Background: Mutations in CNNM2 cause severe dominant hypomagnesemia.
Results: Structure of CNNM2 consists of an extracellular N terminus and intracellular C terminus containing CBS domains, which are affected by the identified mutations.
Conclusion: CNNM2 is intensively processed before being expressed in its final structure at the plasma membrane.
Significance: CNNM2 structure analysis will aid to elucidate CNNM2 function in renal magnesium transport.
Keywords: Kidney, Magnesium Homeostasis, Membrane Trafficking, Protein Processing, Protein Structure, CNNM2, Distal Convoluted Tubule, Hypomagnesemia
Abstract
Recently, mutations in the cyclin M2 (CNNM2) gene were identified to be causative for severe hypomagnesemia. In kidney, CNNM2 is a basolaterally expressed protein with predominant expression in the distal convoluted tubule. Transcellular magnesium (Mg2+) reabsorption in the distal convoluted tubule represents the final step before Mg2+ is excreted into the urine, thus fine-tuning its final excretion via a tightly regulated mechanism. The present study aims to get insight in the structure of CNNM2 and to characterize its post-translational modifications. Here, membrane topology studies using intramolecular epitopes and immunocytochemistry showed that CNNM2 has an extracellular N terminus and an intracellular C terminus. This suggests that one of the predicted transmembrane regions might be re-entrant. By homology modeling, we demonstrated that the loss-of-function mutation as found in patients disturbs the potential ATP binding by the intracellular cystathionine β-synthase domains. In addition, the cellular processing pathway of CNNM2 was exposed in detail. In the endoplasmic reticulum, the signal peptidase complex cleaves off a large N-terminal signal peptide of about 64 amino acids. Mutagenesis screening showed that CNNM2 is glycosylated at residue Asn-112, stabilizing CNNM2 on the plasma membrane. Interestingly, co-immunoprecipitation studies evidenced that CNNM2a forms heterodimers with the smaller isoform CNNM2b. These new findings on CNNM2 structure and processing may aid to elucidate the physiological role of CNNM2 in Mg2+ reabsorption in the kidney.
Introduction
As the second most abundant intracellular cation in humans, magnesium (Mg2+) constitutes an essential co-factor in vital processes such as energy metabolism, DNA transcription, and protein synthesis. Balance between intestinal absorption and renal excretion is tightly regulated to keep the plasma Mg2+ concentration in its physiological range (0.7–1.1 mm) (1). Specifically, bulk Mg2+ reabsorption in kidney from the pro-urine is dependent on passive, paracellular transport in the proximal tubule and the thick ascending limb of Henle's loop, whereas fine-tuning is achieved by active, transcellular transport in the distal convoluted tubule (DCT)2 (1). In the latter segment, tight regulation of Mg2+ transport will determine the final urinary Mg2+ excretion because no Mg2+ reabsorption takes place beyond the DCT. The TRPM6 ion channel, expressed at the apical side of the DCT, constitutes the gatekeeper in active Mg2+ uptake (2), whereas a basolateral Mg2+ extrusion mechanism remains to be identified.
Recently, mutations in the cyclin M2 (CNNM2) gene were described leading to a dominant form of hypomagnesemia caused by renal Mg2+ wasting (3). Patients suffer from muscle weakness, tremor, and headaches accompanied by low Mg2+ serum concentrations (0.3–0.5 mm) (3, 4). Interestingly, no other electrolyte disturbance was detected (3). CNNM2, formerly known as ancient conserved domain protein 2 (ACDP2), belongs to the CNNM family consisting of four proteins (CNNM1–4) that share homology to cyclins, although no cyclin-related function has been described (5). Two predicted CNNM2 protein isoforms are conserved between humans and mouse. Although the 875-amino acid CNNM2a is encoded by splice variant 1 (Cnnm2v1, NM_017649), the 22-residue-shorter isoform b is translated from splice variant 2 (Cnnm2v2, NM_199076), which lacks exon 6. CNNM2 expressed in Xenopus laevis oocytes and in the Mg2+-deficient Salmonella enterica strain MM281 has been suggested to be involved in Mg2+ transport (6, 7). However, electrophysiological analysis using mammalian cells expressing CNNM2 showed Mg2+-sensitive Na+ currents instead of Mg2+ currents (3). This last finding could indicate that instead of transporting Mg2+ itself, CNNM2 might be involved in a Mg2+-sensing mechanism that, in turn, could regulate renal Mg2+ uptake.
At present, no studies have been reported showing the structure of CNNM2 or any of the other CNNM family members. The aim of this study was to elucidate the membrane topology of CNNM2 and to provide a molecular characterization of the intracellular processing mechanisms.
EXPERIMENTAL PROCEDURES
Expression Profiling
Three C57BL/6 mice were sacrificed; kidney, duodenum, ileum, jejunum, colon, cecum, brain, lung, liver, spleen, testis, muscle, and heart tissues were collected. The transgenic parvalbumin-EGFP mice were a gift kind from Dr. Monyer (University of Heidelberg, Germany (8)), and TRPV5-GFP mice were kindly provided by Dr. Praetorius from the University of Aarhus, Denmark (9). They were used to isolate the DCT and connecting tubule (CNT), respectively. Mice were anesthetized by a mixture injection of ketamine (0.1 mg/g of body weight) and xylazine (0.01 mg/g of body weight) and perfused with 20 ml of PBS (in mm: 137 NaCl, 2.7 KCl, 10 Na2HPO4, 1.76 KH2PO4) through the heart. The kidneys were removed, minced, and digested in collagenase (1 mg/ml collagenase A (Worthington), 1 mg/ml hyaluronidase) in Krebs buffer (in mm: 145 NaCl, 5 KCl, 10 HEPES, 1 NaH2PO4, 2.5 CaCl2, 1.8 MgSO4, 5 glucose) at 37 °C for 18 min. The collagenase-digested tubules sized between 40 and 100 μm were sorted based on GFP fluorescence by COPAS (Complex Object Parametric Analysis and Sorting, Union Biometrica).
Total RNA was isolated using TRIzol total RNA isolation agent (Invitrogen). Obtained RNA was treated with DNase (Promega) to remove genomic DNA. Subsequently, reverse transcription of the RNA by M-MLV reverse transcriptase (Invitrogen) was performed for 1 h at 37 °C. Gene expression levels were determined by quantitative real-time PCR on a Bio-Rad analyzer and normalized for Gapdh expression levels. Primer sequences are provided in supplemental Table 1.
Immunohistochemistry
Immunohistochemistry was performed as described previously (10). In short, co-staining for CNNM2 with Na+-K+-Cl− cotransporter (NKCC2), thiazide-sensitive Na+-Cl− cotransporter (NCC), calbindin-D28k, and aquaporin-2 (AQP2) was performed on 7-μm sections of fixed frozen mouse kidney samples. The sections were incubated for 16 h at 4 °C with the following primary antibodies: guinea pig anti-CNNM2 (1:100 3)), rabbit anti-NKCC2 (1:100 (39)), rabbit anti-NCC (1:100 40)), rabbit anti-calbindin-D28k (1:500 (41)), and rabbit anti-AQP2 (1:300, kindly provided by Dr. Deen, Nijmegen, The Netherlands). For detection, kidney sections were incubated with Alexa Fluor-conjugated secondary antibodies. Images were taken with an AxioCam camera (Zeiss) and AxioVision software (Zeiss).
DNA Constructs
Mouse Cnnm2 was cloned into the pCINeo HA IRES GFP vector as described previously (3). To obtain a C-terminal vesicular stomatitis virus epitope, an XbaI restriction site was introduced before the stop codon using the QuikChange site-directed mutagenesis kit (Agilent, Amstelveen, The Netherlands) according to the manufacturer's protocol. Subsequently, oligonucleotides (forward, 5′-GAATCGCTTGGGGAAGTAGCTCGAGAATTCCGCCCC-3′, reverse, 5′-GGGGCGGAATTCTCGAGCTACTTCCCCAAGCGATTC-3′) encoding the vesicular stomatitis virus epitope were hybridized and ligated into the pCINEO HA-CNNM2 IRES GFP vector using the XbaI and XhoI restriction sites. CNNM2 mutations were inserted in the construct using the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol. Human CNNM4 full-length cDNA clone (GenBankTM accession number BC063295) was amplified using Phusion polymerase (Finnzymes, Vantaa, Finland) and primers (forward, 5′-CGGCTAGCGCCACCATGGCGCCGGTGG-3′, reverse, 5′-CGACCGGTCTATGCGTAGTCTGGCACGTCGTATGGGTAGATGGCATTCTCGTGGGAG-3′) to introduce an in-frame HA tag before the stop codon and AgeI and NheI restriction sites. The amplicons were cloned into the pCINeo IRES GFP expression vector using the AgeI and NheI restriction sites. All constructs were verified by sequence analysis.
Mouse Cnnm2 was cloned into pcDNA3 (Invitrogen) as described previously (3). To obtain splice variant 2 with a 66-nucleotide deletion corresponding to exon 6, site-directed mutagenesis (QuikChange, Agilent) was used, according to the two-stage PCR mutagenesis protocol described by Wang and Malcolm (11). The same protocol was used to delete the C-terminal tag and to insert internal HA tags with an additional serine residue (NH2-YPYDVPDYAS-COOH) (12) after residues Thr-24, Gly-210, or Pro-744 or to replace the C-terminal HA tag with a FLAG sequence. Mutagenesis primer sequences are available upon request. To obtain an N-terminal HA-tagged expression clone, the ORF in pFLCI-mCnnm2WT (3) was amplified using Expand polymerase (Roche Applied Science) using forward primer 5′-ATGGGGTACCATGGCTTACCCATATGATGTTCCAGATTACGCTATTGGCTGTGGCGCTTGTG-3′ containing an in-frame HA tag after the Kozak sequence and reverse primer 5′-ATCTTCTAGAGGTCAGACCAAGTCCAGGAG-3′ and subsequently cloned in pcDNA3 as described previously (13). The complete ORFs of all expression constructs were sequence-verified.
Cell Culture and Transfection
HEK293 cells were grown at 37 °C in DMEM (Biowhittaker Europe, Vervier, Belgium) supplemented with 10% (v/v) FCS (PAA Laboratories, Linz, Austria), nonessential amino acids, and 2 mm l-glutamine in a humidified 5% (v/v) CO2 atmosphere. A few hours before transfection with Lipofectamine 2000 (Invitrogen), cells were seeded in 12-well plates and subsequently transiently transfected with 0.5 μg of pCINeo-IRES-GFP constructs encoding HA-tagged wild type or mutated CNNM2 proteins at a 1:2 DNA to Lipofectamine ratio. COS-7 cells (ATCC CRL-1651) were grown in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Biochrom, Berlin, Germany) in a humidified environment of 5% (v/v) CO2 at 37 °C.
Immunocytochemistry
COS-7 cells were seeded on glass coverslips and transfected using Lipofectamine 2000 according to the manufacturer's protocol with constructs expressing epitope-tagged CNNM2 or CNNM4. After 2 days of growth in medium supplemented with 20 mm MgCl2, the cells were rinsed with PBS and fixed for 10 min with 4% (w/v) methanol-free formaldehyde solution (Thermo Scientific) in PBS. In a control experiment (supplemental Fig. S1, lower panels), living cells were incubated with primary antibody rat anti-HA (Roche Applied Science, high affinity 3F10, 1:250) for 60 min at 4 °C before formaldehyde fixation. After rinses in PBS, some of the cells were incubated for 10 min with 0.2% (v/v) Triton X-100 in PBS for permeabilization. After PBS rinses, the cells were incubated with blocking buffer (5% (v/v) donkey serum (Sigma) in PBS) for 1 h and immunolabeled with the primary antibodies rat anti-HA (1:250), mouse anti-FLAG M2 (Sigma, 1:250), rabbit anti-calnexin (C5C9, Cell Signaling Technology, 1:50), or rabbit anti-CNNM2 (Abcam, Ab111351, 1:50) in blocking buffer for 90 min. After rinses in PBS, cells were incubated with Alexa Fluor 488-conjugated donkey anti-rabbit or anti-mouse IgG or Alexa Fluor 594-conjugated donkey anti-rat IgG secondary antibodies (Invitrogen, 1:500) and DAPI (2 μg/ml) for 45 min. After PBS and ethanol rinses, the cells were mounted on slides using ProLong Gold antifade reagent (Invitrogen). Fluorescence microscopy was performed with a Leica DMIRE2 inverted microscope, and images were taken with the Openlab software.
Homology Modeling
A homology model was built using the modeling script in the WHATIF and YASARA Twinset with standard parameters (14, 15). We used the structure of a cystathionine B-synthase (CBS) domain pair structure of the Mg2+ and Co2+ efflux protein CorC from Bordetella parapertussis (Protein Data Bank (PDB) file 3jtf) as a template for modeling.
Cell Surface Biotinylation, PNGase Treatment, and Immunoblotting
Biotinylation experiments were performed as described previously (16). In short, HEK293 cells were transfected for 48 h, and cell surfaces were biotinylated at 4 °C for 1 h by adding sulfo-NHS-LC-LC-biotin. Subsequently, protein lysates were incubated overnight with NeutrAvidin-agarose beads (Pierce).
Protein lysates were denatured in Laemmli containing 100 mm DTT for 30 min at 37 °C and subsequently subjected to SDS-PAGE. Then, immunoblots were incubated with mouse anti-HA (Cell Signaling Technology) primary antibodies and peroxidase conjugated sheep anti-mouse secondary antibodies (Sigma). PNGase F (New England Biolabs) was added to the protein lysates in Laemmli-DTT buffer and incubated at 37 °C for 1 h before loading the protein samples on gel.
Co-immunoprecipitation
COS-7 cells were cultured and transfected as described above with expression vectors for HA- and FLAG-tagged wild type CNNM2 or CNNM4 isoforms. 48 h after transfection, cells were scraped from dishes in ice-cold 50 mm Tris (pH 8.0) followed by centrifugation at 5000 × g. The pellets were lysed in lysis buffer (150 mm NaCl; 0.1% (v/v) Nonidet P-40; 50 mm Tris, pH 7.5; and Complete protease inhibitor mixture (Roche Applied Science)) by repeated passage through a 26-gauge needle. Protein concentration measurement was performed using a BCA assay (Pierce). The next incubation steps were all done under rotary agitation at 4 °C. Protein aliquots of 100 μg in lysis buffer were precleared by incubation with protein G-Sepharose (Sigma-Aldrich) for 2 h after which the precleared lysate was incubated for 18 h with 2.5 μg of anti-HA antibody (Roche Applied Science, high affinity 3F10). The antibody-lysate mixture was added to 50 μl of fresh protein G-Sepharose beads and incubated for 1 h. The beads were collected by centrifugation at 1000 × g for 1 min at 4 °C and washed four times with lysis buffer. The proteins were separated from the beads by boiling for 5 min in 1× Laemmli sample buffer and detected by immunoblotting using a mouse monoclonal antibody against FLAG M2 (Sigma-Aldrich, 1:1000).
In Vitro Transcription/Translation
The cDNA template for in vitro transcription contained the Cnnm2-HA sequence downstream of a T7 promoter sequence in a pT7Ts vector. The coupled in vitro transcription/translation reaction was incubated at 30 °C for 90 min. The reaction mixture contained 2 μg of cDNA template, 2 μl of reaction buffer, 1 μl of T7 RNA polymerase, 1 μl of amino acid mix without methionine, 3 μl of [35S]Met (1 μCi/μl), RNasin, and 25 μl of rabbit reticulocyte lysate (Promega). The reaction was performed with or without signal peptidase inhibitor MeoSuc-Ala-Ala-Pro-Val-chloromethylketone (AAPV-CMK, Sigma). The protein mixture was separated on an 8% (w/v) SDS-PAGE gel. The gel was fixed in a mixture of acetic acid, methanol, and H2O (20:10:70) and incubated in 20% (w/v) 2,5-diphenyloxazole in dimethyl sulfoxide (DMSO) to intensify the radioactive signal. The proteins were visualized using CL-XPosure films (Pierce, Etten-Leur, The Netherlands).
Statistical Analysis
In all experiments, data are expressed as means ± S.E. Statistical significance was determined using an unpaired Student's t test, and differences with p < 0.05 were regarded as statistically significant.
RESULTS
Expression Profile of CNNM Family
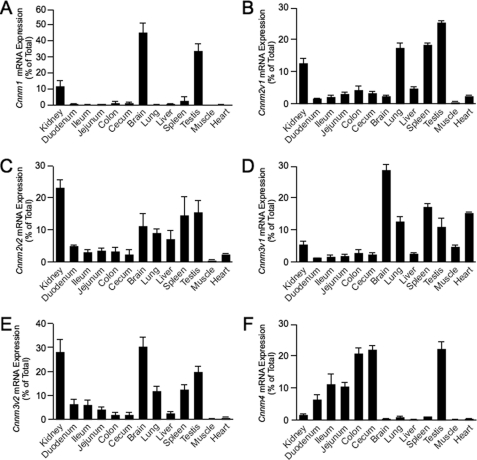
To examine the transcript expression levels for CNNM1–4 in various tissues, a mouse tissue cDNA panel was constructed, and Cnnm mRNA expressions were quantified by real-time PCR analysis. Gapdh expression was used to normalize values. Cnnm3v1 and Cnnm3v2 (containing an alternative last exon) show a ubiquitous expression pattern with highest expression in kidney, brain, lung, spleen, and heart (Fig. 1, D and E). Cnnm1 expression is predominantly found in brain and testis (Fig. 1A), whereas Cnnm4 shows highest expression along the gastrointestinal tract (Fig. 1F). Interestingly, Cnnm2 shows a ubiquitous expression pattern with highest expression in kidney, lung, spleen, and testis (Fig. 1, B and C). CNNM2A and CNNM2B (encoded by Cnnm2v1 and Cnnm2v2, lacking exon 6) share a similar pattern; only minor differences between the tissue expression levels were measured.
FIGURE 1.
Tissue expression pattern of CNNM isoform-encoding transcripts. A–F, the mRNA expression levels of Cnnm1 (A), Cnnm2 splice variant 1 (B), Cnnm2 splice variant 2 (C), Cnnm3 splice variant 1 (D), Cnnm3 splice variant 2 (E), and Cnnm4 (F) in a panel of mouse tissues were measured by quantitative RT-PCR and normalized for Gapdh expression. Data represent the mean of three individual experiments ± S.E. and are expressed as the percentage of the total tissue expression.
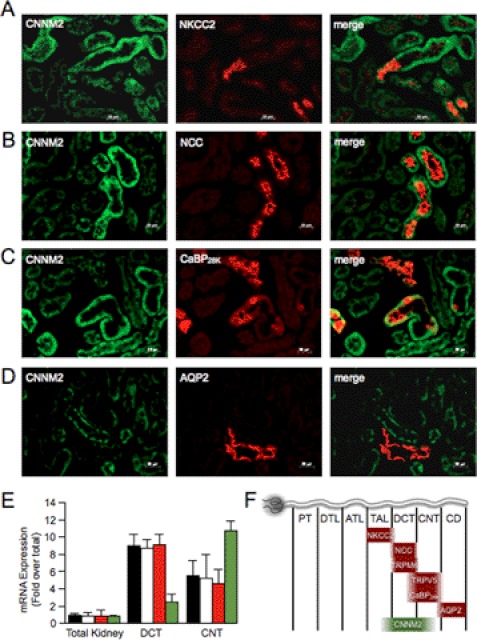
Because CNNM2 has been described to be causative for hypomagnesemia due to excessive renal Mg2+ wasting (3), its kidney localization was investigated in detail. Using immunohistochemistry, co-expression of CNNM2 with markers of different distal nephron segments was examined: the Na-K-Cl cotransporter NKCC2 (thick ascending limb of Henle's loop), the thiazide-sensitive Na-Cl cotransporter NCC (DCT), calbindin-D28k (CNT and cortical collecting duct), and aquaporin-2 (collecting duct). CNNM2 was expressed on the basolateral side of the cells (Fig. 2, A–D, green), opposed to the luminal expression of the marker proteins (Fig. 2, A–D, red). CNNM2 immunopositive tubules were negative for aquaporin-2 staining, but did co-stain with NCC-positive tubules. Only a part of the tubules positive for calbindin-D28k showed CNNM2 staining. Although most of NKCC2-positive tubules are negative for CNNM2, only a few NKCC2-positive tubules show faint CNNM2 staining. These results indicate that CNNM2 is predominantly expressed in DCT. mRNA expression levels measured by real-time PCR on GFP-sorted DCT and CNT tubules showed an equivalent pattern; expression of Cnnm2v1 and Cnnm2v2 was high in DCT and to a lesser extent also in the CNT (Fig. 2E). As a control for DCT and CNT specificity of the mRNA, we examined the expression of the transcripts for the marker proteins TRPM6 and TRPV5, respectively (Fig. 2E). These and previous results (3) indicate that CNNM2 is predominantly expressed basolaterally in DCT and also, but in lower amounts, present in thick ascending limb of Henle's loop and CNT (Fig. 2F).
FIGURE 2.
Kidney expression pattern of CNNM2 shows highest expression in DCT. A–D, double immunofluorescence staining of mouse kidney cortex sections for CNNM2 (in green) and NKCC2 (A, red), NCC (B, red), calbindin-D28k (CaBP28K) (C, red), or aquaporin-2 (D, red). Bars represent 20 μm. E, the mRNA expression levels of Cnnm2v1 (black bars), Cnnm2v2 (white bars), Trpm6 (red bars), and Trpv5 (green bars) in COPAS-selected mouse DCT, CNT, and control (none-selected) kidney tubules were measured by quantitative RT-PCR and normalized for Gapdh expression. Data represent the mean of three individual experiments ± S.E. and are expressed as -fold difference when compared with the expression in none-selected tubules. F, a schematic overview of marker protein kidney expression. PT, proximal tubule; DTL, descending thin limb of Henle's loop; ATL, ascending thin limb of Henle's loop; TAL, thick ascending limb of Henle's loop; CD, collecting duct.
CNNM2 Has an Intracellular C Terminus Containing CBS Domains
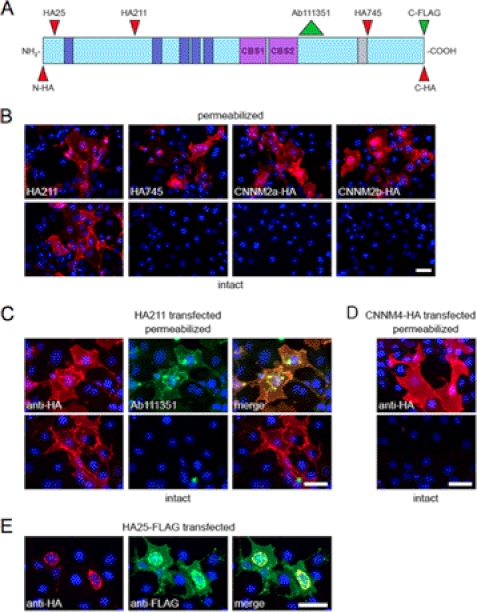
The consensus topology prediction for the plasma membrane protein CNNM2 indicates five potential transmembrane (TM) domains and an extracellular C terminus (3), which would locate the CBS domains (see below) in an unprecedented, extracellular position. We decided to investigate the topology in detail using an intramolecular epitope strategy by expression of various epitope-tagged CNNM2 proteins (Fig. 3A) in COS-7 cells with subsequent immunocytochemistry. A similar approach was used successfully to aid in the topology determination of other plasma membrane transporters such as multidrug resistance proteins (17) and an iron transporter (18). Immunocytochemistry of cells overexpressing CNNM2a proteins with HA epitopes at positions 211 (HA211), 745 (HA745), or at the extreme C terminus revealed these proteins to be targeted to the plasma membrane of the cells (Fig. 3B, upper panels), similar to the results obtained in MDCK-C7 cells (3). Importantly, C-terminally HA-tagged CNNM2b showed a localization that is undistinguishable from that of CNNM2a, indicating that the absence of the amino acids translated from exon 6 does not influence sorting to the plasma membrane.
FIGURE 3.
CNNM2 topology determination. A, linear representation of the CNNM2 protein with domain structures based on UniProt Q3TWN3. Predicted TM domains are indicated in dark blue, CBS domains are in purple, and the exon 6 coding region, which is absent in CNNM2b, is in gray. The position of the epitope tags and the epitope recognized by the Abcam 111351 antibody are indicated by narrow and wide arrowheads, respectively. N-HA, N-terminal HA tag; C-HA, C-terminal HA tag. B–E, immunofluorescence images of transiently transfected COS-7 cells. Bars represent 50 μm in each panel. Red signal represents immunodetected HA epitopes. Nuclei stained with DAPI are shown in blue. B, permeabilized (upper panels) and intact (lower panels) cells expressing, from left to right: CNNM2a with an HA tag at positions 211 or 745 or at the C terminus and CNNM2b with a C-terminal HA tag. C, cells, expressing CNNM2a with an HA tag at position 211, were additionally stained with Ab111351 (in green) under permeabilized and nonpermeabilized conditions (upper and lower panels, respectively). D, permeabilized (top) and intact (bottom) cells transfected with C-terminally HA-tagged CNNM4. E, cells transfected with C-terminally FLAG-tagged CNNM2a with an intramolecular HA epitope at position 25. Immunodetection of permeabilized cells for the HA tag (left), FLAG tag (middle, in green) and the merged image on the right are indicated.
To determine whether the epitopes are located on the inside or the outside of the cells, formaldehyde-fixed cells, both intact and permeabilized, were immunostained for the HA epitope. The HA tag at position 211 could be detected on intact cells (Fig. 3B, lower panels), indicating extracellular localization. On the contrary, detection of HA745 or the epitope at the C terminus required cell permeabilization, suggesting that these latter two are located intracellularly. Similar results were obtained when living cells were incubated with anti-HA antibodies prior to fixation (supplemental Fig. S1).
As an alternative approach to confirm that the C terminus is located intracellularly, cells that expressed HA211 were immunostained with Abcam antibody 111351, raised against amino acids 590–640, located just after the pair of CBS domains. Also here, the epitope could only be detected after permeabilization (Fig. 3C), indicative for an intracellular position of the C terminus that importantly includes the two CBS domains.
To investigate whether the intracellular localization of the C terminus is shared with other members of the CNNM family, the same permeabilization assay was performed on cells transfected with C-terminally HA-tagged CNNM4 (Fig. 3D). Again, the plasma membrane staining was only observed after permeabilization, indicating that also for CNNM4, the C terminus resides inside the cell. In conclusion, the CNNM2 protein has a membrane topology with its C terminus inside of the cell.
CNNM2 N Terminus Contains a Large Signal Peptide and a Glycosylation Site
We observed that the HA epitope at the extreme N terminus or at position 25 of CNNM2a was not detected at the plasma membrane, but rather in distinct perinuclear structures (supplemental Fig. S2), which suggested that the N terminus might contain a signal peptide and, therefore, ultimately results in cleavage of the HA epitope. To exclude incorrect processing of the recombinant protein, a construct was expressed with the HA epitope at position 25 of CNNM2a and a FLAG epitope at the C terminus. We observed that the FLAG epitope was correctly targeted at the plasma membrane, but the HA epitope was retained in perinuclear structures (Fig. 3E). Colocalization studies with an antibody against calnexin (supplemental Fig. S2) showed that these perinuclear structures are part of the endoplasmic reticulum (ER), which is in line with the presence of a signal peptide.
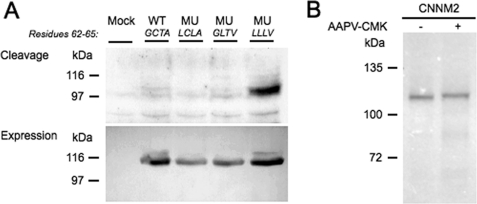
Signal peptide prediction software (SignalP (19)) predicts a long N-terminal signal peptide of 64 amino acids. Examination of the N-terminally HA-tagged CNNM2 construct by immunoblotting confirmed that the CNNM2 N terminus is cleaved (Fig. 4A, upper panel) because the HA tag was undetectable. Mutation of the predicted cleavage region (62GCTA65) to hydrophobic residues 62LLLV65 marred this cleavage event, evidenced by detection of an HA-positive band in the mutant (Fig. 4A, upper panel, right). To identify the exact cleavage site, constructs were generated in which the Cys-63/Ala-65 and the Gly-62/Thr-64 positions were mutated to hydrophobic amino acids. However, none of these two mutations were able to impair the cleavage of CNNM2 (Fig. 4A, upper panel).
FIGURE 4.
SPC cleaves the CNNM2 signal peptide. A, cleavage analysis of the CNNM2 protein by immunoblotting of double-tagged CNNM2 transfected HEK293 cells against the N-terminal HA tag (upper blot). As a control for CNNM2 expression, the noncleavable CNNM2 C terminus was targeted by immunoblotting against the vesicular stomatitis virus tag (lower blot). The blot shown is representative of five independent experiments. WT, wild type; MU, mutant. B, coupled in vitro transcription/translation protein synthesis of the CNNM2-HA protein in the presence (+) and absence (−) of SPC inhibitor AAPV-CMK. The gel shown is representative of three independent experiments.
To confirm that the signal peptidase complex (SPC, a protein complex expressed on the ER membrane known to be involved in signal peptide cleavage) facilitates cleavage, an in vitro protein translation assay in the presence of SPC inhibitor AAPV-CMK was performed. SPC inhibition by AAPV-CMK prevented signal peptide cleavage, as shown by a higher molecular weight on gel (Fig. 4B). Together, we conclude that SPC is responsible for CNNM2 signal peptide cleavage.
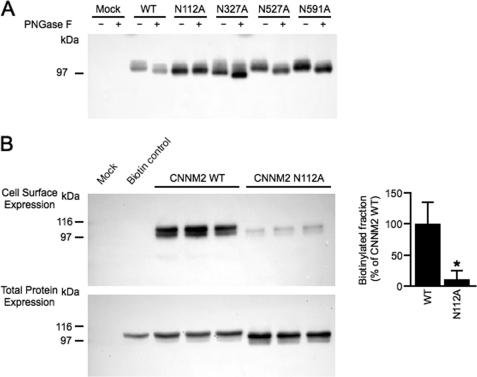
In eukaryotic cells, trafficking of membrane proteins to the cell surface is often regulated by glycosylation of asparagine residues present in extracellular domains. To determine the CNNM2 glycosylation profile, four potential candidate asparagine residues were mutated to alanine, namely Asn-112, Asn-327, Asn-527, and Asn-591 (based on prediction software NetNGlyc 1.0, Center for Biological Sequence Analysis (CBS)). Subsequently, the presence of N-linked glycans in the mutants was discerned by immunoblotting the proteins after treatment with PNGase F, an enzyme causing N-glycan cleavage. The glycosylated wild type CNNM2 runs at ∼105 kDa, but PNGase F treatment reduced its molecular mass to ∼96 kDa (Fig. 5A). Of all the candidates, only residue Asn-112 was identified as a glycosylation target in CNNM2 because the N112A mutation resulted in a molecular mass of 96 kDa (Fig. 5A). PNGase F treatment did not cause a further reduction of the molecular mass, indicating that no N-glycan is present in this mutant. To determine the effect of the N-glycan at position 112 on membrane expression of the CNNM2 protein, cell surface biotinylation studies of the N112A mutant were performed (Fig. 5B, left panel). A reduction of 90% in plasma membrane expression of the CNNM2-N112A mutant was observed when compared with the wild type CNNM2 protein (Fig. 5B, right panel), suggesting that Asn-112 glycosylation is necessary for CNNM2 membrane stability. Taken together, the CNNM2 extracellular N terminus provides important sites for post-translational modifications necessary for proper plasma membrane expression.
FIGURE 5.
Glycosylation on residue Asn-112 regulates CNNM2 membrane trafficking. A, immunoblot of C-terminally HA-tagged wild type CNNM2 (WT) and CNNM2 glycosylation mutants treated with (+) or without (−) PNGase F. B, cell surface biotinylation of HEK293 cells expressing HA-tagged wild type CNNM2 or glycosylation mutant N112A. Upper and lower blots, representative immunoblots showing that glycosylation at position Asn-112 is necessary for CNNM2 membrane expression (upper blot) and a CNNM2 expression control (lower blot). On the right, a quantification by blot intensities is shown of cell surface expression of CNNM2-WT (n = 6) and CNNM2-N112A (n = 6) corrected for total protein expression. Error bars represent S.E.; *, p < 0.05 versus CNNM2-WT.
C-terminal CBS Domains Are Disrupted by T568I Mutation
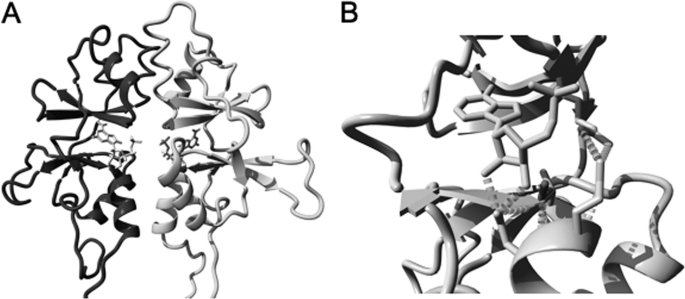
One of the mutations recently described to be causative for hypomagnesemia is located in the CBS domains within the C terminus (3). Therefore, emphasis was placed on determining the structure of the intracellular CBS domains (Fig. 3A) that have been associated with ATP binding, among other functions (20). Here, we provide a homology model of the CNNM2 CBS domains based on the B. parapertussis Mg2+/Co2+ CorC protein (Fig. 6A). Analysis of this model showed conservation of the potential ATP-binding site in the CBS dimer of CNNM2 (Fig. 6A). Interestingly, the T568I mutation, identified in a Czech family to be causative for hypomagnesemia (3), is lining the ATP-binding site in the CBS domain (Fig. 6B). In our model, mutation of the threonine into the bigger isoleucine residue causes steric bumps with the ATP molecule. Besides that, Thr-568 also forms a hydrogen bond that stabilizes the position of the Glu-570 and Asp-571 residues (Fig. 6B). These two residues form important hydrogen bonds and ionic interactions to the ATP molecule and residues lining the ATP-binding pocket, respectively. According to the model, the T568I mutation of the patient will also affect the position of the Glu-570 and Asp-571 residues and could, therefore, severely affect ATP binding. In conclusion, this model might explain why the T568I mutation of the patient is causative for nonfunctional CNNM2.
FIGURE 6.
The CBS domains are potentially involved in ATP binding. A, homology model of the CBS domain pair of CNNM2, modeled using the structure of the CorC Mg2+ and Co2+ efflux protein from B. parapertussis (PDB file 3jtf). The two CBS domain monomers are shown with the Thr-568 highlighted. ATP is depicted in the center of the model. B, detail of the ATP-binding region in the CBS domain. ATP is shown in this model. The wild type Thr-568 residue is highlighted, and the extra side-chain of the mutant Ile-568 is indicated. Hydrogen bonds with the surrounding Glu-570 and Asp-571 residues are demonstrated.
CNNM2a and CNNM2b Form Multimers
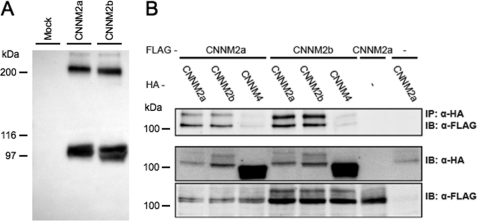
In GenBank, two splice variants exist that are conserved between human and mouse, the first being full length, the second lacking exon 6. Interestingly, splice variant 2 was consistently found in conjunction with that of splice variant 1 in all tissues examined, including kidney (Fig. 1). Further examination of potential interactions between CNNM2a and CNNM2b was performed by studying the dimerization of CNNM2 isoforms. Interestingly, besides the CNNM2 monomer (with an expected molecular mass of 105 kDa) (5), additional complexes were visualized in the immunoblot at ∼200 kDa (Fig. 7A). This molecular mass matches the mass of a hypothetical CNNM2 dimer. To confirm CNNM2 oligomerization events, co-immunoprecipitation assays of CNNM2 isoforms were performed. CNNM2a-HA and CNNM2b-HA were each able to co-immunoprecipitate both CNNM2a and C-terminally FLAG-tagged CNNM2b. CNNM4-HA only weakly co-immunoprecipitated, although it was highly expressed, indicating the specificity of these interactions (Fig. 7B). These data show that CNNM2a and CNNM2b are able to form specific homo- and heteromultimeric complexes.
FIGURE 7.
CNNM2a and CNNM2b form homo- and heterotypic interactions. A, Western blot analysis of CNNM2 protein showing CNNM2a and CNNM2b monomers at ∼100 kDa and potential CNNM2 dimers at ∼200 kDa. B, co-immunoprecipitation (IP) studies of CNNM proteins in COS-7 cells. C-terminal FLAG-tagged CNNM2a or CNNM2b were co-expressed with C-terminal HA-tagged CNNM2a, CNNM2b, or CNNM4 as indicated in the two top rows. The upper blot shows the detection of the FLAG-tagged proteins in anti-HA precipitated cell lysates. The lower two blots show input controls (10%) of HA-tagged and FLAG-tagged proteins, respectively. The results shown are representative of three individual experiments. IB, immunoblot.
DISCUSSION
Mutations in CNNM2 have, recently, been described to be causative for a severe form of hypomagnesemia (3). Initially described as nuclear proteins involved in cell cycle regulation (5), the family of CNNM proteins has now been shown to reside at the plasma membrane (3, 6). Although their physiological role is still poorly understood, all members are likely involved in electrolyte homeostasis. CNNM1 has been identified as a cytosolic copper chaperone, CNNM2 was shown to mediate Mg2+-sensitive Na+ currents in HEK293 cells, and Cnnm2, Cnnm3, and Cnnm4 have been linked with serum Mg2+ levels (3, 21, 22). Furthermore, Cnnm2 has been associated with coronary artery disease and hypertension (23–25). Finally, mutations in Cnnm4 were shown to be causative for recessive cone-rod dystrophy with amelogenesis imperfecta (26, 27). In the present study, we aimed to elucidate the CNNM2 protein topology and characterize its post-translational modifications.
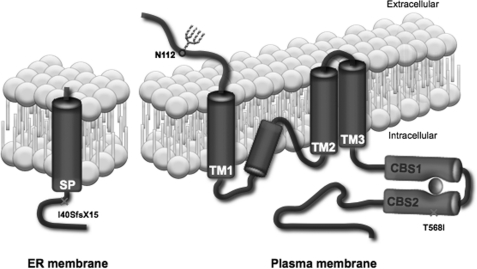
Although Gómez García et al. (28) have been working on structural elucidation of the CBS domains of CNNM4, complete structural information for any of the CNNM proteins is presently unknown. Using immunocytochemical methods, we examined the membrane topology of CNNM2. Based on our results showing an extracellular N terminus and an intracellular C terminus (Fig. 3), we propose a structure comparable with that of the glutamate receptor (GluR) ion channels (29), containing three full membrane-spanning regions and an additional re-entrant loop. In silico analysis of the CNNM2 protein sequence showed that of the four hydrophobic regions, the second one is the shortest and the least hydrophobic, indicating that the second hydrophobic region might not be completely membrane-spanning, but instead forms a re-entrant loop (Fig. 8).
FIGURE 8.
Schematic model of CNNM2 structure. A model representing the structure of the CNNM2 protein at the plasma membrane and showing the signal peptide cleavage in the endoplasmic reticulum is illustrated. Crosses represent the locations of the mutations. The glycosylation site at position Asn-112 is shown in the extracellular N terminus. The CBS domains are represented in purple, and binding ATP is in red. SP, signal peptide; TM, transmembrane helix; N112, Asn-112.
Similar to the glutamate receptor family, CNNM2 contains an N-terminal signal peptide that is cleaved in the ER. Here, the SPC seems to be the protease complex responsible for CNNM2 cleavage (Fig. 4B). The human SPC is a complex of five proteins expressed on the ER membrane and recruited post-translationally to the signal recognition particle to perform signal peptide cleavage (30). Our results do not identify the exact position of the cleavage site. Signal peptide recognition is known to be dependent on the residues at positions −1 and −3 of the actual cleavage site (31). Nevertheless, conversion of the −1 and −3 position of the predicted site (Gly-62/Thr-64) to hydrophobic residues did not prevent cleavage. Likewise, the cleavage was also not impaired when the alternative cleavage site (Cys-63/Ala-65) was mutated. Only mutation of both sites prevented CNNM2 cleavage, suggesting that the SPC can cleave CNNM2 at the preferential site between Thr-64 and Ala-65, but also at an alternate site between Ala-65 and Ala-66. Thus, CNNM2 contains a remarkable lengthy signal peptide of ∼64 amino acids. Long signal peptides have been associated with multiple functions, but might serve as a mechanism to achieve proper membrane insertion of the protein (32). This also suggests that the first predicted transmembrane domain (Fig. 3A) actually represents the hydrophobic core of the signal peptide that, after cleavage, will remain at the ER membrane (Fig. 8).
After cleavage in the ER, CNNM2 is further processed by glycosylation in the Golgi apparatus. N-Glycosylation is known to facilitate plasma membrane trafficking for several membrane proteins (33). Here, Asn-112 was identified as the residue that is glycosylated and implicated in CNNM2 membrane stability (Fig. 5, A and B). This can be interpreted as an additional argument showing that the N terminus is located on the extracellular side of the membrane. Interestingly, our immunoblotting data showed a shift of the molecular weight of the CNNM2 protein when the alternative residue Asn-327 was mutated. However, PNGase F treatment still lowers the molecular weight of the N327A mutant, indicating that the protein is not deglycosylated (Fig. 5A). Because Asn-327 is predicted to be located in the second TM domain, we suspect that mutation of this residue instigates an altered protein folding that runs different on gel, explaining the observed shift.
Once at the plasma membrane, the functional CNNM2 unit likely represents a multimer. Interestingly, we showed that both CNNM2a and CNNM2b can form homomultimers as well as heteromultimers (Fig. 7B). This interaction is specific, as evidenced by the fact that CNNM2a and CNNM2b hardly dimerize with CNNM4, which shares the highest sequence homology with CNNM2 (96.7% similarity and 81.4% identity at the protein level) (5).
In magnesium research, CNNM2 function is heavily debated. CNNM2 was first reported as a Mg2+ transporter (6, 7), but recent findings questioned the ability of CNNM2 to transport Mg2+ and proposed a Mg2+-sensing function (3). We, therefore, examined whether our topological findings could further elucidate the function of CNNM2. We reasoned that although CNNM2 might contain a re-entrant region, it is unlikely that this region forms a pore for Mg2+ transport. The re-entrant loop does not include negatively charged residues that could provide Mg2+ specificity. Furthermore, the protein topology of only three transmembrane segments would be small in comparison with known eukaryotic Mg2+ transporters such as TRPM6/7 (tetramers of six TM proteins) (2) and SLC41A1 (10/11 TM domains) (34). Thus, our topological findings strengthen the previously reported patch clamp data (3), suggesting that CNNM2 might be a Mg2+ sensor indirectly regulating Mg2+ transport.
To further substantiate this hypothesis, we focused on the CBS domains during analysis of the CNNM2 structure. CBS domains are found in 100–200 mammalian proteins and play a role in a variety of functions such as adenosine phosphate binding and ionic strength sensing (35, 36). The CBS domains in CNNM2 are highly homologous to those found in the B. parapertussis Co2+/Mg2+ protein CorC, of which the structure has been crystallized recently (PDB 3jtf). As shown by homology modeling based on the CorC structure, the CBS domains in the CNNM2 C terminus contain a potential ATP-binding domain (Fig. 4B). ATP binding in CBS domains has been reported to activate or further inhibit protein function in eukaryotic CBS domain-containing proteins (20).
Our model does not allow discrimination between ATP and Mg-ATP binding, but we expect that the CBS domains of CNNM2 are also able to bind Mg-ATP. This theory is strengthened by the recently elucidated crystal structure of the bacterial Mg2+ transporter MgtE in which its CBS domains are involved in Mg2+ binding (PDB 2yvy (37, 38). As the majority of intracellular Mg2+ is ATP bound, the intracellular Mg-ATP concentrations are representative for the cellular Mg2+ content. The suggestion that CNNM2 binds Mg-ATP implies that rather than ATP being an energy source for active transport, the Mg-ATP binding would provide a Mg2+-sensing mechanism. Given that the CNNM2 T568I loss-of-function mutation found in patients (3) is located at the region predicted to form the ATP-binding pocket (Fig. 6B), the (Mg-)ATP binding is likely of major importance for CNNM2 function. Because the T568I mutation is specifically disturbing the ATP-binding pocket, Mg2+ alone will possibly not be able to activate CNNM2.
In conclusion, we propose a protein topology of CNNM2 consisting of three membrane-spanning domains and a re-entrant loop (Fig. 8). CNNM2 is extensively post-translationally modified by signal peptide cleavage and glycosylation before being functional in multimers at the plasma membrane. Rather than transporting Mg2+ itself, we hypothesize that CNNM2 would sense intracellular Mg2+ concentrations and regulate other Mg2+ transporters. Future research should focus on the identification of CNNM2 protein partners to further elucidate its role in Mg2+ homeostasis.
Supplementary Material
Acknowledgments
In particular, we thank Eric Jansen for supplying us materials and excellent technical suggestions for the in vitro translation experiments. We thank Femke van Zeeland, AnneMiete van der Kemp, Michelle Damen, Kelly Menting, Evelien Oerlemans, and Kerstin Sommer for technical support.
This work was supported by the Netherlands Organization for Scientific Research Grants ZonMw 9120.8026 and EURYI 2006 (to J. G. J. H.) and by grants from the European Community, FP7 (EUNEFRON 201590) (to I. C. M. and D. M.).

This article contains supplemental Table S1 and Figs. S1 and S2.
- DCT
- distal convoluted tubule
- CNT
- connecting tubule
- NCC
- Na+-Cl− cotransporter
- NKCC2
- Na+-K+-Cl− cotransporter
- IRES
- internal ribosomal entry site
- TM
- transmembrane
- CBS
- cystathionine B-synthase
- ER
- endoplasmic reticulum
- AAPV-CMK
- MeoSuc-Ala-Ala-Pro-Val-chloromethylketone
- PNGase F
- N-glycosidase F
- SPC
- signal peptidase complex
- PT
- proximal tubule
- TAL
- thick ascending limb of Henle's loop.
REFERENCES
- 1. Glaudemans B., Knoers N. V., Hoenderop J. G., Bindels R. J. (2010) New molecular players facilitating Mg2+ reabsorption in the distal convoluted tubule. Kidney Int. 77, 17–22 [DOI] [PubMed] [Google Scholar]
- 2. Voets T., Nilius B., Hoefs S., van der Kemp A. W., Droogmans G., Bindels R. J., Hoenderop J. G. (2004) TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J. Biol. Chem. 279, 19–25 [DOI] [PubMed] [Google Scholar]
- 3. Stuiver M., Lainez S., Will C., Terryn S., Günzel D., Debaix H., Sommer K., Kopplin K., Thumfart J., Kampik N. B., Querfeld U., Willnow T. E., Němec V., Wagner C. A., Hoenderop J. G., Devuyst O., Knoers N. V., Bindels R. J., Meij I. C., Müller D. (2011) CNNM2, encoding a basolateral protein required for renal Mg2+ handling, is mutated in dominant hypomagnesemia. Am. J. Hum. Genet. 88, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meij I. C., van den Heuvel L. P., Hemmes S., van der Vliet W. A., Willems J. L., Monnens L. A., Knoers N. V. (2003) Exclusion of mutations in FXYD2, CLDN16, and SLC12A3 in two families with primary renal Mg2+ loss. Nephrol. Dial. Transplant 18, 512–516 [DOI] [PubMed] [Google Scholar]
- 5. Wang C. Y., Shi J. D., Yang P., Kumar P. G., Li Q. Z., Run Q. G., Su Y. C., Scott H. S., Kao K. J., She J. X. (2003) Molecular cloning and characterization of a novel gene family of four ancient conserved domain proteins (ACDP). Gene 306, 37–44 [DOI] [PubMed] [Google Scholar]
- 6. Goytain A., Quamme G. A. (2005) Functional characterization of ACDP2 (ancient conserved domain protein), a divalent metal transporter. Physiol. Genomics. 22, 382–389 [DOI] [PubMed] [Google Scholar]
- 7. Sponder G., Svidova S., Schweigel M., Vormann J., Kolisek M. (2010) Splice-variant 1 of the ancient domain protein 2 (ACDP2) complements the magnesium-deficient growth phenotype of Salmonella enterica sv. typhimurium strain MM281. Magnes. Res. 23, 105–114 [DOI] [PubMed] [Google Scholar]
- 8. Meyer A. H., Katona I., Blatow M., Rozov A., Monyer H. (2002) In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J. Neurosci. 22, 7055–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hofmeister M. V., Fenton R. A., Praetorius J. (2009) Fluorescence isolation of mouse late distal convoluted tubules and connecting tubules: effects of vasopressin and vitamin D3 on Ca2+ signaling. Am. J. Physiol. Renal Physiol. 296, F194–F203 [DOI] [PubMed] [Google Scholar]
- 10. Hoenderop J. G., Dardenne O., Van Abel M., Van Der Kemp A. W., Van Os C. H., St -Arnaud R., Bindels R. J. (2002) Modulation of renal Ca2+ transport protein genes by dietary Ca2+ and 1,25-dihydroxyvitamin D3 in 25-hydroxyvitamin D3-1α-hydroxylase knockout mice. Faseb. J. 16, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 11. Wang W., Malcolm B. A. (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 12. Kast C., Gros P. (1997) Topology mapping of the amino-terminal half of multidrug resistance-associated protein by epitope insertion and immunofluorescence. J. Biol. Chem. 272, 26479–26487 [DOI] [PubMed] [Google Scholar]
- 13. Günzel D., Stuiver M., Kausalya P. J., Haisch L., Krug S. M., Rosenthal R., Meij I. C., Hunziker W., Fromm M., Müller D. (2009) Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J. Cell Sci. 122, 1507–1517 [DOI] [PubMed] [Google Scholar]
- 14. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, Suppl. 9, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 16. van de Graaf S. F., Hoenderop J. G., Gkika D., Lamers D., Prenen J., Rescher U., Gerke V., Staub O., Nilius B., Bindels R. J. (2003) Functional expression of the epithelial Ca2+ channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. EMBO J. 22, 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kast C., Canfield V., Levenson R., Gros P. (1995) Membrane topology of P-glycoprotein as determined by epitope insertion: transmembrane organization of the N-terminal domain of mdr3. Biochemistry 34, 4402–4411 [DOI] [PubMed] [Google Scholar]
- 18. Czachorowski M., Lam-Yuk-Tseung S., Cellier M., Gros P. (2009) Transmembrane topology of the mammalian Slc11a2 iron transporter. Biochemistry 48, 8422–8434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 20. Baykov A. A., Tuominen H. K., Lahti R. (2011) The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem. Biol. 6, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 21. Alderton A., Davies P., Illman K., Brown D. R. (2007) Ancient conserved domain protein-1 binds copper and modifies its retention in cells. J. Neurochem. 103, 312–321 [DOI] [PubMed] [Google Scholar]
- 22. Meyer T. E., Verwoert G. C., Hwang S. J., Glazer N. L., Smith A. V., van Rooij F. J., Ehret G. B., Boerwinkle E., Felix J. F., Leak T. S., Harris T. B., Yang Q., Dehghan A., Aspelund T., Katz R., Homuth G., Kocher T., Rettig R., Ried J. S., Gieger C., Prucha H., Pfeufer A., Meitinger T., Coresh J., Hofman A., Sarnak M. J., Chen Y. D., Uitterlinden A. G., Chakravarti A., Psaty B. M., van Duijn C. M., Kao W. H., Witteman J. C., Gudnason V., Siscovick D. S., Fox C. S., Köttgen A. (2010) Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six loci influencing serum magnesium levels. PLoS Genet. 6, e1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schunkert H., König I. R., Kathiresan S., Reilly M. P., Assimes T. L., Holm H., Preuss M., Stewart A. F., Barbalic M., Gieger C., Absher D., Aherrahrou Z., Allayee H., Altshuler D., Anand S. S., Andersen K., Anderson J. L., Ardissino D., Ball S. G., Balmforth A. J., Barnes T. A., Becker D. M., Becker L. C., Berger K., Bis J. C., Boekholdt S. M., Boerwinkle E., Braund P. S., Brown M. J., Burnett M. S., Buysschaert I., Carlquist J. F., Chen L., Cichon S., Codd V., Davies R. W., Dedoussis G., Dehghan A., Demissie S., Devaney J. M., Diemert P., Do R., Doering A., Eifert S., Mokhtari N. E., Ellis S. G., Elosua R., Engert J. C., Epstein S. E., de Faire U., Fischer M., Folsom A. R., Freyer J., Gigante B., Girelli D., Gretarsdottir S., Gudnason V., Gulcher J. R., Halperin E., Hammond N., Hazen S. L., Hofman A., Horne B. D., Illig T., Iribarren C., Jones G. T., Jukema J. W., Kaiser M. A., Kaplan L. M., Kastelein J. J., Khaw K. T., Knowles J. W., Kolovou G., Kong A., Laaksonen R., Lambrechts D., Leander K., Lettre G., Li M., Lieb W., Loley C., Lotery A. J., Mannucci P. M., Maouche S., Martinelli N., McKeown P. P., Meisinger C., Meitinger T., Melander O., Merlini P. A., Mooser V., Morgan T., Mühleisen T. W., Muhlestein J. B., Münzel T., Musunuru K., Nahrstaedt J., Nelson C. P., Nöthen M. M., Olivieri O., Patel R. S., Patterson C. C., Peters A., Peyvandi F., Qu L., Quyyumi A. A., Rader D. J., Rallidis L. S., Rice C., Rosendaal F. R., Rubin D., Salomaa V., Sampietro M. L., Sandhu M. S., Schadt E., Schäfer A., Schillert A., Schreiber S., Schrezenmeir J., Schwartz S. M., Siscovick D. S., Sivananthan M., Sivapalaratnam S., Smith A., Smith T. B., Snoep J. D., Soranzo N., Spertus J. A., Stark K., Stirrups K., Stoll M., Tang W. H., Tennstedt S., Thorgeirsson G., Thorleifsson G., Tomaszewski M., Uitterlinden A. G., van Rij A. M., Voight B. F., Wareham N. J., Wells G. A., Wichmann H. E., Wild P. S., Willenborg C., Witteman J. C., Wright B. J., Ye S., Zeller T., Ziegler A., Cambien F., Goodall A. H., Cupples L. A., Quertermous T., März W., Hengstenberg C., Blankenberg S., Ouwehand W. H., Hall A. S., Deloukas P., Thompson J. R., Stefansson K., Roberts R., Thorsteinsdottir U., O'Donnell C. J., McPherson R., Erdmann J. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeuchi F., Isono M., Katsuya T., Yamamoto K., Yokota M., Sugiyama T., Nabika T., Fujioka A., Ohnaka K., Asano H., Yamori Y., Yamaguchi S., Kobayashi S., Takayanagi R., Ogihara T., Kato N. (2010) Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation 121, 2302–2309 [DOI] [PubMed] [Google Scholar]
- 25. Newton-Cheh C., Johnson T., Gateva V., Tobin M. D., Bochud M., Coin L., Najjar S. S., Zhao J. H., Heath S. C., Eyheramendy S., Papadakis K., Voight B. F., Scott L. J., Zhang F., Farrall M., Tanaka T., Wallace C., Chambers J. C., Khaw K. T., Nilsson P., van der Harst P., Polidoro S., Grobbee D. E., Onland-Moret N. C., Bots M. L., Wain L. V., Elliott K. S., Teumer A., Luan J., Lucas G., Kuusisto J., Burton P. R., Hadley D., McArdle W. L., Brown M., Dominiczak A., Newhouse S. J., Samani N. J., Webster J., Zeggini E., Beckmann J. S., Bergmann S., Lim N., Song K., Vollenweider P., Waeber G., Waterworth D. M., Yuan X., Groop L., Orho-Melander M., Allione A., Di Gregorio A., Guarrera S., Panico S., Ricceri F., Romanazzi V., Sacerdote C., Vineis P., Barroso I., Sandhu M. S., Luben R. N., Crawford G. J., Jousilahti P., Perola M., Boehnke M., Bonnycastle L. L., Collins F. S., Jackson A. U., Mohlke K. L., Stringham H, M., Valle T. T., Willer C. J., Bergman R. N., Morken M. A., Döring A., Gieger C., Illig T., Meitinger T., Org E., Pfeufer A., Wichmann H. E., Kathiresan S., Marrugat J., O'Donnell C. J., Schwartz S. M., Siscovick D. S., Subirana I., Freimer N. B., Hartikainen A. L., McCarthy M. I., O'Reilly P. F., Peltonen L., Pouta A., de Jong P. E., Snieder H., van Gilst W. H., Clarke R., Goel A., Hamsten A., Peden J. F., Seedorf U., Syvänen A. C., Tognoni G., Lakatta E. G., Sanna S., Scheet P., Schlessinger D., Scuteri A., Dörr M., Ernst F., Felix S. B., Homuth G., Lorbeer R., Reffelmann T., Rettig R., Völker U., Galan P., Gut I. G., Hercberg S., Lathrop G. M., Zelenika D., Deloukas P., Soranzo N., Williams F. M., Zhai G., Salomaa V., Laakso M., Elosua R., Forouhi N. G., Völzke H., Uiterwaal C. S., van der Schouw Y. T., Numans M. E., Matullo G., Navis G., Berglund G., Bingham S. A., Kooner J. S., Connell J. M., Bandinelli S., Ferrucci L., Watkins H., Spector T. D., Tuomilehto J., Altshuler D., Strachan D. P., Laan M., Meneton P., Wareham N. J., Uda M., Jarvelin M. R., Mooser V., Melander O., Loos R. J., Elliott P., Abecasis G. R., Caulfield M., Munroe P. B. (2009) Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 41, 666–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parry D. A., Mighell A. J., El-Sayed W., Shore R. C., Jalili I. K., Dollfus H., Bloch-Zupan A., Carlos R., Carr I. M., Downey L. M., Blain K. M., Mansfield D. C., Shahrabi M., Heidari M., Aref P., Abbasi M., Michaelides M., Moore A. T., Kirkham J., Inglehearn C. F. (2009) Mutations in CNNM4 cause Jalili syndrome, consisting of autosomal-recessive cone-rod dystrophy and amelogenesis imperfecta. Am. J. Hum. Genet. 84, 266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polok B., Escher P., Ambresin A., Chouery E., Bolay S., Meunier I., Nan F., Hamel C., Munier F. L., Thilo B., Mégarbané A., Schorderet D. F. (2009) Mutations in CNNM4 cause recessive cone-rod dystrophy with amelogenesis imperfecta. Am. J. Hum. Genet. 84, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gómez García I., Oyenarte I., Martínez-Cruz L. A. (2011) Purification, crystallization and preliminary crystallographic analysis of the CBS pair of the human metal transporter CNNM4. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Traynelis S. F., Wollmuth L. P., McBain C. J., Menniti F. S., Vance K. M., Ogden K. K., Hansen K. B., Yuan H., Myers S. J., Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paetzel M., Karla A., Strynadka N. C., Dalbey R. E. (2002) Signal peptidases. Chem. Rev. 102, 4549–4580 [DOI] [PubMed] [Google Scholar]
- 31. Nielsen H., Engelbrecht J., Brunak S., Von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein. Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 32. Hiss J. A., Schneider G. (2009) Architecture, function and prediction of long signal peptides. Brief Bioinform. 10, 569–578 [DOI] [PubMed] [Google Scholar]
- 33. Vagin O., Kraut J. A., Sachs G. (2009) Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am. J. Physiol. Renal Physiol. 296, F459–F469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wabakken T., Rian E., Kveine M., Aasheim H. C. (2003) The human solute carrier SLC41A1 belongs to a novel eukaryotic subfamily with homology to prokaryotic MgtE Mg2+ transporters. Biochem. Biophys. Res. Commun. 306, 718–724 [DOI] [PubMed] [Google Scholar]
- 35. Ignoul S., Eggermont J. (2005) CBS domains: structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 289, C1369–C1378 [DOI] [PubMed] [Google Scholar]
- 36. Biemans-Oldehinkel E., Mahmood N. A., Poolman B. (2006) A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. U.S.A. 103, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hattori M., Tanaka Y., Fukai S., Ishitani R., Nureki O. (2007) Crystal structure of the MgtE Mg2+ transporter. Nature 448, 1072–1075 [DOI] [PubMed] [Google Scholar]
- 38. Ishitani R., Sugita Y., Dohmae N., Furuya N., Hattori M., Nureki O. (2008) Mg2+-sensing mechanism of Mg2+ transporter MgtE probed by molecular dynamics study. Proc. Natl. Acad. Sci. U.S.A. 105, 15393–15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loffing J., Vallon V., Loffing-Cueni D., Aregger F., Richter K., Pietri L., Bloch-Faure M., Hoenderop J. G., Shull G. E., Meneton P., Kaissling B. (2004) Altered renal distal tubule structure and renal Na+ and Ca2+ handling in a mouse model for Gitelman's syndrome. J. Am. Soc. Nephrol. 15, 2276–2288 [DOI] [PubMed] [Google Scholar]
- 40. Nijenhuis T., Hoenderop J. G., Loffing J., van der Kemp A. W., van Os C. H., Bindels R. J. (2003) Thiazide-induced hypocalciuria is accompanied by a decreased expression of Ca2+ transport proteins in kidney. Kidney Int. 64, 555–564 [DOI] [PubMed] [Google Scholar]
- 41. Bindels R. J., Hartog A., Timmermans J., Van Os C. H. (1991) Active Ca2+ transport in primary cultures of rabbit kidney CCD: stimulation by 1,25-dihydroxyvitamin D3 and PTH. Am. J. Physiol. 261, F799–F807 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.