Background: Monomeric ephrinA1 exhibits anti-tumor activity and possesses a class-characteristic G-H loop for receptor binding.
Results: Alanine-scanning mutagenesis of the G-H loop revealed amino acids with specific contributions to the function of the ligand.
Conclusion: Our results document a critical importance of the G-H loop for ephrinA1 tumor-suppressing activity.
Significance: More potent anti-tumor isoforms of ephrinA1 can be generated.
Keywords: Akt, Anticancer Drug, Brain Tumors, Cancer Therapy, Cell Signaling, Cell Surface Receptor, Drug Action, Glioblastoma, Molecular Markers, Alanine-scanning
Abstract
The EphA2 receptor is overexpressed in glioblastoma multiforme and has been to shown to contribute to cell transformation, tumor initiation, progression, and maintenance. EphrinA1 (eA1) is a preferred ligand for the receptor. Treatment with monomeric eA1, the form of eA1 found in the extracellular environment, causes receptor phosphorylation, internalization, and down-regulation with subsequent anti-tumor effects. Here, we investigated the structure-function relationship of a monomeric eA1 focusing on its G-H loop (108FQRFTPFTLGKEFKE123G), a highly conserved region among eAs that mediates binding to their receptors. Alanine substitution mutants of the G-H loop amino acids were transfected into U-251 MG glioblastoma multiforme cells, and functional activity of each mutant in conditioned media was assessed by EphA2 down-regulation, ERK and AKT activation and cellular response assays. Alanine substitutions at positions Pro-113 Thr-115, Gly-117, Glu-122, and also Gln-109 enhanced the EphA2 receptor down-regulation and decreased p-ERK and p-AKT. Substitution mutants of eA1 at positions Phe-108, Arg-110, Phe-111, Thr-112, Phe-114, Leu-116, Lys-118, Glu-119, and Phe-120 had a deleterious effect on EphA2 down-regulation when compared with eA1-WT. Mutants at positions Phe-108, Lys-18, Lys-121, Gly-123 retained similar properties to eA1-WT. Recombinant eA1-R110A, -T115A, -G117A, and -F120A have been found to exhibit the same characteristics as the ligands contained in the conditioned media mainly due to the differences in their binding to the receptor. Thus, we have identified variants of eA1 that possess either superagonistic or antagonistic properties. These new findings will be important in the understanding of the receptor/ligand interactions and in further design of anti-cancer therapies targeting the eA/EphA system.
Introduction
The Eph receptors constitute the largest classified family of vertebrate receptor-tyrosine kinases. The interaction between the Eph receptors and their ligands, the ephrins, regulates cellular repulsion and adhesion, an effect that plays a major instructive role in tissue patterning, neuronal targeting, and vascular and neuronal development during embryogenesis (1–3) as well as neuronal plasticity and regeneration in the adult central nervous system (4–7). Similar to other receptor-tyrosine kinases, Ephs and ephrins have also been implicated in carcinogenesis, assuming critical roles in oncogenic transformation, metastasis, and angiogenesis (8–10). Accordingly, Ephs and ephrins have been shown to be overexpressed in a variety of solid tumors (11–13), including breast (14, 15), pancreas (16), gastric (17), colorectal (18), prostate (19), and brain (20, 21).
Eph receptors and ephrin ligands are classified into A and B classes according to their sequence homology. There are nine EphA receptors encoded in the human genome that bind to five glycosylphosphatidylinositol-anchored class A ephrins (eAs)5 and five EphB receptors that bind to three transmembrane-tethered ephrinB ligands (2, 3). In general, binding within each class is prominent; however, interclass interactions such as the binding of eA5 to EphB2 or ephrinB2 to EphA4 (22, 23) have been documented. Eph receptors and ephrin ligands are unique in that their binding results in bidirectional signaling that affects the Eph-expressing cell (forward signaling) and the ephrin-expressing cell (reverse signaling) and often involves signaling pathways with opposite effects, e.g. cellular adhesion versus repulsion (3, 24).
Our laboratory has documented that the EphA2 receptor, but not its preferred ligand eA1, is prominently overexpressed in the highly aggressive and lethal primary brain tumor glioblastoma multiforme (GBM). Moreover, EphA2 overexpression in GBM has been significantly linked to malignancy grade (20, 21), malignant progression (25), and patient survival (26, 27). Thus, the EphA2 receptor represents an attractive molecular target for therapeutic and diagnostic/imaging applications for GBM. We have also shown that eA1-Fc, a recombinant soluble homodimer of eA1, induces EphA2 phosphorylation, internalization, and subsequent down-regulation that mediates the reversal of several of the malignant properties of GBM cells, i.e. invasion, migration, and anchorage-independent growth (20).
This effect was also observed when eA1 was ectopically expressed in GBM cells; however, in this case eA1 was being released into the extracellular environment as a soluble, monomeric protein capable of mediating its effects in a paracrine manner (28). Importantly, this finding supports the development of a soluble, monomeric eA1-based ligand that may be used as a vector for targeted delivery of therapeutic or imaging agents to GBM tumors. With that in mind, we have recently conjugated a bacterial toxin to eA1-Fc and demonstrated remarkable EphA2-specificity and cytotoxic potency of this conjugate on GBM cells (29). Taken together, these observations prompted us to study the structure-function relationship of wild type (WT) eA1 by generating structural mutants by site-directed mutagenesis and analyzing their effect on EphA2 down-regulation as a readily measured functional end point of their activity.
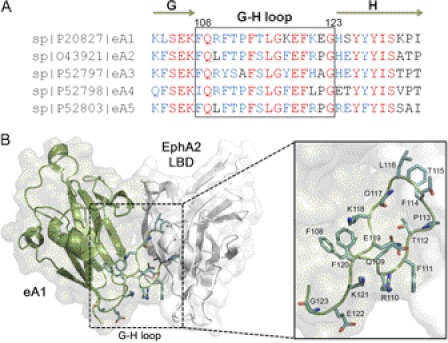
In general, the structure of eA ligands is a variation of the Greek key β-barrel-fold with parallel and anti-parallel β-strands that are connected by several highly flexible loops (30–32). Of particular importance for receptor binding is the conserved G-H loop (Fig. 1A). This loop for eA1 packs within a surface pocket of the N-terminal ligand binding domain of EphA2 (Fig. 1B). In this work the G-H loop of eA1 (residues 108–123) was subjected to alanine-scanning mutagenesis in an effort to identify variants that alter cellular effects. Most variants had either no or deleterious consequences on EphA2 binding. At least three variants within the turn of the loop (P113A, T115A, and G117A) had increased activity when compared with eA1-WT. These improved variants should help in the design of more potent anti-tumor agents based on eA1.
FIGURE 1.
The eA1 G-H loop and its interaction with EphA2. A, sequence alignment of the G-H loop of human eA ligands. The residues indicated in red are conserved. Blue shading indicates the most common residue or residues with comparable biophysical properties. The G and H β-strands are indicated by green arrows. B, structure model of the eA1-EphA2 complex. The G-H loop of eA1 (green) docks within a surface cavity of the ligand binding domain of the EphA2 receptor (white). The side chains of the G-H loop are highlighted in light blue (PDB 3CZU (31)). The figure was produced using PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC.).
EXPERIMENTAL PROCEDURES
Cell Culture
GBM cell lines U-251 MG, U-373, and SNB-19 were obtained from the American Type Culture Collection (Manassas, VA). U-251 MG cells were grown in DMEM + glutamine, 10% (v/v) FBS, 0.1 mmol/liter nonessential amino acids, and 1% (v/v) penicillin/streptomycin. eA1-mutant cell lines, which are derived from the U-251 MG parental cell line, were grown in the above medium supplemented with 200 μg/ml Geneticin. U-373 cells were grown in MEM, 10% (v/v) FBS, and 1% (v/v) penicillin/streptomycin, and SNB-19 cells were grown in RPMI, 10% (v/v) FBS, 0.1 mmol/liter nonessential amino acids, sodium pyruvate, and 1% (v/v) penicillin/streptomycin. All cultures were maintained at 37 °C and 5% CO2.
Alanine-scanning Mutagenesis
eA1 mutants were prepared using a transformer site-directed mutagenesis kit from Clontech (Mountain View, CA) and an eA1-pcDNA3.1+ plasmid generated from full-length human ephrin mRNA obtained from human umbilical vein endothelial cells using specific primers for the eA1 gene. Mutagenesis and selection primers used are listed in supplemental Table S1. BMH71-18 mutS chemically competent Escherichia coli cells were then transformed with mutant ephrinA1 plasmids and cultured overnight at 37 °C with shaking at 280 rpm; plasmid DNA was subsequently isolated with a Qiaprep miniprep kit (Qiagen, Valencia, CA). Sequence-verified eA1-mutant plasmids (Wake Forest University DNA Sequencing Laboratory) were then transfected into U-251 MG cells as described below.
Transfection Experiments
Mutant plasmids were transfected into U-251 MG glioma cells in Opti-MEM (Invitrogen) using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. After 24 h, Opti-MEM media was replaced with growth medium containing 20% FBS and then again, 24 h later, with 10% FBS growth medium. Forty-eight hours later cells were split into 100-mm2 dishes and selected with growth medium containing 800 μg/ml Geneticin. Individual clones were isolated with cloning rings, transferred into a 24-well plate, and maintained in growth medium with 200 μg/ml Geneticin. Mutant eA1 expression in the cell media of clones was verified by eA1 immunoblotting, and high-expressing clones were further cultured in growth medium without Geneticin for at least 48 h to obtain conditioned media for EphA2 down-regulation assays.
EphA2 Down-regulation Assay
U-251 MG cells were grown to 60% confluence in 60-mm dishes at 37 °C, 5% CO2, and subsequently dosed with conditioned media containing equivalent amounts of wild type eA1 or mutant eA1 as evaluated by eA1 Western blotting and densitometry analysis. For determination of the effect of all the mutants together on EphA2 down-regulation and activation of AKT and ERK as well as in the migration/cell morphology assays, eA1 was measured by ELISA (see below). Negative controls for these experiments included fresh media and conditioned media from vector-transfected U-251 MG cells, whereas positive controls were recombinant homodimeric eA1-Fc at 1 μg/ml (R&D Systems, Minneapolis, MN) and wild type eA1-conditioned media. U-251 MG cells were incubated for 24 h at 37 °C, 5% CO2 with the corresponding conditioned media or controls before obtaining cell lysates.
Sandwich ELISA Analysis of eA1 in Conditioned Media (CM)
eA1 in CM was determined using a sandwich ELISA assay. CMs were collected from subconfluent monolayers of U-251 MG parental and vector-, WT-, and mutant eA1-transfected cells. Media were centrifuged for 5 min ×1000 rpm to pellet any insoluble debris, and supernatant was collected. A 96-well plate was coated with 5 μg/ml polyclonal goat anti-mouse human eA1 (R&D Systems), blocked in 5% milk, and incubated with serial concentrations of eA1-Fc (R&D Systems) or CM. Detection was achieved using a primary antibody against human eA1 (clone V18, Santa Cruz Biotechnology, Santa Cruz, CA) followed by HRP-conjugated anti-rabbit secondary antibody and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS). Detection was performed at 405 nm. Both primary and secondary antibodies were raised against epitopes outside of the eA1 GH-loop and thus enable detection of eA1 GH-loop mutants. A standard curve was constructed using the signals corresponding to the eA1-Fc serial dilutions, and mutant eA1 content in CM was calculated.
Conditioned Media Treatment of U-251 MG Cells
Subconfluent cultures of U-251 MG cells were grown in 60-mm2 dishes. Cells were treated with conditioned media dilutions based on the results from the sandwich ELISA assay, recombinant mouse eA1/Fc chimera (R&D Systems), or conditioned growth media control for the indicated times. Cells were photographed by phase contrast microscopy with a 20× objective lens, and images were processed using Photoshop (Adobe Systems). Cell lysates were collected and used for Western blotting.
Western Blotting
Cell lysates were prepared by washing treated U-251 MG cells with PBS and lysed in radioimmunoprecipitation assay buffer (0.5% (w/v) sodium deoxycholate, 0.1% (w/v) sodium dodecyl sulfate, 0.5% (w/v) Igepal in 10 mm phosphate-buffered saline) containing mammalian protease inhibitor mixture and 0.5% v/v sodium vanadate (Sigma). Cell lysates were separated by SDS-PAGE in 10% polyacrylamide gels (EphA2/actin immunoblotting) or 12% polyacrylamide gels (ephrinA1-immunoblotting for conditioned media) and then transferred to a polyvinylidene difluoride membrane (Pierce). Membranes were subsequently blocked (5% (w/v) milk in PBS containing 0.05% (v/v) Tween 20 (Sigma)) for at least 1 h at room temperature and then incubated overnight at 4 h with shaking with the appropriate primary antibodies including anti-EphA2 clone D7 at 1:1000 (Santa Cruz Biotechnology), anti-β-actin diluted at 1:50,000 (Sigma), anti-ephrinA1 clone V18 at 1:300 (Santa Cruz Biotechnology), anti-phospho-p44/42 MAPK (ERK1/2) (Thr-202/Tyr-204) XPTM, anti-p44/42 MAPK (Erk1/2), anti-phospho-AKT (Se-r473) XPTM, anti-Akt (Cell Signaling Technology, Danvers, MA), and anti-α tubulin (Thermo Scientific, Fremont, CA). After three 5-min washes in 0.05% (v/v) Tween 20 in PBS, membranes were incubated with secondary antibody conjugated to horseradish peroxidase (goat anti-mouse IgG or goat anti-rabbit IgG) at a dilution of 1:5000 in blotto for 1 h at room temperature with shaking. Membranes were then again washed three times with 0.05% (v/v) Tween 20 in PBS, and proteins were detected using the Enhanced Chemiluminescence Plus Western blotting detection system (Amersham Biosciences). Membranes were subsequently exposed to BioMax XAR autoradiographic film (Eastman Kodak Co.) for 10–20 s. Films were scanned at a resolution of 600 dpi using a HP ScanJet3979 and Adobe Photoshop 5.0 Software. Densitometry analysis was conducted using Image J Software (National Institutes of Health).
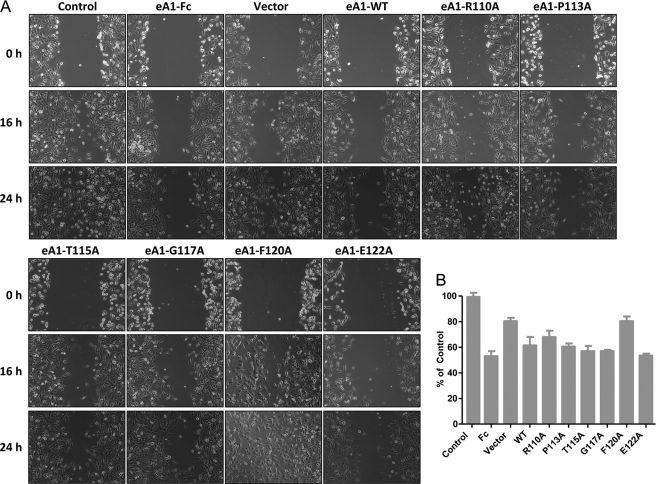
Migration Assay
Wounds were made in a confluent monolayer of U-251 MG cells with a sterile 200-μl tip, and growth media containing WT and mutant eA1 was added. Conditioned growth medium from untransfected cells was used as the negative control, whereas eA1-Fc (1 μg/ml) was added as a positive control. Phase contrast microscopy pictures were taken of the same field at 0, 8, 16, and 24 h. Distance of the wound in μm was measured in five places for each of three wounds for each treatment or cell type at each time point using ImagePro Plus software, and the percent wound closure over 24 h was calculated for graphical representation.
Production of Recombinant Variants of eA1
Recombinant eA1-WT and its variants R110A, T115A, G117A, and F120A were produced using the baculovirus expression vector system from BD Biosciences. eA1 (19–175) WT, one of the forms of eA1 that we identified in the media of cancer cells (unpublished) and the four eA1 mutants were amplified by PCR using the forward primer 5′-TATAGGATCCCATCACCATCACCATCACGATCGCCACACCGTC-3′ (BamHI restriction site and the coding sequence for the N terminus Histidine tag), and the reverse primer 5′-CACGAATTCCTATTATTAAACCCGCACCTCTGGGTCATC-3′ (EcoRI restriction site and stop codons). The amplified fragments were cloned into BamHI-EcoRI sites in the Baculovirus transfer vector pAcGP67-B (BD Biosciences) and sequenced. This vector carries the gp67 Baculovirus-encoded secretion signal sequence upstream the MCS forcing the secretion of the recombinant proteins. The generated recombinant bacmids pBeA1, pBeA1-110, pBeA1-115, pBeA1-117, and pBEe1-120, respectively, were amplified in E. coli DH5α cells.
Sf9 insect cells were co-transfected with pBEA1, pBeA1-110, pBEA1-115, pBEA1-117, and pBEA1-120 recombinant bacmids and the linearized BaculogoldTM Baculovirus DNA (BD Biosciences) using the BaculogoldTM Transfection kit (BD Biosciences) following the instructions of the supplier. Infectious recombinant baculoviruses were amplified two times in BD BaculogoldTM MaX-XP Serum-free Insect Cell Medium (BD Biosciences) in Sf9 serum-free media-adapted cells at 27 °C for 5 days to obtain high titer virus stocks. Sf9 cells, serum-free media-adapted, were infected with high titer virus stocks and grown at 27 °C for 5 days to produce the recombinant proteins.
Sf9-baculovirus infected media containing the recombinant proteins were collected, and floating cells were removed by centrifugation (at 3000 × g for 10 min). Supernatant was filtered through a 0.22-μm pore filter, 0.1 m urea was added, and the pH was adjusted to 7.4. The His6-tagged recombinant proteins were purified by affinity chromatography with HisTrap HP affinity column (GE Healthcare). The column was equilibrated in buffer A (50 mm NaH2PO4·H2O, 150 mm NaCl, pH 7.4). After loading, the column was washed with 10 column volumes of buffer A, and the recombinant proteins were eluted with a step gradient with buffer B (50 mm NaH2PO4·H2O, 300 mm NaCl, 250 mm imidazole, pH 7.4). Recombinant proteins were filtered (0.22 μm) and stored at −20 °C.
Surface Plasmon Resonance Assay
Binding interactions between eA1 (wild type and mutants) and EphA2 were measured by surface plasmon resonance in a BIAcore T100 instrument (33, 34). Recombinant human EphA2 receptor (R&D Systems) was immobilized on the dextran matrix of a CM5 biosensor chip using amine coupling chemistry (29 μg/ml at pH 4) to achieve a sparse monolayer in the sample chamber (5991 ± 139 response units (RU), n = 3) and a blank immobilization in the reference chamber. Unreacted sites in both chambers were blocked with ethanolamine. All surface plasmon resonance procedures were carried out at 25.0 °C.
Binding curves were obtained with eA1 monomer (0–3000 nm) and dimer (0–2400 nm) by delivering aliquots of each protein in HEPES-buffered saline, pH 7.4, to both sample and reference chambers at 30 μl/min. RU versus time profiles were monitored for 700 s during the binding step; dissociation was then monitored for 1500 s as buffer was delivered. Bound ligands were removed by regeneration with 1 m NaCl followed by 0.1% SDS. Time-dependent changes obtained with buffer delivery were subtracted from each protein binding profile (sample − reference RU signals) to obtain double-corrected kinetic traces.
Anchorage-independent Growth
U-251 MG cells (2 × 103) were plated in 6-well plates in growth medium plus 0.35% agar on a base layer of BactoTMAgar, BD Biosciences, growth medium plus 0.5% agar. Cells were supplemented with eA1-WT, eA1-R110A, eA1-T115A, eA1-G117A, and eA1-F120A at 1.0 and 0.1 μg/ml or vehicle alone. Fresh media-eA1 was added to the cells 3 days and 1 week after plating. Colonies were counted after 14 days. Clusters of colonies greater than 50 cells were counted in 10 random fields at low power. Each experimental condition was done in triplicate for every assay.
Statistical Analysis
Probability (p) values were calculated using the analysis of variance one-way test using MS Excel; p values ≤0.05 were considered to be statistically significant.
RESULTS
EphA2 Is Differentially Down-regulated by G-H Loop eA1 Alanine Mutants
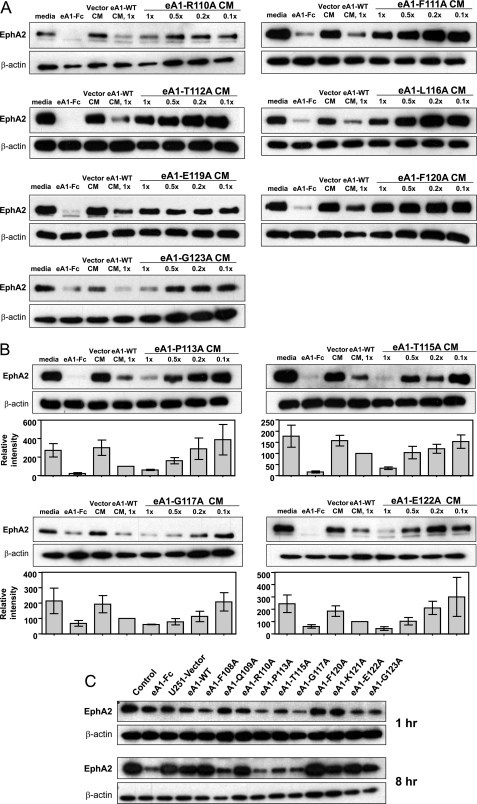
We demonstrated that EphA2 is differentially down-regulated in U-251 MG cells treated with an equimolar amount of CM obtained from U-251[eA1](+) and various U-251[G-H loop mutant eA1s](+) cells. First, we determined that there are three principal groups of alanine-mutated eA1 variants. In the first group a diminished EphA2 down-regulation was observed among CM from R110A, F111A, T112A, L116A, E119A, F120A, and G123A eA1 mutants when compared with eA1-WT CM (Fig. 2A). In the second group three substitution mutations, namely F108A, K118A, and K121A yielded activity similar to those obtained with U-251[eA1](+) CM (supplemental Fig. S1). Interestingly, the third group of variants (Pro-113, Thr-115, Gly-117, and Glu-122) exhibited a 39, 67, 39, and 57% (Fig. 2B) enhanced down-regulation of EphA2, respectively. Q109A demonstrated a similar effect (not shown). It is noteworthy that T115A and E122A are two of few other residues that are unique to eA1 G-H loop. Further experiments revealed that non-conservative substitutions at position 115, such as T115R, T115D, and T115G, not only abolished the enhancing effect on EphA2 down-regulation that had been obtained with T115A CM, but they made the variants inactive (supplemental Fig. S2). One substitution, F114A, was the only mutant of eA1 found in small amounts in the cell lysate and not released into the media and, thus, of unknown activity (not shown).
FIGURE 2.
EphA2 down-regulation ability of alanine point-mutants of human ephrinA1. A, eA1 G-H loop mutants with diminished ability to down-regulate EphA2. Shown are Western blots of EphA2 immunoreactivity in U-251 MG cells treated with CM from eA1-R110A, eA1-F111A, eA1-T112A, eA1-L116A, eA1-E119A, eA1-F120Am and eA1-G123A for 24 h. Equivalent dosing (1.0× to 0.1×) was verified by Western blotting; eA1-Fc (1 mg/ml) and eA1-WT CM were used as positive controls. B, eA1 G-H loop mutants with enhanced effect on EphA2 receptor down-regulating ability. Shown is a Western blot of EphA2 immunoreactivity in U-251 MG cells treated with CM from eA1-P113A, eA1-T115A, eA1-G117A, and eA1-E122A for 24 h. C, temporal analysis of EphA2 down-regulation in response to the treatment with eA1-Fc (1 μg/ml), U-251 vector, eA1-WT, or eA1-mutant CM. Western blot of EphA2 immunoreactivity in U-251 MG cells treated for 1 h and for 8 h are shown. ELISA was used to confirm equivalent dosing of eA1 mutants.
G-H Loop Mutant CMs Promote Cell Rounding and Decrease Migration of GBM Cells
Next, U-251 MG parental and G48a GBM cells were treated with equal concentrations of the different mutant eA1 CMs as measured directly by ELISA. We investigated an ability of the mutants to elicit a characteristic morphological response to eA1 in a form of cell rounding (20, 35). Cells treated with the alanine mutant CMs became rounded within 20 min of treatment as did cells exposed to U-251[eA1](+) CM or homodimeric eA1-Fc (not shown; more detailed analysis of cell rounding was performed with recombinant forms of eA1 as shown in Fig. 5E). Cell rounding was accompanied by EphA2 receptor down-regulation (Fig. 2C). Interestingly, P113A, T115A, and G117A CM induced rapid (1 h) EphA2 receptor down-regulation (Fig. 2C). This prominent effect of the three mutants was sustained for 8 h post-treatment when also the Q109A mutant showed an increased activity. Furthermore, the treatment of U-251 MG cells with similar concentrations of CM from P113A, T115A, G117A, K121A, and E122A mutants also decreased the migration of U-251 MG cells (Fig. 3).
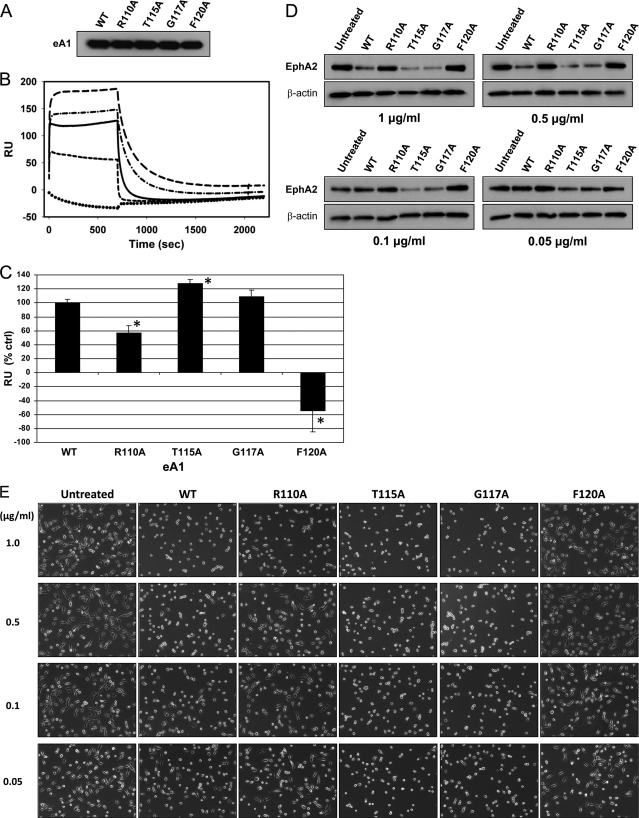
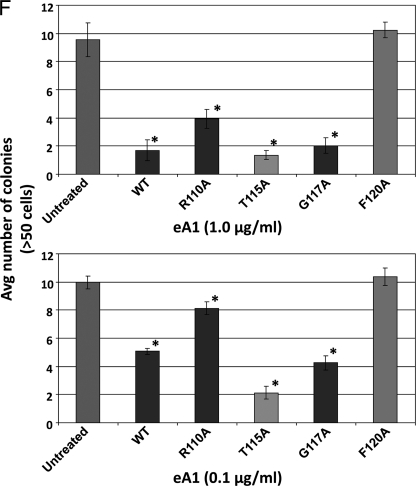
FIGURE 5.
Binding and biological activity of recombinant eA1 G-H loop mutants. A, Western blot of recombinant forms of eA1 mutants. B, surface plasmon resonance analysis of the binding of eA1-WT and recombinant eA1 mutants to immobilized EphA2. Each protein was delivered at 1 μm for 700 s followed by a 1500-s dissociation step. Solid line, eA1-WT; long dash, eA1-T115A; dash-dot, eA1-G117A; short dash, eA1-R110A; dotted line, eA1-F120A. C, quantification of binding of recombinant eA1 mutants as assayed by surface plasmon resonance. Significant differences (p < 0.05) versus eA1-WT are indicated by asterisks. D, down-regulation of the EphA2 receptor in GBM cells by recombinant eA1 mutants as shown by EphA2 western blotting. E, cell rounding of GBM cells in response to various concentrations of recombinant eA1 mutants. F, quantification of the effect of recombinant eA1 mutants on anchorage-independent growth of GBM cells. Significant differences (p < 0.05) versus untreated group are indicated by asterisk.
FIGURE 3.
Effect of eA1-WT and eA1-mutant CMs on the migration of GBM cells. A, phase contrast micrographs of a wound on confluent U-251 MG cells (20× magnification) treated with control, eA1-Fc (1 μg/ml), eA1-WT, and mutant eA1 CM over a period of 24 h. B, shown is quantification of migration as the percentage of wound closure at 24 h of U251-MG cells treated with control, eA1-Fc (1 mg/ml), eA1-WT, and mutant eA1 CM. ELISA was used to confirm equivalent dosing of eA1 mutants.
Ras-MAPK and PI3K-AKT Pathways Are Differentially Affected by eA1 G-H Loop Mutants
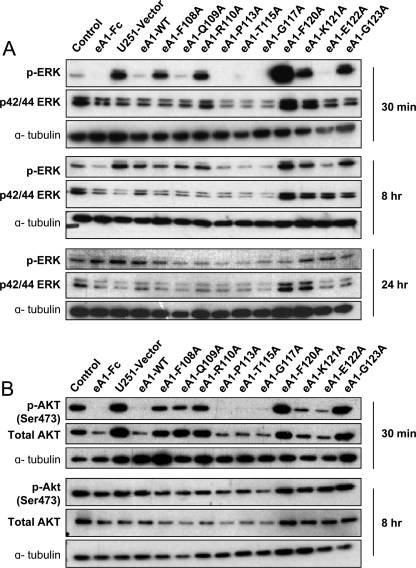
We next investigated the effect of WT and mutant eA1s on intracellular signaling, specifically on classical oncogenic pathways such as Ras-MAPK and the phosphoinositide 3-kinase/AKT pathways known to be responsive to eA1 stimulation. Dimeric eA1-Fc suppresses signaling through the Ras-MAPK pathway (36–38) as does soluble monomeric eA1 (28); hence, we examined whether or not an altered suppression of this pathway would occur after treatment with the mutant eA1 CM. Indeed, treatment with Q109A, P113A, T115A, G117A, and E122A CM led to a decrease in phosphorylation of ERK at 30 min (Fig. 4A). Surprisingly, this effect was sustained for 8 h and less so at 24 h post-treatment (Fig. 4A). It appears that eA1-F120A actually activated p-ERK.
FIGURE 4.
Effect of eA1 G-H loop point mutants on oncogenic signaling. A, Western blots of p-ERK and p42/p44 MAPK immunoreactivity in U-251 MG cells treated with control, eA1-Fc (1 μg/ml), eA1-WT, and eA1-mutant CMs for 30 min, 8 h, and 24 h. B, Western blots of p-AKT and total AKT immunoreactivity in U-251 MG cells treated with control CM, eA1-Fc (1 μg/ml), eA1-WT, and eA1-mutant CMs for 30 min and 8 h. ELISA was used to confirm equivalent dosing of eA1 mutants in both assays.
Decreased phosphorylation of AKT was shown to follow EphA2 receptor activation by eA1-Fc (39). Therefore, we investigated the effect of mutant monomeric eA1s on the PI3K/AKT pathway. The treatment of U-251 MG cells with P113A, T115A, G117A, and E122A CM led to a decrease in p-AKT at Ser-473 30 min after the treatment (Fig. 4B). This effect was at least partially sustained by P113A, T115A, and G117A mutants for 8 h (Fig. 4B).
eA1 and Its Variants Bind to Immobilized EphA2 with Different Affinities
Purified recombinant monomeric eA1-WT and its four variants (Fig. 5A), chosen on the basis of varying activities identified in the experiments with CM, were further characterized for their binding abilities and biological properties. In the binding assays using BIAcore system, plotting the maximum signal changes versus eA1 concentration yielded hyperbolic binding profiles (supplemental Fig. S3). Analysis by nonlinear regression (SigmaPlot 11, SYSTAT Software, Santa Fe, CA) with a single site-saturable model yielded Bmax = 229 ± 7 RU and Kd = 89 ± 21 nm for eA1-Fc (dimer); eA1 monomer yielded Bmax = 163 ± 26 RU and Kd = 580 ± 240 nm.
EphA2 binding experiments with wild type eA1 and mutants carried out at 1 μm yielded rapid signal changes during the binding step that approached a plateau by 30–60 s; dissociation rates exhibited half-times <50 s (Fig. 5B). Plotting the maximum binding signals observed in replicate experiments with eA1-WT (n = 11) and the mutants T115A (n = 4), F120A (n = 4), R110A (n = 3) and G117A (n = 3) yielded a consistent pattern of tighter binding by eA1-T115 and eA1-G117A variants when compared with eA1-WT, and a partial (eA1-R110) or complete loss (eA1-F120) of the binding by the others (Fig. 5C).
Recombinant Variants of eA1 Display Enhanced Biological Properties Relative to Wild Type eA1
The recombinant variants of eA1 were then tested for their ability to down-regulate the EphA2 receptor, to induce cell rounding, and to affect anchorage-independent growth of GBM cells as an in vitro anti-tumor activity measure. The most active variants in all these assays were eA1-T115A and eA1-G117A, whereas eA1-R110A and eA1-F120A were either significantly less active or lost the activity altogether when compared with eA1-WT. For example, eA1-T115A and -G117A down-regulated the EphA2 receptor at concentrations as low as 0.05 μg/ml whereas eA1-WT lacked activity at 0.1 μg/ml (Fig. 5D). Furthermore, the same was observed in a cell-rounding assay when these two super agonistic variants of eA1 were still able to change the GBM cells morphology at the lowest concentrations used (Fig. 5E). In the anchorage-independent growth assay, eA1-T115A and eA1-G117A showed significant inhibition of this growth comparable with eA1-WT at 1.0 μg/ml, but they demonstrated more pronounced inhibition than eA1-WT at the lower 0.1 μg/ml concentration (Fig. 5F and supplemental Fig. S4, A and 4B).
DISCUSSION
In this work we have directly documented a critical role of the eA1 G-H loop in mediating functional soluble monomeric eA1 activity. Importantly, the P113A, T115A, G117A, E122A, and to some extent Q109A mutants of the eA1 ligand exhibited an enhanced down-regulation of the EphA2 receptor, the typical morphological changes of cancer cell-rounding, and altered cell migration, all at significantly lower concentrations (Table 1). In addition, the treatment with mutant eA1 ligands had profound and sustained effects on classical oncogenic pathways in which the eA1/EphA2 system is ultimately involved. These results were recapitulated with the use of recombinant purified variants of eA1. Thus, eA1-T115A and eA1-G117A bind tighter to the immobilized EphA2 receptor than eA1-WT whereas A1-R110A and eA1-F120A bind less tightly or lose the binding completely. The two superagonistic variants of eA1 have also significantly more pronounced biological activities in down-regulation of EphA2, cell rounding, and anchorage-independent growth assays when compared with eA1-WT or eA1-R110A or eA1-F120A (Fig. 5).
TABLE 1.
Activity of eA1 G-H loop mutants in GBM cells
+ to +++, arbitrary degree of potentiation of eA1 and its substitution mutants activity. - to —, arbitrary degree of alteration of eA1 mutants activity. NA, not done.
| Mutant | EphA2 down-regulation | p-ERK down-regulation | p-Akt down-regulation |
|---|---|---|---|
| WT | ++ | ++ | + |
| F108A | ++ | - | + |
| Q109A | +++ | +++ | ++ |
| R110A | – | – | - |
| F111A | – | NA | NA |
| T112A | – | NA | NA |
| P113A | +++ | +++ | +++ |
| F114A | NA | NA | NA |
| T115A | +++ | +++ | +++ |
| L116A | – | NA | NA |
| G117A | +++ | +++ | +++ |
| K118A | ++ | NA | NA |
| E119A | – | NA | NA |
| F120A | – | — | – |
| K121A | ++ | - | – |
| E122A | +++ | +++ | +++ |
| G123A | – | – | + |
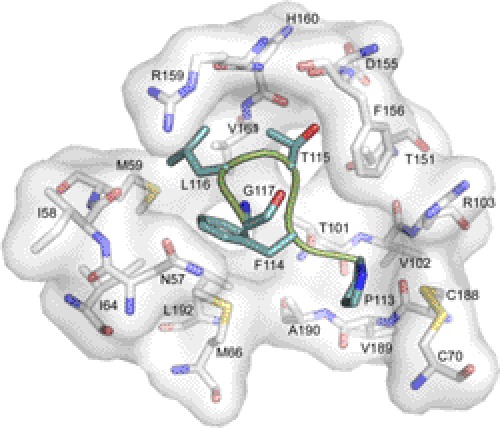
The G-H loop of eA1 packs into a surface pocket of the EphA2 receptor (Fig. 1). Several of the variants lost part or all of the ability to interact with the EphA2 (R110A, F111A, T112A, L116A, E119A, and G123A). The most likely scenarios for the observed loss in activity are that the structure of loop may have been distorted or altered in its conformation dynamics. For example, L116 fills completely a pocket primary generated by the side chains of Met-59 and Val-61 (Fig. 6). A mutation to Ala would result in a loss of this hydrophobic interaction. Mutation of eA1 at position 119 was detrimental in terms of EphA2 receptor down-regulation, a result that is also in line with its role as the “latch” that strengthens the eA1/EphA2 interaction (31). The F108A, K118A, and K121A variants exhibited WT levels of activity.
FIGURE 6.
Molecular interactions of the G-H loop near eA1 residues 113–117. The coloring scheme is the same as Fig. 1 (PDB 3CZU (31)).
In contrast, the P113A, T115A, G117A, E122A, and Q109A mutants had improved interaction with EphA2, as supported by their ability to bind at significantly lower concentrations (Fig. 5C). A closer inspection at the details of the interactions of P113A, T115A, and G117A with EphA2 (Fig. 6) provides some rationale for the consequences of the mutations. Pro-113 packs against the disulfide bond between residues Cys-70 and Cys-188 and the backbone atoms of residues Val-89 and Ala-190 of EphA2. Perhaps the removal of the conformational rigidity of Pro-113 enabled the loop to have more facile binding. Thr-115 fills a pocket lined by several residues including Thr-101, Thr-151, Phe-156, Asp-155, and Val-61. It is unclear why an Ala substitution would be more favorable, but the requirement for a small, uncharged amino acid at this position is supported by the complete loss of EphA2 binding by the T115D and T115R variants (supplemental Fig. S2). It is also unclear why the mutation of Gly-117, Glu-122, and Gln-109 to Ala would be favorable. Clearly more work will be required to ascertain the molecular basis for the observed improvements. Nonetheless, it is encouraging to find that single mutations can lead to significant improvements in the eA1-EphA2 interaction. Further optimization of the G-H loop and perhaps others may ultimately prove to yield eA1 variants that will be useful for cancer therapy or diagnosis.
Mutation of ephrinA1 at the five critical residues led to sustained EphA2 receptor down-regulation in GBM cells. It had been previously documented that upon exposure to dimeric eA1-Fc, the EphA2 receptor present on tumor cells undergoes tyrosine phosphorylation and is down-regulated (40–42). The effects of binding of eA1 to EphA2 have been hypothesized to constitute a dual process comprised of direct signaling via ligand-receptor interaction and the effects resulting from the down-regulation of the receptor. The five mutants showed an enhanced ability to down-regulate the receptor when compared with eA1-WT treatment.
Binding of eA1 to EphA2 leads to SHP-2 recruitment and subsequent FAK dephosphorylation that suppresses integrin function and diminishes cell adhesion to the extracellular matrix (35). The effects of this signaling cascade are visible in a short period of time after eA1 administration, when tumor cell rounding ensues most likely from the formation of the kinase-active Src-FAK complex, which leads to contraction of the cytoskeleton via myosin II and RhoA activity (43). Mutant eA1 CMs corresponding to the four critical residues cause a rapid and profound cell rounding (Fig. 5E) (28). In addition, administration of eA1-Fc, eA1-WT, or the four mutants CM led to decreased migration of GBM cells, in line with the result obtained in breast and prostate cancer cells (37, 44) and in contrast with data obtained in osteosarcoma (45).
Previous reports indicated that eA1-Fc suppressed signaling through the Ras-MAPK pathway (28, 36–38). In contrast, other reports indicate that ERK1/2 activity is actually increased after treatment with eA1-Fc (44, 45). The treatment of U-251 MG cells with eA1-P113A, -T115A, -G117A, and -E122A CM decreased phosphorylation of extracellular signal-regulated kinase beyond to what was seen with eA1-WT CM. Of particular interest is the fact that this effect was sustained for an extended period of time, as decreased p-ERK was observed at 8 h of post-treatment in cells treated with these four mutants.
AKT activation in the majority of GBM has been reported to result from PTEN inactivation, receptor-tyrosine kinase activation, or AKT amplification (46, 47). Recent reports also indicate the existence of a negative regulatory loop between ligand-activated EphA2 and AKT (39, 48). In accordance with these reports, treatment of U-251 MG cells with eA1-WT and mutant eA1 CM corresponding to the critical residues in the G-H loop led to a significant reduction in AKT phosphorylation. The effect of the four eA1 mutant CM on the p-AKT levels was greater than that of the eA1-WT. The results further support the importance of eA1 signaling axis and its involvement in GBM progression/maintenance.
Our previous work had determined that EphA2 represents a promising target for new therapeutics to combat GBM (21). The construction of a targeted cytotoxin wherein eA1-Fc was conjugated to the truncated form of Pseudomonas exotoxin A yielded a highly potent and specific agent that kills GBM, breast, and prostate cancer cells at very low concentrations (29). The results of the work presented here should prove helpful in further rational design of these and similar cytotoxins. Being that eA1 is a tumor-suppressing factor in GBM by itself, finding an optimal variant(s) will have an important impact on the design and production of novel anti-tumor agents.
Supplementary Material
Acknowledgment
We thank Dr. Young A. Choi for help with experiments involving recombinant eA1s.
This work was supported, in whole or in part, by National Institutes of Health Grant CA 74145 (NCI; to W. D.). This work was supported by the Brain Tumor Center of Excellence (to W. D.) and Institutional Development Grant 2006-IDG-1004 from the North Carolina Biotechnology Center (to R. H.).

This article contains supplemental Table S1 and Figs. S1–S4.
- eA1
- ephrinA1
- GBM
- glioblastoma multiforme
- CM
- conditioned media
- RU
- response units.
REFERENCES
- 1. Merlos-Suárez A., Batlle E. (2008) Eph-ephrin signaling in adult tissues and cancer. Curr Opin Cell Biol. 20, 194–200 [DOI] [PubMed] [Google Scholar]
- 2. Pasquale E. B. (2005) Eph receptor signaling casts a wide net on cell behavior. Nat. Rev. Mol. Cell Biol. 6, 462–475 [DOI] [PubMed] [Google Scholar]
- 3. Pasquale E. B. (2008) Eph-ephrin bidirectional signaling in physiology and disease. Cell 133, 38–52 [DOI] [PubMed] [Google Scholar]
- 4. Drescher U., Kremoser C., Handwerker C., Löschinger J., Noda M., Bonhoeffer F. (1995) In vitro guidance of retinal ganglion cell axons by RAGS, a 25-kDa tectal protein related to ligands for Eph receptor-tyrosine kinases. Cell 82, 359–370 [DOI] [PubMed] [Google Scholar]
- 5. Knöll B., Drescher U. (2002) Ephrin-As as receptors in topographic projections. Trends Neurosci 25, 145–149 [DOI] [PubMed] [Google Scholar]
- 6. Lai K. O., Ip N. Y. (2009) Synapse development and plasticity. Roles of ephrin/Eph receptor signaling. Curr. Opin. Neurobiol. 19, 275–283 [DOI] [PubMed] [Google Scholar]
- 7. Nakamoto M., Cheng H. J., Friedman G. C., McLaughlin T., Hansen M. J., Yoon C. H., O'Leary D. D., Flanagan J. G. (1996) Topographically specific effects of ELF-1 on retinal axon guidance in vitro and retinal axon mapping in vivo. Cell 86, 755–766 [DOI] [PubMed] [Google Scholar]
- 8. Surawska H., Ma P. C., Salgia R. (2004) The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 15, 419–433 [DOI] [PubMed] [Google Scholar]
- 9. Wimmer-Kleikamp S. H., Lackmann M. (2005) Eph-modulated cell morphology, adhesion, and motility in carcinogenesis. IUBMB Life 57, 421–431 [DOI] [PubMed] [Google Scholar]
- 10. Beauchamp A., Debinski W. (2012) Ephs and ephrins in cancer. Ephrin-A1 signaling. Semin. Cell Dev. Biol. 23, 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ireton R. C., Chen J. (2005) EphA2 receptor-tyrosine kinase as a promising target for cancer therapeutics. Curr. Cancer Drug Targets 5, 149–157 [DOI] [PubMed] [Google Scholar]
- 12. Kinch M. S., Carles-Kinch K. (2003) Overexpression and functional alterations of the EphA2-tyrosine kinase in cancer. Clin. Exp. Metastasis 20, 59–68 [DOI] [PubMed] [Google Scholar]
- 13. Walker-Daniels J., Hess A. R., Hendrix M. J., Kinch M. S. (2003) Differential regulation of EphA2 in normal and malignant cells. Am. J. Pathol. 162, 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Q., Suo Z., Risberg B., Karlsson M. G., Villman K., Nesland J. M. (2004) Expression of Ephb2 and Ephb4 in breast carcinoma. Pathol. Oncol. Res. 10, 26–33 [DOI] [PubMed] [Google Scholar]
- 15. Zelinski D. P., Zantek N. D., Stewart J. C., Irizarry A. R., Kinch M. S. (2001) EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 61, 2301–2306 [PubMed] [Google Scholar]
- 16. Mudali S. V., Fu B., Lakkur S. S., Luo M., Embuscado E. E., Iacobuzio-Donahue C. A. (2006) Patterns of EphA2 protein expression in primary and metastatic pancreatic carcinoma and correlation with genetic status. Clin. Exp. Metastasis 23, 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura R., Kataoka H., Sato N., Kanamori M., Ihara M., Igarashi H., Ravshanov S., Wang Y. J., Li Z. Y., Shimamura T., Kobayashi T., Konno H., Shinmura K., Tanaka M., Sugimura H. (2005) EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci. 96, 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kataoka H., Igarashi H., Kanamori M., Ihara M., Wang J. D., Wang Y. J., Li Z. Y., Shimamura T., Kobayashi T., Maruyama K., Nakamura T., Arai H., Kajimura M., Hanai H., Tanaka M., Sugimura H. (2004) Correlation of EPHA2 overexpression with high microvessel count in human primary colorectal cancer. Cancer Sci. 95, 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng G., Hu Z., Kinch M. S., Pan C. X., Flockhart D. A., Kao C., Gardner T. A., Zhang S., Li L., Baldridge L. A., Koch M. O., Ulbright T. M., Eble J. N., Cheng L. (2003) High level expression of EphA2 receptor-tyrosine kinase in prostatic intraepithelial neoplasia. Am. J. Pathol. 163, 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wykosky J., Debinski W. (2008) The EphA2 receptor and ephrinA1 ligand in solid tumors. Function and therapeutic targeting. Mol. Cancer Res. 6, 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wykosky J., Gibo D. M., Stanton C., Debinski W. (2005) EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol. Cancer Res. 3, 541–551 [DOI] [PubMed] [Google Scholar]
- 22. Qin H., Noberini R., Huan X., Shi J., Pasquale E. B., Song J. (2010) Structural characterization of the EphA4-Ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity. J. Biol. Chem. 285, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Himanen J. P., Chumley M. J., Lackmann M., Li C., Barton W. A., Jeffrey P. D., Vearing C., Geleick D., Feldheim D. A., Boyd A. W., Henkemeyer M., Nikolov D. B. (2004) Repelling class discrimination. Ephrin-A5 binds to and activates EphB2 receptor signaling. Nat. Neurosci. 7, 501–509 [DOI] [PubMed] [Google Scholar]
- 24. Pasquale E. B. (2010) Eph receptors and ephrins in cancer. Bidirectional signaling and beyond. Nat. Rev. Cancer 10, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Wang Y., Wang Y., Zhen H., Yang H., Fei Z., Zhang J., Liu W., Wang Y., Zhang X. (2007) Expression of EphA2 in human astrocytic tumors. Correlation with pathologic grade, proliferation, and apoptosis. Tumour Biol. 28, 165–172 [DOI] [PubMed] [Google Scholar]
- 26. Liu F., Park P. J., Lai W., Maher E., Chakravarti A., Durso L., Jiang X., Yu Y., Brosius A., Thomas M., Chin L., Brennan C., DePinho R. A., Kohane I., Carroll R. S., Black P. M., Johnson M. D. (2006) A genome-wide screen reveals functional gene clusters in the cancer genome and identifies EphA2 as a mitogen in glioblastoma. Cancer Res. 66, 10815–10823 [DOI] [PubMed] [Google Scholar]
- 27. Wang L. F., Fokas E., Bieker M., Rose F., Rexin P., Zhu Y., Pagenstecher A., Engenhart-Cabillic R., An H. X. (2008) Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol Rep 19, 151–156 [PubMed] [Google Scholar]
- 28. Wykosky J., Palma E., Gibo D. M., Ringler S., Turner C. P., Debinski W. (2008) Soluble monomeric EphrinA1 is released from tumor cells and is a functional ligand for the EphA2 receptor. Oncogene 27, 7260–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wykosky J., Gibo D. M., Debinski W. (2007) A novel, potent, and specific ephrinA1-based cytotoxin against EphA2 receptor-expressing tumor cells. Mol. Cancer Ther. 6, 3208–3218 [DOI] [PubMed] [Google Scholar]
- 30. Qin H., Lim L., Song J. (2012) Protein dynamics at Eph receptor-ligand interfaces as revealed by crystallography, NMR and MD simulations. BMC Biophys. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Himanen J. P., Goldgur Y., Miao H., Myshkin E., Guo H., Buck M., Nguyen M., Rajashankar K. R., Wang B., Nikolov D. B. (2009) Ligand recognition by A-class Eph receptors. Crystal structures of the EphA2 ligand binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 10, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Himanen J. P., Yermekbayeva L., Janes P. W., Walker J. R., Xu K., Atapattu L., Rajashankar K. R., Mensinga A., Lackmann M., Nikolov D. B., Dhe-Paganon S. (2010) Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. U.S.A. 107, 10860–10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hantgan R. R., Stahle M. C., Lord S. T. (2010) Dynamic regulation of fibrinogen. Integrin αIIbβ3 binding. Biochemistry 49, 9217–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hantgan R. R., Stahle M. C. (2009) Integrin priming dynamics. Mechanisms of integrin antagonist-promoted αIIbβ3:PAC-1 molecular recognition. Biochemistry 48, 8355–8365 [DOI] [PubMed] [Google Scholar]
- 35. Miao H., Burnett E., Kinch M., Simon E., Wang B. (2000) Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2, 62–69 [DOI] [PubMed] [Google Scholar]
- 36. Guo H., Miao H., Gerber L., Singh J., Denning M. F., Gilliam A. C., Wang B. (2006) Disruption of EphA2 receptor-tyrosine kinase leads to increased susceptibility to carcinogenesis in mouse skin. Cancer Res. 66, 7050–7058 [DOI] [PubMed] [Google Scholar]
- 37. Macrae M., Neve R. M., Rodriguez-Viciana P., Haqq C., Yeh J., Chen C., Gray J. W., McCormick F. (2005) A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell 8, 111–118 [DOI] [PubMed] [Google Scholar]
- 38. Miao H., Wei B. R., Peehl D. M., Li Q., Alexandrou T., Schelling J. R., Rhim J. S., Sedor J. R., Burnett E., Wang B. (2001) Activation of EphA receptor-tyrosine kinase inhibits the Ras/MAPK pathway. Nat. Cell Biol. 3, 527–530 [DOI] [PubMed] [Google Scholar]
- 39. Miao H., Li D. Q., Mukherjee A., Guo H., Petty A., Cutter J., Basilion J. P., Sedor J., Wu J., Danielpour D., Sloan A. E., Cohen M. L., Wang B. (2009) EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 16, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duxbury M. S., Ito H., Zinner M. J., Ashley S. W., Whang E. E. (2004) Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem. Biophys. Res. Commun. 320, 1096–1102 [DOI] [PubMed] [Google Scholar]
- 41. Shao H., Pandey A., O'Shea K. S., Seldin M., Dixit V. M. (1995) Characterization of B61, the ligand for the Eck receptor protein-tyrosine kinase. J. Biol. Chem. 270, 5636–5641 [DOI] [PubMed] [Google Scholar]
- 42. Walker-Daniels J., Riese D. J., 2nd, Kinch M. S. (2002) c-Cbl-dependent EphA2 protein degradation is induced by ligand binding. Mol. Cancer Res. 1, 79–87 [PubMed] [Google Scholar]
- 43. Parri M., Taddei M. L., Bianchini F., Calorini L., Chiarugi P. (2009) EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 69, 2072–2081 [DOI] [PubMed] [Google Scholar]
- 44. Pratt R. L., Kinch M. S. (2002) Activation of the EphA2-tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene 21, 7690–7699 [DOI] [PubMed] [Google Scholar]
- 45. Fritsche-Guenther R., Noske A., Ungethüm U., Kuban R. J., Schlag P. M., Tunn P. U., Karle J., Krenn V., Dietel M., Sers C. (2010) De novo expression of EphA2 in osteosarcoma modulates activation of the mitogenic signaling pathway. Histopathology 57, 836–850 [DOI] [PubMed] [Google Scholar]
- 46. Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A., Jr., Hartigan J., Smith D. R., Strausberg R. L., Marie S. K., Shinjo S. M., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang N. Y., Fernandez C., Richter M., Xiao Z., Valencia F., Tice D. A., Pasquale E. B. (2011) Cross-talk of the EphA2 receptor with a serine/threonine phosphatase suppresses the Akt-mTORC1 pathway in cancer cells. Cell. Signal. 23, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.