
The mechanism by which GRIN connects the Go signaling pathway to the receptor tyrosine kinase-activated MAPK depends on the ability of GRIN to bind Sprouty. Based on our results, we propose a modulatory role for GRIN (Fig. 9). In the absence of Go activity, GRIN binds and sequesters Sprouty (Fig. 9A). GRIN-bound Sprouty is unable to inhibit the MAPK cascade. Upon activation of Go, by activation of the CB1R, the activated Gαo subunit interacts with GRIN, thus releasing Sprouty, which can then be activated. Sprouty2 has been shown to inhibit FGF activation of MAPK through a number of loci on the FGF pathway. Sprouty contains a PXXPXR motif that competes with Sos for binding to the SH3 domain of Grb2 (24). In addition, phosphorylation of the conserved tyrosines Tyr-55 and Tyr-221 has been shown to be essential for the inhibitory activity of Sprouty (23, 25, 39). GRIN binds Sprouty within the region of 250–279 aa. By binding to that region, GRIN may inhibit Sprouty2 binding of Grb2. GRIN2 expression also appears to decrease the tyrosine phosphorylation of Sprouty2 after FGF stimulation. In this model, Sprouty upon dissociation from GRIN and activation by tyrosine phosphorylation is able to inhibit the FGF-ERK1,2 pathway (Fig. 9B). Thus, GRIN functions as a mediator of Go-coupled receptor function to modulate the inhibitory effects of Sprouty2 on growth factor stimulation of MAPK activation. This model is supported by our observation that exogenous GRIN expression in Neuro-2A cells that have endogenous cannabinoid receptors and FGF receptor results in potentiation of MAPK activation by FGF stimulation (Fig. 9), whereas knocking down native GRIN proteins using siRNA in the same cell line attenuates FGF activation of MAPK (Fig. 9).
FIGURE 9.
Proposed mechanism of Go-GRIN mediated modulation of Sprouty2 inhibitory effects on MAPK activation. A, in the absence of Hu210, Gαo is inactive, and GRIN sequesters Sprouty and prevents it from inhibiting FGF-induced MAPK activation. B, upon stimulation of Go/i-coupled receptors, Gαo is activated and binds to GRIN. Sprouty is released and is able to attenuate FGF activated MAPK. Sprouty2 can potentially inhibit at the level of Grb2 and/or at c-Raf.
In examining the potential role of GRIN in neuronal processes, GRIN proteins, such as Gαo, have been implicated in neurite outgrowth (5, 40). Overexpression of Gαo and GRIN1 in Neuro-2A cells resulted in neurite extension, and both proteins are expressed at the growth cone of neuronal cells (5). GRIN2, a related family member, is also expressed in the brain, particularly enriched in the cerebellum. Its expression in the neurites and role in neurite outgrowth is still under investigation. In addition, Sprouty2 has also been implicated in regulating neurite extension and neuronal development (7, 8, 25, 38, 41). Expression of a nonphosphorylated Sprouty2 mutant promoted FGF-induced outgrowth of neurites in PC12 cells, thus suggesting that Sprouty2 negatively regulates neurite outgrowth (9, 25). GRIN2 has been shown to inhibit tyrosine phosphorylation of Sprouty2 and thus may be a potential mechanism to negatively regulate Sprouty2 and promote neurite outgrowth as well. GRIN1 can also interact with Sprouty2 and perhaps, like GRIN2, may modulate the inhibitory activity of Sprouty2.
Our laboratory has shown that activation of the Go/i-coupled cannabinoid CB1 receptors in Neuro-2A promotes the proteasomal degradation of Rap1GAPII, which subsequently activates Rap1 to induce neurite outgrowth. Whether Gαo signaling through the inhibition of the FGF pathway by GRIN is another mechanism that promotes neurite outgrowth in Neuro-2A cells will be addressed in future studies.
Supplementary Material
Acknowledgment
The cDNA clone KIAA0514 was generously supplied by Dr. T. Nagase (Kazusa DNA Research Institute).
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM54508 (to R. I.) and 5R01DK087650 (to R. I. and S. R. N.) and RO1 CA59998 (to J. D. L.). This work was also supported by the Systems Biology Center of New York (Grant PSDGM071558).

This article contains supplemental Figs. S1–S3.
- GAP
- GTPase-activating protein
- GRIN
- G protein-regulated inducer of neurite outgrowth
- bFGF
- basic fibroblast growth factor
- NHEI
- Na+/H+ exchanger
- DMSO
- dimethyl sulfoxide
- aa
- amino acids
- m
- mouse
- h
- human
- CB1R
- cannabinoid receptor 1.
REFERENCES
- 1. Strittmatter S. M., Valenzuela D., Kennedy T. E., Neer E. J., Fishman M. C. (1990) G0 is a major growth cone protein subject to regulation by GAP-43. Nature 344, 836–841 [DOI] [PubMed] [Google Scholar]
- 2. Neer E. J., Lok J. M., Wolf L. G. (1984) Purification and properties of the inhibitory guanine nucleotide regulatory unit of brain adenylate cyclase. J. Biol. Chem. 259, 14222–14229 [PubMed] [Google Scholar]
- 3. Jordan J. D., Carey K. D., Stork P. J., Iyengar R. (1999) Modulation of rap activity by direct interaction of Gαo with Rap1 GTPase-activating protein. J. Biol. Chem. 274, 21507–21510 [DOI] [PubMed] [Google Scholar]
- 4. Luo Y., Denker B. M. (1999) Interaction of heterotrimeric G protein Gαo with Purkinje cell protein-2: evidence for a novel nucleotide exchange factor. J. Biol. Chem. 274, 10685–10688 [DOI] [PubMed] [Google Scholar]
- 5. Chen L. T., Gilman A. G., Kozasa T. (1999) A candidate target for G protein action in brain. J. Biol. Chem. 274, 26931–26938 [DOI] [PubMed] [Google Scholar]
- 6. Nakata H., Kozasa T. (2005) Functional characterization of Gαo signaling through G protein-regulated inducer of neurite outgrowth 1. Mol. Pharmacol. 67, 695–702 [DOI] [PubMed] [Google Scholar]
- 7. Gross I., Armant O., Benosman S., de Aguilar J. L., Freund J. N., Kedinger M., Licht J. D., Gaiddon C., Loeffler J. P. (2007) Sprouty2 inhibits BDNF-induced signaling and modulates neuronal differentiation and survival. Cell Death Differ. 14, 1802–1812 [DOI] [PubMed] [Google Scholar]
- 8. Ishida M., Ichihara M., Mii S., Jijiwa M., Asai N., Enomoto A., Kato T., Majima A., Ping J., Murakumo Y., Takahashi M. (2007) Sprouty2 regulates growth and differentiation of human neuroblastoma cells through RET tyrosine kinase. Cancer Sci. 98, 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gross I., Bassit B., Benezra M., Licht J. D. (2001) Mammalian Sprouty proteins inhibit cell growth and differentiation by preventing Ras activation. J. Biol. Chem. 276, 46460–46468 [DOI] [PubMed] [Google Scholar]
- 10. Crosby M. E., Almasan A. (2004) Opposing roles of E2Fs in cell proliferation and death. Cancer Biol. Ther. 3, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Persengiev S. P., Kondova I. I., Kilpatrick D. L. (1999) E2F4 actively promotes the initiation and maintenance of nerve growth factor-induced cell differentiation. Mol. Cell. Biol. 19, 6048–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuroda S., Oyasu M., Kawakami M., Kanayama N., Tanizawa K., Saito N., Abe T., Matsuhashi S., Ting K. (1999) Biochemical characterization and expression analysis of neural thrombospondin-1-like proteins NELL1 and NELL2. Biochem. Biophys. Res. Commun. 265, 79–86 [DOI] [PubMed] [Google Scholar]
- 13. Watanabe T. K., Katagiri T., Suzuki M., Shimizu F., Fujiwara T., Kanemoto N., Nakamura Y., Hirai Y., Maekawa H., Takahashi E. (1996) Cloning and characterization of two novel human cDNAs (NELL1 and NELL2) encoding proteins with six EGF-like repeats. Genomics 38, 273–276 [DOI] [PubMed] [Google Scholar]
- 14. Nelson B. R., Claes K., Todd V., Chaverra M., Lefcort F. (2004) NELL2 promotes motor and sensory neuron differentiation and stimulates mitogenesis in DRG in vivo. Dev. Biol. 270, 322–335 [DOI] [PubMed] [Google Scholar]
- 15. Matsuyama S., Doe N., Kurihara N., Tanizawa K., Kuroda S., Iso H., Horie M. (2005) Spatial learning of mice lacking a neuron-specific epidermal growth factor family protein, NELL2. J. Pharmacol. Sci. 98, 239–243 [DOI] [PubMed] [Google Scholar]
- 16. Putney L. K., Denker S. P., Barber D. L. (2002) The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42, 527–552 [DOI] [PubMed] [Google Scholar]
- 17. Slepkov E., Fliegel L. (2002) Structure and function of the NHE1 isoform of the Na+/H+ exchanger. Biochem. Cell Biol. 80, 499–508 [DOI] [PubMed] [Google Scholar]
- 18. Baumgartner M., Patel H., Barber D. L. (2004) Na+/H+ exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am. J. Physiol. Cell Physiol. 287, C844–C850 [DOI] [PubMed] [Google Scholar]
- 19. Cardone R. A., Casavola V., Reshkin S. J. (2005) The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer 5, 786–795 [DOI] [PubMed] [Google Scholar]
- 20. Kim H. J., Bar-Sagi D. (2004) Modulation of signaling by Sprouty: a developing story. Nat. Rev. Mol. Cell Biol. 5, 441–450 [DOI] [PubMed] [Google Scholar]
- 21. Mason J. M., Morrison D. J., Basson M. A., Licht J. D. (2006) Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 16, 45–54 [DOI] [PubMed] [Google Scholar]
- 22. Lim J., Wong E. S., Ong S. H., Yusoff P., Low B. C., Guy G. R. (2000) Sprouty proteins are targeted to membrane ruffles upon growth factor receptor tyrosine kinase activation: identification of a novel translocation domain. J. Biol. Chem. 275, 32837–32845 [DOI] [PubMed] [Google Scholar]
- 23. Mason J. M., Morrison D. J., Bassit B., Dimri M., Band H., Licht J. D., Gross I. (2004) Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol. Biol. Cell 15, 2176–2188 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Lao D. H., Chandramouli S., Yusoff P., Fong C. W., Saw T. Y., Tai L. P., Yu C. Y., Leong H. F., Guy G. R. (2006) A Src homology 3-binding sequence on the C terminus of Sprouty2 is necessary for inhibition of the Ras/ERK pathway downstream of fibroblast growth factor receptor stimulation. J. Biol. Chem. 281, 29993–30000 [DOI] [PubMed] [Google Scholar]
- 25. Hanafusa H., Torii S., Yasunaga T., Nishida E. (2002) Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signaling pathway. Nat. Cell Biol. 4, 850–858 [DOI] [PubMed] [Google Scholar]
- 26. Rubin C., Litvak V., Medvedovsky H., Zwang Y., Lev S., Yarden Y. (2003) Sprouty fine-tunes EGF signaling through interlinked positive and negative-feedback loops. Curr. Biol. 13, 297–307 [DOI] [PubMed] [Google Scholar]
- 27. Fong C. W., Leong H. F., Wong E. S., Lim J., Yusoff P., Guy G. R. (2003) Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J. Biol. Chem. 278, 33456–33464 [DOI] [PubMed] [Google Scholar]
- 28. Crespo P., Xu N., Simonds W. F., Gutkind J. S. (1994) Ras-dependent activation of MAP kinase pathway mediated by G protein βγ subunits. Nature 369, 418–420 [DOI] [PubMed] [Google Scholar]
- 29. Faure M., Voyno-Yasenetskaya T. A., Bourne H. R. (1994) cAMP and βγ subunits of heterotrimeric G proteins stimulate the mitogen-activated protein kinase pathway in COS-7 cells. J. Biol. Chem. 269, 7851–7854 [PubMed] [Google Scholar]
- 30. Koch W. J., Hawes B. E., Allen L. F., Lefkowitz R. J. (1994) Direct evidence that Gi-coupled receptor stimulation of mitogen-activated protein kinase is mediated by Gβγ activation of p21ras. Proc. Natl. Acad. Sci. U.S.A. 91, 12706–12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Biesen T., Hawes B. E., Luttrell D. K., Krueger K. M., Touhara K., Porfiri E., Sakaue M., Luttrell L. M., Lefkowitz R. J. (1995) Receptor-tyrosine kinase- and Gβγ-mediated MAP kinase activation by a common signaling pathway. Nature 376, 781–784 [DOI] [PubMed] [Google Scholar]
- 32. Inglese J., Koch W. J., Touhara K., Lefkowitz R. J. (1995) Gβγ interactions with PH domains and Ras-MAPK signaling pathways. Trends Biochem. Sci. 20, 151–156 [DOI] [PubMed] [Google Scholar]
- 33. Jordan J. D. (2002) Gαo/i targeting of direct interactors for degradation by the ubiquitin-dependent proteasome system: a mechanism for signal integration. in Pharmacology and Biological Chemistry, Mount Sinai School of Medicine of New York University, New York, NY [Google Scholar]
- 34. Yurchak L. K., Sefton B. M. (1995) Palmitoylation of either Cys-3 or Cys-5 is required for the biological activity of the Lck tyrosine protein kinase. Mol. Cell. Biol. 15, 6914–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osterhout J. L., Waheed A. A., Hiol A., Ward R. J., Davey P. C., Nini L., Wang J., Milligan G., Jones T. L., Druey K. M. (2003) Palmitoylation regulates regulator of G protein signaling (RGS) 16 function. II. Palmitoylation of a cysteine residue in the RGS box is critical for RGS16 GTPase accelerating activity and regulation of Gi-coupled signaling. J. Biol. Chem. 278, 19309–19316 [DOI] [PubMed] [Google Scholar]
- 36. Hiol A., Davey P. C., Osterhout J. L., Waheed A. A., Fischer E. R., Chen C. K., Milligan G., Druey K. M., Jones T. L. (2003) Palmitoylation regulates regulators of G protein signaling (RGS) 16 function. I. Mutation of amino-terminal cysteine residues on RGS16 prevents its targeting to lipid rafts and palmitoylation of an internal cysteine residue. J. Biol. Chem. 278, 19301–19308 [DOI] [PubMed] [Google Scholar]
- 37. Acconcia F., Ascenzi P., Bocedi A., Spisni E., Tomasi V., Trentalance A., Visca P., Marino M. (2005) Palmitoylation-dependent estrogen receptor α membrane localization: regulation by 17β-estradiol. Mol. Biol. Cell 16, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chandramouli S., Yu C. Y., Yusoff P., Lao D. H., Leong H. F., Mizuno K., Guy G. R. (2008) Tesk1 interacts with Spry2 to abrogate its inhibition of ERK phosphorylation downstream of receptor tyrosine kinase signaling. J. Biol. Chem. 283, 1679–1691 [DOI] [PubMed] [Google Scholar]
- 39. Rubin C., Zwang Y., Vaisman N., Ron D., Yarden Y. (2005) Phosphorylation of carboxyl-terminal tyrosines modulates the specificity of Sprouty2 inhibition of different signaling pathways. J. Biol. Chem. 280, 9735–9744 [DOI] [PubMed] [Google Scholar]
- 40. Strittmatter S. M., Fishman M. C., Zhu X. P. (1994) Activated mutants of the α subunit of Go promote an increased number of neurites per cell. J. Neurosci. 14, 2327–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taketomi T., Yoshiga D., Taniguchi K., Kobayashi T., Nonami A., Kato R., Sasaki M., Sasaki A., Ishibashi H., Moriyama M., Nakamura K., Nishimura J., Yoshimura A. (2005) Loss of mammalian Sprouty2 leads to enteric neuronal hyperplasia and esophageal achalasia. Nat. Neurosci. 8, 855–857 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.