Background: Unphosphorylated STAT3 (U-STAT3) regulates gene expression, but the mechanisms of its DNA binding are not fully understood.
Results: U-STAT3 binds to the same γ-activated sequence (GAS) DNA-binding site as phosphorylated STAT3. It also binds to AT-rich DNA structures.
Conclusion: U-STAT3 regulates gene expression by binding to GAS and influencing chromatin organization.
Significance: Our data provide an explanation of mechanisms of U-STAT3 binding to DNA.
Keywords: Atomic Force Microscopy, DNA Structure, STAT Transcription Factor, STAT3, Transcription Factors, DNA Binding, Microscale Thermophoresis
Abstract
Phosphorylation of signal transducer and activator of transcription 3 (STAT3) on a single tyrosine residue in response to growth factors, cytokines, interferons, and oncogenes activates its dimerization, translocation to the nucleus, binding to the interferon γ (gamma)-activated sequence (GAS) DNA-binding site and activation of transcription of target genes. STAT3 is constitutively phosphorylated in various cancers and drives gene expression from GAS-containing promoters to promote tumorigenesis. Recently, roles for unphosphorylated STAT3 (U-STAT3) have been described in response to cytokine stimulation, in cancers, and in maintenance of heterochromatin stability. However, the mechanisms underlying U-STAT3 binding to DNA has not been fully investigated. Here, we explore STAT3-DNA interactions by atomic force microscopy (AFM) imaging. We observed that U-STAT3 molecules bind to the GAS DNA-binding site as dimers and monomers. In addition, we observed that U-STAT3 binds to AT-rich DNA sequence sites and recognizes specific DNA structures, such as 4-way junctions and DNA nodes, within negatively supercoiled plasmid DNA. These structures are important for chromatin organization and our data suggest a role for U-STAT3 as a chromatin/genome organizer. Unexpectedly, we found that a C-terminal truncated 67.5-kDa STAT3 isoform recognizes single-stranded spacers within cruciform structures that also have a role in chromatin organization and gene expression. This isoform appears to be abundant in the nuclei of cancer cells and, therefore, may have a role in regulation of gene expression. Taken together, our data highlight novel mechanisms by which U-STAT3 binds to DNA and supports U-STAT3 function as a transcriptional activator and a chromatin/genomic organizer.
Introduction
Signal transducer and activator of transcription 3 (STAT3) belongs to a family of DNA-binding proteins that are activated in response to extracellular ligands such as cytokines, growth factors, and hormones (1). Seven mammalian STAT proteins, STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6, share conserved structural and functional domains: the N-terminal, coiled-coil, a DNA binding, a Src homology 2, and a C-terminal domain containing a conserved tyrosine phosphorylation site (2). STAT proteins are required for diverse biological processes including embryonic development and adult homeostasis, as well as differentiation, proliferation, survival, and apoptosis (3). Cytokines or growth factors binding to their cognate receptors result in STAT recruitment to receptors and phosphorylation by members of the Janus family of kinases (JAKs) or Src kinases on a single tyrosine residue (Tyr705 in STAT3) (1, 2). This process is named STAT “activation,” triggering the dimerization and nuclear translocation of the STAT proteins. In the nucleus, phosphorylated STATs (P-STAT) bind to DNA-response elements named interferon γ (gamma)-activated sequence (GAS)3 in the promoters of target genes and activate specific gene expression programs (1). Two major STAT isoforms were identified for STATs 1, 3, 4, 5A, and 5B: full-length transcription factor α-isoforms and alternatively spliced β-isoforms that lack the transactivation domain (4). STAT isoforms, so-called STATγ, could also be generated by proteolysis (4). Both the alternatively spliced STATβ and proteolytically cleaved STATγ lack a transactivation domain and are thought to negatively regulate transcription (5).
In normal cells and in tissues, ligand-dependent activation of the STATs is a transient process, lasting from several minutes to several hours (2). However, in many cancerous cells, with dysregulated growth factor signaling, STAT proteins are constitutively activated by tyrosine phosphorylation (2). In this respect, STAT3α stands out, based on its constitutive phosphorylation in the majority of human neoplasms and its capacity to induce cell transformation and tumorigenesis (6). It is believed that P-STAT3α mediates its oncogenic effects through transcriptional activation of target genes to enhance proliferation (CCND1 and c-MYC), angiogenesis (VEGF, ADM, and ANGPTL4), invasion (FGA, FGB, CTSB, and SERPINE2), and suppression of apoptosis (BCL-xL, BCL-2, MCL-1, and BIRC5) (7). In addition, P-STAT3α stimulates its own transcription causing an increase in unphosphorylated STAT3α (U-STAT3) (8–10).
U-STAT proteins, have been reported to shuttle between cytoplasmic and nuclear compartments (11), and to influence gene transcription, both in response to cytokines and in cancer cells by mechanisms distinct from those underlying P-STAT3 (10). Furthermore, studies in Drosophila have demonstrated that U-STAT92E is localized in the nucleus in association with heterochromatin-associated protein HP1, and is essential for maintaining heterochromatin stability. Activation of STAT92E by phosphorylation causes STAT dispersal from heterochromatin leading to HP1 dissociation from DNA and heterochromatin destabilization (12, 13). Therefore, unphosphorylated STAT3 may be an important regulator of gene expression, underscoring the need to understand the mechanisms underlying DNA-binding and regulation of gene expression by U-STAT3.
Here, we used atomic force microscopy (AFM) imaging to directly evaluate U-STAT3 binding to DNA in a model system. We observed that U-STAT3 can bind GAS motif as a monomer and as a dimer. In addition, we observed that U-STAT3 binds to AT-rich DNA sequences and recognizes specific DNA structures, such as 4-way junctions and DNA nodes. Our data support U-STAT3 function as an architectural protein and regulator of gene expression.
EXPERIMENTAL PROCEDURES
STAT3-DNA Binding Reactions for AFM Imaging
Reaction mixtures containing 20 ng of plasmid DNA and 100 ng of STAT3 recombinant protein molecules (Active Motif, Carlsbad, CA) were incubated on ice for 15 min and then cross-linked by 0.5% glutaraldehyde. After completion of the fixation step, the complexes were purified using Montage PCR filter units (Millipore Corp., Bedford, MA). The final samples were re-suspended in 20 μl of 10 mm Tris-HCl (pH 7.4), 5 mm NaCl buffer. Appropriate dilutions were made for deposition of the samples onto mica to provide an even distribution of DNA on the mica surface for AFM imaging.
AFM Imaging and Protein Volume Measurement
Freshly cleaved muscovite mica was incubated in a mixture of a 1-(3-aminopropyl)silatrane (APS) solution for 30 min to prepare APS-mica, as described (14). The sample droplets (5 μl) were deposited on APS-mica for 2 min, then washed with de-ionized water and dried with nitrogen gas. Images were acquired in tapping mode using a Multimode SPM Nanoscope IIIa system (Veeco/Digital Instruments, Santa Barbara, CA). Silicon tapping mode probes (Hi'Res DP14; MicroMasch, Estonia) with a curvature radius of 1 nm were used. The nominal spring constants were ∼5.0 newtons/m and the resonant frequency was ∼160 Hz.
Contour length measurements of the DNA molecules were made by tracing the image DNA backbone using the curve tool software of the image module (Digital Instruments, Santa Barbara, CA). Protein volume measurements were determined using the section tool software (Digital Instruments). Perpendicular cross-sections were in two dimensions: the height (h) of the protein was measured by the difference in intensity of the protein compared with background noise; the width was measured at a position corresponding to half-maximal protein height. The volume (V) of the protein was determined as described (15, 16).
Statistical Analysis
The number of complexes was plotted in histograms. Respective density fits were calculated by means of Kernel Density Estimation with Gaussian kernels. The calculations and density fits were performed using the R Statistical Package (R Foundation for Statistical Computing, Vienna, Austria) (17). Bootstrap Kolmogorov-Smirnov testing was employed to compare the distribution of monomer and dimer volumes at the sites of DNA binding for GAS and 4-way junctions.
Cell Lines, Cell Fractionation, and Western Blot Analysis
MCF-7, DU145, PC-3, and LNCaP cell lines were obtained from the American Type Culture Collection (ATCC). Cells were cultured accordingly to the manufacturer's recommendations. For cell fractionation, cells were plated in 100-mm dishes, harvested in 2 days, and cytoplasmic and nuclear extracts were prepared as described (18). Western blot was performed with anti-STAT3 (H-190) antibody from Santa Cruz (Santa Cruz, CA) as described (19).
DNA Affinity Precipitation Assays
DNA affinity precipitation assays (DAPA) was performed as described (20). Briefly, cells were lysed in cell lysis buffer (50 mm Tris-HCl (pH 8), 0.5% Igepal, 150 mm NaCl, 1 mm EDTA, and 10% glycerol, supplemented with protease and phosphatase inhibitors) to generate total cell extracts. Biotinylated DNA oligonucleotides listed in Table 1 were incubated with streptavidin-agarose beads (Chemicon) in IP buffer (cell lysis buffer containing 15 mm NaCl) at 4 °C for 2 h. The oligo-bead complex was washed and incubated with cell extract in IP buffer at 4 °C overnight. The beads were washed two times, resuspended in SDS-PAGE loading buffer, boiled, and run on SDS-PAGE gels before immunoblotting with anti-STAT3 antibody.
TABLE 1.
AFM-identified sequences bound by STAT3 in the pGAS-TA-Luc plasmid
| Site | Distance from NotI site | Corresponding sequences |
|---|---|---|
| S100 | 85–110 bp | (S + 100) 5′-AAAACAAAACGAAACAAACAAACTA |
| (S-100) 5′-CGCAACTGTTGGGAAGGGCGATCG | ||
| S333 | 320–350 bp | (S + 330) 5′-CGCCAAAAACATAAAGAAAG |
| (S-330) 5′-GTTTTTCGCCCTTTGACGTTG | ||
| GAS | 167–195 bp | (GAS) 5′- GTTTCCGGGAAAGCAGTAGGTTTCCGGGAAAG-3′ |
| S+100 | S + 100 mut#1 AAAACAGAACGAGACAGACAAACTA | |
| mutantsa | S + 100 mut#2 AAGGCAGGACGAGGCAGGCAGGCTA |
a S+100 mutants were generated by substitution of A nucleotides within AT-rich hexanucleotides with G nucleotides. The positions of mutated bases are in bold and underlined.
Microscale Thermophoresis DNA Binding Studies
Binding of STAT3 protein to oligonucleotides was measured by microscale thermophoresis (21–23). HEK293 cells were transfected with the STAT3-GFP expression vector as described (19). Cell lysates prepared using 1× RIPA buffer (Millipore Corp., Bedford, MA) (20 × 106 cells/ml) were used as a source of fluorescence-labeled STAT3. The lysates were diluted 150 times with microscale thermophoresis DNA-binding buffer to provide the optimal level of the fluorescent STAT3 proteins (20 nm) in the binding reactions. Nontransfected HEK293 cells were used to evaluate background fluorescence, which turned out to be nondetectable even in nondiluted lysate. Titration series (16 binding mixtures) have been prepared containing constant amounts of cell lysate in each sample and varying concentrations of the oligonucleotides. The final buffer composition contained 25 mm HEPES (pH 7.2), 50 mm NaCl, 2.5 mm MgCl2, and 0.025% Nonidet P-40. Measurements were taken in standard treated capillaries on a Monolith NT.115 instrument (NanoTemper Technologies GmbH, Germany) using 50% IR-laser power and LED excitation source with λ = 470 nm at ambient temperature. NanoTemper Analysis 1.2.20 software was used to fit the data and determine the apparent KD values.
DNA Heteroduplex Formation and Isolation
Plasmid pUC8F14C containing a 106-bp inverted repeat insert has been previously described (24). The 301-bp fragments of pUC8 and similar fragments of insert-containing plasmid were amplified using upstream (5′-biotin-CTGGCACGACAGGTTTCCCGACTG-3′) and downstream (5′-biotin-CTGGCGAAAGGGGGATGTGCTGCA-3′) modified PCR primers (Sigma Genosys). DNA strands were annealed with their complementary counterparts to form heteroduplexes (25). Briefly, each 25-μl hybridization mixture containing 1 pmol of DNA fragments in 200 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1 mm dithiothreitol, 10 mm MgCl2 was incubated stepwise at 95 (5 min), 85 (10 min), and 70 °C (60 min) and then cooled to room temperature. The hybridization products were separated in a 5% native polyacrylamide gel in TBE buffer (90 mm Tris borate, pH 8.3, 2.5 mm EDTA), and bands of heteroduplex fragments that migrate slower than correctly annealed parental fragments were excised. After purification using a GeneClean gel extraction kit (Q-biogen, Carlsbad, CA), the isolated heteroduplexes were resuspended in 40 μl of TE (10 mm Tris-HCl, 1 mm EDTA, pH 7.8), and aliquots were taken for STAT3 binding reactions.
STAT3 Binding Reactions
STAT3 binding assays were performed using STAT3 recombinant protein molecules obtained from Active Motif at various the DNA-protein molar ratios (indicated in the figure legends) in buffer containing 20 mm Tris-HCl, 50 mm KCl, 1 mm EDTA, 4 mm MgCl2, 0.2% Nonidet P-40, and 7% glycerol (pH 7.9). The products were analyzed on a 4–12% TBE gel (Novex, Invitrogen) run at 100 V for 2 h. All of the binding reactions (total volume 30 μl), containing equimolar amounts of DNA probes (1.7 nm) and varying amounts of proteins were incubated for 30 min at 37 °C. The reaction products were loaded on 3.5% native polyacrylamide gels containing 5% glycerol and separated by electrophoresis at 100 V in TBE buffer for 2 h. DNA was visualized as described (25).
RESULTS
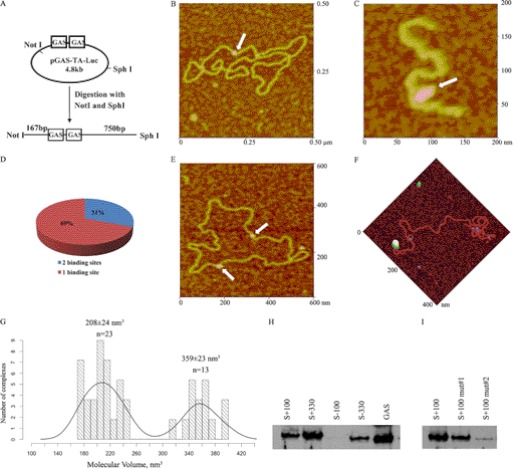
To examine U-STAT3 binding to the GAS motif, recombinant STAT3 proteins were incubated with the plasmid pGAS-TA-Luc for AFM imaging. This plasmid contains two neighboring GAS motifs separated by 10 bp (TTCCGGGAAN10TTCCGGGAA) (Fig. 1A) and is routinely used in luciferase reporter assays to study STAT3 transcriptional activity in response to cytokine stimulation and/or drugs targeting STAT3 dimerization and DNA binding. The purified DNA-protein complexes were deposited on APS-mica and imaged using the AFM tapping mode, which minimizes any lateral force applied to the protein (Fig. 1, B–F). Estimated sizes of STAT3 proteins assembled on DNA, determined from AFM volume measurements, were 359 and 208 nm3, suggesting that U-STAT3 is capable of binding to DNA as a dimer or monomer (Fig. 1G). The expected (theoretical) volumes of the 176-kDa STAT3α dimer are 332 nm3, and the 88-kDa STAT3α monomer are 166 nm3. The discrepancies between theoretical and estimated STAT3 volumes may relate to the protein collapse, flattening, or distortion due to sample drying prior to AFM imaging. However, since the object of this study was to distinguish between dimers and monomers, the relative values are informative.
FIGURE 1.
AFM imaging of STAT3 complexes with pGAS-TA-Luc plasmid. A, experimental design for AFM analysis of STAT3 binding to GAS element within pGAS-TA-Luc. B, representative AFM image of full-length STAT3 (white arrow) binding as a monomer to pGAS-TA-Luc. C, full-length STAT3 binding to the GAS element as a dimer (white arrow). D, percent of protein complexes bound to a single site or to two sites on the pGAS-TA-Luc plasmid. E, representative AFM image of STAT3 bound to two sites (white arrows). F, STAT3 binding to the site located at 100 bp from a NotI-digested end. G, histogram of volume distribution of full-length STAT3 measured by AFM. H, STAT3-DNA interactions analyzed by DAPA with a biotinylated double-stranded DNA oligonucleotides corresponding to each of the four potential STAT3 binding sites in the pGAS-TA-Luc plasmid. I, STAT3 binding to AT-rich sequences is decreased by A to G mutations.
To confirm STAT3 binding to the GAS element within pGAS-TA-Luc, we digested the plasmid bound to STAT3 with NotI and SphI. Digestion of the 4.8-kb pGAS-TA-Luc plasmid results in two DNA fragments: a short 0.9-kb DNA fragment containing a GAS DNA-binding motif and a long 3.9-kb DNA fragment without a STAT3 binding site. The purified short DNA fragments and protein complexes were deposited on APS-mica for AFM imaging. The distances from the NotI site to the GAS element within the short DNA fragments is 167 bp, and from the GAS element to the SphI site is 750 bp (Fig. 1A). Fig. 1C demonstrates that the full-length U-STAT3 protein binds to the GAS element as a dimer.
Our observation that 20 of 62 (31%) analyzed STAT3-plasmid DNA complexes contained STAT3 bound at two sites (Fig. 1D) was an unexpected finding. The second binding site was clearly not a consensus GAS element, because it was separated from the first binding site by more than 10 bp (Fig. 1E). This suggests that U-STAT3 is capable of binding to sequences that may underlie alternative mechanisms for regulation of gene expression by U-STAT3. To characterize the second binding site we digested pGAS-TA-Luc bound to STAT3 at a single site with NotI to obtain a linear 4.8-kb fragment, as shown on Fig. 1A. The measured distances from the ends of the plasmids showed that STAT3 binds to DNA sites located 100 and 323 bp from the ends of the pGAS-TA-Luc linearized plasmid (Fig. 1F). The sequences that correspond to these potential binding sites are listed in Table 1.
To validate AFM observations and confirm that U-STAT3 binds to identified DNA sequences, we performed DAPA by incubating nuclear extracts from DU145 prostate cancer cells with double-stranded biotin-labeled oligonucleotides corresponding to the GAS motif and the novel AFM-identified binding sites. Because AFM measurements do not discriminate the ends generated by NotI, to further determine regions of STAT3 binding, we synthesized double-stranded oligonucleotides corresponding to DNA at positions +S100, −S100, +S333, and −S333. Scrambled DNA sequences were used as negative controls. The DAPA assays demonstrated that STAT3 preferentially binds to S+100 and S+330 sequences (Fig. 1H). In addition, weaker binding was observed to the S−330 sequence. Notably, there were no similarities in DNA sequences S+100, S+330, and S−330, except that they are enriched for AT hexanucleotides (5′-AAACAA, 5′-AACAAA, and 5′-CAAAAA) (Table 1). These are represented among hexanucleotides sequences frequently found in scaffold/matrix-attached regions (S/MARs) (26, 27). Mutations in AT-rich oligonucleotides carrying substitution of A to G abrogated STAT3 binding to the identified sequences (Fig. 1I).
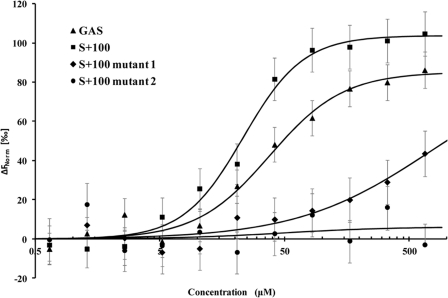
We used microscale thermophoresis to determine relative affinities of STAT3 to GAS and AT-rich oligonucleotide S+100 (Fig. 2). The method is based on the ligand binding-induced change in movement of fluorescent molecules along a temperature gradient (21–23). We used lysates of HEK293 cells expressing STAT3-GFP as a source of fluorescence-labeled STAT3 for the DNA binding assay. Because the U-STAT3-GFP fusion protein is the only fluorescent molecule in the lysates, the interactions of the fusion protein could be studied without purification. Fluorescence of the control lysates of nontransfected cells was undetectable. Binding of highly charged oligonucleotides resulted in significant changes in STAT3 mobility in the temperature gradient and allowed for determination of apparent dissociation constants. The apparent dissociation constants were 37.9 ± 1.0 and 23.3 ± 0.6 μm for GAS and S+100, respectively (Fig. 2). Because binding assays have been performed under identical conditions using the same lysate preparations for all oligonucleotides, this allowed conclusions that U-STAT3-GFP binds to AT-rich (S+100) sequences with 1.6 higher affinity compared with GAS element. Specificity of STAT3 binding to the AT-rich DNA was confirmed by substitutions of A to G that resulted in a dramatic decrease in affinity of S+100 mutant 1, and inability to bind to S+100 mutant 2 (Table 1 and Fig. 2). We interpret the data obtained by AFM, DAPA, and microscale thermophoresis assays as supporting U-STAT3 binding to identified AT-rich and GAS DNA sequences.
FIGURE 2.
STAT3 binding to DNA. Microscale thermophoresis binding measurements of STAT3-EGFP to GAS (KD = 37.9 ± 1.0 μm), S+100 (KD = 23.3 ± 0.57 μm), S+100 mutant 1 (KD = 740 ± 21 μm), and S+100 mutant 2 (no binding). The data suggests comparable affinities of STAT3 interactions with GAS and AT-rich sequences, whereas A to G substitutions in the S+100 sequence (S+100 mutants 1 and S+100 mutant 2, Table 1) result in dramatically reduced affinity. The lines through the points are the best-fit curves using the Hill equation. The error bars represent standard deviation from three independent measurements.
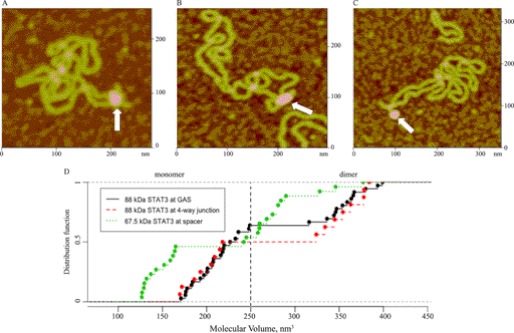
Proteins that bind to AT-rich DNA often bend DNA or preferentially interact with bent DNA or DNA crossings (28, 29). These specific DNA structures have a role in chromatin organization and gene transcription by distant mechanisms. To determine whether U-STAT3 recognizes specific structures within DNA sequences, we used the previously characterized supercoiled pUC8F14C plasmid. This plasmid does not have a GAS element, but contains a 106-base pair palindrome with AT-rich hexanucleotides adapted in the cruciform structure with a 4-way junction region (14, 24). We observed that STAT3 binds to the 4-way junction region (Fig. 3A) and DNA nodes (Fig. 3B), supporting a potential chromatin-organizing role by bringing distal DNA regions together.
FIGURE 3.
STAT3 binding to DNA structures in the defined model plasmids. A, AFM image of a full-length STAT3 dimer bound to a 4-way junction region in a cruciform structure (white arrow). B, AFM image of full-length STAT3 dimer bound to a node region (white arrow). C, AFM image of a 67.5-kDa STAT3 monomer bound to the end of the hairpin arm in the cruciform structure (white arrow). D, the empirical cumulative distribution functions (distribution function) of the volumes are depicted for full-length STAT3 bound to pGAS-TA-Luc and pUC8F14C plasmids, and the C-terminal truncated 67.5-kDa STAT3 bound to pUC8F14C plasmid. The increased value of ECDF at each filled circle on the graph is the empirical probability (i.e. relative frequency) of the corresponding STAT3-DNA complex.
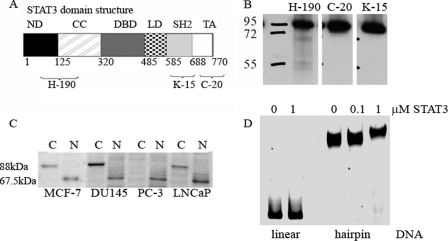
The volumes for full-length STAT3 (88-kDa monomer and 176-kDa dimer) bound to pUC8F14C were slightly different from those determined for pGAS-TA-Luc (Table 2 and Fig. 1G). We tested the null hypothesis, that mono and dimer volumes for 88-kDa STAT3 binding at the GAS site and at the 4-way junction are drawn from the same distribution. The Bootstrap Kolmogorov-Smirnov test yielded p = 0.846. (The number of bootstraps was set at 1000; the naive Kolmogorov-Smirnov p value is 0.918.) We interpret these data in support of the null hypothesis that full-length STAT3 forms similar complexes on both plasmids. However, we also observed binding of the protein with measured volumes of 144 and 267 nm3 to the pUC8F14C suggesting that an isoform shorter than full-length STAT3 binds to the hairpin spacer (Table 2 and Fig. 3C). To determine whether this shorter isoform and full-length STAT3 are drawn from the same distribution, the Bootstrap Kolmogorov-Smirnov test yielded p = 0.001 and p = 0.017 for full-length STAT3 bound to GAS and pUC8F14C, respectively (Fig. 3D). Analysis of the STAT3 recombinant protein sample confirmed the presence of a shorter isoform with a molecular mass of 67.5-kDa that likely resulted from partial proteolysis of STAT3. The use of antibodies for the various domains (N-terminal, Src homology 2, and C-terminal) in combination with mass spectrometry analysis demonstrated that this isoform did not contain the C-terminal domain but retained the DNA-binding motif (Fig. 4, A and B). Therefore, we view these AFM observations as demonstrating the truncated STAT3 isoform binding specifically to a hairpin spacer. Notably, the number of complexes formed by the C-terminal truncated isoform is comparable with the number of complexes formed by the full-length STAT3 protein, despite the lower concentration of this isoform in the protein sample, suggesting that this isoform binds with higher affinity to the hairpin spacer DNA.
TABLE 2.
Analysis of U-STAT3 species bound to pUC8F14C plasmid in AFM images
| Names of the proteins | Molecular mass | No. of complexes | Theoretical (T) volume | Experimental (E) mean volume | Ratio E/T volume |
|---|---|---|---|---|---|
| 67.5-kDa monomer | 67.5 kDa | 12 | 128 nm3 | 144 ± 16 nm3 | 1.13 |
| 88-kDa monomer | 88 kDa | 8 | 166 nm3 | 196 ± 19 nm3 | 1.18 |
| 67.5-kDa dimera | 135 kDa | 14 | 256 nm3 | 267 ± 20 nm3 | 1.04 |
| 88-kDa dimerb | 176 kDa | 9 | 332 nm3 | 357 ± 22 nm3 | 1.08 |
a Ratio of 67.5 dimer/monomer experimental volumes is 1.85.
b Ratio of 88kDa dimer/monomer experimental volumes is 1.82 (theoretical volumes ratio is 2).
FIGURE 4.
Characteristics of the 67.5-kDa STAT3 isoform. A, STAT3 domain structure and antibody recognition sites. B, Western blot analysis of the recombinant STAT3 protein with antibodies against various domains. C, Western blot analysis of STAT3 with antibody against N-terminal domain (H-190) in cancer cell cytoplasmic (C) and nuclear (N) extracts. D, EMSA demonstrates that STAT3 binds to a heteroduplex containing 106-bp hairpin structures, but not to corresponding linear DNA.
To validate AFM observations of STAT3 binding to hairpin structures within the pUC8F14C plasmid DNA, we also performed EMSA with recombinant STAT3 proteins. DNA fragments amplified from pUC8F14C and parental plasmid pUC8 were used for heteroduplex formation as previously described (25). The heteroduplex and linear DNA fragments were incubated with recombinant STAT3 protein. Data presented in Fig. 4D demonstrate that recombinant STAT3 binds to the heteroduplex containing hairpin, but not to the linear DNA.
The truncated isoforms, STAT3γ, have been previously observed in specific cell types (4). We fractionated breast and prostate cancer cells into cytoplasmic and nuclear protein extracts to determine the presence of C-terminal truncated isoforms using an antibody against the N-terminal domain of STAT3. The 67.5-kDa isoform was barely detected in the cytoplasmic protein extracts, but was readily detected in nuclear extracts of MCF-7, PC3, and DU145 cancer cells (Fig. 4C). The presence of nuclear 67.5-kDa STAT3 that is capable of DNA binding suggests that this isoform may play a regulatory role in cancer cells. Taken together, our data demonstrate that U-STAT3 may recognize specific DNA structural elements, suggesting an architectural role for this protein.
DISCUSSION
Our experimental data provide direct evidence that U-STAT3 is capable of binding to DNA. We observed that U-STAT3 binds to GAS sequences as a dimer or monomer, consistent with previous observations by Yang et al. (8–10) supporting a role for U-STAT3 in regulation of gene expression. Our data show that U-STAT3 behaves similarly to U-STAT1 that binds to DNA as a monomer by contacting one-half of a palindromic GAS motif (30), or form dimers through N-domain interactions that also allow them to bind GAS sequences in vitro, although with much lower affinity, compared with the phosphorylated STAT1 dimer (31, 32). Microscale thermophoresis confirms that U-STAT3 binds to DNA with low affinity (the apparent KD is 37.9 μm). This is consistent with KD values reported for other transcription factors (33). Our data suggest that U-STAT3 acts as a weak, but potentially biologically relevant transcription factor through its direct binding to a responsive GAS promoter.
However, we observed that U-STAT3 also binds to AT-rich DNA sequences. These data are in agreement with a report by Yoo et al. (34) demonstrating that AT-rich sequences within the α2-macroglobulin promoter are necessary for STAT3- and c-Jun-dependent synergistic gene activation. AT-rich regions play important roles in regulation of gene expression and/or chromatin organization due to their special structure with a narrow minor groove that can be recognized by proteins (35–38). Using microscale thermophoresis, we found that U-STAT3 binds to AT-rich sequences with slightly higher affinity than to the GAS element. Mutation of A to G nucleotides within hexanucleotides 5′-AAACAA, 5′-AACAAA, and 5′-CAAAAA, frequently found in S/MARs (26, 27), decreased STAT3-DNA binding. S/MARs DNA elements activate or inhibit gene expression indirectly by regulating chromatin loop organization and functioning as a “platform” for the aggregation of chromatin-remodeling proteins (38). Our data provide evidence suggestive of an architectural role of U-STAT3 in cells through its binding to the 4-way junctions and DNA crossings. Future studies will determine the mechanism for the U-STAT3 architectural role in chromatin organization through binding to AT-rich DNA sequences.
Previous reports have demonstrated that seemingly unrelated proteins, which strongly bind in vitro to the 4-way junction DNA, appear to function as architectural proteins (39, 40). The ability to bind AT-rich DNA sequences and recognize DNA structures may be a constitutive trait of architectural proteins. The most common function for the proteins demonstrating binding to crossed and/or bent DNA is to promote the formation of higher order protein-DNA or protein-protein complexes (38, 39, 41). A well studied example is the DNA bending caused by LEF-1 binding to the center of the enhancer of the T cell receptor α gene, thus allowing interactions among several transcription factors (41). Another lies in SATB1, a MAR-binding protein, that regulates genes by folding chromatin into a loop domain and attracting other regulatory proteins (38). Recent reports by Shi et al. (12, 13) on noncanonical JAK-STAT signaling in Drosophila demonstrated that dissociation of U-STAT from DNA in response to cytokine signaling triggers changes in chromatin dynamics. To date, there is little evidence for this mechanism in mammalian cells, although a role for STAT family members in chromatin remodeling in mammalian cells has been suggested (42–44). Christova et al. (43) showed that transcriptional induction of the human major histocompatibility complex requires JAK-STAT signaling to trigger higher-order chromatin remodeling of the entire major histocompatibility complex locus preceding its transcriptional activation. Shi et al. (44) observed that activation of JAK3-STAT5 during T-helper cell differentiation causes chromatin remodeling at the interferon-γ (Ifng) locus resulting in Ifng accessibility and histone acetylation. Based on our data, it is tempting to speculate that the ability of U-STAT3 to directly bind to AT-rich DNA regions and/or to crossed or bent DNA, bringing together distant DNA regions (DNA looping) and other proteins, may underlie the mechanism by which STAT3 controls chromatin organization. Indeed, we observed the situation in vitro when full-length U-STAT3 proteins bring together distant regions of plasmid DNAs similar to the DNA looping mechanism induced by MAR proteins. In Drosophila, phosphorylation of STAT92E triggers its dissociation from heterochromatin and stimulates binding to its own DNA binding motif. Consistent with findings in Drosophila, we propose that phosphorylation of STAT3 may trigger its dissociation from AT-rich DNA sites and stimulate binding to the GAS motif. This would result in changes in DNA looping and chromatin reorganization that takes place after JAK-STAT pathway activation. Further studies are needed to explore this hypothesis in vivo.
Finally, our AFM imaging data has led us to identify a C-terminal truncated 67.5-kDa isoform present in nuclear extracts of cancer cells. Previous reports have shown that limited proteolytic processing by serine proteases, calcium-dependent cysteine protease calpain, or by caspases generate STAT-truncated proteins, designated STATγs (45–48). Post-translation truncation of the C-terminal domain and tyrosine phosphorylation site makes STATγ isoforms distinct from STAT3α isoforms structurally and functionally. Because STATγ molecules retain the DNA-binding domain, they are viewed as dominant-negative regulators of STAT-dependent transcription (46, 48) that may have entirely different functions from full-length STAT3α that was a primary object of our study. Our data shows that the 67.5-kDa isoform binds to the regions of single-stranded DNA within a cruciform formed by a palindrome sequence. Palindrome sequences, which are often capable of forming an alternative DNA structure (cruciform) have been implicated in various DNA-mediated processes including replication, transcription, and genomic instability (49). Palindromes often act as symmetrical recognition sites for the binding of regulatory factors, especially dimeric transcription factors (40, 49–51). The GAS element itself is an imperfect palindrome (TTCNNNGAA), although rather short, providing STAT3 with secondary structure recognition, as well as the primary DNA sequences (52, 53). Therefore, the C-terminal truncated 67.5-kDa isoform may directly regulate gene transcription through binding to palindrome sequences within gene promoters. Moreover, because this C-terminal truncated isoform cannot undergo phosphorylation, it may function independently of JAK activation and interfere with functions of full-length STAT3.
In conclusion, investigations of DNA binding by U-STAT3 suggest that U-STAT3 may be involved in the regulation of gene activity through binding to AT-rich DNA sequences, regulation of chromatin structure, and recognition of secondary structures in addition to transcriptional activator functions. The expression of the C-terminal truncated isoform and its binding to palindrome structures in DNA may interfere with functions of full-length U-STAT3 and effect a biological role in cancer cells. Enhanced understanding of these additional DNA binding mechanisms of STAT3 offers a broadened view of potential molecular targets for specific functions of these molecules.
Acknowledgments
We thank LCCC Proteomics and Tissue Cores for excellent assistance. Shared resources of the Lombardi Comprehensive Cancer Center are partially supported by National Institutes of Health Grant CA56036-08 (Cancer Center Support Grant, to Lombardi Comprehensive Cancer Center).
This work was supported, in whole or in part, by the National Institutes of Health Grant CA56036-08, American Cancer Society Grant IRG 97-152-17 (to O. T.) and by the National Institutes of Health Intramural Research Program of the NCI Center for Cancer Research.
- GAS
- γ-activated sequence
- U-STAT3
- unphosphorylated STAT3
- AFM
- atomic force microscopy
- APS
- 1-(3-aminopropyl)silatrane
- DAPA
- DNA affinity precipitation assays
- S/MAR
- scaffold/matrix-attached region.
REFERENCES
- 1. Darnell J. E., Jr. (1997) STATs and gene regulation. Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 2. Bromberg J., Chen X. (2001) STAT proteins. Signal tranducers and activators of transcription. Methods Enzymol. 333, 138–151 [DOI] [PubMed] [Google Scholar]
- 3. Levy D. E., Darnell J. E., Jr. (2002) Stats, transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 4. Lim C. P., Cao X. (2006) Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2, 536–550 [DOI] [PubMed] [Google Scholar]
- 5. Ramos H. L., O'Shea J. J., Watford W. T. (2007) STAT5 isoforms. Controversies and clarifications. Biochem. J. 404, e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Stat3 as an oncogene. Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 7. Turkson J., Jove R. (2000) STAT proteins, novel molecular targets for cancer drug discovery. Oncogene 19, 6613–6626 [DOI] [PubMed] [Google Scholar]
- 8. Yang J., Chatterjee-Kishore M., Staugaitis S. M., Nguyen H., Schlessinger K., Levy D. E., Stark G. R. (2005) Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 65, 939–947 [PubMed] [Google Scholar]
- 9. Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. (2007) Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Stark G. R. (2008) Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 11. Reich N. C., Liu L. (2006) Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 12. Shi S., Calhoun H. C., Xia F., Li J., Le L., Li W. X. (2006) JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 38, 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi S., Larson K., Guo D., Lim S. J., Dutta P., Yan S. J., Li W. X. (2008) Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 10, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chasovskikh S., Dimtchev A., Smulson M., Dritschilo A. (2005) DNA transitions induced by binding of PARP-1 to cruciform structures in supercoiled plasmids. Cytometry A 68, 21–27 [DOI] [PubMed] [Google Scholar]
- 15. Pietrasanta L. I., Thrower D., Hsieh W., Rao S., Stemmann O., Lechner J., Carbon J., Hansma H. (1999) Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 96, 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schneider S. W., Lärmer J., Henderson R. M., Oberleithner H. (1998) Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 435, 362–367 [DOI] [PubMed] [Google Scholar]
- 17. R Development Core Team (2011). R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 18. Timofeeva O. A., Plisov S., Evseev A. A., Peng S., Jose-Kampfner M., Lovvorn H. N., Dome J. S., Perantoni A. O. (2006) Serine-phosphorylated STAT1 is a prosurvival factor in Wilms' tumor pathogenesis. Oncogene 25, 7555–7564 [DOI] [PubMed] [Google Scholar]
- 19. Timofeeva O. A., Gaponenko V., Lockett S. J., Tarasov S. G., Jiang S., Michejda C. J., Perantoni A. O., Tarasova N. I. (2007) Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem. Biol. 2, 799–809 [DOI] [PubMed] [Google Scholar]
- 20. Yu Q., Thieu V. T., Kaplan M. H. (2007) Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Rα gene during Th1 differentiation. EMBO J. 26, 2052–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100. [DOI] [PubMed] [Google Scholar]
- 23. Zillner K., Jerabek-Willemsen M., Duhr S., Braun D., Längst G., Baaske P. (2012) Microscale thermophoresis as a sensitive method to quantify protein. Nucleic acid interactions in solution. Methods Mol. Biol. 815, 241–252 [DOI] [PubMed] [Google Scholar]
- 24. Oussatcheva E. A., Shlyakhtenko L. S., Glass R., Sinden R. R., Lyubchenko Y. L., Potaman V. N. (1999) Structure of branched DNA molecules. Gel retardation and atomic force microscopy studies. J. Mol. Biol. 292, 75–86 [DOI] [PubMed] [Google Scholar]
- 25. Lonskaya I., Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 280, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 26. Liebich I., Bode J., Frisch M., Wingender E. (2002) S/MARt DB, a database on scaffold/matrix-attached regions. Nucleic Acids Res. 30, 372–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liebich I., Bode J., Reuter I., Wingender E. (2002) Evaluation of sequence motifs found in scaffold/matrix-attached regions (S/MARs). Nucleic Acids Res. 30, 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen B., Young J., Leng F. (2010) DNA bending by the mammalian high-mobility group protein AT hook 2. Biochemistry 49, 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lobov I. B., Tsutsui K., Mitchell A. R., Podgornaya O. I. (2001) Specificity of SAF-A and lamin B binding in vitro correlates with the satellite DNA bending state. J. Cell Biochem. 83, 218–229 [DOI] [PubMed] [Google Scholar]
- 30. Chatterjee-Kishore M., Wright K. L., Ting J. P., Stark G. R. (2000) How Stat1 mediates constitutive gene expression. A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19, 4111–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E., Jr., Kuriyan J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 [DOI] [PubMed] [Google Scholar]
- 32. Vinkemeier U., Cohen S. L., Moarefi I., Chait B. T., Kuriyan J., Darnell J. E., Jr. (1996) DNA binding of in vitro activated Stat1α, Stat1β, and truncated Stat1. Interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15, 5616–5626 [PMC free article] [PubMed] [Google Scholar]
- 33. Fordyce P. M., Gerber D., Tran D., Zheng J., Li H., DeRisi J. L., Quake S. R. (2010) De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol. 28, 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoo J. Y., Wang W., Desiderio S., Nathans D. (2001) Synergistic activity of STAT3 and c-Jun at a specific array of DNA elements in the α2-macroglobulin promoter. J. Biol. Chem. 276, 26421–26429 [DOI] [PubMed] [Google Scholar]
- 35. Moreau J., Marcaud L., Maschat F., Kejzlarova-Lepesant J., Lepesant J. A., Scherrer K. (1982) A+T-rich linkers define functional domains in eukaryotic DNA. Nature 295, 260–262 [DOI] [PubMed] [Google Scholar]
- 36. Oosumi T., Garlick B., Belknap W. R. (1995) Identification and characterization of putative transposable DNA elements in solanaceous plants and Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 92, 8886–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patikoglou G. A., Kim J. L., Sun L., Yang S. H., Kodadek T., Burley S. K. (1999) TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 [DOI] [PubMed] [Google Scholar]
- 39. Zlatanova J., van Holde K. (1998) Binding to four-way junction DNA. A common property of architectural proteins? FASEB J. 12, 421–431 [PubMed] [Google Scholar]
- 40. Brázda V., Laister R. C., Jagelská E., Arrowsmith C. (2011) Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Love J. J., Li X., Case D. A., Giese K., Grosschedl R., Wright P. E. (1995) Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376, 791–795 [DOI] [PubMed] [Google Scholar]
- 42. Li W. X. (2008) Canonical and noncanonical JAK-STAT signaling. Trends Cell Biol. 18, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Christova R., Jones T., Wu P. J., Bolzer A., Costa-Pereira A. P., Watling D., Kerr I. M., Sheer D. (2007) P-STAT1 mediates higher-order chromatin remodeling of the human MHC in response to IFNγ. J. Cell Sci. 120, 3262–3270 [DOI] [PubMed] [Google Scholar]
- 44. Shi M., Lin T. H., Appell K. C., Berg L. J. (2008) Janus kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity 28, 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hevehan D. L., Miller W. M., Papoutsakis E. T. (2002) Differential expression and phosphorylation of distinct STAT3 proteins during granulocytic differentiation. Blood 99, 1627–1637 [DOI] [PubMed] [Google Scholar]
- 46. Azam M., Lee C., Strehlow I., Schindler C. (1997) Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity 6, 691–701 [DOI] [PubMed] [Google Scholar]
- 47. Oda A., Wakao H., Fujita H. (2002) Calpain is a signal transducer and activator of transcription (STAT) 3 and STAT5 protease. Blood 99, 1850–1852 [DOI] [PubMed] [Google Scholar]
- 48. Darnowski J. W., Goulette F. A., Guan Y. J., Chatterjee D., Yang Z. F., Cousens L. P., Chin Y. E. (2006) Stat3 cleavage by caspases. Impact on full-length Stat3 expression, fragment formation, and transcriptional activity. J. Biol. Chem. 281, 17707–17717 [DOI] [PubMed] [Google Scholar]
- 49. Humphrey-Dixon E. L., Sharp R., Schuckers M., Lock R. (2011) Comparative genome analysis suggests characteristics of yeast inverted repeats that are important for transcriptional activity. Genome 54, 934–942 [DOI] [PubMed] [Google Scholar]
- 50. LeBlanc J. F., McLane K. E., Parren P. W., Burton D. R., Ghazal P. (1998) Recognition properties of a sequence-specific DNA binding antibody. Biochemistry 37, 6015–6022 [DOI] [PubMed] [Google Scholar]
- 51. Lu L., Jia H., Dröge P., Li J. (2007) The human genome-wide distribution of DNA palindromes. Funct. Integr. Genomics 7, 221–227 [DOI] [PubMed] [Google Scholar]
- 52. Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. (2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276, 6675–6688 [DOI] [PubMed] [Google Scholar]
- 53. Seidel H. M., Milocco L. H., Lamb P., Darnell J. E., Jr., Stein R. B., Rosen J. (1995) Spacing of palindromic half-sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 92, 3041–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]