Background: Talin regulates integrin affinity and nucleates the integrin-cytoskeleton linkage at the plasma membrane.
Results: Specific inter-domain interactions between the talin head and two rod regions block interaction with actin or plasma membrane localization.
Conclusion: Autoinhibitory interactions between the talin head and rod domains maintain cytosolic talin.
Significance: Structurally defined inter-domain interactions regulate talin localization and function.
Keywords: Actin, Integrins, Membrane, NMR, Subcellular Fractionation, Talin
Abstract
Talin, which is composed of head (THD) and rod domains, plays an important role in cell adhesion events in diverse species including most metazoans and Dictyostelium discoideum. Talin is abundant in the cytosol; however, it mediates adhesion by associating with integrins in the plasma membrane where it forms a primary link between integrins and the actin cytoskeleton. Cells modulate the partitioning of talin between the plasma membrane and the cytosol to control cell adhesion. Here, we combine nuclear magnetic resonance spectroscopy (NMR) with subcellular fractionation to characterize two distinct THD-rod domain interactions that control the interaction of talin with the actin cytoskeleton or its localization to the plasma membrane. An interaction between a discrete vinculin-binding region of the rod (VBS1/2a; Tln1(482–787)), and the THD restrains talin from interacting with the plasma membrane. Furthermore, we show that vinculin binding to VBS1/2a results in talin recruitment to the plasma membrane. Thus, we have structurally defined specific inter-domain interactions between THD and the talin rod domain that regulate the subcellular localization of talin.
Introduction
Integrin adhesion receptors are composed of type I transmembrane α and β subunits (1). Affinity modulation of integrins for their ligands and the connection of most integrins to the actin cytoskeleton are mediated by adaptors that bind to the integrin cytoplasmic domains (1–4). Among the numerous proteins that associate with integrin cytoplasmic domains, talin has emerged as a central player in both affinity modulation and integrin-actin cytoskeleton linkage (5–13).
In platelets, cells in which integrin affinity is tightly regulated, full-length (FL)2 talin remains in the cytosol until the cells are stimulated by agonists, resulting in talin redistribution to the plasma membrane (14) and increased integrin affinity (15, 16). Furthermore, recent studies using model cellular systems have established that talin is cytosolic until it interacts with an activated Rap1-RIAM complex that recruits FL talin to the plasma membrane (17–19). These data suggest that talin might be autoinhibited to regulate its localization and thus interactions with integrins and resulting biological functions.
Talin consists of a globular head domain of ∼50 kDa (THD; Tln1(1–433)) and a rod domain of ∼220 kDa (Tln1(482–2541)) connected by a short, flexible linker region (Tln1(434–481)) (6, 20). THD contains a FERM (band 4.1/ezrin/radixin/moesin) domain comprised of the F1 (Tln1(86–205)), F2 (Tln1(206–305)), and F3 (Tln1(309–405)) subdomains, but it is atypical in that it contains an additional N-terminal domain F0 (Tln1(1–85)) (21), and has an extended structure (22) rather than the clover leaf structure adopted by most FERM domains (23). The PTB-like F3 domain contains a major integrin-binding site (24, 25) that interacts with both the membrane-proximal (26, 27) and -distal regions (24, 25, 28) of the integrin β cytoplasmic tails. Perturbation of either of these interactions compromises talin-mediated integrin activation (24–26, 28–30). In addition to the integrin-binding sites, THD contains lipid-binding sites in the F1, F2, and F3 domains (21, 26, 31). These lipid-binding sites, which are aligned along one surface of the extended FERM domain, make extensive contacts with the plasma membrane (22) and play a critical role in integrin activation (21, 26, 31). The rod domain of talin consists of 62 amphipathic α-helices that are assembled into a series of α-helical bundles (33, 34), and autoinhibitory interactions between THD and the rod domain regulate the functions of talin including its interactions with integrins (17, 35–40).
Here we have combined nuclear magnetic resonance spectroscopy (NMR) with subcellular fractionation to map the THD-rod domain interactions that regulate subcellular localization of talin. We report that THD associates with the plasma membrane to a much greater extent than FL talin. We characterize two distinct THD-rod domain interactions that control the interaction of talin with the actin cytoskeleton or localization to the plasma membrane. We find that five acidic residues within Domain E (Tln1(1655–1822)) of the rod that make electrostatic interactions with the F3 domain of THD (37) inhibit the interaction of talin with actin; however, disruption of the Domain E-F3 interaction does not cause plasma membrane localization. NMR analysis revealed a second inter-domain interaction between THD and a pair of vinculin-binding α-helical bundles in the talin rod termed VBS1/2a (Tln1(482–787)); mutations that disrupt the VBS1/2a-F2F3 interaction led to plasma membrane localization of talin. Furthermore, expression of the vinculin head domain (VinHD; vinculin(2–258)) that binds to VBS1/2a (41–44) results in talin recruitment to the plasma membrane, accounting for the capacity of VinHD to induce talin-dependent integrin activation (43, 45). Thus, specific contacts between THD and helical bundles in the talin rod control the subcellular localization of talin.
EXPERIMENTAL PROCEDURES
Antibodies and cDNAs
Anti-HA mouse monoclonal antibody (MMS101P) was purchased from Covance Research Products Inc. Anti-RhoGDI rabbit polyclonal antibody was from Santa Cruz Biotechnology, Inc. Anti-GFP (Living Colors® Full-length) rabbit polyclonal antibody was obtained from Clontech. Ab2308 (46) is a rabbit polyclonal antibody against anti-αIIb integrin.3 Anti-talin mouse monoclonal antibody, 8D4, was obtained from Sigma. Anti-calnexin rabbit polyclonal antibody and anti-LAMP1 (LY1C6) mouse monoclonal antibody were purchased from Abcam. Anti-lamin A/C rabbit polyclonal antibody was obtained from Cell Signaling Technology.
HA-tagged mouse FL wild type talin in mammalian expression vector, pcDNA3.1, has been previously described (17). HA-tagged mouse THD (Tln1(1–433)) was amplified by PCR and subcloned into the mammalian expression pcDNA3-HA vector. cDNAs encoding mouse talin1 truncation mutants 465 (Tln1(1–465)), Tln1(1–787), Tln1(1–1654), Tln1(1–1822), and Tln1(1–2298) were amplified by PCR and subcloned into pcDNA3.1 vector. Each truncation mutant carries a stop codon immediately after the indicated residue number; e.g. Tln1(1–465) has a stop codon at residue 466. Residue numbers correspond to those of NP_035732.2 in the NCBI Protein database. The 5′-primer was designed to introduce an N-terminal HA tag during expression in mammalian cells. Deletion of Domain E (Tln1(1655–1822)) in FL talin was introduced by PCR using a fragment of FL talin flanked by PmlI restriction enzyme sites as a template. When the deletion was confirmed, the PmlI fragment carrying the Domain E deletion was ligated back into the PmlI site of FL talin to produce FL ΔE. Point mutations in FL 5K, FL M319A, and FL 4A4K, and FL 5K4A4K were introduced using the QuikChange II XL site-directed mutagenesis kit from Stratagene with FL talin as a template and with mutagenic primers.
RIAM176-CAAX, a talin-binding fragment of RIAM (RIAM(1–176)) fused to GFP and a Rap1 membrane targeting site (18) was described previously. cDNA encoding chicken VinHD (vinculin(2–258)) was amplified by PCR and subcloned into the mammalian expression pEGFP-C1 vector. Point mutation in VinHD-A50I was introduced using the QuikChange II XL site-directed mutagenesis kit from Stratagene with VinHD as a template and with mutagenic primers.
Cell Culture
All the experiments in this study were carried out using CHO cells stably expressing αIIbβ3 integrins (A5 cells) (47). A5 cells were cultured in Dulbecco's modified Eagle's medium supplemented with nonessential amino acids, l-glutamine, antibiotics, and 10% fetal bovine serum.
Subcellular Fractionation
A5 cells, transiently transfected with the indicated cDNAs for 24 h, were subjected to subcellular fractionation as described before (17). In brief, cells were harvested in fractionation buffer (20 mm HEPES-KOH, pH 7.5, 1.5 mm MgCl2, 5 mm KCl, 0.2 mm Na3VO4, 10 μg/ml of leupeptin, 10 μg/ml of aprotinin, 1 mm PMSF, and Complete mini protease inhibitor tablet (Roche Applied Bioscience)) and incubated on ice for 10 min. Swollen cells were disrupted by shearing through 27-gauge needles, and a fraction of the total cell lysate was saved for analysis of total protein expression. The remaining lysates were centrifuged at 425 × g for 10 min in an Eppendorf microcentrifuge model 5417R to pellet nuclei and unbroken cells. The supernatant was further centrifuged at 20,817 × g for 30 min to precipitate the membrane- and cytoskeleton-containing fraction, denoted here as “high-speed pellet.” The high-speed pellet was washed and then solubilized in fractionation buffer containing 1% Nonidet P-40 on ice. Each sample was then mixed with 5× sample buffer containing SDS and β-mercaptoethanol and boiled at 95 ºC for 5 min. Samples were run on SDS-PAGE gels (Invitrogen), and expression of talin in total lysates, cytosolic, and high-speed pellet fractions were analyzed by Western blotting using anti-HA antibody. Integrin αIIb-specific Rb2308 and anti-RhoGDI antibodies were used as membrane and cytosolic markers, respectively. The bands corresponding to the high-speed pellet and cytosolic fractions were scanned and quantified with Odyssey infrared imaging system from Li-Cor Biosciences, and represented as percent of total (cytosolic + high-speed pellet).
Purification of Plasma Membranes
Twenty four h after transfection with the indicated cDNAs, A5 cells were detached with 5 mm EDTA/PBS and subjected to surface biotinylation with 3 mm EZ-Link Sulfo-NHS-Biotin (Thermo Fisher Scientific) dissolved in 0.1 m sodium phosphate buffer, pH 8.0, for 30 min. Cells were then washed extensively with PBS and subjected to subcellular fractionation as described above. The sedimented high-speed pellet was then resuspended in fractionation buffer and incubated with BcMagTM Streptavidin Magnetic Beads (Bioclone Inc., San Diego, CA) for 1 h at room temperature with rotation. The beads were magnetically captured and washed to separate the plasma membrane from intracellular organelle membranes. Whole cell, nuclear/intact cell, cytosolic, high-speed pellet, and plasma membrane fractions (WCL, N/IC, C, P, and PM, respectively) were saved for subsequent Western blot analysis. Samples were run on SDS-PAGE gels, and expression of recombinant HA-talin in each fraction was analyzed by Western blotting using anti-HA antibody. Endogenous FL talin was detected with anti-talin 8D4 antibody. RIAM176-CAAX was detected by anti-GFP antibody. Integrin αIIb-specific Rb2308, anti-calnexin, anti-lamin A/C, anti-LAMP1, and anti-RhoGDI antibodies were used as plasma membrane, ER membrane, nuclear membrane, lysosomal membrane, and cytosolic markers, respectively. The bands corresponding to talin in each fraction were scanned and quantified with the Odyssey infrared imaging system from Li-Cor Biosciences, and talin associated with the plasma membrane (PM) fraction was represented as percent of total (cytosolic + high-speed pellet).
F-Actin Depolymerization
Recombinant DNase I from bovine pancreas (Roche Applied Bioscience) was treated with 2 mm PMSF in fractionation buffer for 1 h prior to use. To depolymerize F-actin, we incubated homogenized cell lysates with DNase I at a final concentration of 3 mg/ml for 2 h (48). Subsequent fractionation steps were performed as described above.
Actin Co-sedimentation Assay
Assay was performed using 2 μm purified, recombinant talin with or without 10 μm polymerized human platelet non-muscle actin (Cytoskeleton Inc., Denver, CO). The mixture was incubated for 1 h at room temperature and centrifuged at 541,000 × g for 30 min at 22 ºC in a Beckman Ultracentrifuge TL100 equipped with TLA100.3 rotor (Beckman Coulter). Supernatants and pellets were run on SDS-PAGE gels, which were stained with Coomassie Blue and scanned with the Odyssey infrared imaging system (Li-Cor Biosciences). Bands corresponding to each talin were quantified, and actin binding of talin is represented as: percent of total (pellet + supernatant) in the presence of actin − percent of total in the absence of actin.
Expression and Purification of Recombinant Talin Proteins
The cDNAs encoding mouse talin F2F3 (Tln1(196–405)), F2 (Tln1(196–309)), F3 (Tln1(309–400)), vinculin-binding site (VBS) 1 (Tln1(482–655)), VBS2a (Tln1(655–787)), VBS2b (Tln1(787–911)), VBS1/2a (Tln1(482–787)), and VBS1/2a/2b (Tln1(482–911)) were synthesized by PCR using a mouse talin1 cDNA as template and cloned into the expression vector pET-151TOPO (Invitrogen). These talin polypeptides were expressed in Escherichia coli BL21 STAR (DE3) cultured in either LB for unlabeled protein or in M9 minimal media for preparation of isotopically labeled samples for NMR analysis. Recombinant polyhistidine (His)-tagged talin polypeptides were purified by nickel-affinity chromatography following standard procedures. The His tag was removed by cleavage with AcTEV protease (Invitrogen), and the proteins were further purified by anion exchange or cation exchange depending on the construct.
The cDNAs encoding human Tln1(1–1654) and Tln(1–1822) were generated by PCR with human talin1 as template and cloned into pET28 bacterial expression vector that attaches an N-terminal His tag (EMD Chemicals). Residue numbers for human talin1 are based on NP_006280.3 on the NCBI Protein database. These proteins were then expressed in E. coli BL21-DE-pLys and purified with His-binding beads according to the manufacturer's instructions (EMD Chemicals). After the purified proteins were dialyzed against TBS, they were run through a HiLoad 16/60 Superdex 200 size exclusion column with TBS to remove smaller fragments generated by proteolytic cleavages during purification.
Protein concentrations were determined using their respective extinction coefficient at 280 nm. Protein concentrations were based on absorption coefficients calculated from their amino acid compositions using ProtParam (www.expasy.org).
NMR Spectroscopy
NMR experiments for the resonance assignment of Tln1(482–655), Tln1(655–787), and Tln1(787–911) were carried out with 1 mm protein in 20 mm sodium phosphate, pH 6.5, 50 mm NaCl, 2 mm DTT, 10% (v/v) 2H2O. NMR spectra of all the proteins were obtained at 298 K using Bruker AVANCE DRX 600 or AVANCE AVII 800 spectrometers both equipped with CryoProbes (Bruker, Coventry, UK). Proton chemical shifts were referenced to external DSS, and 15N and 13C chemical shifts were referenced indirectly using the recommended gyromagnetic ratios (49). Spectra were processed with TopSpin (Bruker) and analyzed using Analysis (50). Three-dimensional HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB experiments were used for the sequential assignment of the backbone NH, N, CO, Cα, and Cβ resonances. The resonance assignments of Tln1(482–655), Tln1(655–787), and Tln1(787–911) have been deposited in the BioMagResBank (BMRB) with accession numbers 17555, 17350, and 17332, respectively. The NMR assignments of the F2 and F2F3 domains were deposited in the BMRB data base and published previously (51); F2 (Tln1(196–309)), BMRB code 16930; F3 (Tln1(309–405)), BMRB code 7150; F2F3 (Tln1(196–405)), BMRB code 16932. Resonance assignments were then transferred to the double domain Tln1(482–787) using the single domains assignments and confirmed using a HNCA collected on Tln1(482–787).
NMR Interaction Studies
All experiments were carried out in 20 mm phosphate, pH 6.5, 50 mm NaCl, 2 mm DTT. Initially, 15N-labeled F2F3 was screened against the five large fragments of talin Tln1(1–433), Tln1(434–911), Tln1(913–1653), Tln1(1655–2294), and Tln1(2300–2541). 1H,15N-HSQC spectra were measured for the 15N-labeled protein alone and then in the presence of 1:1 and 1:3 molar ratios with unlabeled fragments of talin. For the Domain E-F3 interaction, spectra were recorded at ratios of 1:0, 1:4, and 1:6.
Phospholipid Co-sedimentation Assay
Large multilamellar vesicles were prepared essentially as described earlier (31). Briefly, films of dried phospholipids (Sigma) were swollen at 5 mg/ml in 20 mm HEPES, pH 7.4, 0.2 mm EGTA for 3 h at 42 ºC. The vesicles were then centrifuged at 20,000 × g for 20 min at 4 ºC, and the pellet was resuspended in the same buffer at 5 mg/ml. Protein samples were diluted into 20 mm Tris-HCl, pH 7.4, 0.1 mm EDTA, 15 mm β-mercaptoethanol. After centrifugation at 20,000 × g for 20 min at 4 ºC, proteins at 0.15 mg/ml were incubated for 30 min at 25 ºC in the absence or presence of phospholipid vesicles at 0.5 mg/ml (200 μl total volume), followed by centrifugation at 25,000 × g for 20 min at 4 ºC. Pellet and supernatant fractions were analyzed on a 10–20% gradient gel (Expedeon, Cambridge, UK) and proteins were detected by Coomassie Blue staining.
Statistics
Statistical analyses were performed with a paired t test using GraphPad Software. The resulting statistics are indicated in each figure as follows: N.S., not significant (p > 0.05), and significant: *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
RESULTS
Discrete Region of Rod Domain Maintains a Cytosolic Pool of Talin
As noted above, THD contains multiple lipid-binding sites that could support membrane interactions (21, 22, 26, 31), and previous studies raised the possibility that such sites within FL talin might be masked by the talin rod domain (17–19).
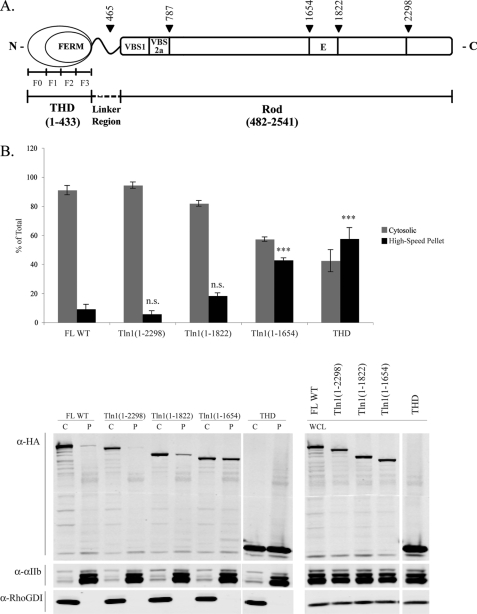
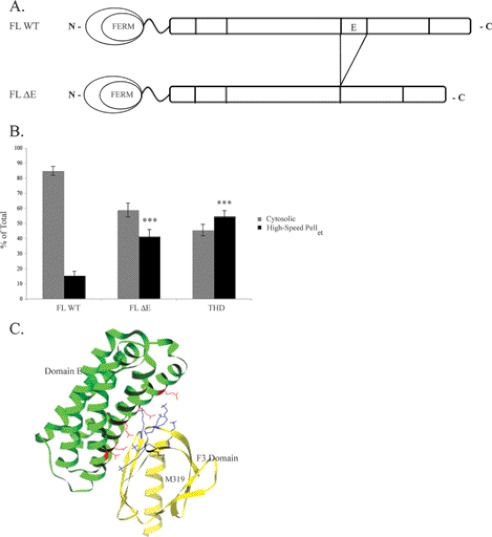
To localize potential inhibitory site(s) in the rod domain, we constructed a series of C-terminal truncations of talin and examined the partitioning of these mutants by subcellular fractionation. The rod domain of talin comprises 62 amphipathic α-helices that are assembled into a series of α-helical bundles (33, 34). Therefore, to avoid disrupting the tertiary structure of the resulting protein, we introduced stop codons between α-helical bundles as designated by arrowheads in Fig. 1A. Talin1(1–1654) (Tln1(1–1654)) showed markedly increased association with the membrane- and cytoskeleton-containing fraction, obtained after 20,817 × g centrifugation for 30 min and denoted here as high-speed pellet, approaching that of THD (Fig. 1B, 43 ± 1.5 and 58 ± 7.6%, respectively). In contrast, Tln1(1–1822) and Tln1(1–2298) associated with the high-speed pellet in similar amounts to that observed for FL talin (Fig. 1B, 18 ± 2.0, 5.7 ± 2.1, and 9.1 ± 3.2%, respectively). The membrane marker, αIIb integrin, and the cytosolic marker, RhoGDI, were detected only in the high-speed pellet and the cytosolic fractions, respectively, validating the subcellular fractionation. These data suggest that deletion of the five-helix bundle contained within Tln1(1655–1822), termed Domain E (37), increases association of talin with the high-speed pellet. To test the effect of loss of Domain E, we deleted it in the context of the FL talin protein (FL ΔE) (Fig. 2A). FL ΔE also exhibited markedly increased high-speed pellet association (Fig. 2B, 41 ± 4.6%). Thus, Domain E is required to keep talin in the cytosol.
FIGURE 1.
Truncations that delete Domain E result in increased association of talin with the high-speed pellet. A, schematic diagram of talin: talin consists of THD (Tln1(1–433)) and a rod domain (Tln1(482–2541)) connected by a linker region. THD contains an atypical FERM domain that can be subdivided into the F0, F1, F2, and F3 domains. The rod domain contains a series of α-helical bundles, one of which is Domain E located within Tln1(1655–1822). The N-terminal rod domains VBS1 and VBS2a corresponding to Tln1(482–787) and Tln1(656–787), respectively, are also shown. Arrowheads indicate the locations of stop codons. B, A5 cells transfected with the indicated talin constructs were subjected to subcellular fractionation, and the distribution of talin was assessed by Western blotting with anti-HA antibody. Integrin αIIb and RhoGDI served as the membrane and cytosolic markers, respectively. Densitometry was used to quantify the blots, and the amount of talin in each fraction is expressed as percent of total (cytosolic + high-speed pellet) recovered. Results represent mean ± S.E. (n ≥ 3). N.S., not significant, p > 0.05; ***, p < 0.001. A representative blot is shown (C, P, and WCL indicate cytosol, high-speed pellet, and whole cell lysates, respectively).
FIGURE 2.
Internal deletion of Domain E increases the amount of talin in the high speed pellet. A, schematic diagram of FL talin and FL talin mutant with Domain E deletion (FL ΔE). B, after 24 h transfection, localization of FL talin, FL ΔE, or THD in A5 cells was assessed by subcellular fractionation, followed by Western blotting with anti-HA antibody. Integrin αIIb and RhoGDI served as membrane and cytosolic markers, respectively. The blots were quantified with densitometric scanning, and the amount of talin in each fraction is expressed as percent of total (cytosolic + high-speed pellet). Results represent mean ± S.E. (n ≥ 3). ***, p < 0.001. C, ribbon representation of the talin F3 domain (yellow) in complex with rod Domain E (green) (Protein Data Bank accession code 2KGX) (37). The residues in Domain E and those in the F3 domain that lie at the interacting interface are highlighted in red and blue, respectively: Asp-1676, Asp-1763, Glu-1770, Glu-1798, and Glu-1805 in Domain E and Lys-316, Lys-318, Lys-320, Lys-322, Lys-324, and Lys-364 in the F3 domain. Met-319 in the F3 domain reportedly involved in an intramolecular interaction with the talin rod (38) is indicated in black.
Domain E-F3 Interaction Maintains Talin in Cytosol
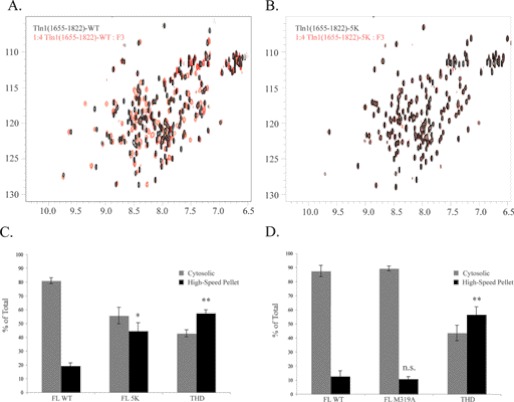
Previous studies used NMR to demonstrate that the talin rod domain interacts with the F3 fragment of the talin FERM domain (37, 38). Goult et al. (37) identified five acidic residues in Domain E (Fig. 2C, Asp-1676, Asp-D1763, Glu-1770, Glu-1798, and Glu-1805, shown in red) that form an interface with a basic surface on the F3 domain. Reverse-charge mutations were made at each of these five residues, which lie at the interface, to disrupt the electrostatic association between Domain E and the F3 domain (FL talin (D1676K, D1763K, E1770K, E1798K, E1805K) denoted FL 5K). Purified, 15N-labeled recombinant Domain E protein bearing these mutations (Tln1(1655–1822) (D1676K, D1763K, E1770K, E1798K, E1805K)) exhibited a well dispersed 1H,15N-HSQC spectrum that closely resembled that of the wild type protein (Fig. 3, A and B), indicating that the 5K mutation did not affect the folding of Domain E. Furthermore, these lysine mutations in Domain E blocked the interaction with the F3 domain as detected by NMR. As reported previously (37), incubation of 15N-labeled Domain E with unlabeled F3 resulted in a large number of chemical shift changes indicative of a direct interaction (Fig. 3A). However, addition of F3 to 15N-labeled Domain E containing the 5K mutation resulted in no chemical shift changes even at a 6-fold molar excess of the F3 (WT Domain E is saturated at a 3.5-fold excess of F3) (Fig. 3B). Thus, the lysine mutations in Domain E inhibit its interaction with the F3 in THD. Basic residues on the F3 domain that are in contact with the above mentioned acidic residues in Domain E are colored in blue (Fig. 2C, Lys-316, Lys-318, Lys-320, Lys-322, Lys-324, and Lys-364). FL talin containing the 5K mutation exhibited increased association with the high-speed pellet (Fig. 3C, 44 ± 6.0%), similar to that seen with Tln1(1–1654) and FL ΔE (Figs. 1B and 2B, respectively). FL 5K mutant was expressed well in cells at a comparable level to FL talin and THD (data not shown).
FIGURE 3.
Domain E-F3 interaction prevents membrane association of talin with the high-speed pellet. A and B, disruption of the Domain E-F3 interaction by the 5K mutations within Domain E was confirmed by NMR. A, 1H,15N-HSQC spectra of 100 μm 15N-labeled Tln1(1655–1822) rod domain in the absence (black) or presence (red) of a 4-fold molar excess of the F3 FERM subdomain. B, 100 μm 15N-labeled Tln1(1655–1822)-5K rod domain mutant in the absence (black) or presence (red) of a 4-fold molar excess of the F3 FERM subdomain. C, all of the residues in Domain E that are at the interface with the F3 were mutated to lysines (FL 5K) and the localization of this mutant was determined in A5 cells by subcellular fractionation followed by Western blotting with anti-HA antibody. The blots were quantified with densitometric scanning, and the amount of talin in each fraction is expressed as percent of total (cytosolic + high-speed pellet) recovered. Results represent mean ± S.E. (n ≥ 3). D, localization of FL talin carrying an M319A point mutation (FL M319A) was also tested, as described above. Results represent mean ± S.E. (n ≥ 3). C and D, N.S., not significant, p > 0.05; *, p = 0.01–0.05; **, p = 0.001–0.01.
Goksoy et al. (38) reported that the THD-talin rod interaction can be disrupted by a single M319A point mutation in the F3 domain. However, residue Met-319, colored in black in Fig. 2C, appears not to be at the interface between Domain E and the talin F3. In contrast to FL ΔE and FL 5K mutants (Figs. 2B and 3C, respectively), the association of FL M319A with the high-speed pellet was at a level similar to that of FL talin (Fig. 3D, 11 ± 1.7 and 13 ± 4.0%, respectively). Thus, specific interactions between Domain E and the F3 domain of THD maintain talin in the cytosol.
Loss of Domain E Is Insufficient for Plasma Membrane Localization
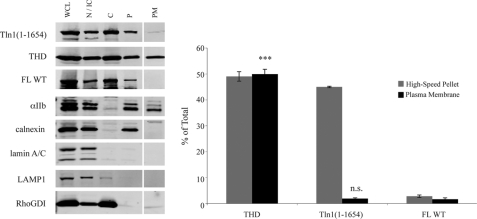
Integrin-activating signals cause talin to translocate from the cytosol to the plasma membrane (14, 52). To test whether the loss of Domain E and the consequent high-speed pellet association might allow talin to localize to the plasma membrane, we modified our fractionation protocol to separate the plasma membrane from intracellular membranes and the cytoskeleton. Cells expressing THD and Tln1(1–1654) were first labeled with cell-impermeable sulfo-biotin. After disruption and subsequent fractionation steps, the plasma membrane was further purified by capture with streptavidin-conjugated magnetic beads, and the association of talin with the recovered plasma membrane was examined. Although initial subcellular fractionation showed similar levels of Tln1(1–1654) and THD in the high-speed pellet (Fig. 4, gray bars), the abundance of Tln1(1–1654) was markedly reduced in the purified plasma membrane fraction relative to that of THD (Fig. 4, black bars). As expected, FL talin was almost undetectable in both the high-speed pellet and the plasma membrane fraction (Fig. 4). The presence of αIIb integrin (plasma membrane marker) and the absence of calnexin (ER membrane marker), lamin A/C (nuclear membrane marker), LAMP1 (lysosomal membrane marker), and RhoGDI (cytosolic marker) confirmed enrichment of the plasma membrane in this fraction (Fig. 4). Thus, although the loss of Domain E-F3 interaction increases abundance of talin in the high-speed pellet, it results in association of talin with a component other than the plasma membrane. Moreover, efficient plasma membrane localization still appears to require relief of additional constraints imposed by residues between Tln1(434) and Tln1(1654).
FIGURE 4.
Disruption of Domain E-F3 interaction is insufficient for plasma membrane localization of talin. Twenty-four h after transfection with THD or Tln1(1–1654), A5 cells were detached and subjected to surface biotinylation. Following subsequent fractionation of the surface-biotinylated cells, plasma membrane was isolated with streptavidin-conjugated magnetic beads. Distribution of the expressed talin was assessed by Western blotting with anti-HA antibody. Endogenous FL talin was detected with anti-talin antibody 8D4. Integrin αIIb, calnexin, lamin A/C, LAMP1, and RhoGDI served as the plasma membrane, ER membrane, nuclear membrane, lysosomal membrane, and cytosolic markers, respectively. A representative blot is shown (WCL, N/IC, C, P, and PM indicate whole cell lysates, nuclear/intact cell, cytosolic, high-speed pellet, and plasma membrane fractions, respectively). The PM fraction was run on the same gel, transferred onto the same blot, and scanned under the same condition as other fractions. The vertical space inserted between P and PM lanes indicates where we removed intervening, irrelevant samples. Densitometry was used to quantify the blots, and talin in high-speed pellet (gray bars) and in the plasma membrane fraction (black bars) are represented as percent of total. Bar graph represents mean ± S.E. (n ≥ 3). N.S., not significant, p > 0.05; ***, p < 0.001.
Loss of Domain E-THD Interactions Lead to Association of Talin with Cytoskeletal Actin
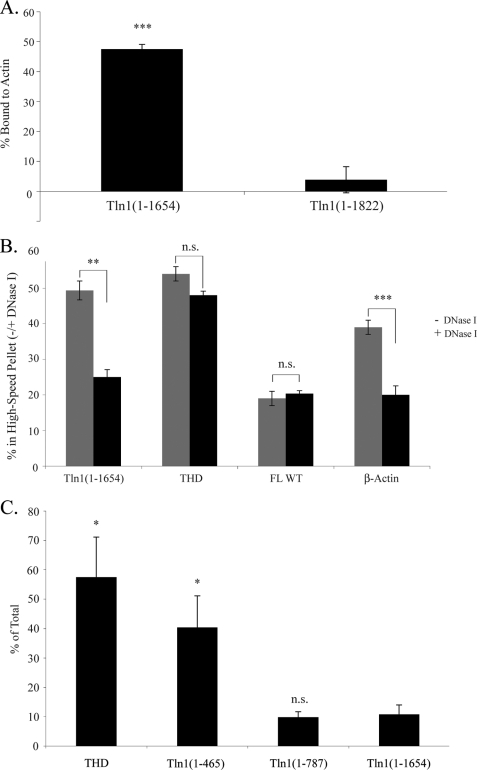
We then asked which component of the high-speed pellet fraction interacted with Tln1(1–1654). As described above, this pellet obtained after high-speed centrifugation, 20,817 × g for 30 min, of the postnuclear supernatant contains F-actin in addition to the plasma membrane and intracellular membranes. Because there are multiple actin-binding sites within talin (53), we tested the possibility that the absence of Domain E allows Tln1(1–1654) to associate with the actin cytoskeleton, rather than directly with the plasma membranes. Purified actin co-sedimentation assays showed that nearly 50% of Tln1(1–1654) bound to F-actin, whereas actin-binding of Tln1(1–1822) was negligible (Fig. 5A). The abundance of Tln1(1–1654) in the high-speed pellet fraction was reduced by half after the treatment of cell lysates with DNase I, which depolymerizes F-actin (48), whereas those of THD and FL WT talin were unaffected (Fig. 5B). Moreover, DNase I treatment reduced β-actin content in the high-speed pellet by half, to the same extent it affected the abundance of Tln1(1–1654), showing partial depolymerization of F-actin (Fig. 5B). Thus, Domain E-THD interaction prevents association of talin with the actin cytoskeleton.
FIGURE 5.
Loss of Domain E leads to association of talin with F-actin, and deletion of VBS1 results in increased association of talin with the plasma membrane. A, actin binding of purified, recombinant Tln1(1–1654) and Tln1(1–1822) were compared. 2 μm talin with or without 10 μm actin was centrifuged, and the supernatant and pellet fractions were analyzed by Coomassie Blue staining of SDS-PAGE gels. Data are represented as percent of talin in the pellet, with mean ± S.E. (n ≥ 3). B, 24 h after transfection, A5 cells expressing THD, Tln1(1–1654), or FL talin were subjected to DNase I treatment to depolymerize F-actin. After subcellular fractionation, talin distribution was assessed by Western blotting with anti-HA antibody. The blots were quantified by densitometric scanning. Abundance of talin in the high-speed pellet, with and without DNase I treatment, is represented as percent of total talin (cytosolic + high-speed pellet) recovered. The efficacy of DNase I treatment to depolymerize F-actin was verified by the measurement of β-actin in the high-speed pellet. Results represent mean ± S.E. (n ≥ 3). C, 24 h after transfection, the localization of recombinant THD, Tln1(1–465), Tln1(1–787), or Tln1(1–1654) in A5 cells was assessed as described in the legend to Fig. 4. Talin in the plasma membrane fraction is represented as percent of total. Bar graph represents mean ± S.E. (n ≥ 3). A–C, N.S., not significant, p > 0.05. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
VBS1/2a Inhibits Plasma Membrane Localization of Talin
The foregoing data indicate that plasma membrane localization of Tln1(1–1654) is restricted by sequences between Tln1(434) and Tln1(1654). To map the region that is involved in the regulation of talin plasma membrane localization further, we created two more truncation mutants (Fig. 1A), Tln1(1–465) and Tln1(1–787), and examined their association with the plasma membrane fraction. The abundance of Tln1(1–787) in the plasma membrane fraction was similar to that of Tln1(1–1654) (Fig. 5C, 10 ± 1.9 and 11 ± 3.2%, respectively) in the plasma membrane fraction. In contrast, Tln1(1–465) exhibited markedly increased association with the plasma membrane similar to that of THD (Fig. 5C, 40 ± 11 and 57 ± 14%, respectively). These data indicate that the loss of the Tln1(466–787) fragment, a region of the talin rod containing two helical bundles (VBS1 and VBS2a) that comprise vinculin-binding sites (44), leads to plasma membrane localization.
VBS1/2a Interacts with THD
To investigate whether VBS1/2a (Tln1(482–787)) limits plasma membrane localization of talin by making inter-domain contacts with THD, we performed NMR spectroscopy using 15N-labeled VBS1/2a in the presence of talin F2F3. VBS1/2a specifically interacted with talin F2F3, whereas it exhibited no interaction with any of the other talin polypeptides, e.g. Tln1(913–1653) and Tln1(2300–2541). Increasing the amounts of talin F2F3 resulted in increased line broadening most likely due to intermediate exchange on the NMR time scale, and consequently it was not possible to determine the affinity of the interaction by NMR. Furthermore, changes in the 1H,15N-HSQC spectra of 15N-labeled F2F3 domain in the presence of VBS1/2a confirmed the inter-domain interaction between VBS1/2a and THD.
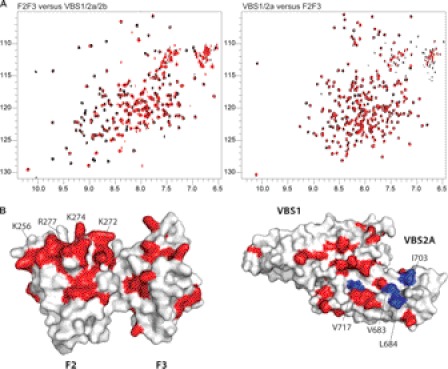
To further characterize this interaction, we assigned the backbone chemical shifts of VBS1, VBS2a, and the adjacent helical bundle, VBS2b (Tln1(787–911)). This enabled us to identify the peaks that are affected by the interaction with the F2F3 and to map them onto the surface of the known structures of the domains involved (Fig. 6A). Interestingly, observed shifts in signals and broadening effects were mapped predominantly over one face of the F2F3 and VBS1/2a (Fig. 6B), implying an interaction between these surfaces of the two domains. Furthermore, the inter-domain interaction with the F2F3 domain appears to require both VBS1 and VBS2a as a single module, because individual VBS1 or VBS2a domains did not cause significant chemical shift changes in 15N-labeled F2F3 (data not shown).
FIGURE 6.
VBS1/2a interacts with the F2F3 domain of talin. A, 1H,15N-HSQC spectra of the 15N-labeled talin polypeptide (100 μm) indicated were collected in the absence (black) or presence (red) of the unlabeled talin polypeptide indicated (molar ratio 1:3). B, molecular surface representation of F2F3 (left) and VBS1/2a (right), residues in 15N-labeled F2F3 whose signals are affected by the addition of unlabeled VBS1/2a are highlighted in red. Residues in the talin F2 MOP (Lys-256, Arg-277, Lys-272, and Lys-274) are indicated. Residues in 15N-labeled VBS1/2a whose signals are perturbed following addition of unlabeled F2F3 are highlighted in red in the molecular surface representation on the right. Some of the residues mutated are highlighted.
Disruption of the VBS1/2a-F2F3 Interaction Targets Talin to the Plasma Membrane
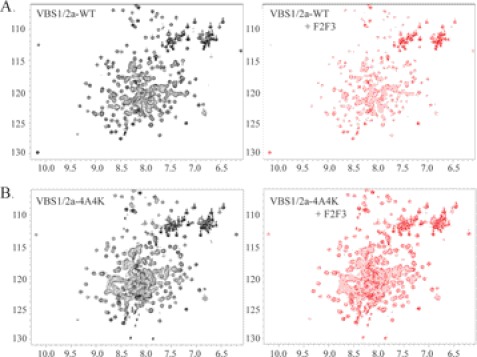
The residues in the VBS1/2a and the F2F3 whose 1H,15N-HSQC signals show large changes upon mixing the two polypeptides may define an interaction surface (Fig. 6B). To assess the effect of disrupting this surface on VBS1/2a, we introduced mutations in FL talin at each of the following residues: VBS1 residues Trp-531, Glu-538, Glu-542, and Glu-586 were substituted with alanines and VBS2a residues Val-683, Leu-684, Ile-703, and Val-717 were replaced with lysines (FL 4A4K). We confirmed that these structure-based mutations in VBS1/2a indeed disrupted the inter-domain interaction with the F2F3 as shown by the marked reduction in broadening and chemical shift changes compared with the wild type protein (Fig. 7, A and B). Importantly, the purified, recombinant VBS1/2a protein carrying the 4A4K mutations showed a well dispersed NMR spectrum similar to that of the wild type protein (Fig. 7, A and B), indicating that 4A4K mutations did not affect the tertiary structure of the protein.
FIGURE 7.
Disruption of the VBS1/2a-F2F3 interaction by the 4A4K mutations within VBS1/2a was confirmed by NMR. A, 1H,15N-HSQC spectra of 15N-labeled talin VBS1/2a rod domain (100 μm) in the presence of the F2F3 FERM subdomain at ratios of 1:0 (left panel) and 1:4 (right panel). Addition of F2F3 to 15N-labeled VBS1/2a (right panel) results in a marked decrease in signal intensity and chemical shift changes, and increased line broadening relative to VBS1/2a-WT alone (left panel). B, 100 μm 15N-labeled talin VBS1/2a-4A4K rod domain mutant in the presence of the F2F3 FERM subdomains at ratios of 1:0 (left panel) and 1:3 (right panel). Here the addition of F2F3 (right panel) has only minor effects on the 5N-labeled VBS1/2a-4A4K spectrum, which is almost identical in both the absence and presence of F2F3.
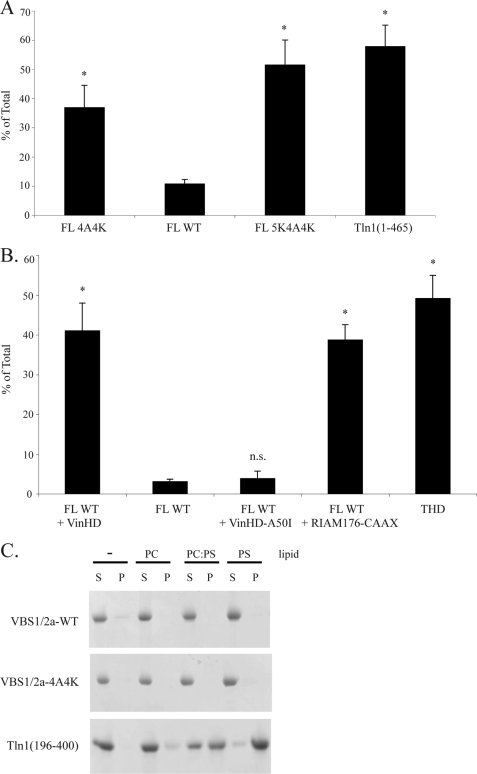
Next, the plasma membrane localization of the FL 4A4K mutant was determined. The same 4A4K mutations in the VBS1/2a were also made in combination with Domain E 5K mutations (FL 5K4A4K). FL 4A4K and FL 5K4A4K showed increased plasma membrane association to levels approaching that seen with Tln1(1–465) (Fig. 8A, 37 ± 7.5, 52 ± 8.4, 58 ± 7.2, and 102 ± 21.0%, respectively), whereas FL talin exhibited ∼10% plasma membrane association (Fig. 8A).
FIGURE 8.
Disruption of the VBS1/2a-F2F3 interaction increases plasma membrane localization of talin, and the expression of the VinHD recruits talin to the plasma membrane. A, residues in VBS1 and VBS2a that are at the interface with the F2F3 were mutated to alanines or lysines, respectively (FL 4A4K), and the plasma membrane localization of the mutant was determined in A5 cells. Localization of a talin mutant carrying VBS1/2a 4A4K mutations in combination with the Domain E 5K mutations was also analyzed (FL 5K4A4K). The localization of FL 4A4K, FL WT talin, FL 5K4A4K, or Tln1(1–465) in A5 cells was assessed as described in the legend to Fig. 4. Bar graph represents mean ± S.E. (n ≥ 3). B, FL WT talin and VinHD were co-expressed in A5 cells, and the plasma membrane localization of talin in A5 cells was assessed as described in the legend to Fig. 4. Localization of THD, FL talin in the presence of VinHD-A50I mutant and FL talin in the presence of RIAM176-CAAX were also studied. Bar graph represents mean ± S.E. (n ≥ 3). A and B, N.S., not significant, p > 0.05; *, p = 0.01–0.05. C, VBS1/2a domain does not interact with negatively charged membrane phospholipids. VBS1/2a-WT or the VBS1/2a-4A4K mutant (0.15 mg/ml) were mixed with vesicles at 0.5 mg/ml consisting of phosphatidylcholine (PC), phosphatidylserine (PS), or a 4:1 ratio of PC:PS and then centrifuged. Tln1(196–400), which binds tightly to negatively charged lipids (31), was used as a positive control.
To test whether this increase in plasma membrane localization was due to the inadvertent introduction of a plasma membrane localization site in VBS1/2a, we performed lipid co-sedimentation experiments. Even using large multilamellar vesicles consisting of 100% phosphatidylserine, we observed no interaction between either VBS1/2a or the VBS1/2a-4A4K mutant and the membrane (Fig. 8C). Therefore, we conclude that the VBS1/2a-F2F3 interaction inhibits plasma membrane localization of talin.
VinHD Promotes Plasma Membrane Localization of Talin
Talin and vinculin both localize to integrin-based adhesion sites (8) where they form a complex that stabilizes the adhesion (43, 54). The talin rod contains multiple vinculin-binding sites (55), three of which are localized in VBS1/2a. VinHD contains the talin-binding site (41, 42, 44, 56). Recently, activated forms of vinculin, including the isolated VinHD, have been reported to activate αIIbβ3 integrins in a talin-dependent fashion (45). To investigate whether VinHD binding leads to plasma localization of talin, we examined the localization of FL talin in the presence of VinHD. Remarkably, expression of the VinHD increased plasma membrane association of FL talin to a level approaching that seen with THD alone (Fig. 8B, 41 ± 6.9, and 49 ± 5.7%, respectively). Importantly, an A50I mutation in VinHD that prevents vinculin from binding to talin (57) failed to increase plasma membrane association of FL talin (Fig. 8B). Moreover, VinHD-induced plasma membrane localization of FL talin was indistinguishable from localization of talin to the plasma membrane mediated by the talin-binding fragment of RIAM fused to a Rap1A membrane targeting site (RIAM176-CAAX) (Fig. 8B). Thus, vinculin binding leads to talin recruitment to the plasma membrane.
DISCUSSION
Talin plays a major role in both integrin affinity modulation and integrin-actin cytoskeleton linkage (5, 7–13), and the subcellular localization of talin is important in controlling these biological functions. Yet, our understanding of how the activity of talin is regulated remains incomplete. Sequential C-terminal truncations of the talin rod domain and subsequent mutational analysis revealed that an α-helical bundle termed Domain E (37), spanning residues Tln1(1655–1822), keeps talin in the cytosol via an inter-domain interaction with the F3 domain of talin. However, whereas disruption of the Domain E-F3 interaction results in talin associating with the actin cytoskeleton, it is insufficient for plasma membrane localization of talin. Subsequent NMR analysis identified another inter-domain interaction between the VBS1/2a module at the N terminus of the talin rod and the F2F3 domain, dissociation of which led to plasma membrane localization of talin. Thus, we have mapped two inhibitory inter-domain interactions within talin that regulate its subcellular localization.
Interactions between Domain E and the F3 domain retain talin in the cytosolic compartment. Disruption of the Domain E-F3 inter-domain interaction increases the abundance of talin in the high-speed pellet fraction, however, this is due to its association with the actin cytoskeleton and not with the plasma membrane. Therefore, our results imply that the presence of Domain E prevents interaction of talin with cytoskeletal actin, although the precise mechanism remains to be resolved. Several regions of talin, including the talin FERM domain and several noncontiguous regions in the rod domain, bind actin (53, 58, 59), and it is reasonable to propose that one or more of these sites may be cryptic in FL talin and masked by the Domain E-F3 interaction.
Disruption of the Domain E-F3 interaction is insufficient for plasma membrane localization of talin. Unlike THD, Tln1(1–1654) fails to associate with the purified plasma membrane fraction, suggesting that exposure of the F3 domain alone is not sufficient for plasma membrane localization of talin. This idea agrees with the presence of lipid-binding sites in other subdomains of the talin head that are also critical for integrin activation. In addition to the F3 domain (26), a series of basic residues in the F1 domain (21) and a series of basic residues in the F2 domain, called the membrane orientation patch (MOP) (31), are all involved in talin-membrane association, integrin activation, and for the ability of talin to support focal adhesion assembly (36). The structure of the THD reveals an open, extended conformation in which all three lipid-binding sites on the surface of THD are orientated toward the membrane (22). Therefore, it is possible that additional membrane interactions via the F1 domain (21) and/or the F2 MOP (31) are required for the effective localization of talin to the plasma membrane and that these lipid-binding sites remain somehow masked in Tln1(1–1654).
Insight into the structural basis of such masking has come from our NMR studies that revealed a second inter-domain interaction between the two helical bundles (VBS1/2a) at the N terminus of the talin rod and the F2F3 domain of talin. Truncation of VBS1/2a and disruption of the VBS1/2a-F2F3 interaction by mutations increased the abundance of talin in the plasma membrane fraction, suggesting that this inter-domain interaction acts as the inhibitor of plasma membrane localization of talin. As noted earlier, the F2F3 contains the lipid binding MOP, and two of the F2 residues that interface with VBS1/2a are important in lipid binding of talin, i.e. Lys-272 and Lys-274 (31). Therefore, by masking one or both of these MOP residues, the VBS1/2a-F2F3 interaction might interfere with efficient plasma membrane localization of talin. The involvement of the F2F3 domain is also consistent with observations by others that the N terminus of THD cooperates with the F3 domain in integrin activation (35).
THD-talin rod domain interactions have also been reported by others (38), in which the authors concluded that a single M319A point mutation in the F3 domain disrupts its interaction with the talin rod, causing constitutive activation of talin and thereby inducing integrin activation (38). However, our examination of FL M319A localization by subcellular fractionation shows that FL M319A is mostly cytosolic, and its distribution was indistinguishable from FL WT talin. These data indicate that Met-319 does not play a direct role in the inter-domain interactions that regulate talin membrane attachment. This conclusion is in agreement with Goult et al. (37) who reported that Met-319 is not directly involved in the Domain E-F3 interaction and that the M319A mutation does not have a significant effect on this inter-domain association. Together, these results suggest that a single point mutation is unlikely to release autoinhibition of talin completely, at least in the context of subcellular localization of talin. Consistent with this concept, when Domain E mutations were tested individually in FL talin, none of the single point mutants exhibited increased association of talin with the high-speed pellet fraction (data not shown).
Several reports suggest that autoinhibition of talin can be released by calpain-mediated proteolytic cleavage (40, 60) or by phosphatidylinositol 4,5-biphosphate binding (38, 39). However, the relative roles of these processes in the regulation of talin and in physiological integrin activation have yet to be elucidated. Moreover, serine/threonine phosphorylation of talin has been shown to increase in thrombin-activated human platelets, although it is inadequate for plasma membrane localization of talin (52). The phosphorylation sites have been mapped in talin isolated from activated human platelets (32), and two of the major sites (Thr-144/Thr-150) are contained within the F1 loop implicated in lipid association (21). Interestingly, substitution of these residues for alanine accelerates talin-dependent focal adhesion assembly (36), suggesting that phosphorylation at these sites negatively regulates talin function, possibly by inhibiting THD-membrane interactions.
The ability of the VinHD to promote plasma membrane localization of talin raises the intriguing possibility that VinHD binding to the VBS1/2a fragment in the talin rod domain out-competes the VBS1/2a-F2F3 inter-domain interaction, releasing the autoinhibitory conformation of talin and resulting in the recruitment of talin to the plasma membrane. The concept certainly agrees with the recent observation that the activated forms of vinculin activate αIIbβ3 integrins in a talin-dependent manner (45). In summary, our work has revealed two regulatory inter-domain interactions that constrain the subcellular localization of talin, and opens the door to future studies aimed at identification of additional biochemical signals that unmask the membrane-binding sites in the head domain of talin.
This work was supported, in whole or in part, by National Institutes of Health Grants U54 GM64346 (to the Cell Migration Consortium), HL078784 and HL57900, a Tobacco-Related Disease Research Program Dissertation Fellowship Award, and the Wellcome Trust, Cancer Research UK.
The resonance assignments of Tln1(482–655), Tln1(655–787), and Tln1(787–911) have been deposited in the BioMagResBank (BMRB) with accession numbers 17555, 17350, and 17332, respectively.
P. E. Hughes, F. Diaz-Gonzalez, L. Leong, C. Wu, J. A. McDonald, S. J. Shattil, and M. H. Ginsberg, unpublished data.
- FL
- full-length
- THD
- talin head domain
- VinHD
- vinculin head domain
- PM
- plasma membrane
- MOP
- membrane orientation patch
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Hynes R. O. (2002) Integrins, bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2. Tamkun J. W., DeSimone D. W., Fonda D., Patel R. S., Buck C., Horwitz A. F., Hynes R. O. (1986) Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 46, 271–282 [DOI] [PubMed] [Google Scholar]
- 3. Legate K. R., Fässler R. (2009) Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 122, 187–198 [DOI] [PubMed] [Google Scholar]
- 4. Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. (2007) Functional atlas of the integrin adhesome. Nat. Cell Biol. 9, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., Fässler R. (2007) Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. 204, 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Critchley D. R. (2009) Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 38, 235–254 [DOI] [PubMed] [Google Scholar]
- 7. Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. (2002) Talin is essential for integrin function in Drosophila. Dev. Cell 3, 569–579 [DOI] [PubMed] [Google Scholar]
- 8. Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., Waterman C. M. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrich B. G., Fogelstrand P., Partridge A. W., Yousefi N., Ablooglu A. J., Shattil S. J., Ginsberg M. H. (2007) The antithrombotic potential of selective blockade of talin-dependent integrin αIIbβ3 (platelet GPIIb-IIIa) activation. J. Clin. Invest. 117, 2250–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petrich B. G., Marchese P., Ruggeri Z. M., Spiess S., Weichert R. A., Ye F., Tiedt R., Skoda R. C., Monkley S. J., Critchley D. R., Ginsberg M. H. (2007) Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J. Exp. Med. 204, 3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calderwood D. A., Zent R., Grant R., Rees D. J., Hynes R. O., Ginsberg M. H. (1999) The Talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274, 28071–28074 [DOI] [PubMed] [Google Scholar]
- 12. Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Talin binding to integrin β tails. A final common step in integrin activation. Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 13. Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., Ginsberg M. H. (2010) Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckerle M. C., Miller D. E., Bertagnolli M. E., Locke S. J. (1989) Activation-dependent redistribution of the adhesion plaque protein, talin, in intact human platelets. J. Cell Biol. 109, 3333–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma Y. Q., Qin J., Plow E. F. (2007) Platelet integrin α(IIb)β(3). Activation mechanisms. J. Thromb. Haemost. 5, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 16. Shattil S. J., Brass L. F. (1987) Induction of the fibrinogen receptor on human platelets by intracellular mediators. J. Biol. Chem. 262, 992–1000 [PubMed] [Google Scholar]
- 17. Han J., Lim C. J., Watanabe N., Soriani A., Ratnikov B., Calderwood D. A., Puzon-McLaughlin W., Lafuente E. M., Boussiotis V. A., Shattil S. J., Ginsberg M. H. (2006) Reconstructing and deconstructing agonist-induced activation of integrin αIIbβ3. Curr. Biol. 16, 1796–1806 [DOI] [PubMed] [Google Scholar]
- 18. Lee H. S., Lim C. J., Puzon-McLaughlin W., Shattil S. J., Ginsberg M. H. (2009) RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284, 5119–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watanabe N., Bodin L., Pandey M., Krause M., Coughlin S., Boussiotis V. A., Ginsberg M. H., Shattil S. J. (2008) Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin αIIbβ3. J. Cell Biol. 181, 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rees D. J., Ades S. E., Singer S. J., Hynes R. O. (1990) Sequence and domain structure of talin. Nature 347, 685–689 [DOI] [PubMed] [Google Scholar]
- 21. Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C., Calderwood D. A., Critchley D. R., Barsukov I. L. (2010) Structure of a double ubiquitin-like domain in the talin head. A role in integrin activation. EMBO J. 29, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elliott P. R., Goult B. T., Kopp P. M., Bate N., Grossmann J. G., Roberts G. C., Critchley D. R., Barsukov I. L. (2010) The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure 18, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bretscher A., Chambers D., Nguyen R., Reczek D. (2000) ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 16, 113–143 [DOI] [PubMed] [Google Scholar]
- 24. Calderwood D. A., Yan B., de Pereda J. M., Alvarez B. G., Fujioka Y., Liddington R. C., Ginsberg M. H. (2002) The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 277, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 25. García-Alvarez B., de Pereda J. M., Calderwood D. A., Ulmer T. S., Critchley D., Campbell I. D., Ginsberg M. H., Liddington R. C. (2003) Structural determinants of integrin recognition by talin. Mol. Cell 11, 49–58 [DOI] [PubMed] [Google Scholar]
- 26. Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. (2007) Structural basis of integrin activation by talin. Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 27. Ulmer T. S., Calderwood D. A., Ginsberg M. H., Campbell I. D. (2003) Domain-specific interactions of talin with the membrane-proximal region of the integrin β3 subunit. Biochemistry 42, 8307–8312 [DOI] [PubMed] [Google Scholar]
- 28. Calderwood D. A., Fujioka Y., de Pereda J. M., García-Alvarez B., Nakamoto T., Margolis B., McGlade C. J., Liddington R. C., Ginsberg M. H. (2003) Integrin β cytoplasmic domain interactions with phosphotyrosine-binding domains. A structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. U.S.A. 100, 2272–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Toole T. E., Ylanne J., Culley B. M. (1995) Regulation of integrin affinity states through an NPXY motif in the β subunit cytoplasmic domain. J. Biol. Chem. 270, 8553–8558 [DOI] [PubMed] [Google Scholar]
- 30. Ulmer T. S., Yaspan B., Ginsberg M. H., Campbell I. D. (2001) NMR analysis of structure and dynamics of the cytosolic tails of integrin αIIbβ3 in aqueous solution. Biochemistry 40, 7498–7508 [DOI] [PubMed] [Google Scholar]
- 31. Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H., Campbell I. D. (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ratnikov B., Ptak C., Han J., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2005) Talin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 118, 4921–4923 [DOI] [PubMed] [Google Scholar]
- 33. Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 34. Patel B., Gingras A. R., Bobkov A. A., Fujimoto L. M., Zhang M., Liddington R. C., Mazzeo D., Emsley J., Roberts G. C., Barsukov I. L., Critchley D. R. (2006) The activity of the vinculin-binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 281, 7458–7467 [DOI] [PubMed] [Google Scholar]
- 35. Bouaouina M., Lad Y., Calderwood D. A. (2008) The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate β1 and β3 integrins. J. Biol. Chem. 283, 6118–6125 [DOI] [PubMed] [Google Scholar]
- 36. Kopp P. M., Bate N., Hansen T. M., Brindle N. P., Praekelt U., Debrand E., Coleman S., Mazzeo D., Goult B. T., Gingras A. R., Pritchard C. A., Critchley D. R., Monkley S. J. (2010) Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. Eur. J. Cell Biol. 89, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goult B. T., Bate N., Anthis N. J., Wegener K. L., Gingras A. R., Patel B., Barsukov I. L., Campbell I. D., Roberts G. C., Critchley D. R. (2009) The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 284, 15097–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., Qin J. (2008) Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell 31, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martel V., Racaud-Sultan C., Dupe S., Marie C., Paulhe F., Galmiche A., Block M. R., Albiges-Rizo C. (2001) Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 276, 21217–21227 [DOI] [PubMed] [Google Scholar]
- 40. Yan B., Calderwood D. A., Yaspan B., Ginsberg M. H. (2001) Calpain cleavage promotes talin binding to the β3 integrin cytoplasmic domain. J. Biol. Chem. 276, 28164–28170 [DOI] [PubMed] [Google Scholar]
- 41. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) The structure and regulation of vinculin. Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 43. Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohmori T., Kashiwakura Y., Ishiwata A., Madoiwa S., Mimuro J., Honda S., Miyata T., Sakata Y. (2010) Vinculin activates inside-out signaling of integrin αIIbβ3 in Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 400, 323–328 [DOI] [PubMed] [Google Scholar]
- 46. Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. (1996) Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 271, 6571–6574 [DOI] [PubMed] [Google Scholar]
- 47. O'Toole T. E., Mandelman D., Forsyth J., Shattil S. J., Plow E. F., Ginsberg M. H. (1991) Modulation of the affinity of integrin αIIbβ3 (GPIIb-IIIa) by the cytoplasmic domain of αIIb. Science 254, 845–847 [DOI] [PubMed] [Google Scholar]
- 48. Painter R. G., Gaarde W., Ginsberg M. H. (1985) Direct evidence for the interaction of platelet surface membrane proteins GPIIb and III with cytoskeletal components. Protein cross-linking studies. J. Cell. Biochem. 27, 277–290 [DOI] [PubMed] [Google Scholar]
- 49. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 50. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 51. Kalli A. C., Wegener K. L., Goult B. T., Anthis N. J., Campbell I. D., Sansom M. S. (2010) The structure of the talin/integrin complex at a lipid bilayer. An NMR and MD simulation study. Structure 18, 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bertagnolli M. E., Locke S. J., Hensler M. E., Bray P. F., Beckerle M. C. (1993) Talin distribution and phosphorylation in thrombin-activated platelets. J. Cell Sci. 106, 1189–1199 [DOI] [PubMed] [Google Scholar]
- 53. Hemmings L., Rees D. J., Ohanian V., Bolton S. J., Gilmore A. P., Patel B., Priddle H., Trevithick J. E., Hynes R. O., Critchley D. R. (1996) Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 109, 2715–2726 [DOI] [PubMed] [Google Scholar]
- 54. Saunders R. M., Holt M. R., Jennings L., Sutton D. H., Barsukov I. L., Bobkov A., Liddington R. C., Adamson E. A., Dunn G. A., Critchley D. R. (2006) Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 85, 487–500 [DOI] [PubMed] [Google Scholar]
- 55. Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) Mapping and consensus sequence identification for multiple vinculin-binding sites within the talin rod. J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 56. Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 58. Gingras A. R., Bate N., Goult B. T., Hazelwood L., Canestrelli I., Grossmann J. G., Liu H., Putz N. S., Roberts G. C., Volkmann N., Hanein D., Barsukov I. L., Critchley D. R. (2008) The structure of the C-terminal actin-binding domain of talin. EMBO J. 27, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee H. S., Bellin R. M., Walker D. L., Patel B., Powers P., Liu H., Garcia-Alvarez B., de Pereda J. M., Liddington R. C., Volkmann N., Hanein D., Critchley D. R., Robson R. M. (2004) Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 343, 771–784 [DOI] [PubMed] [Google Scholar]
- 60. Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. (2004) Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6, 977–983 [DOI] [PubMed] [Google Scholar]