Background: The glypican family of cell-surface proteoglycans regulates growth factor signaling during development through their core proteins and heparan sulfate chains.
Results: The crystal structure of N-glycosylated human glypican-1 is described.
Conclusion: The structure reveals the complete disulfide bond arrangement for the conserved Cys residues in glypicans.
Significance: Increased structural knowledge of glypicans will help elucidate their important functions in shaping animal development.
Keywords: Cell Surface Receptor, Crystal Structure, Disulfide, Glycobiology, Protein Structure, Proteoglycan Structure, Glypican
Abstract
Glypicans are a family of cell-surface proteoglycans that regulate Wnt, hedgehog, bone morphogenetic protein, and fibroblast growth factor signaling. Loss-of-function mutations in glypican core proteins and in glycosaminoglycan-synthesizing enzymes have revealed that glypican core proteins and their glycosaminoglycan chains are important in shaping animal development. Glypican core proteins consist of a stable α-helical domain containing 14 conserved Cys residues followed by a glycosaminoglycan attachment domain that becomes exclusively substituted with heparan sulfate (HS) and presumably adopts a random coil conformation. Removal of the α-helical domain results in almost exclusive addition of the glycosaminoglycan chondroitin sulfate, suggesting that factors in the α-helical domain promote assembly of HS. Glypican-1 is involved in brain development and is one of six members of the vertebrate family of glypicans. We expressed and crystallized N-glycosylated human glypican-1 lacking HS and N-glycosylated glypican-1 lacking the HS attachment domain. The crystal structure of glypican-1 was solved using crystals of selenomethionine-labeled glypican-1 core protein lacking the HS domain. No additional electron density was observed for crystals of glypican-1 containing the HS attachment domain, and CD spectra of the two protein species were highly similar. The crystal structure of N-glycosylated human glypican-1 core protein at 2.5 Å, the first crystal structure of a vertebrate glypican, reveals the complete disulfide bond arrangement of the conserved Cys residues, and it also extends the structural knowledge of glypicans for one α-helix and two long loops. Importantly, the loops are evolutionarily conserved in vertebrate glypican-1, and one of them is involved in glycosaminoglycan class determination.
Introduction
Proteoglycans (PGs)2 consist of a core protein covalently substituted with glycosaminoglycans (GAGs), long unbranched polysaccharide chains. Several genetic diseases in humans and numerous genetic experiments in fruit flies, nematodes, and mice have revealed that GAGs and their core proteins are important in shaping animal development (1, 2). Glypicans are a family of cell-surface PGs substituted with the GAG heparan sulfate (HS). Glypicans regulate growth factor signaling by binding of growth factors to their HS chains or, as has been shown recently, by binding of growth factors to the core proteins. Based on sequence comparisons, glypicans fall into two subfamilies: glypican-1, -2, -4, and -6 and glypican-3 and -5, with ∼25% amino acid identity between the groups. Glypicans are around 550 amino acids in length and all share an N-terminal secretory signal peptide, 14 evolutionarily conserved Cys residues, an HS attachment domain near the C terminus, and a hydrophobic domain for addition of the glycosylphosphatidylinositol (GPI) anchor at the C terminus. Aside from the six members of the glypican family in vertebrates, there are also two glypicans in Drosophila melanogaster (Dally and Dally-like protein) and two in Caenorhabditis elegans (Gpn-1 and Lon-2). Dally-like protein (DLP) is divergent from the other glypicans, in particular by the insertion of 100 amino acids N-terminal to the last two conserved Cys residues (supplemental Fig. S1). Loss-of-function mutations in GPC3 cause Simpson-Golabi-Behmel syndrome, an X-linked condition characterized by prenatal and postnatal overgrowth (3). Mutations in GPC6 cause omodysplasia-1, a congenital disease affecting the skeletal system (4). Clinical features of omodysplasia-1 involve short stature and facial dysmorphism. Also, common variation in the GPC5 gene is associated with acquired nephrotic syndrome (5). GPC1 is expressed in the developing and adult brain, and GPC1 knock-out mice have reduced brain size at birth (6).
The structurally related but functionally distinct GAGs HS and chondroitin sulfate (CS) are added to core proteins that contain a GAG attachment motif consisting of the dipeptide Ser-Gly. Synthesis is initiated by the addition of a xylose residue to the hydroxyl group of the Ser residue in the GAG attachment domain, followed by sequential addition of two galactose (Gal) and one glucuronic acid (GlcA) residue. This forms the linker region common for HS and CS, GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser. Synthesis of HS is initiated by the addition of an N-acetylglucosamine (GlcNAc) residue, whereas CS synthesis begins with the addition of an N-acetylgalactosamine (GalNAc). After the addition of the GlcNAc or GalNAc residues, HS and CS chains are polymerized, forming their respective disaccharide backbones. Exactly what determines whether a GlcNAc residue or a GalNAc is added to the common linker region is not known. It has been suggested that HS assembly is promoted by the repetition of the Ser-Gly dipeptide and by flanking acidic residues (7, 8). Glypicans are substituted with HS and contain GAG attachment domains similar to those suggested to promote HS assembly. However, expression of the rat GPC1 GAG attachment domain alone results in substitution with ∼90% CS (9), strongly suggesting that factors in the glypican core protein further away from the GAG attachment sites promote the assembly of HS.
Recently, it has been shown that functional aspects of glypicans also reside in the protein cores. For example, Hedgehog growth factor signaling is deficient in D. melanogaster animals when the D. melanogaster glypican Dlp has been knocked down, and the Hedgehog response can be rescued by expression of DLP without the HS chains (10, 11). Moreover, other reports have shown that glypican core proteins bind directly to growth factors such as Hedgehog (12), bone morphogenetic protein 4 (13), decapentaplegic, Wnt (14, 15), and insulin-like growth factor II (16) with significant biological function independent of the presence of HS chains.
We have previously shown that anchorless recombinant human GPC1 (rhGPC1) is a stable α-helical protein that resists high concentrations of the chemical denaturants urea and guanidine-HCl (17). Unfolding occurs in a single step, and unfolding data can be closely fitted by a two-state model, indicating that GPC1 is a densely packed globular protein. Recently, the core protein structure of DLP was published, revealing an extended α-helical fold for glypican core proteins (18). Here, we present the independently solved structure of GPC1 core protein using multiple-wavelength anomalous dispersion phasing on selenomethionine (SeMet)-labeled rhGPC1, the first crystal structure of a vertebrate glypican. Structural alignment of GPC1 and DLP reveals a common fold for glypican core proteins. The GPC1 structure presented here also expands our knowledge of glypican core proteins, in particular for the disulfide bond arrangement of the conserved Cys residues, one α-helix and two long loops. One of the loops has been shown to be involved in GAG class determination, but lack of structural data has previously hampered further mutagenic investigations.
EXPERIMENTAL PROCEDURES
Materials
Human GPC1 cDNA clone IMAGE ID 6275649 was obtained from Geneservice (UK). The expression vector pCEP-BM40-HisTEV and 293 human embryonic kidney (HEK293) cells were obtained as described previously (17). All restriction enzymes used were from Fermentas (Canada). Nickel-nitrilotriacetic acid columns were from GE Healthcare. Protein-free medium for CHO cells and Coomassie Blue were from Sigma. Lipofectamine, 4–12% BisTris SDS-polyacrylamide gels, l-methionine-free Dulbecco's modified Eagle's medium, dialyzed fetal bovine serum (FBS), and hygromycin B were from Invitrogen. l-Selenomethionine was from Acros Organics (Belgium). Crystallization plates and polyethylene glycol Mr 6000 (PEG 6000) were obtained from Hampton Research. Other materials related to crystallography were purchased from Molecular Dimensions (UK).
Cloning and Purification of Human GPC1
The cDNA sequence coding for the mature form of human GPC1, amino acids 24–529, i.e. without the sequences for the N- and C-terminal signal peptides, was amplified and introduced into the vector pCEP4-BM40-HisTEV as described previously (17). This vector contains the sequence for an N-terminal BM40 secretion peptide followed by a His6 tag and a tobacco etch virus protease cleavage site. Furthermore, the HS attachment sites were disrupted by site-directed mutagenesis to create a vector expressing GPC1 ΔHS (17). A vector coding for GPC1 ΔC was created by amplifying the sequence for amino acids 24–479 using the forward primer 5′-TTAAGCTTGACCCGGCCAGCAAGAG-3′ and the reverse primer 5′-TTGGATCCTTAGAAGTCCACGTCGTTGCCGTT-3′ and by introducing the PCR product into the pCEP4-BM40-HisTEV plasmid. Restriction enzyme sites are shown in boldface. All constructs were verified by sequencing at Eurofins MWG Operon (Germany). Stable HEK 293 cell clones expressing GPC1 ΔHS and GPC1 ΔC were generated by limiting dilution after several weeks of growth in selective antibiotics. Stable cell clones expressing large amounts of rhGPC1 were grown to confluence in minimal essential medium with Earle's salts supplemented with 10% (v/v) FBS, 2 mm l-glutamine, penicillin (100 units/ml), streptomycin (100 μg/ml), and hygromycin B (200 μg/ml). After extensive washing with PBS, the rhGPC1-expressing cells were maintained for 3 or 4 days in protein-free medium supplemented with 2 mm l-glutamine, penicillin (100 units/ml), streptomycin (100 μg/ml), and hygromycin B (100 μg/ml). Conditioned medium was stored frozen, and rhGPC1 was purified from conditioned medium of ∼200-ml batches using nickel affinity chromatography. Protein purification was analyzed by SDS-PAGE, and purified rhGPC1 was dialyzed to 20 mm Tris-HCl, pH 8.0, concentrated by ultrafiltration, and stored frozen. SeMet labeling of GPC1 ΔC was performed in l-methionine-free Dulbecco's modified Eagle's medium supplemented with 2% dialyzed FBS. After a 6-h preincubation with l-methionine-free medium, the medium was replaced with medium containing 30 mg/liter l-selenomethionine. Cells suffered from SeMet toxicity, which made it difficult to grow the cells in medium supplemented with SeMet for more than 24 h. SeMet-labeled GPC1 ΔC was purified in the presence of 0.1 mm tris(2-carboxyethyl)phosphine hydrochloride to avoid oxidation of SeMet residues (19).
CD Spectroscopy
CD spectra were recorded using a Jasco J-810 spectropolarimeter equipped with a Peltier thermostated cell holder. The cuvette was thermostated at 20 °C. For far-UV CD, a 1-mm quartz cuvette was used, and the measurement range was 190–250 nm. The following parameters were used: sensitivity, 100 millidegrees (mdeg); data pitch, 1 nm; scan rate, 100 nm/min; response time, 4 s; bandwidth, 1 nm; and accumulation, 2. Near-UV CD was measured in a 10-mm quartz cuvette, and the measurement range was 250–340 nm. The following parameters were used: sensitivity, 100 mdeg; data pitch, 0.2 nm; scan rate, 50 nm/min; response time, 4 s; bandwidth, 0.5 nm; and accumulation, 4. Base-line spectra were recorded for pure buffer and subtracted from the corresponding protein spectra. Stock solutions of rhGPC1 were clarified by centrifugation at 15,000 × g for 15 min, and protein concentration was determined by measuring the absorbance at 280 nm using a micro-volume spectrophotometer (GE Healthcare) and the molar extinction coefficient for bovine serum albumin. Samples were prepared by diluting the rhGPC1 stock solution to 0.1 mg/ml for far-UV CD and to 1 mg/ml for near-UV CD. Protein was maintained in 20 mm sodium phosphate buffer, pH 7.4. Mean residue ellipticity was calculated using 110.5 and 112.0 as the mean residue molecular weights of GPC1 ΔHS and GPC1 ΔC, respectively.
Crystallization
Initial high throughput crystallization screens performed with GPC1 ΔHS at a protein concentration of 40 mg/ml were set up in MRC plates using a Mosquito nanoliter pipetting robot (TTP Labtech, UK) at the MAX-lab Crystallization Facility, Lund University. The initial screens, PACT premier (Molecular Dimensions), Crystal Screen 1 and 2 (Hampton Research), and JCSG+ (Qiagen GmbH, Germany), were set up using the vapor diffusion technique in sitting drops where 100 nl of protein solution were mixed with 100 nl of reservoir solution and equilibrated against 80 μl of reservoir solution. After 1 day, needle clusters began to appear under conditions containing polyethylene glycol Mr = 3350 (PEG 3350) with anions such as Br−, F−, and NO3−, pH 6.5. After 1 week, crystal plates appeared in drops with 20% PEG 6000, 0.2 m CaCl2 in 0.1 m Tris-HCl, pH 8.0. After 2 weeks, the same appeared in drops with 20% PEG 6000, 0.2 m NH4Cl in 0.1 m Tris-HCl, pH 8.0. Crystal-like material also appeared in 20% PEG 8000, 0.2 m magnesium acetate, 0.1 m sodium cacodylate trihydrate, pH 6.5, after 2 weeks. Optimization screens were produced from stock solutions using a liquid handling pipetting robot (Tecan Group Ltd., Switzerland) and set up in MRC plates using the nanoliter pipetting robot as for the initial screens. The best results were obtained with the crystals grown in CaCl2 with PEG 6000 as precipitating agent. The optimized conditions were scaled up to microliter drops in 24-well plates using the sitting drop vapor diffusion method at 20 °C with 500 μl of reservoir solution. Typically, crystals of rhGPC1 were grown by mixing 2 μl of protein solution with 2 μl of reservoir solution containing 12–14% PEG 6000, 0.1 m Tris-HCl, 0.2 m CaCl2, pH 8.0. GPC1 ΔC was crystallized at 20 mg/ml. GPC1 ΔHS was crystallized at a higher protein concentration, up to 40 mg/ml, and benefited from low concentrations of dithiothreitol in the protein solution. In general, crystallization of GPC1 ΔC was more easily reproduced in comparison to crystallization of GPC1 ΔHS. Crystallization of SeMet-labeled GPC1 ΔC was also performed using the same conditions but with lower protein concentration, and the protein was maintained in 20 mm Tris-HCl, pH 8.0, supplemented with 0.1 mm tris(2-carboxyethyl)phosphine hydrochloride. Large but thin plate-like crystals with dimensions around 0.6 × 0.3 mm formed within 1 week for GPC1 ΔHS and GPC1 ΔC. Crystals of SeMet-labeled GPC1 ΔC were typically smaller, around 0.2 × 0.2 mm. Crystallization of rhGPC1 was not affected by the presence of the His6 tag.
Data Collection and Processing
Crystals were soaked in 15% ethylene glycol, 18% PEG 6000, 0.1 m Tris-HCl, 0.2 m CaCl2, pH 8.0, and flash-frozen under a stream of liquid nitrogen. X-ray diffraction data were collected on stations I911-2 and I911-3 of the MAX-II synchrotron radiation source, Lund, Sweden, on 165- and 225-mm CCD detectors from MarResearch (Germany). All data sets were integrated and scaled using the XDS suite (20) and further processed using programs from the CCP4 (21) and Phenix (22) suites. Data collection statistics are given in Table 1. Diffraction patterns of GPC1 ΔHS crystals could be indexed in space group P21 and had unit cell dimensions a = 47.2 Å, b = 169.0 Å, c = 151.6 Å, β = 95.0°. Variation in cell dimensions was seen between different crystals, in particular for the c dimension, with lengths ranging from 148 to 154 Å. The crystals contain four GPC1 molecules in the asymmetric unit, giving a Matthews volume of 2.7 Å3/Da and a solvent content of 54%. Crystals of GPC1 ΔC and SeMet GPC1 ΔC also belong to space group P21 with similar unit cell dimensions. The diffraction anisotropy of the data from GPC1 ΔC crystals was analyzed using phenix.xtriage (22) and found to be significant. The B factor was twice as large in the c* direction as in the a* and b* directions. This effectively limited the resolution to around 2.9 Å in the c* direction. However, refinement against data truncated using the UCLA anisotropy server (23) did not result in improved maps, so the original data were retained. The data also show a significant translational noncrystallographic symmetry, with a peak at (0.5, 0.021, 0.5) in the native Patterson of 35% of the height of the origin peak. This translation relates chains A and B to C and D in the asymmetric unit, respectively.
TABLE 1.
Data collection and refinement statistics
The Rfree values were calculated using 5% of the unique data (3753 reflections for the truncated form and 2529 for the full-length protein).
| Native GPC1 ΔC | SeMet GPC1 ΔC (peak) | SeMet GPC1 ΔC (inflection) | Native GPC1 ΔHS | |
|---|---|---|---|---|
| Space group | P21 | P21 | P21 | P21 |
| Unit cell parameters (Å, °) | a = 47.2, b = 168.7, c = 147.8, β = 94.6 | a = 47.1, b = 168.5, c = 147.0, β = 94.6 | a = 47.2, b = 168.6, c = 147.4, β = 94.6 | a = 47.2, b = 169.0 c = 151.6, β = 95.0 |
| Beamline | I911-2 | I911-3 | I911-3 | I911-2 |
| Wavelength | 1.0397 Å | 0.9789 Å | 0.9794 Å | 1.0397 Å |
| Resolution range | 30 to 2.49 Å (2.64 to 2.49 Å) | 30 to 3.3 Å (3.39 to 3.30 Å) | 30 to 3.3 Å (3.39 to 3.30 Å) | 30 to 2.95 Å (3.03 to 2.95 Å) |
| Rmerge (I) | 8.3% (68.3%) | 10.7% (33.0%) | 12.9% (51.1%) | 13.5% (83.2%) |
| Total observations | 328,111 | 347,933 | 381,017 | 440,496 |
| Unique reflections | 79,825 | 66,568 | 66,846 | 49,537 |
| Average redundancy | 4.1 (3.5) | 5.2 (5.3) | 5.7 (5.6) | 8.9 (8.2) |
| Completeness | 98.9% (94.9%) | 98.2% (98.0%) | 98.0% (95.4%) | 99.4% (94.1%) |
| I/σ(I) | 11.6 (2.37) | 11.3 (4.47) | 10.85 (3.29) | 15.4 (3.4) |
| Refinement | ||||
| Rwork/Rfree | 25.3/29.2% | 22.1/27.9% | ||
| No. of atoms | ||||
| Protein/carbohydrate | 12,572 | 12,630 | ||
| Water | 242 | 126 | ||
| Root mean square deviations from ideal geometry | ||||
| Bond lengths | 0.003 Å | 0.004 Å | ||
| Bond angles | 0.72° | 0.84° | ||
| Ramachandran analysis | ||||
| In preferred regions | 98.6% | 98.2% | ||
| In allowed regions | 1.4% | 1.8% | ||
| Outliers | 0% | 0% | ||
Structure Determination and Refinement
X-ray data were collected to 3.3 Å resolution from SeMet GPC1 ΔC crystals at selenium peak and inflection wavelengths at station I911-3 of MAX-II. The processed data sets were sent to the AutoRickshaw server at the European Molecular Biology Laboratory in Hamburg, Germany (24, 25), for structure determination using the two-wavelength multiple anomalous dispersion method. In AutoRickshaw, the data sets were prepared using the CCP4 programs CAD and SCALEIT (21, 26). Furthermore, heavy atoms were found using SHELXD (27), and heavy atom refinement and phase calculation were performed using SHARP (28) and MLPHARE (21). Initial density modification and phase extension were performed using DM (29) and further explored using RESOLVE (30). Finally, the electron density map was interpreted using HELICAP (31), and model building was performed by BUCCANEER (32). This resulted in a more than 90% complete model built at 3.8 Å that was partially refined by CNS (33) and Refmac5 (34) at 3.3 Å (Rmodel = 23% and Rfree = 32%). The selenium positions were confirmed by calculating an anomalous difference map at peak wavelength using phases from a partially refined model. The initial structure was improved by extensive manual rebuilding in Coot (35) followed by many rounds of refinement using the diffraction data to 2.55 Å from a native GPC1 ΔC crystal. Phenix.refine (22) with Ramachandran restraints was used for the final model refinement, and Molprobity (36) was used for validation. Structural alignments were made using the secondary structure matching algorithm as implemented in Coot. All graphical representations of the GPC1 structure were generated using the PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger.
Structural Conservation Analysis
Alignments of glypican amino acid sequences were performed using ClustalW2 (37). The amino acid sequences used are given in Fig. 5 figure legend. DNA sequences were translated using the ExPASy Translate Tool. Structural conservation analysis was performed using the ConSurf server (38).
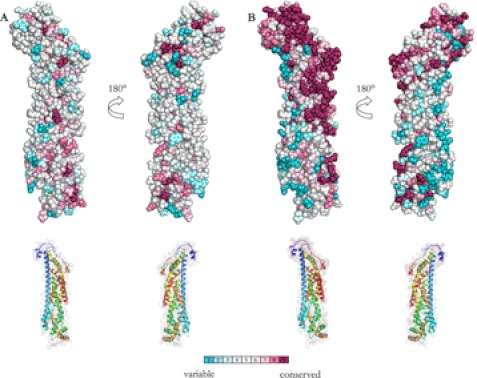
FIGURE 5.
Structural conservation of surface-exposed amino acids in GPC1 homologs (A) and in GPC1 orthologs (B). Multiple sequence alignments were performed using ClustalW (37), and conservation scores were calculated with the Bayesian method using the ConSurf server (38). Images of structural conservation of surface-exposed amino acids were generated using Consurf output in combination with PyMOL. Amino acid conservation is shown in colors from turquoise (variable) to purple (conserved). The inlaid smaller figures show the structural conservation space fill model in transparency aligned with a ribbon representation of the structure. A, multiple sequence alignment and amino acid conservation were calculated using GPC1 homologs. The following amino acid sequences were used (UniProt accession numbers): GPC1 human, P35052; GPC2 human, Q8N158; GPC3 human, P51654; GPC4 human O75487; GPC5 human, P78333; GPC6 human, Q9Y625; Dally D. melanogaster, Q24114; DLP D. melanogaster, Q9GLP5. B, following amino acid sequences were used to visualize structural conservation in GPC1 orthologs (i.e. GPC1 vertebrate homologs): GPC1 human, P35052; GPC1 mouse, Q9QZF2; GPC1 rat, P35052; GPC1 bovine, Q2KJ65; GPC1 chick, P50593; Gpc1 zebrafish, Q1LXM6; gpc1 western clawed frog, Q0V9W0. Also included were the translated mRNA sequences (given by NBCI nucleotide accession numbers) for GPC1 genes: Gpc1 short-tailed opossums, XM_001377888; Gpc1 rhesus macaque, XM_001089255; Gpc1 common chimpanzee, XM_001156197.
RESULTS
Expression and Characterization of Anchorless GPC1
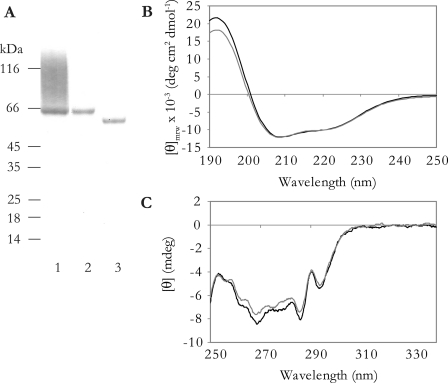
We expressed His6-tagged full-length rhGPC1 without the GPI anchor signal peptide in 293 human embryonic kidney cells. This secreted rhGPC1 (protein referred to as GPC1 WT) was purified under native conditions, and analysis on SDS-PAGE of the purified protein showed a core protein band of ∼60 kDa and a PG smear of ∼60–120 kDa (Fig. 1A, lane 1). Degradation with HS lyase and peptide:N-glycosidase F showed that this protein was substituted exclusively with HS and also contained N-linked glycans (17, 39). To produce rhGPC1 for conformational and structural studies, the HS attachment sites were disrupted by site-directed mutagenesis, resulting in expression of core protein containing N-linked glycans with a molecular mass of ∼60 kDa (protein referred to as GPC1 ΔHS, Fig. 1A, lane 2). Also, the HS attachment site in GPC1 is regarded, by secondary structure and disorder predictions, as adopting a random coil conformation despite the fact that it contains two Cys residues. Therefore, rhGPC1 was truncated C-terminally before the HS attachment domain, resulting in a core protein band (GPC1 ΔC) of smaller size (∼55 kDa) on SDS-PAGE (Fig. 1A, lane 3). We have previously shown that the PG form of GPC1 WT and GPC1 ΔHS is identical in folding, as judged by far-UV CD and fluorescence emission spectroscopy (17). To ensure that GPC1 ΔHS and GPC1 ΔC are also structurally similar, we subjected the two forms of rhGPC1 to far- and near-UV CD spectroscopy. Far-UV CD reports on the peptide bond conformation (i.e. secondary structure content) of the investigated protein and near-UV CD reports on the immediate environment of Trp, Tyr, and Phe residues (i.e. the tertiary structure). Both protein forms exhibited very similar far- and near-UV CD spectra with peaks and troughs located at identical wavelengths, indicating that removal of the HS attachment domain affects neither the secondary nor the tertiary structure of GPC1 (Fig. 1, B and C). Also, expression levels were similar for GPC1 ΔHS and GPC1 ΔC. Together, this suggests that the C-terminal HS attachment site in GPC1 does not contribute to the folding and that the two Cys residues in the HS attachment domain either form a disulfide bond together or are free.
FIGURE 1.
SDS-PAGE and spectroscopic analysis of rhGPC1. A, SDS-PAGE and Coomassie Blue staining of nickel-nitrilotriacetic acid-purified full-length rhGPC1 expressed without the GPI anchor signal peptide (lane 1). Expression of full-length rhGPC1 with disrupted HS attachment site (GPC1 ΔHS) resulted in expression of core protein only (lane 2). GPC1 with the C-terminal HS attachment site removed by truncation resulted in expression of core protein of smaller size (lane 3). Three μg of purified protein were added to the well of lane 1, and 1 μg was loaded in the wells of lanes 2 and 3. B and C, far- and near-UV-CD spectra, respectively, of GPC1 ΔHS (black, solid line) and GPC1 ΔC (gray, solid line) in 20 mm sodium phosphate, pH 7.4, were recorded at 20 °C.
Overall Three-dimensional Structure of GPC1 ΔC
GPC1 ΔHS and GPC1 ΔC both crystallized in space group P21 with four molecules in the asymmetric unit. Crystals of GPC1 ΔHS diffracted x-rays to 2.95 Å and crystals of GPC1 ΔC diffracted to 2.5 Å. Aside from the improved diffraction from GPC1 ΔC crystals, reproducibility of crystallization was greatly improved for GPC1 ΔC compared with GPC1 ΔHS. SeMet derivative crystals of GPC1 ΔC also crystallized in space group P21 with unit cell parameters similar to crystals of native GPC1 ΔC and diffracted to 3.3 Å. The structure of GPC1 ΔC was solved by the anomalous dispersion technique using the peak and inflection wavelengths for selenium. Data collection and processing statistics are presented in Table 1. The initial auto-built model was refined against a native data set diffracting to 2.5 Å. The final Rmodel and Rfree values are 25.3 and 29.2%, respectively. These somewhat high values can be explained partly by the significant anisotropy in the diffraction data. Analysis of the dataset for GPC1 ΔC using the UCLA Anisotropy Server (23) revealed strong anisotropy, with a difference of ∼40 Å2 between the B-factors for the a*/b* directions and the c* direction. This effectively restricted diffraction data to 2.9 Å in the c* direction. We refined the structure against ellipsoidally truncated data from the UCLA server, but this resulted in no improvement in model or map quality, so we reverted to refinement against the full dataset.
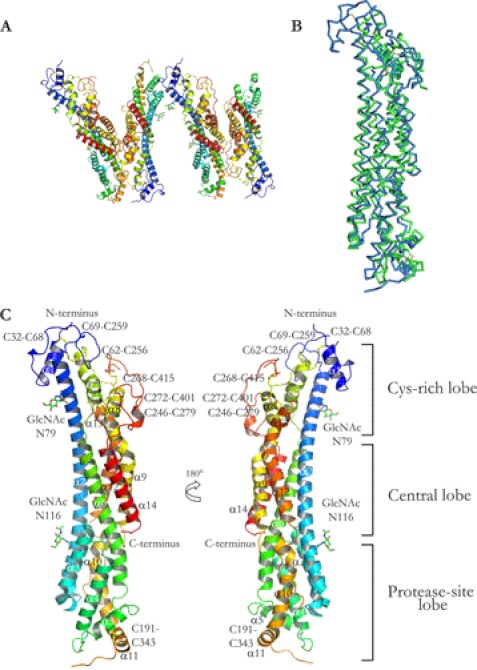
The final model encompasses residues Ser27–Asp475 of GPC1 ΔC and the first GlcNAc residues on Asn79 and Asn116. A major gap in the model occurs between residues 350 and 361 and a smaller one between 406 and 412. No electron density was observed for the His6 tag or the protease cleavage site. The asymmetric unit of the GPC1 ΔC consists of four monomers apparently organized as two dimers (Fig. 2A). The individual monomers in the dimer pairs are facing in opposite directions. Some variation in the quality of the electron density was seen for the different monomers. The most complete model was obtained with the monomer designated D; the other chains suffer from increased gap lengths. In particular, a major gap between residues 180 and 200 is seen in the other monomers. The missing amino acids in monomer D are charged residues present in extended loops. The highly extended GPC1 molecules pack in layers with extensive contacts in two dimensions but only limited contact in the third dimension (supplemental Fig. S2). This is consistent with the anisotropy observed in the diffraction datasets.
FIGURE 2.
Overview of GPC1 ΔC structure and comparison with DLP. A, overview of the asymmetric unit of GPC1 ΔC with two dimer pairs. B, ribbon representation of α-carbon alignment of GPC1 ΔC monomer (blue) and DLP (green, Protein Data Bank code 3ODN). Alignment was performed in PyMOL, version 1.2r3pre, Schrödinger, LLC. C, overview of the GPC1 ΔC structure represented as schematic in rainbow colors (N terminus in blue, C terminus in red) with loops 1–3 (L1–L3) and α-helices (α1–α14). Also indicated are the disulfide bonds (yellow solid lines), the first residues of the N-glycans (GlcNAc), and the assignment of different lobes in the GPC1 ΔC structure.
The structure of GPC1 is similar to the recently published structure of the D. melanogaster glypican DLP, with similarly positioned α-helices (Fig. 2B). The GPC1 structure thus reveals a cylindrical (dimensions 120 × 30 × 30 Å) all α-helical fold for glypican core proteins that is structurally conserved between D. melanogaster and humans, although GPC1 and DLP only share 25% sequence identity (as determined by FASTA (40)). The overlap between GPC1 and DLP structures is shown as α-carbon traces in Fig. 2B. The root mean square deviation in α-carbon positions between monomer D of GPC1 ΔC and DLP is 2.6 Å for 331 positions when superimposed by the secondary structure matching algorithm in Coot (35). The α-helical fold is also consistent with the far-UV CD spectra in Fig. 1B, displaying one peak at 195 nm and two troughs at 209 and 222 nm, which is typical for α-helical proteins. The GPC1 structure consists of 14 α-helices and three major loops, termed α1–α14 and L1–L3 (Fig. 2C). In comparison with the DLP structure, the GPC1 structure extends structural knowledge of glypicans, especially for the N-terminal loops (L1 and L3) and helix α1. The 60-residue, 83-Å-long helix α2 (71–130) spans the whole protein, with N-linked glycans near either end (on Asn79 and Asn116). Out of 14 evolutionarily conserved Cys residues in GPC1, 12 are located in a Cys-rich region near the N terminus. This region was termed the N-lobe (18) based on the observation that it contains the N terminus of the α-helix (α2 in GPC1) that spans the entire protein. We propose that this region should instead be termed the Cys-rich lobe (Fig. 2C), as the lobe is not entirely contained within the N-terminal part of the sequence. The Cys-rich lobe is followed by a middle segment containing five helices, α2, α6, α9, α12, and α14, forming the heart of the structure. This region is stabilized by two evolutionarily conserved hydrophobic centers that connect the α-helices and also contain the C terminus of GPC1 ΔC. This lobe was termed the M-lobe in Ref. 18. Based on the conserved hydrophobic centers in this lobe, we propose that this part of GPC1 should be called the central lobe. The last part of the GPC1 structure contains a long loop, L2, stretching from Cys343 to Glu376. This loop is processed by furin proteases in many of the glypican core proteins (41, 42); however, only minor processing of GPC1 occurs in our hands. Because of the disorder of the loop, residues 350–361 are not modeled. This lobe was termed the C-lobe in Ref. 18; an alternative name could be the protease site lobe.
Analysis of Crystal Contacts
The interface between pairs of GPC1 molecules in the crystal is located in the central lobe and involves α9 and α12 in both interacting monomers. Analysis of the crystal packing reveals at least two additional crystal contacts, one involving the end of α2 interacting with α14 in another monomer and the other involving the N-terminal parts of different monomers (supplemental Fig. 2). The interface involving α2 and α14 is the largest, with a buried surface area of ∼500 Å2. However, calculations of the stabilities of the interfaces using PISA software (43) indicate that the interactions are too weak to form oligomeric states of GPC1 in solution. Preliminary dynamic light scattering experiments confirm these calculations, with profiles exhibiting single peaks for both GPC1 ΔHS and GPC1 ΔC (data not shown).
Evolutionarily Conserved Cysteine Residues in Anchorless GPC1
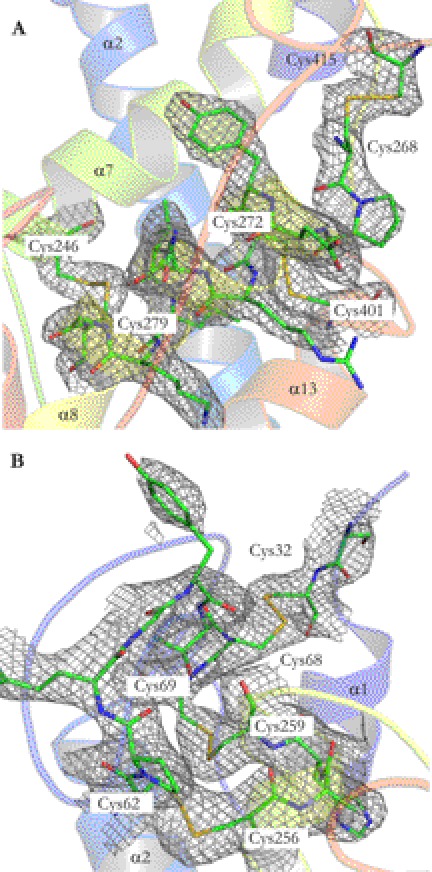
All of the 14 Cys residues evolutionarily conserved in the glypican family are clearly defined in the electron density and all are clearly involved in disulfide bonds. This gives a high degree of confidence to the presented model. The disulfide bonds and their connections are presented in Table 2, and a detailed view of the model and electron densities for the disulfide bonds is shown in Fig. 3. Six disulfide bonds connect the structure in the Cys-rich lobe. Disulfides Cys268–Cys415 and Cys272–Cys401 connect the structure between the furin-like convertase site, consistent with the two fragments that appear when running reducing SDS-PAGE on glypicans processed by furin-like convertases (41, 42). Together with Cys246–Cys279, this forms three disulfide bonds in or closely situated to α8 in the Cys-rich lobe (Fig. 3A). Cys32–Cys68, Cys62–Cys256, and Cys69–Cys259 form three longitudinally placed disulfide bonds in the N-terminal part of the Cys-rich lobe (Fig. 3B). All detailed pictures of the GPC1 structure are presented with the Cys-rich lobe facing upwards. The remaining disulfide bond is located in the protease lobe, being formed by Cys191–Cys343. The structure of DLP (18) lacked three out of seven disulfide bonds, because the electron density was not clear in the Cys-rich region. However, the four disulfide bonds present in the DLP structure are consistent with those presented here.
TABLE 2.
Disulfide bonds in GPC1
| Cysteine residues | Connected structural elements |
|---|---|
| Cys32–Cys68 | α1-L1 |
| Cys62–Cys256 | L1-α7 |
| Cys69–Cys259 | L1-α7 |
| Cys191–Cys343 | α5-α11 |
| Cys246–Cys279 | α7-α8 |
| Cys268–Cys415 | Turn between α7 and α8-L3 |
| Cys272–Cys401 | α8-α13 |
FIGURE 3.
Structure model and electron densities of the six disulfide bonds in the N-terminal Cys-rich lobe of GPC1. Red, oxygen; blue, nitrogen; green, carbon; yellow, sulfur. Model of the three disulfide bonds (Cys415–Cys268; Cys272–Cys401; Cys246–Cys279) in α8 in the Cys-rich region (A) and of the three longitudinally placed disulfide bonds in the L1 loop (Cys32–Cys68; Cys69–Cys259, and Cys62–Cys256) (B). Electron densities, contoured at 2.0 σ in A and 1.0 σ in B, were calculated as 2|Fo|−|Fc| maps using the program FFT in CCP4 (21, 55). Figures were generated using PyMOL.
GPC1 also contains two additional Cys residues in the HS attachment region, shared with GPC4 and GPC6. These Cys residues are not part of the presented GPC1 model and can either be free or form a disulfide bond. Crystallization of GPC1 ΔHS is facilitated by the addition of low concentrations of reducing agent, suggesting that the Cys residues are not bound together.
Other Evolutionarily Conserved Residues in Anchorless GPC1
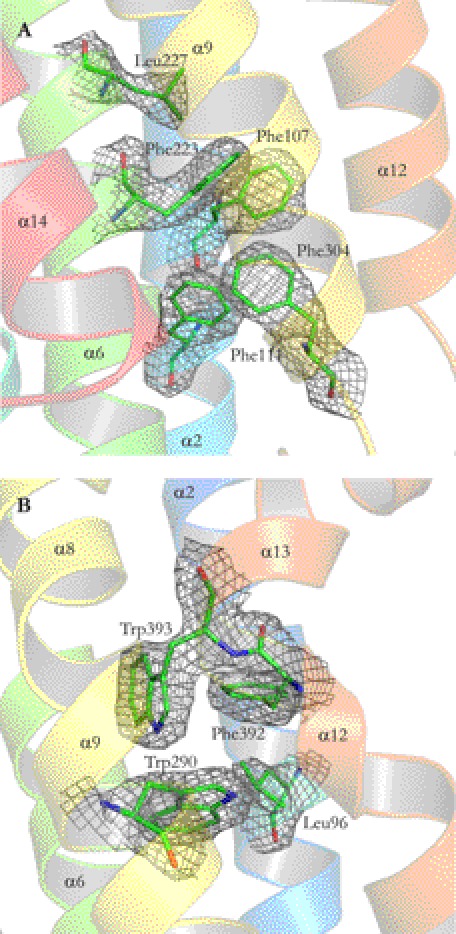
A multiple sequence alignment of GPC1 through GPC6, Dally, and DLP is shown in supplemental Fig. S1. The alignment reveals that, aside from the 14 conserved Cys residues, other evolutionarily conserved amino acids are mostly of nonpolar character. In particular, two hydrophobic centers are formed in the central lobe. Phe107 and Leu227 are identical in human and D. melanogaster glypicans (supplemental Fig. S1) and form a tight hydrophobic region with the conserved or conservatively substituted Phe111, Phe223, and Phe304 (Fig. 4A). The other conserved hydrophobic region in the central lobe is built from the conserved Trp290 and the conservatively substituted Phe392, Trp393, and Leu96 (Fig. 4B). A Trp to Arg mutation in GPC3 of the amino acid corresponding to Trp290 in Gpc1 has been found in a Simpson-Golabi-Behmel syndrome patient, and it results in poor substitution of HS chains to the core protein (44). The mutation would result in a buried positive charge, which would either disrupt or prevent proper folding of GPC3.
FIGURE 4.
Structure model and electron densities of two hydrophobic centers conserved in glypicans. Red, oxygen; blue, nitrogen; green, carbon; Phe107 and Leu227 are identical in human and D. melanogaster glypicans and form a tight hydrophobic core in the central lobe of GPC1 with the conservatively substituted Phe111, Phe223, and Phe304 (A). Another hydrophobic region is also present in the central lobe, built from the conserved Trp290 and the conservatively substituted Phe392, Trp393, and Leu96 (B). Presented in mesh are 2|Fo|−|Fc| electron density maps generated as in Fig. 3. The maps are contoured at 1.0 σ. Figures were generated using PyMOL.
Structural Conservation of the C-terminal Segment in GPC1 Orthologs
Evolutionary conservation of surface-exposed amino acids suggests that these exposed amino acids are involved in some interaction with other macromolecules relating to the function of the protein. To investigate in detail if surface-exposed amino acids are evolutionarily conserved in glypicans, we performed multiple sequence alignment of GPC1 through GPC6, as well as the D. melanogaster glypicans and calculated amino acid conservation using the ConSurf Server. Fig. 5A shows the conservation of glypican amino acids as projected onto a space-filling model of GPC1. This model suggests that few surface-exposed amino acids are conserved between the evolutionarily distant glypicans. However, we also performed the same analysis using multiple sequence alignment of GPC1 orthologs, i.e. GPC1 from different vertebrates. Multiple sequence alignment was performed using GPC1 sequences from Homo sapiens (human), Mus musculus (mouse), Rattus norvegicus (rat), Bos taurus (bovine), Gallus gallus (chicken), Danio rerio (zebrafish), Xenopus tropicalis (western clawed frog), Monodelphis domestica (short-tailed opossums), Maccaca mulatta (rhesus macaque), and Pan troglodytes (common chimpanzee). As shown in Fig. 5B, L1 and large parts of L3 are evolutionarily conserved within vertebrate GPC1 sequences, together with the C-terminal α14. Interestingly, L3 has been shown by mutagenesis to be involved in GAG class determination in rat GPC1 (9).
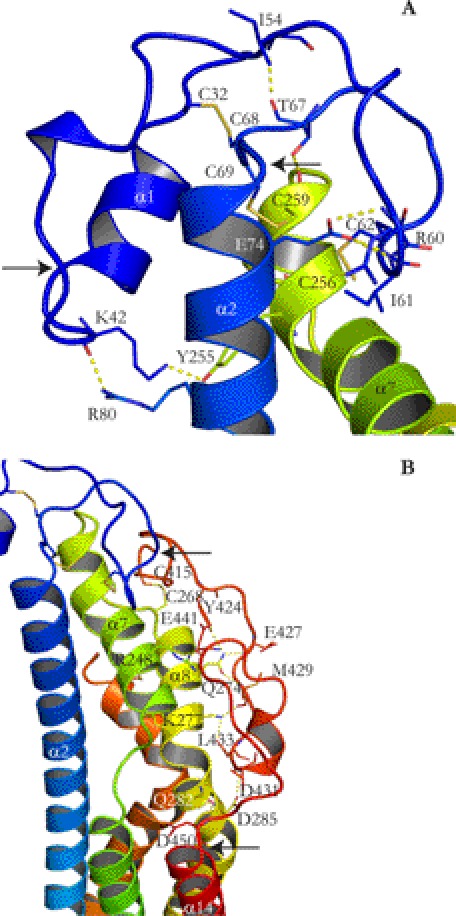
The L1 loop connects α1 and α2 (Fig. 6A). Rather than being extended, the loop is folded upon itself and is stabilized by internal hydrogen bonds. The loop is tethered to the rest of the structure via three disulfide bonds. Cys68 is connected to α1 through Cys32, and Cys62 and Cys69 are connected to α7 through Cys256 and Cys259, respectively. The L3 loop (Fig. 6B) connects α13 and α14 and is formed by amino acids Cys401 to Glu450. Missing in the structure are amino acids 406–412. The L3 loop is stabilized by forming a disulfide bond, Cys268–Cys415, where Cys268 is located in a turn between α7 and α8. The L3 loop is built onto α7, α8, and α9 and forms a number of hydrogen bonds with these α-helices (Fig. 6B). Comparison of the temperature factors for L1 and L3 suggest that L3 is far less flexible than L1, consistent with the hydrogen bonds between L3 and α7, α8, and α9 (data not shown).
FIGURE 6.
Structure of evolutionarily conserved L1 and L3. A, L1 (blue) with start and end marked by arrows is folded upon itself with hydrogen bonds (dotted yellow lines) and is tethered to the rest of the structure by several disulfide bonds (yellow solid lines). B, L3 (orange and red) with start and end marked by arrows is connected to α7, α8, and α9 with several hydrogen bonds (dotted yellow lines).
The structural conservation observed in GPC1 suggests that different structural elements important for specific functions of individual vertebrate glypicans can be identified using similar approaches for the other members in the glypican family. However, our attempts to do so have so far failed, perhaps in part because the sequence of GPC1 is known for many species, contrary to other members of the glypican family.
HS Attachment Site in GPC1
The structure of GPC1 was solved by anomalous dispersion phasing on crystals of SeMet-labeled GPC1 ΔC. To build a model of full-length GPC1 in which the HS attachment site was included, we performed model refinement using data from a native GPC1 ΔHS crystal. However, no additional electron density was observed, and Rfree and Rmodel values were similar to values obtained using GPC1 ΔC (Table 1). Thus, it was not possible to obtain information regarding the C-terminal HS attachment site in GPC1 using the GPC1 ΔHS crystals. This also suggests that the C-terminal HS attachment domain in GPC1 is disordered.
DISCUSSION
In a previous study, we investigated the in-solution conformation and stability of GPC1 and showed that the GPC1 core protein is a stable α-helical protein (17). Thermal and chemical unfolding of GPC1 ΔHS showed that high temperatures (∼70 °C) or high concentrations of urea or guanidine HCl (5.8 or 2.6 m, respectively) are required to denature the protein. Chemical denaturation occurred in a single step for GPC1 ΔHS for both secondary and tertiary structure, and unfolding data closely fitted a two-state model. The stability of GPC1 ΔHS, ΔGNU, was estimated to be ∼60 kJ/mol, which is in the higher range of what is usually seen for globular proteins (45). Together, these data suggested that GPC1 is a densely packed one-domain globular protein. These previous results support the crystal structure of GPC1, which is α-helical, consists of a single domain and, at least as far as the core helices are concerned, is apparently quite rigid. In particular, the disulfide bonds and the long helix α2 possibly contribute to the cooperative unfolding of GPC1.
During refinement of the GPC1 structure, the core protein structure of the D. melanogaster glypican DLP was published (18). This structure is similar to the GPC1 core protein structure, with similarly placed α-helices (Fig. 2B). The two structures were created independently, giving strength for both models as the correct overall fold for glypican core proteins. It should be noted that the GPC1 structure was determined using crystals of N-glycosylated GPC1, whereas DLP was crystallized after mutagenic disruption of both N-glycosylation sites in DLP. N-Linked glycans are involved in protein folding, quality control, sorting, degradation, and secretion (46). Given the similarities of the structures of N-glycosylated GPC1 and unglycosylated DLP, it is clearly possible to obtain the correct glypican fold in the absence of N-glycosylation. This is also supported by our previous study, investigating the importance the N-glycosylation for HS substitution, folding, and stability of GPC1, where we show that the correct folding is obtained for GPC1 even when the two N-glycosylation sites have been disrupted (39). Also, the N-linked glycans on glypicans are most likely involved in increasing the solubility of the proteins, as N-glycosylated GPC1 was crystallized at a protein concentration of 20 mg/ml in comparison with nonglycosylated DLP at 3.5 mg/ml.
The core proteins of proteoglycans have been regarded as inert, with all function of the PGs related to the GAG chains. This view arose from a wealth of genetic experiments implicating GAGs as regulators of morphogenesis and because GAGs are fairly easy to label and purify. The PG core proteins, however, are often large, heavily glycosylated, and difficult to express in bacteria and have therefore been more difficult to study. Moreover, although GPC1 is a fairly large protein with longest dimension around 120 Å, this is rather small in comparison with the HS chains that become attached to glypicans. An HS chain can consist of up to 50–100 repeating disaccharide units of GlcNAc and GlcA. From the solution structure of HS, the length of an HS chain consisting of eight disaccharide units has been estimated to be around 70 Å (47). Thus, a full HS chain can be ∼500 Å long, as compared with the 100 Å for the folded glypican core protein. However, in recent years many interactions with PG core proteins have been found. For example, the ectodomains of syndecan core proteins have been shown to interact directly with integrins (48, 49). Glypican core proteins have been suggested to have functional roles, by interacting with a number of growth factors, including hedgehog, bone morphogenetic protein 4, decapentaplegic, insulin-like growth factor II, and Wnt. Recently, a region was identified in DLP as being important for hedgehog signaling, although hedgehog itself does not seem to bind and the interaction partner for this region remains elusive (18). This region is localized to α4 and the first part of α5 (in our numbering α5 and α6, because the first α-helix is missing in the DLP structure). This region contains a number of amino acids conserved in GPC1 through GPC6 as well as Dally and Dlp that are involved in the folding of the protein. Among the amino acids in this region is the GPC1 Cys191–Cys343 disulfide bond. The structural similarity in this region is confirmed by our GPC1 ΔC structure, with two α-helices connected by a turn. The possibility arises that the interaction is governed by charged residues in this region and that mutagenesis of GPC1 to mimic the DLP region important for hedgehog signaling may confer hedgehog signaling effects on GPC1 core protein.
Amino acids in protein loops display higher sequence variation than residues that form the core of a structure, unless the loops are involved in the function of the protein. Also, protein loops can adopt a more unique folding than the common protein secondary structure elements α-helix and β-sheet. This is perhaps best exemplified by the hypervariable loops on antibodies, which are responsible for the unique binding of antibodies to their antigens. By our analysis of GPC1 vertebrate orthologs, the L1 and L3 loops of GPC1 are evolutionarily conserved, suggesting that they are involved in the function of GPC1. No interaction partner has been found for GPC1 core protein, contrary to some of the other glypican core proteins. GPC1 knock-out mice are viable and fertile but have reduced brain size at birth (6). This has been suggested to be an effect of a transient reduction in HS-dependent fibroblast growth factor signaling during embryogenesis. Moreover, heparan sulfate and chondroitin sulfate proteoglycans (HSPGs and CSPGs, respectively) serve different functions during brain development, with CSPGs inhibiting and HSPGs promoting axon extension (50). GPC1 is exclusively substituted with HS, whereas GPC5, for example, can exist as an HS CS/DS (where DS is dermatan sulfate) hybrid. However, expression of the GPC1 GAG attachment domain alone results in addition of ∼90% CS (9), strongly suggesting that factors in the GPC1 core protein further away from the GAG attachment sites promote the assembly of HS. Mutagenesis of amino acids in the L3 loop resulted in increased proportion of CS substituted on GPC1, but mutagenesis of the entire loop resulted in no protein expression (9). The structure of the L3 loop presented here should facilitate further mutagenic studies. It should also be noted that the information regarding HS assembly enzymes has increased during recent years. HS polymerization and initiation are performed by the exostosin and exostosin-like (EXTL) families, respectively. EXT1 and EXT2 function as an HS co-polymerase and were first described as tumor suppressor genes, as they were found to be the cause of hereditary multiple exostoses, a disease characterized by the formation of multiple cartilaginous tumors (51, 52). The EXTL family consists of three members, EXTL1–3, where EXTL2 and EXTL3 harbor HS initiating α1,4-GlcNAc transferase activity and are ubiquitously expressed in the human body (53). Only EXTL3 has a D. melanogaster ortholog, suggesting that this is the functional HS initiation enzyme, whereas structural data are available for EXTL2, the smallest member in the EXTL family. Given the requirement for the glypican core protein to ensure exclusive HS attachment on GPC1, it is reasonable to suggest that EXTL2 or EXTL3 interact not only with the HS attachment site but also with the core protein. Mutagenesis of the L3 loop suggests that this is the interacting surface for EXTL and glypicans. It is also possible that disruption of the L3 loop results in displacement of the HS attachment site. An alternative explanation could also be that the glypican core protein provides steric hindrance for the CS initiation enzyme, chondroitin N-acetylgalactosaminyltransferase (54), to ensure exclusive HS attachment. Unfortunately, no structural information is available for the CS initiating enzymes. The L1 loop is also evolutionarily conserved as shown by our analysis of structural conservation of GPC1 vertebrate orthologs, but thus far, no function has been suggested for this loop.
The glypican core proteins have long been assumed to adopt a globular fold, but the structures of GPC1 and DLP are cylindrical in shape. The C-terminal α14 is placed on the long side of the protein, making it difficult to predict how the core proteins are situated in the membranes. The first Ser residue in the HS attachment site is located 10 amino acids from the C terminus of the GPC1 ΔC structure, placing the HS attachment close to the folded core protein. We have previously shown that the HS chains on GPC1 do not affect its secondary or tertiary structure (17). In fact, results obtained by differential scanning calorimetry suggest that the HS chains cause the core protein monomers to repel one another, possibly by electrostatic repulsion of the negatively charged HS chains. Others have reported that glypican core protein binds weakly to heparin-agarose (18). The C-terminal Ser residue responsible for GPI attachment is located 50 amino acids downstream of the visible ends of the models of GPC1 and DLP, which is long enough that the C terminus of the protein could be placed in a wide variety of orientations with respect to the core protein. Thus, when the GPI anchor is anchored in the membrane, it may allow great freedom of orientation of GPC1 and DLP with respect to the membrane surface. Our attempts to locate the GPC1 HS attachment domain and C terminus using crystallography have failed, which together with the presented in-solution data on the GPC1 conformation indicate that the HS attachment site is a disordered tail in the protein. Experimental approaches other than x-ray crystallography may be needed to finally answer the important question of the placement of the C-terminal part in glypicans, how the HS chains are extended from the core protein, and how glypicans are situated in the membranes. Moreover, the presented GPC1 structure provides new information that will be important in determining the mechanism behind exclusive HS attachment on glypicans.
Supplementary Material
Acknowledgments
We thank the beamline staff at MAX-lab for help during data collection and Dr. Mattias Hansson at the MAX-lab Crystallization Facility for help with setting up high throughput crystallization plates. The technical assistance of Sol Da Rocha is greatly appreciated. We are also grateful to Axel Hyrenius Wittsten for help with CD.
This work was supported by Swedish Research Council Grants 2006-4387 (to D. L.) and 2005-6749 and 2010-3914 (to K. M.), the Swedish Cancer Foundation, the Medical Faculty of Lund University, the Royal Physiographic Society in Lund, and the Crafoord, Wiberg, Jeanson, Segerfalk, and Kock Foundations.

This article contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (codes 4ACR and 4AD7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PG
- proteoglycan
- HS
- heparan sulfate
- GAG
- glycosaminoglycan
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SeMet
- selenomethionine
- CS
- chondroitin sulfate
- GPI
- glycosylphosphatidylinositol
- rh
- recombinant human
- mdeg
- millidegrees
- EXTL
- exostosin-like.
REFERENCES
- 1. Bülow H. E., Hobert O. (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 [DOI] [PubMed] [Google Scholar]
- 2. Häcker U., Nybakken K., Perrimon N. (2005) Heparan sulfate proteoglycans. The sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- 3. Pilia G., Hughes-Benzie R. M., MacKenzie A., Baybayan P., Chen E. Y., Huber R., Neri G., Cao A., Forabosco A., Schlessinger D. (1996) Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 12, 241–247 [DOI] [PubMed] [Google Scholar]
- 4. Campos-Xavier A. B., Martinet D., Bateman J., Belluoccio D., Rowley L., Tan T. Y., Baxová A., Gustavson K. H., Borochowitz Z. U., Innes A. M., Unger S., Beckmann J. S., Mittaz L., Ballhausen D., Superti-Furga A., Savarirayan R., Bonafé L. (2009) Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 84, 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okamoto K., Tokunaga K., Doi K., Fujita T., Suzuki H., Katoh T., Watanabe T., Nishida N., Mabuchi A., Takahashi A., Kubo M., Maeda S., Nakamura Y., Noiri E. (2011) Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat. Genet. 43, 459–463 [DOI] [PubMed] [Google Scholar]
- 6. Jen Y. H., Musacchio M., Lander A. (2009) Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neural Dev. 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang L., Esko J. D. (1994) Amino acid determinants that drive heparan sulfate assembly in a proteoglycan. J. Biol. Chem. 269, 19295–19299 [PubMed] [Google Scholar]
- 8. Zhang L., David G., Esko J. D. (1995) Repetitive Ser-Gly sequences enhance heparan sulfate assembly in proteoglycans. J. Biol. Chem. 270, 27127–27135 [DOI] [PubMed] [Google Scholar]
- 9. Chen R. L., Lander A. D. (2001) Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J. Biol. Chem. 276, 7507–7517 [DOI] [PubMed] [Google Scholar]
- 10. Williams E. H., Pappano W. N., Saunders A. M., Kim M. S., Leahy D. J., Beachy P. A. (2010) Dally-like core protein and its mammalian homologs mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc. Natl. Acad. Sci. U.S.A. 107, 5869–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan D., Wu Y., Yang Y., Belenkaya T. Y., Tang X., Lin X. (2010) The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development 137, 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capurro M. I., Xu P., Shi W., Li F., Jia A., Filmus J. (2008) Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell 14, 700–711 [DOI] [PubMed] [Google Scholar]
- 13. Kirkpatrick C. A., Knox S. M., Staatz W. D., Fox B., Lercher D. M., Selleck S. B. (2006) The function of a Drosophila glypican does not depend entirely on heparan sulfate modification. Dev. Biol. 300, 570–582 [DOI] [PubMed] [Google Scholar]
- 14. Capurro M. I., Xiang Y. Y., Lobe C., Filmus J. (2005) Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 65, 6245–6254 [DOI] [PubMed] [Google Scholar]
- 15. Ohkawara B., Yamamoto T. S., Tada M., Ueno N. (2003) Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development 130, 2129–2138 [DOI] [PubMed] [Google Scholar]
- 16. Cheng W., Tseng C. J., Lin T. T., Cheng I., Pan H. W., Hsu H. C., Lee Y. M. (2008) Glypican-3-mediated oncogenesis involves the insulin-like growth factor-signaling pathway. Carcinogenesis 29, 1319–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Svensson G., Linse S., Mani K. (2009) Chemical and thermal unfolding of glypican-1. Protective effect of heparan sulfate against heat-induced irreversible aggregation. Biochemistry 48, 9994–10004 [DOI] [PubMed] [Google Scholar]
- 18. Kim M. S., Saunders A. M., Hamaoka B. Y., Beachy P. A., Leahy D. J. (2011) Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 13112–13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu H., Lustbader J. W., Liu Y., Canfield R. E., Hendrickson W. A. (1994) Structure of human chorionic gonadotropin at 2.6 Å resolution from MAD analysis of the selenomethionyl protein. Structure 2, 545–558 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) Integration, scaling, space-group assignment, and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collaborative Computational Project No. 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes. Crystal structure of a PE-PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Auto-rickshaw. An automated crystal structure determination platform as an efficient tool for the validation of an x-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 25. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2009) On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D Biol. Crystallogr. 65, 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howell P. L., Smith G. D. (1992) Identification of heavy atom derivatives by normal probability methods. J. Appl. Crystallogr. 25, 81–86 [Google Scholar]
- 27. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 28. de La Fortelle E., Bricogne G., Carter C. W., Jr. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 29. Cowtan K. (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D Biol. Crystallogr. 55, 1555–1567 [DOI] [PubMed] [Google Scholar]
- 30. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) Breaking good resolutions with ARP/wARP. J. Synchrotron Radiat. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 32. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 33. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Svensson G., Hyrenius Wittsten A., Linse S., Mani K. (2011) The structural role of N-linked glycans on human glypican-1. Biochemistry 50, 9377–9387 [DOI] [PubMed] [Google Scholar]
- 40. Pearson W. R., Lipman D. J. (1988) Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 85, 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Cat B., Muyldermans S. Y., Coomans C., Degeest G., Vanderschueren B., Creemers J., Biemar F., Peers B., David G. (2003) Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J. Cell Biol. 163, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Capurro M. I., Shi W., Sandal S., Filmus J. (2005) Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma growth. J. Biol. Chem. 280, 41201–41206 [DOI] [PubMed] [Google Scholar]
- 43. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 44. Veugelers M., Cat B. D., Muyldermans S. Y., Reekmans G., Delande N., Frints S., Legius E., Fryns J. P., Schrander-Stumpel C., Weidle B., Magdalena N., David G. (2000) Mutational analysis of the GPC3/GPC4 glypican gene cluster on Xq26 in patients with Simpson-Golabi-Behmel syndrome. Identification of loss-of-function mutations in the GPC3 gene. Hum. Mol. Genet. 9, 1321–1328 [DOI] [PubMed] [Google Scholar]
- 45. Makhatadze G. I., Privalov P. L. (1995) Energetics of protein structure. Adv. Protein Chem. 47, 307–425 [DOI] [PubMed] [Google Scholar]
- 46. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 47. Khan S., Rodriguez E., Patel R., Gor J., Mulloy B., Perkins S. J. (2011) The solution structure of heparan sulfate differs from that of heparin: implications for function. J. Biol. Chem. 286, 24842–24854 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Whiteford J. R., Couchman J. R. (2006) A conserved NXIP motif is required for cell adhesion properties of the syndecan-4 ectodomain. J. Biol. Chem. 281, 32156–32163 [DOI] [PubMed] [Google Scholar]
- 49. Beauvais D. M., Burbach B. J., Rapraeger A. C. (2004) The syndecan-1 ectodomain regulates αvβ3 integrin activity in human mammary carcinoma cells. J. Cell Biol. 167, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsumoto Y., Irie F., Inatani M., Tessier-Lavigne M., Yamaguchi Y. (2007) Netrin-1/DCC signaling in commissural axon guidance requires cell-autonomous expression of heparan sulfate. J. Neurosci. 27, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stickens D., Clines G., Burbee D., Ramos P., Thomas S., Hogue D., Hecht J. T., Lovett M., Evans G. A. (1996) The EXT2 multiple exostoses gene defines a family of putative tumor suppressor genes. Nat. Genet. 14, 25–32 [DOI] [PubMed] [Google Scholar]
- 52. Lind T., Tufaro F., McCormick C., Lindahl U., Lidholt K. (1998) The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 273, 26265–26268 [DOI] [PubMed] [Google Scholar]
- 53. Kitagawa H., Shimakawa H., Sugahara K. (1999) The tumor suppressor EXT-like gene EXTL2 encodes an α1,4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 [DOI] [PubMed] [Google Scholar]
- 54. Uyama T., Kitagawa H., Tamura Ji. J., Sugahara K. (2002) Molecular cloning and expression of human chondroitin N-acetylgalactosaminyltransferase. The key enzyme for chain initiation and elongation of chondroitin/dermatan sulfate on the protein linkage region tetrasaccharide shared by heparin/heparan sulfate. J. Biol. Chem. 277, 8841–8846 [DOI] [PubMed] [Google Scholar]
- 55. Read R. J., Schierbeek A. J. (1988) A phased translation function. J. Appl. Crystallogr. 21, 490–495 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.