Background: MDM2 uses a dual-site mechanism to ubiquitinate p53, involving the acidic domain.
Results: MDM2 acid domain binds NUMB and is critical for NUMB ubiquitination.
Conclusion: Small molecule ligands that target the acid domain of MDM2 stabilize Numb and activate tumor suppressor activities of p53 in cells.
Significance: NUMB regulates steady-state levels of p53 in cells by disrupting p53-MDM2 acidic domain interactions.
Keywords: Cancer, Cell growth, Cell-penetrating peptides, E3 ubiquitin ligase, p53, MDM2, NUMB, Ubiquitination
Abstract
The E3 ubiquitin ligase, MDM2, uses a dual-site mechanism to ubiquitinate and degrade the tumor suppressor protein p53, involving interactions with the N-terminal hydrophobic pocket and the acidic domain of MDM2. The results presented here demonstrate that MDM2 also uses this same dual-site mechanism to bind to the cell fate determinant NUMB with both the N-terminal hydrophobic pocket and the acidic domain of MDM2 also involved in forming the interaction with NUMB. Furthermore, the acidic domain interactions are crucial for MDM2-mediated ubiquitination of NUMB. Contrary to p53, where two separate domains form the interface with MDM2, only one region within the phosphotyrosine binding domain of NUMB (amino acids 113–148) mediates binding to both these regions of MDM2. By binding to both domains on MDM2, NUMB disrupts the MDM2-p53 complex and MDM2-catalyzed ubiquitination of p53. Therefore, we have identified the mechanism NUMB uses to regulate the steady-state levels of the p53 in cells. By targeting the acidic domain of MDM2 using acid domain-binding ligands we can overcome MDM2-mediated ubiquitination and degradation of NUMB impacting on the stabilization of p53 in cells. Furthermore, delivery of MDM2 acid domain-binding ligands to cancer cells promotes p53-dependent growth arrest and the induction of apoptosis. This highlights the dual-site mechanism of MDM2 on another physiological substrate and identifies the acid domain as well as N terminus as a potential target for small molecules that inhibit MDM2.
Introduction
Murine double minute 2 (MDM2)2 is a major regulator of the p53 tumor suppressor protein (1). MDM2 attenuates the transcriptional activity of p53 (2, 3) and regulates the levels of p53 in cells by acting as an E3 ligase for the ubiquitination and proteosomal degradation of p53 (4, 5). More recent evidence suggests that MDM2 also functions as an ATP-dependent molecular chaperone for p53 (6) and regulates the translation of p53 (7). The complex interaction between MDM2 and p53 involves two distinct binding sites on p53. By binding to the transactivation domain of p53 (BOX-I) via its N-terminal hydrophobic domain, MDM2 disrupts the transcriptional activity of p53 (3, 8). This interaction also results in conformational changes that facilitate binding between the S9-S10 linker/BOX-V region in the core DNA binding domain of p53 and the acid domain of MDM2 (9). The interaction with the acid domain is a lower affinity interaction but importantly comprises the ubiquitination signal. Mutation of either the BOX-V region of p53 (9) or the acid domain of MDM2 results in a decrease in the ubiquitination and degradation of p53 (9–11). Furthermore, ligands that disrupt the p53-BOX-V:MDM2-acid domain interaction are potent E3-ligase inhibitors in vitro and in cells (9). The discovery of the role of the acid domain of MDM2 in ubiquitination of p53 has opened up new avenues for manipulating MDM2 E3-ligase activity in cells.
MDM2 has multiple p53-independent functions that contribute to its activity as an oncogene (12). The interferon regulatory factor (IRF)-2 transcription factor was recently identified as a novel substrate for MDM2-mediated ubiquitination (13). MDM2 was found to bind to IRF-2 using a dual-site mechanism in a similar manner to p53, and acid domain interactions were found to be crucial for MDM2-mediated ubiquitination, suggesting a common mechanism for MDM2-catalyzed ubiquitination (13). Furthermore, the acid domain of MDM2 is important in forming the interaction with a number of other proteins, including Arf (14), the retinoblastoma tumor suppressor protein (Rb) (15), and p21(Waf1) (16).
The cell fate determinant NUMB was identified as an MDM2 binding partner using the N-terminal hydrophobic pocket domain of MDM2 as bait in a yeast two-hybrid screen (17). NUMB was subsequently shown to be ubiquitinated and degraded by MDM2 (18). NUMB is best known for its role as an antagonist of the Notch signaling pathway (19, 20). NUMB interacts with the active intracellular domain of Notch (NICD) promoting degradation of Notch, preventing its localization to the nucleus and its activity as a transcription factor (19, 20). However, NUMB has also recently been shown to play a role in the stabilization and activation of p53 in cells, which was shown to be dependent on MDM2 (21). However, the exact mechanism by which NUMB overcomes MDM2 inhibition of p53 is still unclear. In this report, we investigate the interaction between MDM2 and NUMB in detail and investigate the role of the acid domain of MDM2 in the regulation of NUMB. In contrast to p53 where two separate domains form the interface with MDM2, we demonstrate here that one region within the phosphotyrosine binding (PTB) domain of NUMB interacts with both the N terminus and the acid domain of MDM2, highlighting the significance of the dual-site binding mechanism. By interacting with both these regions of MDM2, NUMB can disrupt the p53-MDM2 complex and prevent ubiquitination of p53. In addition, we show the importance of acidic domain interactions for ubiquitination and degradation of NUMB. Using peptides derived from the PTB domain of NUMB and other small molecule ligands that bind to the acid domain of MDM2, we can reverse the effects of MDM2 on degradation of NUMB in cancer cells. Furthermore, these acid domain-binding ligands prevent MDM2-mediated ubiquitination resulting in stabilization of p53 and the induction of growth arrest and apoptosis in cells.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Peptides
pCMV-His-ubiquitin, pcDNA3.1-MDM2, and pT7.7/MDM2 were gifts from Prof. David Lane. pcDNA3.1-NUMB and pGEX2T-NUMB were gifts from Prof. M. Oren. Full-length NUMB, PTB domain, and ΔPTB domain of NUMB were subcloned into pFN2A and pcDNA3.1vectors (Promega, Madison, WI) using the following primers: pcDNA3.1-NUMB PTB forward (CCGGAATTCATGCAGTGGCAGACAGATG) and reverse (CAACTCGACTTACCGCTTCTGCTTGCG), pcDNA3.1- NUMB ΔPTB forward (CCGGAATTCATGGAGAAGGAATGTGGAG) and reverse (CAACTCGAGCGTTTAAAGTTCAATTTC), pFN2A-NUMB forward (GTTCGCGATCGCCAACAAATTACGGCAAAGTTTTAG) and reverse (ACAGGTTTAAACAAGTTCAATTTCAAACGTCTTC), pFN2A-NUMB PTB forward (CAAGCAGAAGCGGTGAGAGAAGGAATGTG) and reverse (CACATTCCTTCTCTCACCGCTTCTGCTTG), and pFN2A-NUMB ΔPTB forward (CACCGCGATCGCCGAGAAGGAATGTGGAGTGACTGC) and reverse (ATTAGTTTAAACAAGTTCAATTTCAAACGTCTTCTGTAAGTCA). GST-MDM2, GST-ΔN MDM2, GST-ΔAD MDM2, GST-AD MDM2, and GST-N MDM2 have been previously described (9). Goat anti-NUMB antibody (Abcam Ab 4147) was used to detect NUMB. Anti-Notch 1, MAB5352 (Millipore), and anti-Hes-1 (Santa Cruz Biotechnology, H140) were used to detect NICD and Hes1, respectively. Monoclonal antibodies 4B2 and 2A10 were used to detect MDM2, and DO-1 antibody was used to detect p53 (gifts from Prof. T. Hupp). Anti-GST and β-actin antibodies were from Sigma. p21 antibody (Ab1) was from Calbiochem. Biotin-labeled peptides were synthesized by Mimotopes (Australia). HIV-TAT Fluorescein-labeled peptides were from Mimotopes (NUMB peptide 7 was VVDEKTDKDLIVDQTIEKVSFYGRKKRRQRRR, NUMB peptide 8 was KVSFCAPDRNFDRAFSYICRYGRKKRRQRRR, NUMB peptide 9 was YICRDGTTRRWICHCFMAVKYGRKKRRQRRR, and Rb1 was DQIMMCSMYGICKVKNIDLKYGRKKRRQRRR).
Cell Culture and Immunoblots
H1299 and MD MB-468 were maintained in RPMI (Invitrogen) supplemented with 5% FCS (Biosera) and 1% penicillin/streptomycin at 37 °C and 5% CO2. MCF7, ZR75, and A375 were maintained in DMEM (Invitrogen) supplemented with 5% FCS (Biosera) and 1% penicillin/streptomycin at 37 °C and 5% CO2. The cells were transfected at 80% confluency using Lipofectamine 2000 (Invitrogen) or Attractene (Qiagen) following the manufacturer's instructions, and DNA was equalized by addition of empty vector control. Peptides were added to the cells as indicated in the figure legends and incubated overnight. Cells were treated with cisplatin (12 μg/ml) for 24 h as indicated in the figures. Cells were lysed, and immunoblots were carried out as previously described (22).
Protein Purification
MDM2 was expressed and purified from Escherichia coli (E. coli) as previously described (6). GST-MDM2 and GST-NUMB constructs were expressed in E. coli and purified on glutathione-Sepharose beads (GE Healthcare) following the manufacturer's instructions. The GST tag was cleaved using Thrombin (Sigma), PreScission Protease (GE Healthcare), or TEV Protease (Expedeon), following the manufacturer's instructions.
Ubiquitination Assays
Cell-based ubiquitination assays were carried out in H1299 cells following transfection of cells with pcDNA3.1-NUMB, pCMV-his ubiquitin, and pcDNA3.1-MDM2 as previously described (9). The p53 ubiquitination assays were carried out as previously described (9). For NUMB the in vitro ubiquitination reactions contained 25 mm Hepes (pH 8.0), 10 mm MgCl2, 4 mm ATP, 0.5 mm DTT, 0.05% (v/v) Triton X-100, 0.25 mm benzamidine, 10 mm creatine phosphate, 3.5 units/ml creatine kinase, ubiquitin (2 μg), E1-UBE-1 (100 nm), E2-UbcH5 (1 μm), and GST-NUMB (250–500 ng). Reactions were started using purified MDM2 (50–100 ng), incubated for 3 h at 30 °C, and analyzed using 7% Tris acetate NuPAGE gels in a Novex Tris acetate buffer system (Invitrogen) followed by immunoblot with anti-NUMB antibody. Where peptides were used they were added to the reaction mix prior to addition of MDM2 as indicated in the figure legends.
ELISA Assays
For peptide ELISA, biotin-labeled peptides were captured onto streptavidin-coated plates (25 μg/ml, Sigma) as detailed in the figure legends. Non-reactive sites were blocked using PBS containing 3% BSA. The wells were incubated with full-length MDM2, mini-domain MDM2, or mutant MDM2 proteins (as indicated in the figure legends) in PBS-0.1% BSA for 1 h at room temperature. The plates were washed extensively with PBS-0.2% Tween, and MDM2 binding was detected using anti-MDM2 antibodies: 2A10 or 4B2 or anti-GST antibody and enhanced chemiluminescence (as indicated in figure legends). For protein ELISA, 96-well microtiter plates were coated with the protein (as detailed in the figure legends) (MDM2 or MDM2 mutants (Fig. 1, B–E), NUMB or NUMB mutants (Fig. 2G, p53; (Fig. 5, A and B)) in 0.1 m sodium borate. Non-reactive sites were blocked using PBS containing 3% BSA, and the wells were incubated with MDM2 (Figs. 2G and 5A), NUMB (Fig. 1, B–E), in PBS, 0.1% BSA for 1 h at room temperature. Binding was detected using the appropriate primary antibodies and enhanced chemiluminescence as detailed in the figure legends. Competition ELISA assays were carried out as above, but peptides were preincubated with MDM2 on ice for 30 min prior to adding to the plate. Readings were performed using Victor3 1420 Multilabel Counter (Perkin-Elmer) or Fluoroscan Ascent FL (Labsystems).
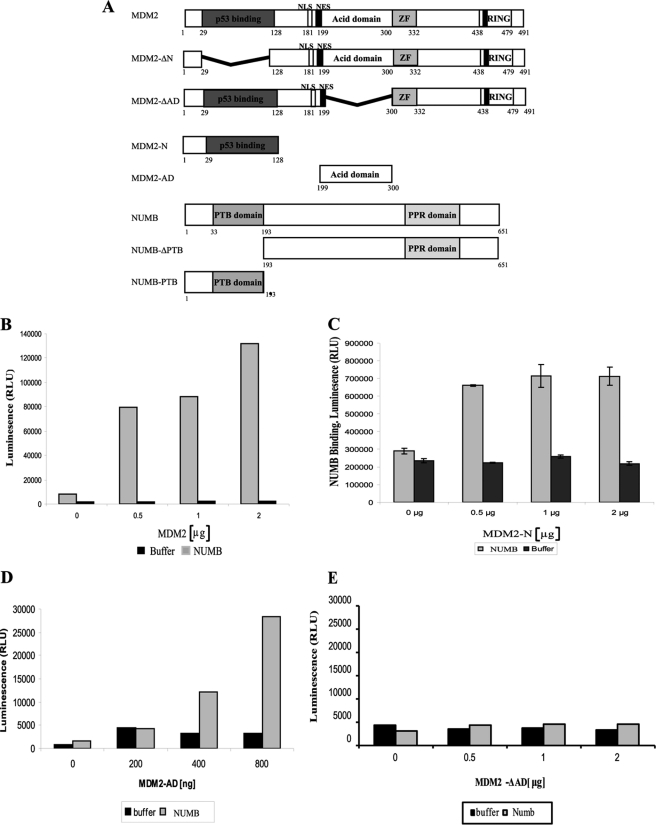
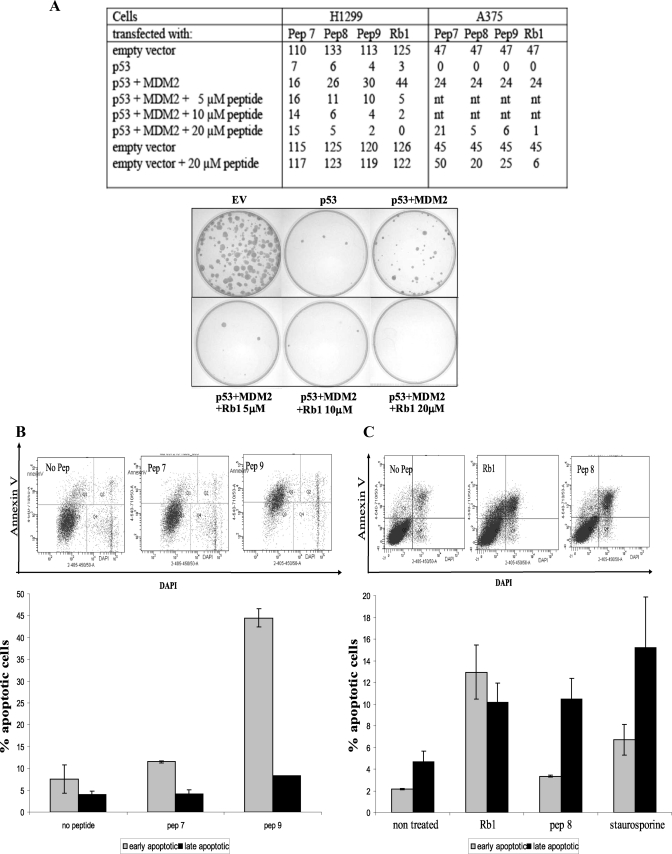
FIGURE 1.
NUMB binds to the Acid Domain of MDM2. A, schematic depicting full-length, mutant, and mini-domain MDM2 and NUMB proteins. ZF represents the zinc finger domain of MDM2. MDM2-ΔN is a mutant MDM2 missing the N-terminal hydrophobic pocket, which binds p53 BOX-I domain. MDM2 ΔAD is missing the acidic domain. NUMB ΔPTB is missing the PTB domain of NUMB. B–E, 0–2 μg of purified MDM2-FL (B), MDM2-N (C), MDM2-AD (D), or MDM2-ΔAD (E) were bound to a 96-well microtiter plate and incubated with full-length NUMB, with the GST tag cleaved. Binding was detected with anti-NUMB antibody followed by chemiluminescence with ECL reagent.
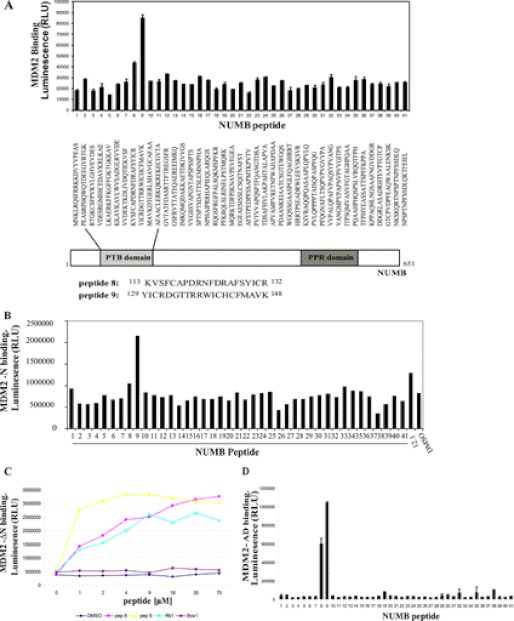
FIGURE 2.
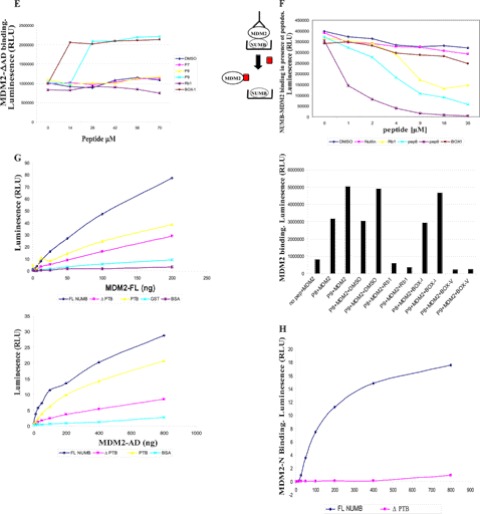
PTB domain of NUMB binds to the N terminus and acidic domain of MDM2. A, B, and D, biotin-tagged peptides to full-length NUMB (1–41) (3.5 μm) captured on a streptavidin-coated microtiter plate and incubated with purified full-length MDM2 (200 ng) (A), the isolated N-terminal hydrophobic pocket (MDM2-N) (200 ng) (B), or the isolated acid domain (MDM2-AD) (200 ng) (D) MDM2 binding detected with anti-MDM2 or anti-GST antibodies, followed by chemiluminescence with ECL reagent. C and E, increasing concentrations of biotin-tagged peptides as indicated (0 to 70 μm) were bound to a streptavidin-coated microtiter plate and incubated with MDM2 missing the N terminus (MDM2-ΔN) (200 ng) (C) or MDM2 missing the acid domain (MDM2-ΔAD) (200 ng) (E). MDM2 binding was detected with anti-GST antibody. F, upper panel: Full-length GST-NUMB protein (500 ng) captured onto a microtiter plate and incubated with full-length MDM2 (200 ng), which had been preincubated with DMSO control or increasing concentrations of peptides as indicated (0 to 35 μm). MDM2 binding was detected with anti-MDM2 antibody. The diagram to the left illustrates the experiment: when competing peptide bound to MDM2, then no NUMB-MDM2 binding was detected. F, lower panel: Biotin-tagged NUMB peptides 8 and 9 (3.5 μm) were bound to a streptavidin-coated plate followed by incubation with MDM2 (200 ng), which had been preincubated with DMSO control or peptides as indicated (35 μm). MDM2 binding was detected with anti-MDM2 antibody. G, full-length NUMB (FL NUMB), the PTB domain of NUMB (PTB), NUMB missing the PTB domain (Δ PTB), or control GST-only and BSA proteins were immobilized on a 96-well microtiter plate and incubated with either full-length MDM2 (upper graph) or the acidic domain of MDM2 (MDM2-AD) (lower graph). Binding was detected with anti-MDM2 antibody followed by chemiluminescence with ECL reagent. H, the N terminus of MDM2 (MDM2-N) was immobilized on a microtiter plate and incubated with full-length NUMB or NUMB missing the PTB domain (Δ PTB). Binding was detected with anti-NUMB antibody followed by chemiluminescence with ECL reagent. The experiments shown in this figure represent at least three separate experiments.
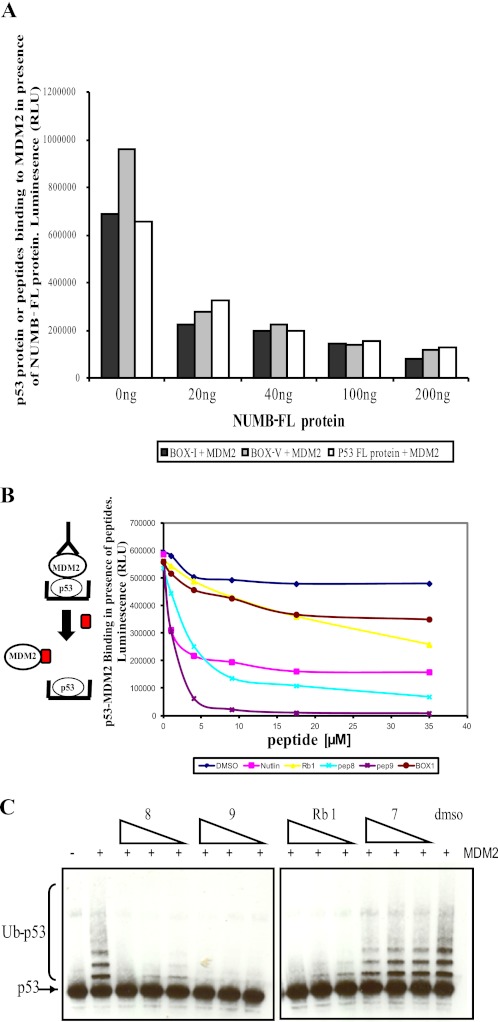
FIGURE 5.
Acid domain-binding ligands inhibit MDM2-mediated ubiquitination of p53 by disrupting the MDM2-p53 complex. A, full-length p53 protein (50 ng) or p53 peptides (BOX-I or BOX-V, 3.5 μm) were captured onto a microtiter plate and incubated with full-length MDM2 (200 ng), which had been preincubated with increasing concentrations of NUMB as indicated (0 to 200 ng). Bound MDM2 was detected with anti-MDM2 antibody. B, full-length p53 protein (50 ng) captured onto a microtiter plate and incubated with full-length MDM2 (200 ng), which had been preincubated with DMSO or increasing concentrations of peptides as indicated (0 to 35 μm). Bound MDM2 was detected with anti-MDM2 antibody. The diagram to the left illustrates the experiment: when competing peptide binds MDM2, then no p53-MDM2 binding was detected. C, full-length p53 protein (50 ng) incubated with ubiquitin (2 μm), E1 (100 nm), and E2 (1 μm) in the presence or absence of MDM2 (100 ng) and increasing amounts of peptides (10, 20, and 40 μm) or DMSO. Samples were analyzed by immunoblotting with anti-p53 antibody, where Ub-p53 represents ubiquitinated p53.
Colony Formation Assay
H1299 or A375 cells were transfected at 90% confluency with pCDNA3.1-p53 (150 ng), pCDNA3.1-MDM2 (250 ng), and DNA equalized by addition of empty vector. Peptides were added to the cells 4 h post transfection. After 24 h cells were then trypsin-treated and re-plated at different concentrations into 10-cm dishes with RPMI or DMEM containing 1.5 mg/ml Geneticin (Invitrogen). Media was replaced after 3 days and then weekly with media plus Geneticin. Colonies were counted after 15 days by fixing in methanol for 30 min following staining with 10% v/v Giemsa (Sigma).
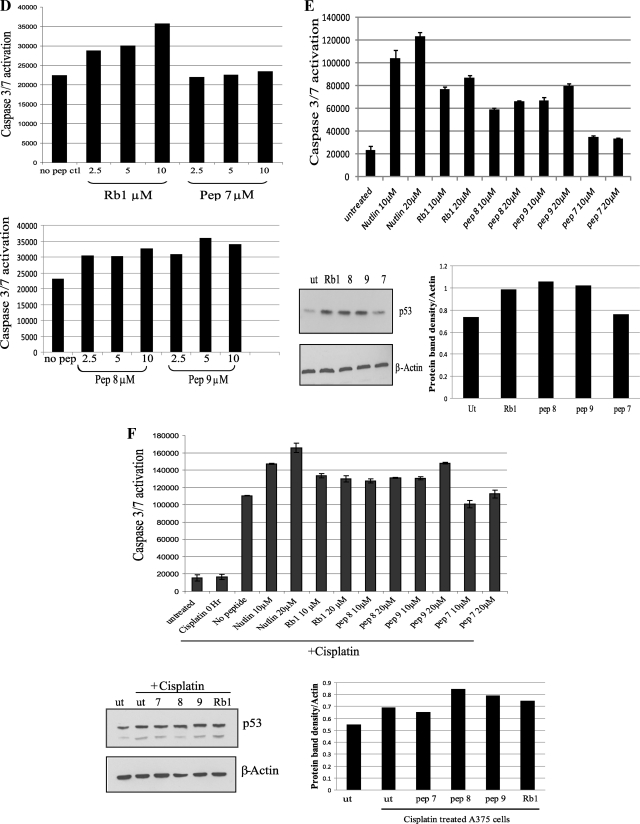
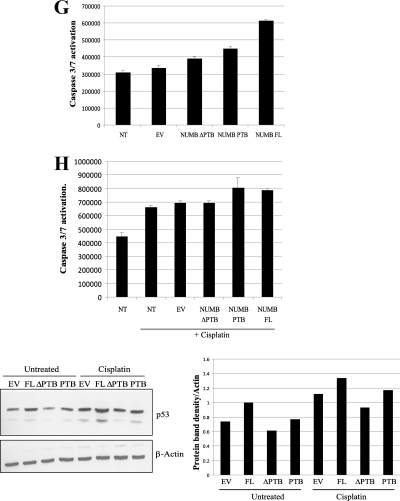
Caspase-Glo 3/7 Assay
The Caspase-Glo 3/7 assay was performed using ZR75 and A375 cells at ∼80% confluency, in a 96-well format. Cells were transfected with plasmids or treated with peptides for 18–24 h (as detailed in figure legend (Fig. 7, D–H)) and then treated with cisplatin (12 μg/ml) for a further 24 h where indicated in the figures. Cells were then processed according to the Caspase-Glo 3/7 kit manufacturer's protocol (Promega).
FIGURE 7.
Acid domain-binding ligands overcome MDM2 inhibition of p53. A, table depicting colony formation assays carried out in H1299 and A375 cells transfected with p53 (150 ng) and MDM2 (250 ng) and compared with empty vector control as indicated. Increasing amounts of peptides (5, 10, and 20 μm) were added to H1299 cells and 20 μm of peptides to A375 cells 4 h post transfection as indicated. After 24 h, cells were then trypsin-treated and re-plated. After 15 days colonies were fixed, stained with Giemsa (Sigma), and counted. Nt indicates not treated. The photographic results with one of the peptides Rb1, in H1299 cells is shown below. B and C, MCF7 cells treated in duplicate with 25 μm peptides for 24 h as indicated. Cells were trypsinized and Annexin V-stained with Allophycocyanin (APC)-tagged antibody. Flow cytometry was performed after the addition of DAPI. Results are also presented graphically as percentage of apoptotic cells. D, ZR75 cells treated with peptides at 2.5, 5, and 10 μm as indicated. Caspase activation was analyzed after 18-h incubation with the peptides. E: Upper panel: A375 cells left untreated or treated with 10 or 20 μm peptides as indicated. Caspase activation was analyzed after 18-h incubation with the peptides. Lower panel: A375 cells were left untreated (ut) or treated with peptides (20 μm) as indicated for 18 h and then analyzed by immunoblotting with p53 and β-actin antibodies. Densitometry of the immunoblots is shown on the right. F: Upper panel, A375 cells left untreated or treated with 10 or 20 μm peptides for 18 h as indicated. Cells were then left untreated as a control or treated with cisplatin (12 μg/ml) for 0 or 24 h as indicated. Caspase activation was then analyzed. Lower panel, A375 cells were left untreated (ut) or treated with peptides as indicated for 18 h followed by treatment with or without cisplatin (12 μg) for 24 h as indicated. Cells were collected and analyzed for levels of p53 and β-actin by Western blotting. Densitometry of the immunoblots is shown on the right. G, A375 cells were untransfected (nt) or transfected with 400 ng of either empty vector (EV), full-length NUMB (NUMB FL), truncated NUMB missing the PTB domain (NUMB ΔPTB), or the isolated PTB domain of NUMB (NUMB PTB) as indicated. Caspase activation was analyzed 24 h post transfection. H: Upper panel, A375 cells were transfected as in G, but cells were then left untreated or treated with cisplatin (12 μg/ml) for 24 h, as indicated, followed by analysis for caspase activation. Lower panel, A375 cells treated as above in G and H, but cells were collected and analyzed for levels of p53 and β-actin by Western blotting as indicated. Densitometry of the immunoblots is shown on the right. The experiments presented in this figure are representative of at least three separate experiments.
FACS
MCF7 cells were seeded into a 6-well plate and grown for at least 24 h prior to peptide treatment. Cells were incubated with peptides (25 μm) for 24 h as indicated. Cells were then trypsinized and harvested by centrifugation and processed according to the Annexin V Apoptosis Detection Kit APC manufacturer's protocol (eBioscience). Samples were analyzed at the Flow Cytometry Facility, MRC Human Genetics Unit, Western General Hospital, Edinburgh.
RESULTS
NUMB Binds to the Acidic Domain of MDM2
To understand the mechanism of action of MDM2-mediated ubiquitination of NUMB and to determine how NUMB might inhibit degradation of p53, we examined the interaction between MDM2 and NUMB in detail. Recent studies into the interaction of the E3 ligase MDM2 with substrates such as p53 and the transcription factor IRF-2 have demonstrated that MDM2 binds to its substrates using a dual-site mechanism (9, 13). However, the salient interaction for p53 ubiquitination is binding of the MDM2 acid domain to the core DNA binding domain of p53, encompassing the BOXV/S9-S10 linker region (9, 23). MDM2-mediated ubiquitination of IRF-2 also requires acid domain interactions (13). NUMB was previously shown to bind to the N-terminal hydrophobic pocket of MDM2 (17, 18), but we wanted to determine if, like p53 and IRF-2, the acidic domain of MDM2 was also important in regulating the ubiquitination and degradation of NUMB.
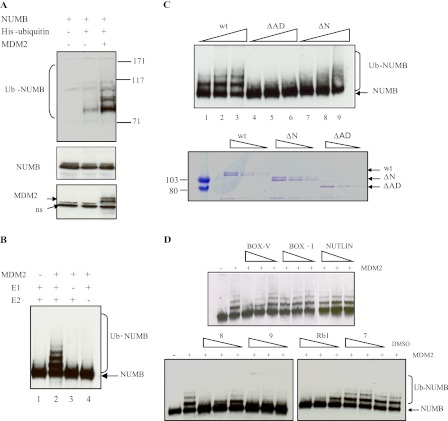
Our initial efforts sought to determine if NUMB could bind to MDM2 or MDM2 mini-domains in vitro using purified proteins (Fig. 1A) in protein-protein interaction assays. Full-length MDM2 or the N terminus of MDM2, purified from E. coli, was immobilized on a plate, and binding to full-length NUMB was detected using an anti-NUMB antibody (Fig. 1, B and C). As expected, NUMB bound to both full-length MDM2 (Fig. 1B) and the N terminus (Fig. 1C). The isolated acidic domain of MDM2 (MDM2-AD) was then examined for NUMB binding (Fig. 1D). Interestingly, NUMB did form an interaction with the acidic domain of MDM2 (Fig. 1D). This result suggests that, in a similar manner to p53, NUMB interacts with two distinct regions of MDM2, the N-terminal hydrophobic pocket and the acidic domain. When MDM2, missing the acidic domain, MDM2-ΔAD, (Fig. 1A) was then examined for NUMB binding (Fig. 1E), we observed a dramatic reduction in binding to full-length NUMB (Fig. 1E). This confirmed the significance of the acidic domain as a major binding interface for NUMB.
The PTB Domain of NUMB Mediates Binding to MDM2
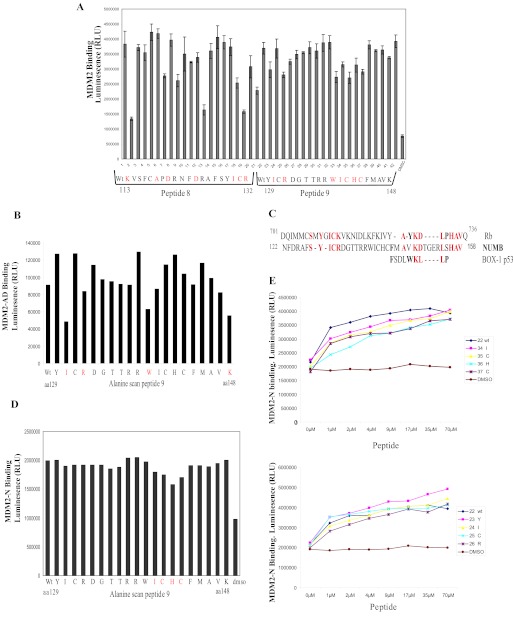
Two separate regions on p53 are involved in dual-site binding to MDM2. Therefore, to establish the regions of NUMB important in forming the interaction with MDM2, we synthesized a panel of overlapping biotin-tagged peptides spanning full-length NUMB and analyzed binding to MDM2 by a peptide-protein interaction assay. The biotin-tagged peptides were immobilized on a streptavidin-coated microtiter plate and incubated with MDM2, and binding was detected by an anti-MDM2 antibody. Two peptides within the PTB domain of NUMB, peptides 8 and 9 (amino acids 113–148), were found to interact with MDM2 (Fig. 2A). The PTB domain has previously been shown to interact with the N terminus of MDM2 (17). Therefore, we analyzed binding further to determine where the PTB domain peptides bound on MDM2 using mini-domain and mutant proteins (Fig. 1A). Peptide 9 of NUMB (amino acids 129–148) could form an interaction with the N-terminal hydrophobic pocket of MDM2 (MDM2-N), similar to BOX-I of p53 or BOX-I-like peptides such as 12.1 (24), which was used as a positive control in this assay (Fig. 2B). Peptide 8 (amino acids 113–132) was unable to bind to this region (Fig. 2B). Although peptide 9 bound the N-terminal hydrophobic pocket of MDM2 when we used MDM2 missing the N terminus (MDM2-ΔN) (Fig. 2C), both peptide 8 or 9 could bind to this truncated form of MDM2 (Fig. 2C), suggesting that regions other than the N terminus of MDM2 also contribute to the interaction with the PTB domain of NUMB.
The PTB domain peptides behaved more like the acidic domain-binding peptide Rb1 than the hydrophobic pocket-binding peptide BOX-1, and we have demonstrated above that purified full-length NUMB could form an interaction with the acid domain of MDM2 (Fig. 1D). Therefore, we wanted to determine if the NUMB PTB domain peptides bound to the acid domain. Indeed NUMB peptides 8 and 9 bound strongly to the acid domain of MDM2 (MDM2-AD) (Fig. 2D). Next mutant MDM2, missing the acidic domain (MDM2-ΔAD), was examined. In a similar manner to BOX-I of p53, peptide 9 was found to bind to MDM2 missing the acidic domain, but no significant binding was observed with peptide 8 (Fig. 2E). The above results suggest that peptide 8 binds only to the acid domain and that peptide 9 can interact with both the N terminus and acid domain of MDM2. Although the N-terminal hydrophobic pocket and acid domain are not directly adjacent to each other, they may be close in the three-dimensional structure of the protein allowing peptide 9 to bind into both regions simultaneously.
Further confirmation of the importance of the acid domain interaction was demonstrated by use of a protein-peptide competition assay. Rb1 peptide, derived from the retinoblastoma tumor suppressor protein, and BOX-V of p53 both bind to the acid domain of MDM2 (9). These peptides and NUMB peptides 8 and 9 were preincubated with MDM2, and then binding to NUMB was analyzed (Fig. 2F, upper panel). We found that, when preincubated with MDM2, these acid domain-binding ligands could inhibit the interaction of full-length NUMB with MDM2. However, hydrophobic pocket-binding ligands BOX-I and Nutlin (25) could not (Fig. 2F, upper panel), confirming the importance of acid domain interactions. Acid domain-binding ligands, Rb1 and BOX-V, could also disrupt the interaction of NUMB PTB domain peptides with MDM2 (Fig. 2F, lower panel). Because BOX-I could not disrupt the interaction between NUMB and MDM2, this result suggests that NUMB may bind to a slightly different region in the N terminus of MDM2 than p53.
To analyze these results further we made truncated NUMB proteins that were terminated after the PTB domain or missing the PTB domain (Fig. 1A). These proteins were then tested for binding to full-length MDM2 and to the isolated N-terminal and acidic domains of MDM2 by protein-protein interaction assays. The isolated PTB domain of NUMB bound to full-length MDM2 and the acidic domain (Fig. 2G), although a reduction in binding was observed compared with the full-length protein. When the truncated NUMB protein missing the PTB domain was examined, there was a dramatic reduction in binding to the acidic domain and also to full-length MDM2 (Fig. 2G). We then examined binding to the N terminus of MDM2 but did not see any binding to the PTB domain in this assay (data not shown). However, this may be due to the fact that the binding site was masked when the PTB domain was bound to the plate, as others have previously shown that the N terminus of MDM2 interacted with the PTB domain of NUMB (17). However, a dramatic reduction in binding to the N terminus of MDM2 was observed with the truncated form of NUMB, missing the PTB domain (Fig. 2H), suggesting that this is also an important binding region for the N terminus of MDM2.
Investigation into the Critical Residues of NUMB PTB Domain Involved in MDM2 Binding
Our results imply that the PTB domain peptides bind to both the N terminus and acidic domains of MDM2. Therefore, to determine the residues in peptides 8 and 9 that were important in forming the interaction with MDM2, we carried out an alanine scan of the peptides, where each residue in turn was mutated to alanine (any alanine residues in the wild-type peptide were mutated to glycine), and then binding to MDM2 was assessed by a peptide-protein interaction assay (Fig. 3A). We observed that a number of residues in peptides 8 and 9 when mutated to alanine had reduced binding to MDM2 (highlighted in red, Fig. 3A). Because peptide 9 of NUMB appeared to bind to both the hydrophobic pocket and the acidic domain of MDM2, we analyzed the alanine scan of peptide 9 further to determine what residues were important for binding to each region of MDM2 by using the isolated N terminus or acidic domain proteins (Fig. 1A). Mutation of residues isoleucine 130, arginine 132, tryptophan 139, and lysine 148 of NUMB showed greatest reduction in binding to the acidic domain (MDM2-AD) compared with wild-type peptide 9; therefore, we propose that these residues are important for mediating binding to the acidic domain of MDM2 (Fig. 3B). When we compared the sequence of the NUMB peptides to the Rb1 peptide (amino acids 701–720 of Rb), which binds to the acidic domain of MDM2, we identified an area of homology between Rb and NUMB at the end of peptide 8 and start of peptide 9, where the peptides overlap: YGICK (amino acids 708–712) in Rb and Y-ICR (amino acids 129–132) in NUMB (Fig. 3C). This correlates with our results demonstrating that these residues were important for MDM2 binding (Fig. 3, A and B). On the other hand, mutated amino acids 140–143 demonstrated slightly reduced binding to the N-terminal hydrophobic pocket compared with wild-type peptide 9 (Fig. 3D). Because peptide 9 binds very strongly to the N-terminal region, we titrated the peptide 9 mutant peptides further and saw a greater reduction in binding to the N-terminal hydrophobic pocket of MDM2, compared with wild-type peptide (Fig. 3E, upper graph). The peptide with a mutated histidine, H142A, in particular showed the greatest reduction in binding. However, the residues important for acidic domain binding above were not significantly reduced compared with the wild-type peptide 9 (Fig. 3E, lower graph). These results demonstrate that, although the PTB domain interacts with two domains on MDM2, different residues within the PTB domain form the interface with the N terminus and the acidic domain of MDM2.
FIGURE 3.
Determination of the critical residues in NUMB that mediate binding to MDM2. A, biotin-tagged NUMB peptides 8 and 9 wild-type (WT) or mutant peptide (7 μm) as indicated were captured onto a streptavidin-coated microtiter plate and incubated with full-length MDM2 (200 ng). MDM2 binding was detected with an anti-MDM2 antibody. The residue that was changed to alanine is shown below the graph, and critical residues for MDM2 binding are highlighted in red. B and D, alanine scan of peptides of biotin-tagged peptide 9 (3.5 μm) bound to a microtiter plate and incubated with GST-tagged MDM2 acidic domain (MDM2-AD) (B) or N terminus (MDM2-N) (D). MDM2 binding was detected with anti-GST antibody. Critical residues for MDM2 binding are highlighted in red. C, alignment of MDM2 binding regions of NUMB, Rb, and BOX-I of p53. Homologous residues are indicated in red, residues similar to BOX-I are in bold. E, upper and lower panel: Increasing amounts of biotin-tagged alanine scan of peptide 9 (0 to 70 μm as indicated) were bound to a streptavidin-coated plate and incubated with the GST-tagged MDM2 N terminus (MDM2-N). Peptide-bound MDM2 was detected with an anti-GST antibody.
Contribution of MDM2 Domains to MDM2-mediated Ubiquitination of NUMB
Because the acidic domain is important for catalyzing ubiquitination of other MDM2 substrates (9, 13), we wanted to determine the significance of the acidic domain for NUMB ubiquitination. Initially, we confirmed MDM2-mediated ubiquitination of NUMB using a well characterized in vivo ubiquitination assay (26). NUMB was transfected into cells in the presence of MDM2 and His-tagged ubiquitin, and a pulldown assay on nickel beads followed by immunoblot analysis was carried out. NUMB was efficiently ubiquitinated in the presence of MDM2 but not in its absence (Fig. 4A). These results confirm earlier reports demonstrating that NUMB is ubiquitinated by MDM2 (18). An in vitro ubiquitination assay was then set up that employed purified components. NUMB, purified from E. coli, was not ubiquitinated in the absence of E1, E2, or MDM2 (Fig. 4B, lanes 1, 3, and 4). However, in the presence of E1, E2, and full-length MDM2, NUMB was subjected to modification producing a characteristic ubiquitin ladder in vitro (Fig. 4B, lane 2).
FIGURE 4.
Contribution of MDM2 domains to the ubiquitination of NUMB. A, H1299 cells transfected with Numb (7.5 μg) in the presence and absence of His-Ubiquitin (10 μg) and Mdm2 (5 μg) as indicated. Samples were supplemented with empty vector, so that the final concentration of DNA was the same in all reactions. Cells were treated for 6 h with 25 μm MG132 before harvesting. 10% of the cell suspension was analyzed for total protein levels. The His-tagged ubiquitinated proteins were isolated using Ni-NTA-agarose (Qiagen) 48 h post transfection and analyzed by immunoblotting on a 7% Tris-acetate gel (Invitrogen) with anti-NUMB antibody. B, NUMB (250 ng) was incubated in the presence of ubiquitin (2 μm) and in the presence or absence of E1 (UBE-1) (100 nm), E2 (UbcH5) (1 μm), and MDM2 (100 ng) as indicated. Ubiquitination was detected by Western blotting with anti-NUMB antibody, where Ub-NUMB represents ubiquitinated NUMB. C, upper panel: NUMB (250 ng) was incubated in the presence of ubiquitin (2 μm), E1 (UBE-1) (100 nm), E2 (UbcH5) (1 μm), and increasing amounts of wild-type MDM2 (lanes 1–3) or MDM2 missing the acidic domain (MDM2-ΔAD) (lanes 4–6) or missing the N terminus (MDM2-ΔN) (lanes 7–9) (100–500 ng). Ubiquitination was detected by Western blotting with anti-NUMB antibody, where Ub-NUMB represents ubiquitinated NUMB. Lower panel: GST-tagged MDM2 wt, ΔAD, and ΔN protein (same amounts as those used for ubiquitination assay) run out on SDS-PAGE and stained with Coomassie Blue reagent. D, NUMB (250 ng) was incubated with ubiquitin (2 μm), E1 (100 nm), and E2 (1 μm) and in the presence or absence of MDM2 (100 ng) and varying amounts of ligand (10, 20, and 40 μm) or DMSO control as indicated. Samples were analyzed by SDS-PAGE and immunoblotting with anti-NUMB antibody, where Ub-NUMB represents ubiquitinated NUMB. Experiments presented in this figure are representative of at least three separate experiments.
Having confirmed that NUMB is ubiquitinated by MDM2 we then determined the contribution of different MDM2 domains to the ubiquitination of NUMB. When MDM2, missing the acidic domain, MDM2-ΔAD (Fig. 4C, upper panel, lanes 4–6), was examined, no ubiquitination was observed compared with wt MDM2 (Fig. 4C, upper panel, lanes 1–3), although mutant MDM2 proteins were present in similar quantities to wild-type protein in the assay (Fig. 4C, lower panel). MDM2, missing the N-terminal domain, MDM2-ΔN (Fig. 1A), also appeared to lose the ability to catalyze ubiquitination of NUMB using low amounts of MDM2 in the assay (Fig. 4C, upper panel, lane 7). However, when higher amounts of MDM2-ΔN were used in the reaction, the distinct mono-ubiquitination ladder observed with the wild-type MDM2 was visible but reduced compared with wt MDM2. However, NUMB appeared to be poly-ubiquitinated, and less un-ubiquitinated NUMB appeared to be present (Fig. 4C, upper panel, lanes 8 and 9). The changes to ubiquitination of NUMB may be due to a conformational change in the MDM2 protein as a result of the loss of the N terminus. However, the results demonstrate that the acid domain of MDM2 is crucial for ubiquitination of NUMB.
Acid Domain-binding Ligands Inhibit MDM2-mediated Ubiquitination of NUMB
We have shown above that acid domain-binding ligands can inhibit the NUMB-MDM2 interaction. Next, we wanted to determine the potential of the acid domain-binding ligands as inhibitors of MDM2-mediated ubiquitination of NUMB. Interestingly, all the acid domain-binding ligands, Rb 1, BOX-V, and peptides 8 and 9 from NUMB, could inhibit MDM2-mediated ubiquitination of NUMB in vitro (Fig. 4D). We have previously demonstrated that hydrophobic pocket-binding ligands like BOX-I and Nutlin are unable to inhibit MDM2-catalyzed ubiquitination of p53 (9) or IRF-2 (13). In a similar manner to p53, ubiquitination of NUMB was not inhibited by BOX-I or Nutlin in this assay (Fig. 4D), which also correlates with earlier results suggesting these N-terminal MDM2-binding peptides do not disrupt the interaction between NUMB and MDM2 (Fig. 2F). Peptide 7 of NUMB was used as a control in this assay, because it did not bind to MDM2 (Fig. 2A), and indeed it did not inhibit ubiquitination (Fig. 4D).
NUMB PTB Domain Peptides Disrupt the MDM2-p53 Interaction and MDM2-mediated Ubiquitination of p53
NUMB is found in a trimeric complex with MDM2 and p53 and has been shown to play a role in the stabilization of p53 in cells (21). However, the mechanism by which NUMB prevents the degradation of p53 is still unclear. Because we found that NUMB binds to the same regions on MDM2 as p53, namely, the N-terminal hydrophobic pocket and the acidic domain, we hypothesized that NUMB could interfere with the MDM2-p53 interaction and thereby disrupt MDM2-mediated ubiquitination.
Initially we sought to determine if the NUMB full-length protein would disrupt the p53-MDM2 complex. We observed binding of purified full-length p53 to MDM2 in a protein-protein interaction assay, however, in the presence of NUMB, MDM2 binding was severely reduced (Fig. 5A). In addition, NUMB protein could also disrupt the interaction of MDM2 with both BOX-I and BOX-V regions of p53 (Fig. 5A). The NUMB PTB domain-derived peptides 8 and 9 were then examined in this assay, and we observed that they could disrupt p53 binding to MDM2 better than the other acid domain-binding peptide Rb1 and even better than Nutlin (Fig. 5B). NUMB was previously shown to bind to p53 in an immunoprecipitation assay in the absence of MDM2, suggesting a direct interaction between NUMB and p53 (21). However, we did not detect p53 binding to any of the NUMB peptides (data not shown).
Because other acid domain-binding ligands, such as Rb1, disrupt the MDM2-p53 complex and also inhibit ubiquitination of p53, the NUMB PTB domain peptides were tested in this assay. Similar to Rb1, both peptides 8 and 9 of NUMB were found to strongly inhibit MDM2-mediated ubiquitination of p53 in vitro (Fig. 5C). The control peptide 7 was unable to inhibit ubiquitination of p53 (Fig. 5C). The results presented suggest that, by binding to similar regions on MDM2 as p53, NUMB can disrupt the MDM2-p53 complex and inhibit ubiquitination of p53.
Acid Domain-binding Ligands Promote Stabilization of NUMB and p53 in Cells
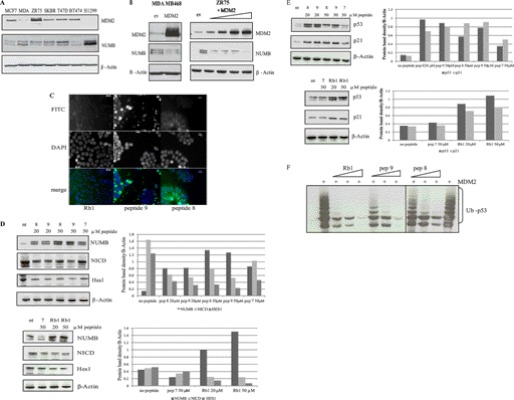
NUMB has previously been shown to be frequently ubiquitinated and degraded in breast cancer cells (27). We and other groups have previously shown that a small peptide could mimic a full-length protein and inhibit MDM2 function (9, 28, 29). Our results above suggest that, by using specific acid-domain-binding inhibitors of MDM2, we may be able to restore NUMB in breast cancer cells and also stabilize p53. Initially we examined a panel of breast cancer cells to determine MDM2 and NUMB protein expression levels. Interestingly we observed an inverse correlation between NUMB and MDM2 (Fig. 6A). Overexpression of MDM2 in osteosarcoma cells has previously been shown to result in degradation of NUMB (17). In addition, we demonstrate that overexpression of MDM2 in MDA MB 468 and ZR75 breast cancer cells resulted in degradation of NUMB (Fig. 6B).
FIGURE 6.
MDM2 acid domain-binding ligands stabilize NUMB and p53 in breast cancer cells. A, panel of breast cancer cells (MCF7, MDA MB 468, ZR75, SKBR, T47D, and BT 474) and a lung cancer cell line, H1299 (used as a control for high NUMB and low MDM2 expression) immunoblotted for protein expression levels of NUMB, MDM2, and β-actin loading control as indicated. B, left panel: MDA-MB 468 cells transiently transfected with MDM2 or empty vector control (ev) (2 μg) as indicated for 24 h followed by immunoblotting with anti-MDM2, anti-NUMB, and β-actin as indicated. Right panel: ZR75 cells were transiently transfected with increasing amounts of MDM2 (0.25, 0.5, 1, and 2 μg). DNA was equalized by addition of empty vector (ev). Samples were analyzed by immunoblotting with anti-MDM2, anti-NUMB antibodies, and β-actin as indicated. C, HIV-TAT-fused peptides 8 and 9 and Rb1 were added to MCF7 cells at 20 μm and analyzed using the FITC and bright-field channels with the Zeiss AxioVision System. Vectashield media with DAPI (Vector) was used to visualize the nuclei. D: Left panel, MCF7 cells treated with 20 or 50 μm of peptides for 24 h as indicated. Cells were analyzed by immunoblotting with antibodies to NUMB, NICD, Hes1, and β-actin loading control as indicated. Right panel, Densitometry of the immunoblots shown on the left. E: Left panel, MCF7 cells treated with 20 and 50 μm of peptides for 24 h as indicated. Cells were analyzed by immunoblotting with antibodies to p53, p21, and β-actin as indicated. Right panel: Densitometry of the immunoblots shown on the right. The experiments presented in this figure are representative of at least three separate experiments. F, H1299 cells transfected with His-ubiquitin (1 μg) and p53 (150 ng) in the presence of MDM2 (1 μg) and HIV-TAT-fused peptides (10, 20, and 50 μm) as indicated. Ubiquitinated p53 was isolated 2 h after treatment with MG132 (20 μm) and 24 h post transfection and analyzed by immunoblotting with anti-p53, where Ub-p53 represents ubiquitinated p53.
The basic domain of HIV-TAT-(49–57) (RKKRRQRRR) has been well used recently as a tool to deliver proteins/small molecule peptides to cells and tissues (30–32). Therefore, the acid domain-binding peptides were synthesized fused to HIV-TAT and with a fluorescent label for intracellular delivery. Fluorescence microscopy was used to determine cellular localization (Fig. 6C). All peptides examined were found to localize to the nucleus and cytoplasm (Fig. 6C), although not all cells examined display bright fluorescent staining.
To examine the effects of the peptides we used MCF7 breast cancer cells, which have wild-type p53 and express high levels of MDM2 and lower levels of NUMB (Fig. 6A). Upon addition of acid domain-binding ligands to MCF7 breast cancer cells, we observed an increase in NUMB protein levels, particularly when 50 μm of the NUMB PTB domain-derived peptides were added (Fig. 6D), although Rb1 peptide was just as good an inhibitor at 20 μm. Furthermore, after addition of the acid domain-binding ligands to cells, expression of both the active intracellular domain of Notch1 (NICD) and its downstream target Hes1 were reduced compared with untreated or control peptide 7-treated cells (Fig. 6D), demonstrating an effect of the stabilization of NUMB on its downstream targets in these cells.
The acid domain-binding ligands were then examined further to determine if they could stabilize p53 in cells. Indeed, 24 h after addition of the peptides to MCF7 breast cancer cells, we observed an increase in protein expression levels of p53 (Fig. 6E), which correlated with the enhancement of NUMB in the presence of these inhibitors that we showed earlier (Fig. 6D). Furthermore, elevated levels of p21, a downstream target of p53, were also observed (Fig. 6E). Along with the stabilization of p53 observed, ubiquitination of p53 was also inhibited in cells after addition of the NUMB PTB domain peptides (Fig. 6F).
Acid Domain-binding Ligands Induce Growth Suppression in Cells
The above results demonstrate that the NUMB PTB domain peptides and other acid domain-binding ligands can increase the stability of p53 in cells and predicts that p53-mediated responses, such as growth suppression and apoptosis, would also be enhanced. Therefore, to test the effect of the acid domain-binding peptides on growth suppression, we set up a colony formation assay in p53-null H1299 cells (Fig. 7A). As expected, when p53 was overexpressed in these cells fewer colonies were observed compared with empty vector control. When low levels of MDM2 were co-transfected with p53, it could overcome the p53-induced suppression of growth. All the acid domain-binding peptides were able to reverse the effects of MDM2 in this assay and restore the p53-induced suppression of growth (Fig. 7A). Peptide 7 of NUMB, which does not bind to MDM2, was used as a control and was unable to overcome the effects of MDM2. This suppression of growth was also observed in the A375, melanoma cell line (Fig. 7A). The effect of the peptides was due to p53, because no effect on growth was observed in the absence of transfected p53 in the p53-null H1299 cells, but some growth arrest was observed, especially with the Rb1 peptide, in A375 cells containing wt p53 (Fig. 7A).
Acid Domain-binding Ligands Induce Apoptosis in Cells
Considering the results of these experiments, we wanted to investigate the effect of the peptides on the induction of apoptosis. Initially, the effect of the acid domain-binding ligands was examined by FACS analysis using Annexin V staining (Fig. 7B). MCF7 breast cancer cells were examined at different time points after addition of the peptides (data not shown), and we found the most dramatic effects were observed at 24 h and; therefore, this time point was used for all further experiments. After addition of NUMB peptide 9 to the cells, we observed a dramatic increase in the percentage of early apoptotic cells, compared with the control peptide 7. The number of late apoptotic cells was also increased compared with controls in the presence of NUMB peptide 9 (Fig. 7B). NUMB peptide 8 and Rb1 peptide were then examined for induction of apoptosis in MCF7 cells (Fig. 7C). A very slight increase in early apoptosis was observed with NUMB peptide 8, but a more dramatic induction of late apoptosis was observed with this peptide. Addition of Rb1 resulted in an increase in the percentage of both early and late apoptotic cells (Fig. 7C).
In addition to annexin V, activation of caspase3/7 was also examined in ZR75 cells (Fig. 7D). After addition of the Rb1 peptide, an increase in caspase activation was observed (Fig. 7D, upper panel), but the negative control peptide 7 was unable to activate the caspases. The NUMB PTB domain peptides 8 and 9 were then examined in this assay to determine if they could induce apoptosis. Peptides 8 and 9 enhanced activation of caspase3/7 in the cells, compared with the control without any peptide (Fig. 7D, lower panel).
This effect was not cell-type-specific or breast cancer-specific, because the peptides also could activate caspases 3/7 in A375 cells (Fig. 7E, upper panel). However, no effect was observed with the peptides in p53-null H1299 cells (data not shown), indicating p53 dependence. The activation of caspases 3/7 correlated with an increase in the stabilization of p53 observed after treatment of A375 cells with the acid domain-binding ligands (Fig. 7E, lower panel).
The PTB Domain of NUMB Enhances p53-induced Apoptosis in Response to DNA Damage
NUMB was previously shown to enhance stabilization of p53 after DNA damage, resulting in enhancement of p53-dependent apoptosis (21). Therefore, activation of caspases 3/7 was examined in A375 cells following treatment with cisplatin. Compared with untreated cells, cisplatin resulted in activation of the caspases, which was further enhanced by addition of the acid domain-binding peptides to the cells (Fig. 7F, upper panel). Nutlin was used as a positive control and Numb peptide 7 as a negative control in these experiments. In addition, we observed stabilization of p53 levels after treatment of the A375 cells with cisplatin, which was enhanced by the acid domain-binding ligands (Fig. 7F, lower panel).
To determine the significance of the PTB domain in stabilization of p53 and activation of apoptosis further, we transfected A375 cells with full-length NUMB, compared with NUMB missing the PTB domain (NUMBΔPTB) or NUMB truncated after the PTB domain (NUMB PTB) in this assay. Full-length NUMB and the PTB domain (NUMB PTB) on its own were capable of stimulating caspase activation (Fig. 7G). However, a reduction in caspase activation was observed with NUMB missing the PTB domain (NUMB ΔPTB) (Fig. 7G). In addition, unlike the mutant NUMB missing the PTB domain, NUMB or the isolated PTB domain of NUMB could enhance caspase activation after cisplatin treatment of A375 cells (Fig. 7H, upper panel). Correlating with these results, p53 was stabilized in the presence of transfected full-length NUMB or slightly by the PTB domain of NUMB but not in the absence of the PTB domain (Fig. 7H, lower panel). These results suggest the importance of the PTB domain in mediating the effect of NUMB on the p53 response to DNA damage.
We have highlighted in this report the importance of the PTB domain of NUMB and the MDM2 dual site-binding mechanism for MDM2-mediated regulation of NUMB. The significance of the PTB domain of NUMB in the regulation of p53 was also demonstrated. By targeting the acid domain of MDM2 with small molecule peptide inhibitors, we can enhance levels of NUMB and restore the levels and tumor suppressor activities of p53 in cells, which identifies the acid domain as a potential target for drug discovery.
DISCUSSION
MDM2 uses a dual-site mechanism to ubiquitinate and degrade substrates such as p53 and IRF-2, involving acidic domain interactions (9, 13). Previous reports from others using mutational analysis have also demonstrated the significance of the acidic domain of MDM2 for its E3 ligase activity (10, 11). In this report we now demonstrate that MDM2 uses this same dual-site mechanism, involving N-terminal hydrophobic pocket and acidic domain interactions, to regulate levels of the cell fate determinant NUMB, another physiological relevant target of MDM2. However, in contrast to p53, which binds to MDM2 through two separate domains, the BOX-I transactivation domain and the BOX-V S9-S10 linker region in the core DNA binding domain, NUMB mediates binding to the two sites in MDM2 through one region within the PTB domain. The PTB domain of NUMB is known to be critical for NUMB function in vivo, and overexpression of rat NUMB PTB domain alone affected neural development (33).
The N terminus of MDM2 was previously shown to interact with the PTB domain of NUMB (17). Our results demonstrate that two 20-residue peptides within the PTB domain of NUMB are able to bind to MDM2. We have narrowed down the interaction between the PTB domain and MDM2 to amino acids 113–148 of NUMB with amino acids 140–143 contributing to the interaction with the N-terminal hydrophobic pocket. Different residues within the PTB domain were involved in acidic domain binding. Although these regions of MDM2 are not adjacent in the protein they may lie beside one another in the three-dimensional conformation of the protein, allowing NUMB to bind simultaneously to both domains. It is not unusual for a short peptide sequence to accommodate binding to more than one region or to more than one protein. The C terminus of p21 contains overlapping recognition sites for both cyclins and proliferating cell nuclear antigen, and a 20-mer peptide is sufficient to mimic the assembly role of full-length p21 (34). The structure of the NUMB PTB domain is similar to other PTB domain- containing proteins such as Shc, X11, and IRS-1 (35) and appears to have a broad binding specificity, capable of binding to multiple peptides that share no strong sequence similarity (35–37). These diverse sequences appear to bind at a common binding site within a large hydrophobic surface groove in the PTB domain. Our results demonstrate that some of the residues exposed in the peptide-binding groove of the NUMB-PTB domain are involved in binding to MDM2. Residues Lys-148, Ala-151, and Arg-165 of dNUMB (Lys-113, Ala-118, and Arg-132 in hNUMB) have all been shown to be involved in binding to the Nak peptide (38), and we found these residues were also important for MDM2 acidic domain binding.
Deletion of the acidic domain resulted in loss of binding to NUMB and loss of MDM2-mediated ubiquitination. These results correlate with our previous findings, demonstrating that acidic domain interactions are critical for MDM2-mediated ubiquitination of p53 and IRF-2 (9, 13). Interestingly, NUMB is thought to play a role in the stabilization of p53 in cells (21, 39), although the mechanism NUMB uses to achieve stabilization of p53 was unclear. We demonstrate here the mechanism whereby NUMB stabilizes p53 in cells. By binding to both the N terminus and the acidic domain of MDM2 PTB domain-derived peptides could physically disrupt the p53-MDM2 complex and inhibit ubiquitination of p53. No real homology was found between PTB domain peptides and MDM2 hydrophobic pocket-binding ligands, such as BOX-I of p53. Also, the BOX-1 peptide or Nutlin could not disrupt the interaction between NUMB and MDM2 or MDM2-mediated ubiquitination. However, MDM2 includes a sequence in the N-terminal hydrophobic pocket (YLGQYI), which resembles the YIGPYF motif. Mutations at Gly-58 of MDM2 (G in above motif) resulted in a decrease in the association of MDM2 with both p53 and NUMB (17), and full-length NUMB could disrupt the BOX-I-MDM2 complex, suggesting there may be some overlap in binding regions. In addition, deletion of the PTB domain resulted in a dramatic decrease in binding to the N terminus and the acidic domains of MDM2.
MDM2 is tightly regulated in the cell, which involves a number of mechanisms, including protein-protein interactions, protein turnover by ubiquitination, and post-translational modification, such as phosphorylation on multiple sites. Both the N terminus and the acidic domain of MDM2 contain residues that are targeted by a number of kinases that phosphorylate MDM2, resulting in changes in the stabilization of p53 (40, 41). This may also be the case for MDM2-mediated regulation of NUMB. CKI phosphorylates the acidic domain of MDM2, and this appears to play two distinct roles in regulating p53. Small molecule inhibitors of CK1α or siRNA to CK1α can stabilize p53 in cells (41). When CK1α was knocked out in cells a change in the stabilization of p53 was observed, although we did not observe any change in stabilization of NUMB in CK1α knock-out cells.3 This indicates that CK1a integration into MDM2 is specific to p53 and does not integrate with NUMB. However, it is highly likely that other kinases may be involved in regulating the interaction with MDM2 and the levels of NUMB in cells, which may impact on the tumor suppressor activities of p53.
The results in this report and our previous studies (9) on the discovery of the acid domain-binding ligands as potential inhibitors of MDM2-mediated ubiquitination lead us to design tools to investigate these interactions further in cells and to determine their effect on the tumor suppressor activities of p53. Using the peptides fused to HIV-TAT, we could deliver them into the cell and demonstrate that the acid domain-binding ligands could enhance levels of NUMB in breast cancer cells. In a similar manner to other acid domain-binding peptides (9), we demonstrated that a small fragment from the NUMB PTB domain could not only disrupt the interaction between p53 and MDM2 but could also inhibit ubiquitination and degradation of p53 in cells. Further analysis of the acid domain-binding ligands revealed that, by targeting the acidic domain of MDM2, the function of p53 as a tumor suppressor protein could be restored in cells with all the acid domain-binding ligands inducing a growth arrest and apoptosis in cells. NUMB has previously been shown to enhance the p53-mediated response to apoptosis after DNA damage (21). Indeed, in this report we demonstrated that the acid domain-binding ligands could enhance apoptosis after treatment of the cells with cisplatin and we also demonstrated the significance of the PTB domain of NUMB for this enhancement of p53-mediated apoptosis.
NUMB is also a major regulator of the Notch signaling pathway, and aberrant activation of Notch is frequently found in breast cancer as a result of ubiquitination and degradation of NUMB. We observed MDM2-mediated degradation of NUMB in breast cancer cells, and it is likely that MDM2 may contribute to the degradation of NUMB and activation of Notch in breast cancer. Therefore, the MDM2-NUMB complex may regulate both the p53 and Notch pathways. This also highlights the importance of targeting the acidic domain of MDM2 for drug discovery. Indeed, treatment of MCF7 cells with the acid domain-binding ligands demonstrated a reduction in the activated form of Notch, NICD, and Hes1, a downstream target of Notch.
Modifying the p53 pathway using drug combinations is an attractive option. A combination of a CDK inhibitor and Nutlin was recently shown to synergize in the activation of p53 (42). Perhaps combining acid domain-binding ligands with Nutlin would also show a synergistic effect on the activation of p53 in cancer cells. Therefore, targeting the acidic domain of MDM2 on its own or in combination with other drugs may be a useful therapeutic strategy for the prevention of cancer, and the data presented provides a good starting point for the design of peptidomimetic inhibitors.
Acknowledgments
We thank Kathryn Ball and Ted Hupp for many reagents and advice. We thank David Lane for providing MDM2 and His-Ub plasmids, Moshe Oren for NUMB constructs.
This work was also supported by Cancer Research UK and the Breast Cancer Campaign.
A. S. Huart and T. Hupp, personal communication.
- MDM2
- murine double minute 2
- IRF
- interferon regulatory factor
- Rb
- retinoblastoma
- NICD
- intracellular domain of Notch
- PTB
- phosphotyrosine binding.
REFERENCES
- 1. Bond G. L., Hu W., Levine A. J. (2005) MDM2 is a central node in the p53 pathway. 12 years and counting. Curr. Cancer Drug Targets 5, 3–8 [DOI] [PubMed] [Google Scholar]
- 2. Oliner J. D., Pietenpol J. A., Thiagalingam S., Gyuris J., Kinzler K. W., Vogelstein B. (1993) Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362, 857–860 [DOI] [PubMed] [Google Scholar]
- 3. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 4. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 5. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 6. Wawrzynow B., Zylicz A., Wallace M., Hupp T., Zylicz M. (2007) MDM2 chaperones the p53 tumor suppressor. J. Biol. Chem. 282, 32603–32612 [DOI] [PubMed] [Google Scholar]
- 7. Yin Y., Stephen C. W., Luciani M. G., Fåhraeus R. (2002) p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 4, 462–467 [DOI] [PubMed] [Google Scholar]
- 8. Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 9. Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 10. Kawai H., Wiederschain D., Yuan Z. M. (2003) Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol. Cell. Biol. 23, 4939–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meulmeester E., Frenk R., Stad R., de Graaf P., Marine J. C., Vousden K. H., Jochemsen A. G. (2003) Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol. Cell. Biol. 23, 4929–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Z., Wang H., Li M., Rayburn E., Agrawal S., Zhang R. (2005) Novel MDM2 p53-independent functions identified through RNA silencing technologies. Ann. N.Y. Acad. Sci. 1058, 205–214 [DOI] [PubMed] [Google Scholar]
- 13. Pettersson S., Kelleher M., Pion E., Wallace M., Ball K. L. (2009) Role of Mdm2 acid domain interactions in recognition and ubiquitination of the transcription factor IRF-2. Biochem. J. 418, 575–585 [DOI] [PubMed] [Google Scholar]
- 14. Bothner B., Lewis W. S., DiGiammarino E. L., Weber J. D., Bothner S. J., Kriwacki R. W. (2001) Defining the molecular basis of Arf and Hdm2 interactions. J. Mol. Biol. 314, 263–277 [DOI] [PubMed] [Google Scholar]
- 15. Sdek P., Ying H., Zheng H., Margulis A., Tang X., Tian K., Xiao Z. X. (2004) The central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2F and cell growth. J. Biol. Chem. 279, 53317–53322 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Z., Wang H., Li M., Agrawal S., Chen X., Zhang R. (2004) MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 279, 16000–16006 [DOI] [PubMed] [Google Scholar]
- 17. Juven-Gershon T., Shifman O., Unger T., Elkeles A., Haupt Y., Oren M. (1998) The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell. Biol. 18, 3974–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yogosawa S., Miyauchi Y., Honda R., Tanaka H., Yasuda H. (2003) Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem. Biophys. Res. Commun. 302, 869–872 [DOI] [PubMed] [Google Scholar]
- 19. Gulino A., Di Marcotullio L., Screpanti I. (2010) The multiple functions of Numb. Exp. Cell Res. 316, 900–906 [DOI] [PubMed] [Google Scholar]
- 20. Pece S., Confalonieri S., R Romano P., Di Fiore P. P. (2011) NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta 1815, 26–43 [DOI] [PubMed] [Google Scholar]
- 21. Colaluca I. N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., Di Fiore P. P. (2008) NUMB controls p53 tumour suppressor activity. Nature 451, 76–80 [DOI] [PubMed] [Google Scholar]
- 22. Pamment J., Ramsay E., Kelleher M., Dornan D., Ball K. L. (2002) Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene 21, 7776–7785 [DOI] [PubMed] [Google Scholar]
- 23. Shimizu H., Burch L. R., Smith A. J., Dornan D., Wallace M., Ball K. L., Hupp T. R. (2002) The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J. Biol. Chem. 277, 28446–28458 [DOI] [PubMed] [Google Scholar]
- 24. Liu W. L., Midgley C., Stephen C., Saville M., Lane D. P. (2001) Biological significance of a small highly conserved region in the N terminus of the p53 tumour suppressor protein. J. Mol. Biol. 313, 711–731 [DOI] [PubMed] [Google Scholar]
- 25. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 26. Treier M., Staszewski L. M., Bohmann D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell 78, 787–798 [DOI] [PubMed] [Google Scholar]
- 27. Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., Di Fiore P. P. (2004) Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown C. J., Cheok C. F., Verma C. S., Lane D. P. Trends Pharmacol. Sci. 32, 53–62 [DOI] [PubMed] [Google Scholar]
- 29. Madhumalar A., Lee H. J., Brown C. J., Lane D., Verma C. (2009) Design of a novel MDM2 binding peptide based on the p53 family. Cell Cycle 8, 2828–2836 [DOI] [PubMed] [Google Scholar]
- 30. Kabouridis P. S. (2003) Biological applications of protein transduction technology. Trends Biotechnol. 21, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harada H., Kizaka-Kondoh S., Hiraoka M. (2006) Antitumor protein therapy. Application of the protein transduction domain to the development of a protein drug for cancer treatment. Breast Cancer 13, 16–26 [DOI] [PubMed] [Google Scholar]
- 32. Harbour J. W., Worley L., Ma D., Cohen M. (2002) Transducible peptide therapy for uveal melanoma and retinoblastoma. Arch. Ophthalmol. 120, 1341–1346 [DOI] [PubMed] [Google Scholar]
- 33. Verdi J. M., Schmandt R., Bashirullah A., Jacob S., Salvino R., Craig C. G., Program A. E., Lipshitz H. D., McGlade C. J. (1996) Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 34. Kontopidis G., Wu S. Y., Zheleva D. I., Taylor P., McInnes C., Lane D. P., Fischer P. M., Walkinshaw M. D. (2005) Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U.S.A. 102, 1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uhlik M. T., Temple B., Bencharit S., Kimple A. J., Siderovski D. P., Johnson G. L. (2005) Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345, 1–20 [DOI] [PubMed] [Google Scholar]
- 36. Dho S. E., Jacob S., Wolting C. D., French M. B., Rohrschneider L. R., McGlade C. J. (1998) The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem. 273, 9179–9187 [DOI] [PubMed] [Google Scholar]
- 37. Li S. C., Zwahlen C., Vincent S. J., McGlade C. J., Kay L. E., Pawson T., Forman-Kay J. D. (1998) Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat. Struct. Biol. 5, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 38. Zwahlen C., Li S. C., Kay L. E., Pawson T., Forman-Kay J. D. (2000) Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb. EMBO J. 19, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carter S., Vousden K. H. (2008) A role for Numb in p53 stabilization. Genome Biol. 9, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meek D. W., Hupp T. R. (2010) The regulation of MDM2 by multisite phosphorylation. Opportunities for molecular-based intervention to target tumours? Semin. Cancer Biol. 20, 19–28 [DOI] [PubMed] [Google Scholar]
- 41. Huart A. S., MacLaine N. J., Meek D. W., Hupp T. R. (2009) CK1α plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J. Biol. Chem. 284, 32384–32394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheok C. F., Dey A., Lane D. P. (2007) Cyclin-dependent kinase inhibitors sensitize tumor cells to nutlin-induced apoptosis. A potent drug combination. Mol. Cancer Res. 5, 1133–1145 [DOI] [PubMed] [Google Scholar]